Abstract
Clostridium difficile-associated disease is increasing in incidence and is costly to treat. Our understanding of how this organism senses its entry into the host and adapts for growth in the large bowel is limited. The small-molecule second messenger cyclic diguanylate (c-di-GMP) has been extensively studied in Gram-negative bacteria and has been shown to modulate motility, biofilm formation, and other processes in response to environmental signals, yet little is known about the functions of this signaling molecule in Gram-positive bacteria or in C. difficile specifically. In the current study, we investigated the function of the second messenger c-di-GMP in C. difficile. To determine the role of c-di-GMP in C. difficile, we ectopically expressed genes encoding a diguanylate cyclase enzyme, which synthesizes c-di-GMP, or a phosphodiesterase enzyme, which degrades c-di-GMP. This strategy allowed us to artificially elevate or deplete intracellular c-di-GMP, respectively, and determine that c-di-GMP represses motility in C. difficile, consistent with previous studies in Gram-negative bacteria, in which c-di-GMP has a negative effect on myriad modes of bacterial motility. Elevated c-di-GMP levels also induced clumping of C. difficile cells, which may signify that C. difficile is capable of forming biofilms in the host. In addition, we directly quantified, for the first time, c-di-GMP production in a Gram-positive bacterium. This work demonstrates the effect of c-di-GMP on the motility of a Gram-positive bacterium and on aggregation of C. difficile, which may be relevant to the function of this signaling molecule during infection.
INTRODUCTION
Cyclic diguanylate (3′,5′-cyclic diguanylic acid) (c-di-GMP) is a second messenger utilized exclusively by prokaryotes to effect global changes in cellular physiology (45). c-di-GMP has been implicated in changes to the cellular envelope in multiple species and mediates the transition from planktonic growth to biofilm formation in many Gram-negative bacteria. c-di-GMP augments biofilm formation by increasing the production of adhesins and extracellular matrix components through altered gene regulation (21, 36, 37, 56, 67, 68) and posttranslational modifications (10, 73). Conversely, c-di-GMP inhibits motility by reducing the transcription or translation of flagellar genes (1, 2, 26, 29, 35), impeding pilus assembly (24, 28), or inhibiting the rotation of assembled flagella (4, 8, 40, 49). In addition to regulating biofilm formation and motility, c-di-GMP coordinates cell-wide responses to lifestyle transitions such as entry into stationary phase (60), expression of virulence genes (22, 30, 64, 69), resistance to antibiotics and host immune responses (22, 33), and cell developmental shifts (13, 41, 70).
The level of c-di-GMP in the cell is controlled by three types of enzymes. c-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGCs) containing a GGDEF domain, named for a conserved motif in the active site (46, 50, 56). c-di-GMP is hydrolyzed by two distinct families of phosphodiesterases (PDEs), known as EAL and HD-GYP domains, which are also named for conserved amino acid residues (6, 9, 48, 51, 65). These are highly modular domains, often coupled to a variety of known sensory and/or regulatory domains, and can occur in multiple proteins within one organism (17, 18). Transcriptional and posttranscriptional control of DGCs and PDEs could allow meticulous temporal and spatial regulation of c-di-GMP in the cell.
The Gram-positive, spore-forming, obligate anaerobe Clostridium difficile causes nosocomial infections that are often recurrent and sometimes fatal. Annual treatment costs are estimated to be in excess of $3 billion in the United States alone (11, 38). C. difficile is transmitted between individuals in the form of metabolically inactive spores that germinate in the gut to produce vegetatively growing bacilli. How C. difficile mediates the adaptation to the intestinal environment and production of cytotoxins is only beginning to be understood. Little is known about how the organism adheres to the intestinal wall and colonizes the host. C. difficile produces flagella that allow swimming motility, which in many bacterial pathogens contributes to pathogenesis. A recent study showed that C. difficile flagellar mutants (fliC and fliD mutants) colonize Caco-2 intestinal epithelial cells in higher numbers than the isogenic wild-type strain (12), while FliC and FliD were dispensable for virulence in hamsters. However, evidence from in vitro and other animal studies indicates that the flagellum is involved in cellular adherence (66), and human patients with C. difficile-associated disease produce antibodies against C. difficile flagellar proteins, indicating that flagella are produced in vivo (66). Furthermore, flagella isolated from C. difficile clinical isolates involved in recent outbreaks show various patterns of protein glycosylation, suggesting that flagellar function and regulation may contribute to differences in transmissibility and virulence observed between strains (71).
Biofilm formation by C. difficile, while not yet demonstrated in vitro (44), has been proposed to occur during colonization of intestinal surfaces. Thick mats of C. difficile have been observed in association with sites of severe inflammation in an infected animal model (31), suggesting that biofilm production occurs in vivo. Other bacteria, including commensal species and pathogens, are known to form multi- or single-species biofilms in the mammalian intestine (16, 32, 72). C. difficile may benefit from passive or active interactions with commensal biofilms or may have distinct mechanisms for development of biofilms in the host.
The sequenced genome of Clostridium difficile 630 encodes 37 different proteins predicted to play a role in c-di-GMP metabolism (52). This is far more than are found in other clostridial genomes such as those of C. perfringens or C. botulinum, which encode 9 and 16 predicted DGCs/PDEs, respectively (55, 58). The enzymatic functionality of several of these C. difficile DGCs and PDEs has been confirmed by expressing them heterologously in Vibrio cholerae, where they result in phenotypes consistent with elevated or lowered levels of intracellular c-di-GMP (5). However, the roles that DGCs and PDEs play in C. difficile biology and in motility in particular are unknown. The identification of a c-di-GMP-binding riboswitch, Cd1, immediately upstream of a flagellar operon strongly suggests that c-di-GMP is involved in regulating flagellar gene expression and cellular motility (63). While Cd1 can regulate the expression of a heterologous reporter gene in response to c-di-GMP (63), neither this riboswitch nor c-di-GMP has been shown to regulate motility or any other process in C. difficile.
In this study, we determined the role of c-di-GMP in regulating the motility of C. difficile. Organisms with multiple DGCs and PDEs exert spatial, temporal, and posttranslational regulation in order to prevent cross talk between pathways, but these signaling networks are often at least partially redundant, and the contribution of any given gene may be masked in a knockout analysis (27, 30, 53). Ectopic expression of a DGC or PDE raises or lowers the global intracellular level of c-di-GMP, respectively, and provides a way to assess the roles of this signaling molecule in regulating pathways of interest (2, 64). In order to apply this approach to the question of C. difficile flagellar motility, we constructed a plasmid for inducible expression of DGC and PDE genes in C. difficile. Here we demonstrate that c-di-GMP negatively regulates motility of C. difficile by repressing expression of flagellar genes. Furthermore, we observed that c-di-GMP promotes aggregation of C. difficile cells, which implies the ability of C. difficile to form biofilms. This study provides the first direct evidence that C. difficile synthesizes c-di-GMP which affects its cellular processes, such as motility and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. C. difficile strains were grown in BHIS medium (59) at 37°C in an anaerobic chamber with an atmosphere of 90% N2, 5% CO2, and 5% H2. Media for growth of C. difficile were supplemented with 10 μg/ml thiamphenicol (BHIS-Tm) and/or 100 μg/ml kanamycin, as appropriate. Escherichia coli strains were propagated in Luria Bertani (LB) medium or on LB agar plates containing 50 μg/ml ampicillin and/or 10 μg/ml chloramphenicol, as needed, at 37°C under aerobic conditions.
Plasmid and strain construction.
All restriction enzymes and DNA polymerase (Phusion) were purchased from New England BioLabs. Oligonucleotide sequences used in this study are listed in Table S2 in the supplemental material. Genomic DNA from C. difficile strain 630 (GenBank accession number AM180355) (52) was used as the template in all PCRs. For expression in E. coli, CD1420 (dccA) was amplified with primers dccAF and dccAR, CD1515 (pdcA) with primers pdcAF and pdcAR, and the EAL domain of CD1515 (pdcA-EAL) with primers pdcAFb and pdcAR. A mutant allele of dccA in which the GGDEF active site is mutated to AADEF (dccAmut) was generated using splicing by overlap extension PCR (SOE-PCR), with primers dccAF and dccAgaR generating the upstream fragment and primers dccAgaF and dccARb generating the downstream fragment. All three resulting fragments were digested with KpnI and PstI and cloned into similarly digested pBAD33 (for dccA and dccAmut) or pMMBneo (for pdcA and pdcA-EAL) (39). The plasmids were confirmed by PCR using gene-specific and plasmid-specific primers (67EHF and 67EHR). The plasmids were introduced by electroporation into E. coli BL21.
For expression of relevant genes in C. difficile, we first generated a vector for inducible expression of these genes. The promoter from the cpr locus (34) was amplified from CD630 genomic DNA using primers PcprF and PcprR. The PCR products were digested with EcoRI and KpnI and cloned into similarly digested pMC123 (34), yielding pMC-Pcpr (see Fig. S1 in the supplemental material). This plasmid was confirmed by PCR with primers pUCmcsF and m13r.
The dccA, dccAmut, and pdcA-EAL genes were cloned into pMC-Pcpr. For dccA and dccAmut, the native ribosomal binding sites (RBS) were retained with primers dccAbamHI and dccApstI. For the truncated pdcA-EAL, an RBS was introduced using the forward primer pdcAEbamHI, which was paired with primer pdcApstI. Each of the 3 fragments was digested with BamHI and PstI and cloned into similarly digested pMC-Pcpr. The plasmids were confirmed by PCR using primers pUCmcsF and m13r and named pDccA, pDccAmut, and pPdcA-EAL. These plasmids, along with the pMC-Pcpr vector, were each transformed by electroporation into HB101(pRK24). The pRK24 plasmid is a broad-host-range plasmid RP4 derivative that mobilizes IncP oriT plasmids. The HB101(pRK24) plasmid donor strains were mated with C. difficile strain 630, resulting in transfer of pDccA, pDccAmut, and pPdcA-EAL into C. difficile. Transconjugants were selected on BHIS-Tm supplemented with kanamycin and appeared after 48 h of growth at 37°C. C. difficile strains containing the desired plasmids were screened by PCR using primers pUCmcsF and m13r to confirm the presence of a plasmid with an insert of the correct size and using primers tcdBF and tcdBR to confirm isolation of C. difficile. At least two isolates were obtained for each conjugation (see Fig. S1 in the supplemental material). Using the same conjugation and screening methods, the pDccA, pDccAmut, and pPdcA-EAL plasmids were moved into C. difficile strain R20291.
Protein purification.
E. coli expression strains were grown at 37°C to an optical density at 600 nm (OD600) of 0.1 to 0.15 (1-cm path length), at which point the temperature was dropped to 25°C and expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h. Cells were lysed by sonication in His6 buffer (10 mM Tris-HCl [pH 7.8], 300 mM NaCl, 50 mM NaH2PO4, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Lysates were clarified by centrifugation and purified on HisPure Ni-nitrilotriacetic acid (Ni-NTA) resin (Thermo Scientific) according to the manufacturer's protocol. DccA was dialyzed overnight against 75 mM Tris (pH 7.8), 250 mM NaCl, 25 mM KCl, 10 mM MgCl2, and 30% glycerol and stored at −20°C. PdcA and PdcAEAL were stored at 4°C and used for enzymatic assays within 5 days of purification.
In vitro enzymatic assays.
Purified DccA and the DccA mutant were assayed for DGC activity using [α-32P]GTP as the substrate as described previously (65). Purified WspR (21) was included as a positive control; buffer only was used as the negative control. The reaction mixtures were treated with 1 μl calf intestinal phosphatase (CIP) (New England BioLabs) for 30 min at 37°C to remove the phosphate radiolabel from unincorporated GTP. Aliquots (0.5 μl) taken before and after CIP treatment were analyzed by thin-layer chromatography as described previously (65). Full-length PdcA, which contains a GGDEF domain with an active-site sequence, suggesting that it lacks DGC activity, was nonetheless assayed for DGC activity as described above but in reaction buffer containing 5 mM CaCl2 to inhibit PDE activity of the EAL domain.
PdcA and PdcA-EAL were assayed for PDE activity by incubating the purified enzymes with 32P-labeled c-di-GMP as described previously (65). The radiolabeled c-di-GMP substrate was synthesized and purified as described previously (43, 65). Buffer-only reaction mixtures were used as a negative control. Aliquots (0.5 μl) of the reaction mixtures were analyzed as described above for the DGC assays.
Measurements of intracellular c-di-GMP levels.
c-di-GMP was measured as described previously (3), with the following modifications. C. difficile strains were grown in 50 ml BHIS-Tm, with or without nisin induction (1 μg/ml), to an OD600 of 0.6; the optical densities of the cultures at time of collection were recorded. Dilutions were plated to determine the number of CFU extracted. The bacteria were collected by centrifugation and washed once with TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA, pH 8), and then nucleotides were extracted with methanol (MeOH)-acetonitrile-distilled water (dH2O) (40:40:20) plus 0.1 N formic acid as described previously (3). Samples (50 μl out of 200 μl total) were analyzed by high-pressure liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) (3). The amount of intracellular c-di-GMP was determined by fitting the peak intensity to a standard curve obtained by analyzing serial dilutions of c-di-GMP and dividing the total amount of c-di-GMP extracted by the total intracellular volume of bacteria extracted. The intracellular volume of bacteria extracted was calculated by multiplying the volume of one bacterial cell by the number of cells extracted, which was based on CFU counts. The volume of one bacterial cell was estimated as a cylinder with a radius of 0.37 μm and a length of 2.235 μm, using ImageJ software to measure cell dimensions from electron micrographs. Figure S3 in the supplemental material shows the measurements obtained from 100 bacterial cells either parallel (for length determination) or perpendicular (for radius determination) to the slide surface. For cell length determination, cells with lengths of greater than 4 μm were presumed to be dividing and were excluded from the analysis.
Motility assays.
Individual colonies were inoculated using sterile toothpicks into 0.3% agar plates containing BHIS-Tm and nisin at 0, 0.01, 0.1, or 1 μg/ml nisin. Plates were incubated at 37°C in an anaerobic chamber for up to 96 h. The diameters of the motility swarms were measured every 24 h. The mean diameters and standard deviations are reported.
RNA isolation and real-time PCR.
RNA was isolated as described previously from C. difficile cultures grown in BHIS-Tm, without or with 1 μg/ml nisin, to an OD600 of 1.0 (34). cDNA samples were prepared from 200 ng RNA using random hexamers and the Tetro cDNA synthesis kit (Bioline). Real-time PCRs were done with 2 ng cDNA template using the SensiMix SYBR and fluorescein kit (Bioline). Primers (see Table 2 in the supplemental material) were designed using the PrimerQuest tool from IDT DNA technologies and detected transcripts of flgB, flgM, fliA, and codY. At least three biological replicates were analyzed. The rpoC transcript was measured and used for normalization (34). The raw threshold cycle (CT) values for rpoC were equivalent in all RNA samples tested, and thus rpoC was not differentially expressed in any of the strains. Controls with no reverse transcriptase were included for all templates and all primer sets. The data were analyzed using the ΔΔCT method and are expressed as the fold change in normalized transcript relative to the average value for the induced vector control.
Detection of flagellin.
Flagellins were assayed as described previously (71). Briefly, C. difficile strains were grown to an OD600 of 1.0. Culture samples were vortexed vigorously for 3 min, and then cells and large debris were removed by centrifugation. The supernatants were analyzed by SDS-PAGE (12%) and Coomassie blue staining. The band corresponding to the 26-kDa protein suspected to be flagellin was identified by using ABI 4800 matrix-assisted laser desorption ionization–time of flight (MALDI TOF/TOF) mass spectrometry at the University of North Carolina Michael Hooker Proteomics Center.
Assessment of aggregation.
Strains were grown in BHIS-Tm containing 0 or 1 μg/ml nisin. After 18 h of growth at 37°C, the optical densities at 600 nm of the unperturbed cultures were measured. The cultures were vortexed vigorously to disrupt the aggregates, and the optical densities were measured again. The assay was performed multiple times, each with three biological replicates. For electron microscopy, the strains were grown as described above, centrifuged for 5 min at 700 × g, suspended in 150 mM sodium phosphate buffer (pH 7.4), and mixed 1:1 with 4% glutaraldehyde in 150 mM sodium phosphate buffer. The cells were dehydrated through increasing concentrations of ethanol and dried using carbon dioxide as the transition solvent in a Samdri-795 critical-point dryer (Tousimis Research Corporation, Rockville, MD). Coverslips were mounted on aluminum stubs with carbon adhesive tabs, followed by sputter coating with a 5-nm thickness of gold-palladium alloy (60 Au-40 Pd) in a Hummer X sputter coater (Anatech USA, Union City, CA). Images were taken using a Zeiss Supra 25 field emission scanning electron microscope (FESEM) operating at 5 kV, with a working distance of 5 mm and a 10-μm aperture (Carl Zeiss SMT Inc., Peabody, MA).
For quantification of aggregation, C. difficile with pDccA or empty vector was grown in BHIS-Tm with 1 μg/ml nisin and then viewed by differential interference contrast (DIC) microscopy at a magnification of ×40. ImageJ was used to identify and count cell aggregates in photographs of 20 randomly selected fields of view for each sample. Aggregates were defined as two or more cells contacting along the long axis or three or more cells contacting at the poles (to exclude cells undergoing division).
RESULTS
Identification of active C. difficile enzymes.
The physiological responses of C. difficile to c-di-GMP are unknown, making characterization of the second-messenger-based regulatory network problematic. Not only is targeted mutagenesis of genes not trivial in this organism, but the large number of C. difficile genes predicted to synthesize or hydrolyze c-di-GMP further complicates attempts to determine their roles, as the effect of disrupting any given gene could be masked by the function of multiple, partially redundant orthologues. To begin to determine the functions of c-di-GMP in C. difficile, we decided to ectopically express an active c-di-GMP synthetase or phosphodiesterase to increase or decrease intracellular c-di-GMP, respectively. We could then examine the cellular responses to changes in the global c-di-GMP concentration, an approach that has been successfully utilized in V. cholerae (2, 64).
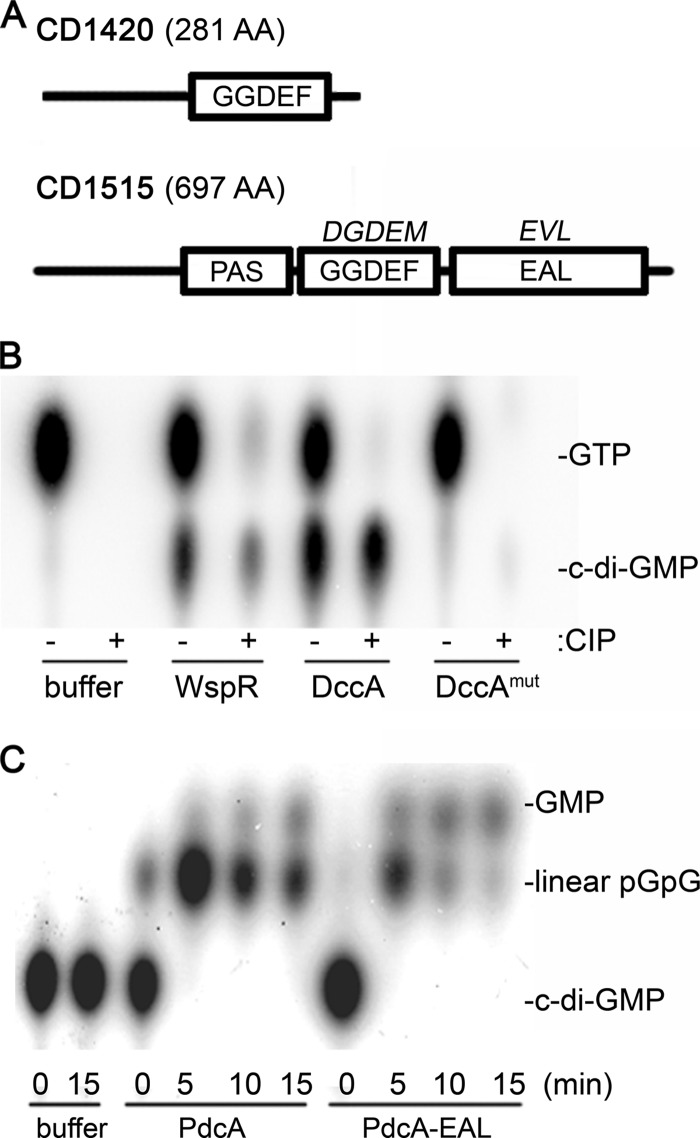
C. difficile encodes 37 putative c-di-GMP DGCs and PDEs (17, 18, 52). CD1420, which encodes the smallest DGC, was previously demonstrated to be a catalytically active enzyme (5). The CD1420 gene product is predicted to be a soluble cytoplasmic protein, with no other sensory or regulatory domains present to modulate its activity (Fig. 1A). Furthermore, CD1420 lacks an I site (5), which should allow unchecked synthesis of c-di-GMP. Thus, we chose to overexpress CD1420, which we have named dccA (diguanylate cyclase from Clostridium A), in C. difficile, and we anticipated that this would result in elevated intracellular c-di-GMP levels.
Fig 1.
Domain architectures and enzymatic activities of the enzymes used in this study. (A) Domain structures of DccA (CD1420) and PdcA (CD1515). The actual amino acid sequences in the GGDEF and EAL motifs are shown above the relevant domain. (B) Purified DccA and DccA with GGDEF-to-AADEF mutations (DccAmut), as well as the WspR DGC (positive control), were assayed in vitro for DGC activity using [α-32P]GTP as the substrate. Reaction products were analyzed before (−) and after (+) treatment with CIP using thin-layer chromatography (TLC). (C) Purified PdcA and its EAL domain alone (PdcA-EAL) were assayed in vitro for c-di-GMP phosphodiesterase activity using 32P-labeled c-di-GMP as the substrate. At the indicated times, reaction products were analyzed by TLC. The identities of spots in panels B and C were assigned based on characteristic relative mobilities, as described previously (47).
There is no single-domain PDE encoded in the C. difficile genome. Eighteen of the 19 EAL-domain containing proteins also contain a GGDEF domain, though in all but one of these cases the GGDEF domain is predicted to be inactive (5). In addition, all but three of the EAL domain proteins have predicted transmembrane domains. The presence of the EAL domain in conjunction with an inactive GGDEF domain or other sensory or regulatory domains could render the protein inactive under certain experimental conditions or could obscure the effect of overexpression. Therefore, we chose to express a truncated phosphodiesterase, consisting only of an EAL domain, to deplete c-di-GMP in C. difficile. We focused on CD1515, which is predicted to encode a soluble PAS-GGDEF-EAL fusion protein. This protein, which we have named PdcA (phosphodiesterase from Clostridium A), has a poorly conserved GGDEF motif that is unlikely to retain any catalytic activity, and a well-conserved EAL domain (Fig. 1A). PdcA is expected to be an active c-di-GMP PDE, as its ectopic expression in a heterologous host, Vibrio cholerae, resulted in reduced biofilm formation and increased motility phenotypes as expected from reduced intracellular c-di-GMP (5).
To confirm the catalytic activities of DccA, full-length PdcA, and the EAL domain of PdcA (PdcA-EAL), we expressed His6-tagged alleles of dccA, pdcA, and pdcA-EAL in E. coli, purified the recombinant proteins, and assayed enzymatic activity in vitro. DccA and the positive control WspR, a DGC from Pseudomonas aeruginosa (21), were tested for DGC activity. Both DccA and WspR produced c-di-GMP when incubated with radiolabeled GTP, demonstrating that both proteins can function as diaguanylate cyclases (Fig. 1B). The ability of DccA to synthesize c-di-GMP was abolished when the GGDEF motif was mutated to AADEF, confirming that DGC activity relies on the GGDEF active site. PdcA and PdcA-EAL were tested for c-di-GMP phosphodiesterase activity. Both the full-length and truncated proteins quickly hydrolyzed c-di-GMP to linear pGpG (Fig. 1C), confirming PDE function and demonstrating that truncation did not affect enzymatic activity of the EAL domain. Indeed, the EAL domain seemed to have higher activity for the secondary hydrolysis of pGpG to GMP, suggesting that the PAS or inactive GGDEF domain may inhibit activity of the full-length protein (Fig. 1C). To avoid any of the experimental or analytical complications that such inhibition might cause in vivo, PdcA lacking the PAS and GGDEF domains, PdcA-EAL, was used for subsequent experiments.
Development of an inducible overexpression system for use in C. difficile.
Few systems are available for inducible expression of genes in C. difficile (15). Therefore, in order to express dccA and pdcA-EAL in C. difficile, we developed a plasmid-based system for the inducible expression of these genes. This plasmid, pMC-Pcpr, derives from pMC123, which is stably maintained extrachromosomally in C. difficile (34) and uses the C. difficile cpr promoter to drive expression of genes (see Fig. S1A in the supplemental material). The clostridial cpr promoter controls the expression of a putative ABC transporter system that enables C. difficile to survive exposure to the cationic antimicrobial peptides nisin and gallidermin (34). The operon under the control of this promoter is upregulated 10- to 20-fold in response to sublethal concentrations of nisin but is not significantly impacted by a number of other stresses, including heat, acid, or oxygen stress (14), confirming that it is induced specifically by nisin and is not part of a general stress response. We cloned dccA, the mutant dccA allele (dccAmut), and pdcA-EAL into pMC-Pcpr to create the expression plasmids pDccA, pDccAmut, and pPdcA-EAL, respectively. The resulting plasmids, as well as pMC-Pcpr, were introduced into C. difficile strain 630 through conjugation. Thiamphenicol-resistant transconjugants were confirmed to contain the desired plasmids and to be C. difficile, as shown by the presence of the chromosomal tcdB gene (see Fig. S1B in the supplemental material). All strains were found to grow equivalently and maximally in BHIS-Tm containing up to 1 μg/ml nisin, with no differences in doubling times observed (data not shown). At nisin concentrations of 5 μg/ml or higher, the growth rates of all strains were slightly impeded, so all subsequent experiments were performed using at most 1 μg/ml nisin.
To test whether growth in the presence of nisin induces expression of the genes placed under the control of the cpr promoter, we performed quantitative reverse transcription-PCR (qRT-PCR) analysis of the dccA and pdcA-EAL transcripts in cultures grown in the presence and absence of 1 μg/ml nisin. In the absence of nisin, the level of dccA transcript in the C. difficile pDccA strain was indistinguishable from that in the control strain carrying the empty vector. When nisin was present in the growth medium, the dccA transcript was induced approximately 6-fold (Fig. 2A). The same results were observed for the dccAmut allele. In the C. difficile pPdcA-EAL strain, the pdcA-EAL transcript was slightly elevated even in the absence of nisin. However, induction with nisin caused a further 7-fold increase in transcript (Fig. 2A).
Fig 2.
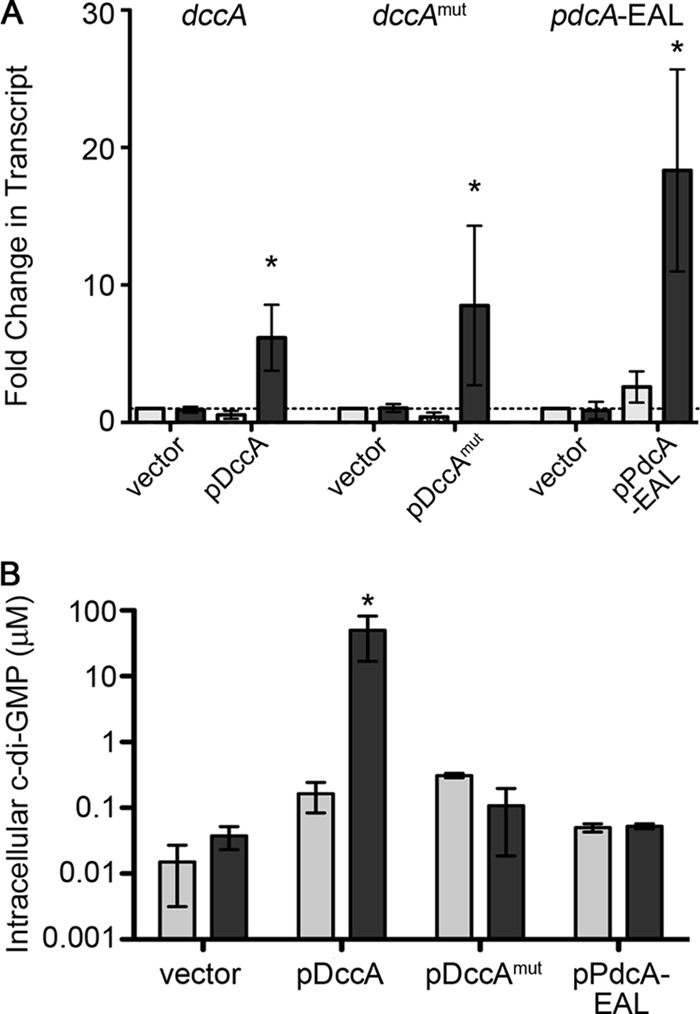
Modulation of intracellular c-di-GMP by ectopic expression of DccA, DccAmut, and PdcA-EAL. (A) Induction of DccA, DccAmut, and PdcA-EAL gene expression by addition of nisin to the growth medium. The levels of the transcripts for the genes indicated at the top were measured by qRT-PCR in the relevant strains grown with (dark gray) or without (light gray) 1 μg/ml nisin. Data were analyzed using the ΔΔCT method and are expressed as the average fold change in transcript compared to that of the induced vector control. (B) Alteration of intracellular c-di-GMP upon expression of DccA, DccAmut, and PdcA-EAL genes. HPLC was used to measure intracellular c-di-GMP in strains containing the indicated plasmids and grown with (dark gray) or without (light gray) 1 μg/ml nisin. Three biological replicates were analyzed. The concentrations shown were calculated as described in Materials and Methods. In panels A and B, data were analyzed by one-way analysis of variance (ANOVA) and Dunnett's posttest, with comparisons to the induced vector control (*, P < 0.05).
Having confirmed that transcription of dccA, dccAmut, and pdcA-EAL is induced by nisin in the respective strains, we further determined the effect of expression on the intracellular c-di-GMP level. The c-di-GMP content of C. difficile with pDccA, pDccAmut, pPdcA-EAL, or the empty vector, each grown with or without 1 μg/ml nisin, was determined by HPLC-tandem mass spectrometry as described previously (3). The estimated intracellular concentration of c-di-GMP in the experimental control strain was determined to be 15 to 50 nM. However, growth of the pDccA strain in the presence of nisin resulted in over a 1,000-fold increase in intracellular c-di-GMP compared to that in the control strain (Fig. 2B). Although there were minor fluctuations in the intracellular c-di-GMP concentration in the PdcA and DccAmut expression strains relative to the control strain, none of the strains showed differences similar to that for induction of DccA. As the samples were near the limit of detection of c-di-GMP (5 to 10 nM), with the exception of DccA induction, it is possible that these differences are due to noise and are not biologically relevant. Surprisingly, induction of pdcA-EAL expression did not detectably reduce the intracellular c-di-GMP content in C. difficile.
Manipulating c-di-GMP levels affects swimming motility of C. difficile.
Elevated c-di-GMP levels are associated with inhibited cellular motility in multiple flagellated species, due to altered gene expression and flagellum assembly and/or function. The presence of a c-di-GMP riboswitch upstream of a flagellar operon suggested that c-di-GMP regulates swimming motility of C. difficile (63), though this has not been demonstrated. We used a soft agar plate assay to monitor and quantify the effect of c-di-GMP on motility, and we observed movement of C. difficile through soft agar which was not impaired by the presence of 1 μg/ml nisin (Fig. 3). In the C. difficile 630 strain background, without nisin, DccA and the enzymatically inactive DccAmut had no impact on motility, while induction of active DccA dramatically decreased motility. The presence of pPdcA-EAL had no significant effect on motility of C. difficile 630, whether or not nisin was present (Fig. 2A).
Fig 3.
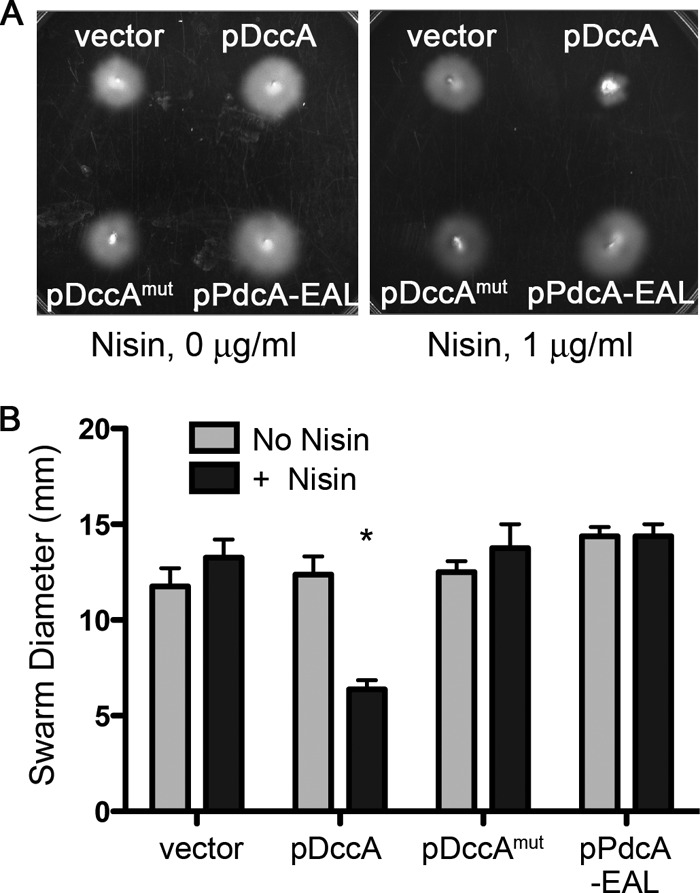
c-di-GMP negatively regulates motility of C. difficile 630. C. difficile strains with pDccA, pDccAmut, pPdcA-EAL, or empty vector were assayed for motility in BHIS-Tm with 0.3% agar and supplemented with 0 or 1 μg/ml nisin. The swarm diameters, indicative of the motility of each strain, were measured daily for 96 h. (A) Representative motility plates after 48 h of incubation at 37°C. (B) Quantitative measurements of motilities of strains grown with or without nisin, after 48 h. Shown are the means and standard deviations of the swarm diameters from four independent experiments. Asterisks indicate statistically significant differences (P < 0.05 by one-way ANOVA and Dunnett's posttest, comparing data to those for the induced vector control.).
To address potential issues due to strain specificity, these plasmids were also introduced into the epidemic C. difficile isolate R20291, where they similarly conferred thiamphenicol resistance and were confirmed by PCR (data not shown). As seen in C. difficile 630, induction of dccA expression with nisin resulted in a dramatic decrease in motility through soft agar (see Fig. S4 in the supplemental material). While expression of pPdcA-EAL in R20291 led to a statistically significant increase in motility, the effect was small and independent of the presence of nisin.
Induction with 1 μg/ml nisin was useful for the identification of motility phenotypes regulated by c-di-GMP but resulted in intracellular c-di-GMP concentrations well above those measured in C. difficile or other bacteria (57). To determine whether intermediate levels of c-di-GMP can affect motility, dccA expression was induced in C. difficile 630 pDccA by the addition of 0, 0.01, 0.1, or 1 μg/ml nisin. The intracellular concentration of c-di-GMP increased with increasing concentrations of nisin in the growth medium (Fig. 4A); as little as 0.01 μg/ml nisin resulted in a significant increase in c-di-GMP. Furthermore, the motility of C. difficile in soft agar medium decreased with increasing intracellular c-di-GMP, and reduced motility could be observed in C. difficile with 100 to 150 nM c-di-GMP (Fig. 4B).
Fig 4.
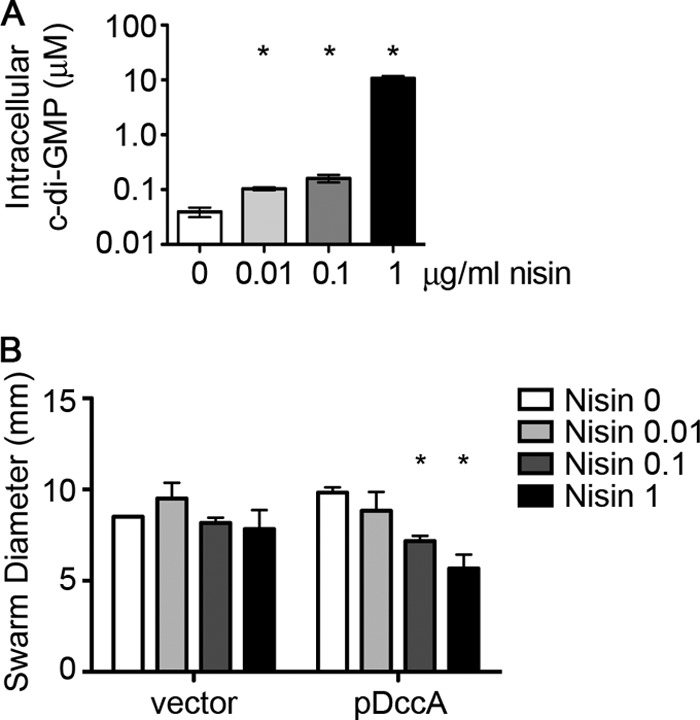
Induction by different concentrations of nisin results in graded effects on motility of C. difficile 630. (A) HPLC-MS/MS was used to determine the amount of c-di-GMP in C. difficile with pDccA grown in BHIS-Tm with 0, 0.01, 0.1, or 1 μg/ml nisin. Three biological replicates were analyzed, and data were normalized to the calculated cell volume extracted, with means and standard deviations shown. Asterisks indicate statistically significant differences from the uninduced sample (P < 0.05 by unpaired t test). (B) C. difficile strains with empty vector or pDccA were assayed for motility in BHIS-Tm with 0.3% agar and supplemented with the indicated concentrations of nisin (μg/ml). Shown are the means and standard deviations of the swarm diameters after 48 h of incubation at 37°C, from four independent experiments. Asterisks indicate statistically significant differences in motility (P < 0.05 by two-way ANOVA and Bonferroni's posttest).
c-di-GMP impedes transcription of flagellar genes.
Previous work has shown that a c-di-GMP riboswitch, named Cd1, lies upstream of the flgB open reading frame and presumably controls expression of flgB and the downstream flagellar genes (63). A transcriptional fusion of the Cd1-containing flgB promoter to a lacZ reporter gene showed that this riboswitch functions as an “off switch” in response to c-di-GMP. Furthermore, in vitro transcription assays demonstrated that addition of c-di-GMP to the reaction mixture led to the appearance of a truncated flagellar transcript, indicating that regulation of flagellar gene expression occurs at the level of transcription rather than translation. These data strongly suggest that c-di-GMP inhibits flagellar gene expression in C. difficile. To test this, C. difficile with pDccA, pDccAmut, pPdcA-EAL, or empty vector was grown in the presence or absence of nisin, and the levels of flagellar gene transcripts in each of these strains were measured by qRT-PCR. We chose three flagellar genes to examine for expression by qRT-PCR: flgB, which is immediately downstream of the Cd1 riboswitch; fliA, which is near the end of the flgB operon and encodes a sigma factor (σ70) predicted to activate flagellar gene expression; and flgM, a gene at the beginning of a second, upstream flagellar operon not predicted to be directly controlled by Cd1. In all cases, increasing intracellular c-di-GMP by inducing dccA expression with nisin dramatically reduced the levels of flgB, fliA, and flgM mRNAs (Fig. 5A); expression of the dccAmut allele had no effect on flgB, fliA, or flgM transcript levels. Furthermore, rpoC transcript levels were equivalent among all strains grown with or without nisin, and the level of codY transcript was unaffected by induction of dccA expression and increased c-di-GMP synthesis. Therefore, dccA expression does not have a global negative effect on transcription. Neither the induction of pdcA-EAL nor dccAmut impacted flagellar gene transcript levels (data not shown), consistent with the wild-type levels of motility of the respective strains.
Fig 5.
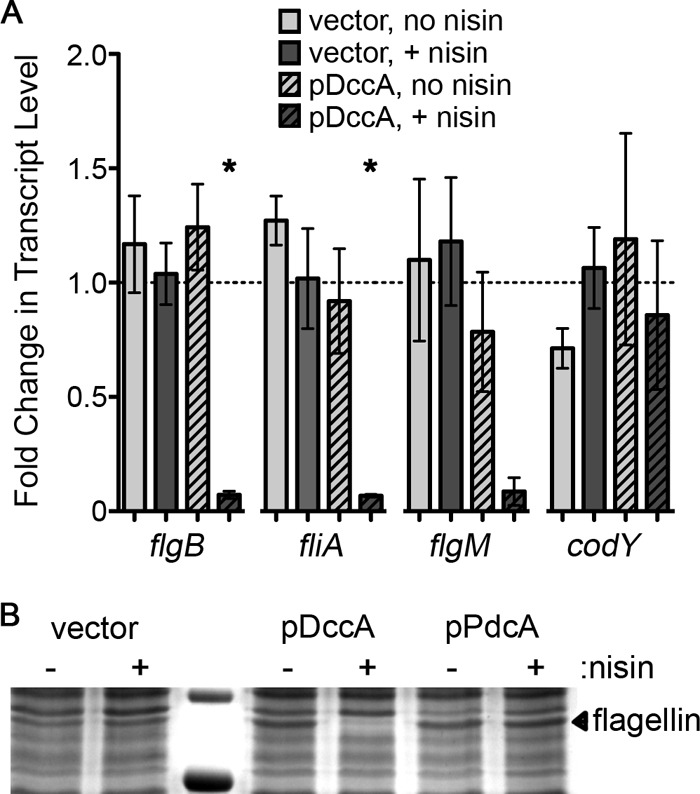
c-di-GMP represses production of flagella by regulating transcription of flagellar genes in C. difficile 630. (A) Using qRT-PCR, transcript levels of rpoC, flgB, flgM, fliA, and codY in the indicated strains grown with or without 1 μg/ml nisin were measured. At least three biological replicates were analyzed. Data were analyzed using the ΔΔCT method with rpoC as the reference transcript and normalized to the induced vector control. Asterisks indicate a P value of <0.05 as determined by one-way ANOVA and Dunnett's posttest, comparing values to that for the induced vector control. (B) The amount of flagellar protein produced was determined by SDS-PAGE of extracellular proteins of uninduced and induced cultures of the indicated strains. The indicated band was determined to be flagellin by mass spectrometry.
Isolation of extracellular proteins from induced and uninduced cultures to enrich for flagellin also showed that cells overexpressing dccA had no appreciable levels of flagellin subunit (Fig. 5B). Induction of dccA expression resulted in the loss of a protein of approximately 30 kDa. This protein band, when excised from the control sample and analyzed by mass spectrometry, contained C. difficile flagellin as the most abundant protein (data not shown). Again consistent with the wild-type motility of C. difficile pPdcA-EAL, induction of pdcA-EAL had no discernible effect on the production or secretion of flagellin (Fig. 3A).
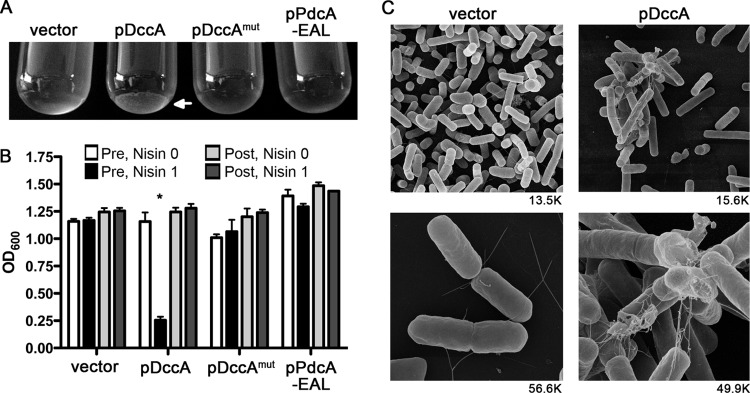
Artificially elevating c-di-GMP levels causes aggregation of cultured cells.
In numerous other bacterial species, increases in c-di-GMP not only impair various types of motility but in many cases upregulate the production of factors that contribute to biofilm production (45). While biofilm formation by C. difficile has not been directly demonstrated and we and others have been unable to observe C. difficile biofilms on abiotic surfaces, it has been proposed that C. difficile may produce such structures on intestinal surfaces (reference 44 and data not shown). We observed that C. difficile containing pDccA formed visible aggregates when the DGC was induced and intracellular c-di-GMP increased. Upon reaching optical densities of above ∼0.8 in broth culture, cells overexpressing dccA began to aggregate and sediment in the culture tube (Fig. 6A). Over time, the optical density of undisturbed cultures expressing induced DccA decreased compared to that of all other strains (Fig. 6B). The aggregates could be dispersed by vigorous vortexing, after which the C. difficile pDccA strain had an optical density comparable to that of the other cultures (Fig. 6B). The optical densities of the cultures containing aggregates, upon vortexing, increased at the same rate as for the other cultures (data not shown), indicating that the aggregated cells remained viable and continued to divide. Further analysis of these aggregates by scanning electron microscopy showed that in cultures of C. difficile pDccA grown with nisin, the cells formed large aggregates visibly bound together by fibrous networks (Fig. 6C). However, the aggregates were completely absent in the vector control strain grown under the same conditions. The control strain contained primarily free-living cells. Using DIC microscopy, we determined the number of aggregates present in stationary-phase cultures of C. difficile with pDccA or empty vector, both grown in the presence of nisin. The vector control strain contained a negligible number of aggregates, all of which consisted of 3 cells or fewer (see Fig. S5 in the supplemental material). In the pDccA strain there were numerous aggregates present, many of which contained 4 or more bacteria. We propose that this c-di-GMP-dependent aggregation phenotype, promoted in this case by the artificial manipulation of c-di-GMP, represents a possible mechanism for biofilm production in the host.
Fig 6.
Elevated c-di-GMP induces aggregation of C. difficile 630 cells. (A) Representative cultures grown in BHIS-Tm with 1 μg/ml nisin. The arrow shows sedimented aggregates in the C. difficile pDccA culture. (B) Quantification of cell aggregation by optical density measurement. The optical densities (600 nm) of the cultures were determined pre- and postvortexing. White bars are for uninduced cultures before vortexing, black bars are for induced cultures before vortexing, light gray bars are for uninduced cultures after vortexing, and dark gray bars are for induced cultures after vortexing. Assays were performed in triplicate. The asterisk indicates a P value of <0.05 as determined using two-way ANOVA and Bonferroni's posttest, comparing each strain to the induced vector control. (C) Scanning electron microscopy of C. difficile pDccA and the vector control strain grown in BHIS-Tm containing 1 μg/ml nisin. The magnification used is shown under each panel.
DISCUSSION
C. difficile infection is a widespread and costly public health problem whose prevalence and severity will likely increase in the coming decades, as an aging population spends more time in hospitals and nursing homes. Characterizing the mechanisms by which C. difficile regulates gene expression while in the host intestinal tract, the only known natural environment in which the bacterium has been shown to grow vegetatively, is critical to understanding the basis for its pathogenicity. This study focused on c-di-GMP, because relatively little is known about the roles of c-di-GMP in Gram-positive bacteria and in C. difficile in particular. Only a few studies have investigated the function of c-di-GMP in Gram-positive species, and to our knowledge, c-di-GMP has never been directly measured in a Gram-positive bacterium. Genetic evidence indicates that in Streptomyces coelicolor c-di-GMP regulates the development of mycelia and the production of the antibiotic actinorhodin (70). In Staphylococcus aureus, a protein with a degenerate GGDEF domain regulates biofilm formation and protein A production, but this is independent of the GGDEF protein's enzymatic activity and cellular c-di-GMP levels (23, 54). Our current work is the first study to directly quantify c-di-GMP production in a Gram-positive bacterium, demonstrate the effect of c-di-GMP on the motility of a Gram-positive species, and show a function for c-di-GMP in C. difficile.
We estimated the intracellular concentration of c-di-GMP in C. difficile to be 15 to 50 nM during exponential growth in rich medium. Compared with that in other bacteria, this is a relatively low concentration of c-di-GMP. Using the data obtained by Simm et al., the concentration of c-di-GMP ranges from 0.1 μM in E. coli to 2 μM in V. cholerae (57). Similarly, we typically observe that the concentration of c-di-GMP in V. cholerae is 1 to 2 μM (unpublished results). However, it is clear that C. difficile has the capacity to generate very high intracellular concentrations of c-di-GMP, as overproduction of DccA increased c-di-GMP levels to approximately 50 μM. Interestingly, this high level of c-di-GMP did not result in the toxicity that has been observed in E. coli (50). In addition, while the numerous GGDEF and EAL domain proteins encoded in the C. difficile genome have been assessed for enzymatic function in a heterologous organism (5), the importance of c-di-GMP in the biology of C. difficile has not been investigated previously. We showed that c-di-GMP represses the motility of C. difficile as it does in other bacteria and that intracellular c-di-GMP is inversely related to motility. Importantly, reduced motility of C. difficile 630 can be observed when the global intracellular c-di-GMP level is ∼100 nM, which represents a 2- to 6-fold increase compared to c-di-GMP levels in wild-type cells grown in vitro. While the physiological levels of c-di-GMP of C. difficile in the host bowel are unknown, our results suggest that relatively small changes in c-di-GMP can alter motility. Determining the natural range of c-di-GMP concentrations in C. difficile during growth in its natural environment will be critical to understanding the impact of c-di-GMP regulation in vivo.
Regulation of C. difficile motility occurs at least in part at the level of flagellar gene expression. This result was anticipated based on the characterization of the Cd1 riboswitch upstream of the operon led by flgB. Interestingly, c-di-GMP also repressed the level of mRNA for flgM, which is carried in a second flagellar operon not under the direct control of Cd1. Both operons contain genes encoding structural and regulatory elements required for proper flagellar function. It is likely that a gene product of the flgB operon regulates expression of the second flagellar operon. For example, expression of fliA, which is presumably cotranscribed with flgB and encodes a RNA polymerase sigma factor (σD), is also repressed by c-di-GMP. In other bacteria, including Bacillus subtilis, FliA is required for flagellar gene expression and motility (7, 25, 62). In C. difficile, repression of fliA transcription by c-di-GMP and the Cd1 riboswitch may in turn reduce transcription of the flgM operon. Such a mechanism would link transcription of both operons to the level of c-di-GMP in the cell, consistent with our expression data (Fig. 6A).
The pPdcA-EAL fragment was enzymatically active in vitro. However, there were no detectable differences in the intracellular levels of c-di-GMP in the pPdcA-EAL strain compared to the vector control strain, nor did we observe increased transcription of flgB, fliA, or flgM. It is possible that our inability to detect decreased c-di-GMP in the pPdcA-EAL strain is due to “buffering” that maintains a minimum level of c-di-GMP in the cell. For example, attempts to reduce c-di-GMP by ectopic expression of PdcA-EAL may lead to increased transcription or activity of one or more of the 18 putative DGCs encoded in the C. difficile genome. Conversely, the expression or activity of PDEs may be downregulated. Either possibility could serve to maintain a minimum level of c-di-GMP in the cell.
There are nearly 20 C. difficile strains for which the full or partial genomes are available. These genomes are highly variable and contain an unusually high number of mobile elements, 11% of the C. difficile 630 genome (52). In order to rule out strain-specific effects, we introduced the empty vector, pDccA, pDccAmut, and pPdcA-EAL into C. difficile isolate R20291, which is considered an epidemic isolate (61). The R20291 strain, of the 027 ribotype, differs genotypically and phenotypically from strain 630 (ribotype 012) (61), including differences in the battery of c-di-GMP signaling factors encoded (5). For example, several c-di-GMP metabolic genes present in 630 are absent or divergent in the R20291 genome (5). As in C. difficile 630, induction of DccA severely inhibited motility of R20291, and this effect was dependent upon an intact GGDEF motif. These results indicate that regulation of motility of C. difficile by c-di-GMP is conserved within the species. Both 630 and R20291 contain the Cd1 riboswitch upstream of the flgB operon, while some C. difficile strains do not (5). It would be interesting to determine if and how c-di-GMP regulates flagellar swimming in C. difficile strains lacking the Cd1 riboswitch. The importance of motility for adherence, colonization, and virulence is uncertain (12, 66), but the production of flagella during infection (42) and variation in flagellar protein glycosylation among strains (71) suggest that flagella may affect immune recognition of the bacteria or variation in virulence between strains.
While C. difficile has not been demonstrated to form biofilms on abiotic surfaces in vitro, it has been suggested that C. difficile forms biofilms in the gut to maintain long-term colonization of the bowel (44). The unexpected observation that C. difficile with elevated c-di-GMP forms visible aggregates suggests that c-di-GMP may augment a biofilm-related process in this bacterium. The ability of C. difficile to aggregate in this manner may be linked to host tissue damage in the mouse model (31). The aggregates observed in cultures of bacteria with high c-di-GMP levels consist of clumps apparently assisted by long, thin fibers. These fibers are approximately 10 nm in thickness, which is consistent with the size of pili. The C. difficile 630 genome contains type IV pilus gene clusters (52), and there is evidence that pili are produced during colonization of the hamster (19). This raises the interesting possibility that c-di-GMP may upregulate production of pili to aid in colonization. Future studies are aimed at determining the composition of these fibers.
The role of c-di-GMP in inversely regulating motility and aggregation in C. difficile, as in many other bacteria, suggests that c-di-GMP may be a key player in pathogenesis. The large number of c-di-GMP regulatory and sensory factors encoded by C. difficile further suggests that this bacterium may carefully modulate the level of c-di-GMP in the cell. The limitation of our strategy of manipulating c-di-GMP by ectopic expression of a DGC and PDE is that it does not address the natural control of intracellular c-di-GMP and its downstream regulatory targets. Future studies will be necessary to understand the complex spatial and temporal regulation of and by c-di-GMP, as well as to identify additional targets of c-di-GMP regulation.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank Abraham L. Sonenshein for helpful advice and critical reading of the manuscript and the Richardson lab for use of a real-time PCR machine. We thank the UNC Michael Hooker Proteomics Center for mass spectrometry services, Victoria Madden and the UNC Microscopy Services Laboratory for electron microscopy assistance, and the MSU Mass Spectrometry Facility for assistance in measuring c-di-GMP.
The project described was supported by NIH grant U54-AI-057157 to R.T. from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense, a UNC University Research Council Small Grant to R.T., NIH grant K01 DK087763 to S.M.M., and NIH grant 2-U54-AI-057153 to C.M.W. from the Region V Great Lakes Regional Center of Excellence for Emerging Infections and Biodefense.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 20 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Albert-Weissenberger C, et al. 2010. Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J. Bacteriol. 192:446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bobrov AG, et al. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehm A, et al. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 5. Bordeleau E, Fortier LC, Malouin F, Burrus V. 2011. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7:e1002039 doi:10.1371/journal.pgen.1002039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang AL, et al. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420–3426 [DOI] [PubMed] [Google Scholar]
- 7. Chen YF, Helmann JD. 1992. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc. Natl. Acad. Sci. U. S. A. 89:5123–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christen M, et al. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829–30837 [DOI] [PubMed] [Google Scholar]
- 10. Da Re S, Ghigo JM. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deneve C, Janoir C, Poilane I, Fantinato C, Collignon A. 2009. New trends in Clostridium difficile virulence and pathogenesis. Int. J. Antimicrob. Agents. 33(Suppl. 1):S24–S38 [DOI] [PubMed] [Google Scholar]
- 12. Dingle TC, Mulvey GL, Armstrong GD. 2011. Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 79:4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duerig A, et al. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emerson JE, Stabler RA, Wren BW, Fairweather NF. 2008. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J. Med. Microbiol. 57:757–764 [DOI] [PubMed] [Google Scholar]
- 15. Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286:27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faruque SM, et al. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galperin MY. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 19. Goulding D, et al. 2009. Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1. Infect. Immun. 77:5478–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hisert KB, et al. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234–1245 [DOI] [PubMed] [Google Scholar]
- 23. Holland LM, et al. 2008. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J. Bacteriol. 190:5178–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang B, Whitchurch CB, Mattick JS. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185:7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hughes KJ, Everiss KD, Kovach ME, Peterson KM. 1994. Sequence analysis of the Vibrio cholerae acfD gene reveals the presence of an overlapping reading frame, orfZ, which encodes a protein that shares sequence similarity to the FliA and FliC products of Salmonella. Gene 146:79–82 [DOI] [PubMed] [Google Scholar]
- 26. Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037 [DOI] [PubMed] [Google Scholar]
- 27. Kader A, Simm R, Gerstel U, Morr M, Romling U. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602–616 [DOI] [PubMed] [Google Scholar]
- 28. Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim YK, McCarter LL. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189:4094–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kulasakara H, et al. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawley TD, et al. 2009. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macfarlane S, Dillon JF. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102:1187–1196 [DOI] [PubMed] [Google Scholar]
- 33. Malone JG, et al. 2010. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000804 doi:10.1371/journal.ppat.1000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun. 79:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. 2006. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J. Biol. Chem. 281:8090–8099 [DOI] [PubMed] [Google Scholar]
- 36. Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 37. Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Brien JA, Lahue BJ, Caro JJ, Davidson DM. 2007. The emerging infectious challenge of clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect. Control Hosp. Epidemiol. 28:1219–1227 [DOI] [PubMed] [Google Scholar]
- 39. Osorio CG, et al. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paul R, et al. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pechine S, et al. 2005. Immunological properties of surface proteins of Clostridium difficile. J. Med. Microbiol. 54:193–196 [DOI] [PubMed] [Google Scholar]
- 43. Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reynolds CB, Emerson JE, de la Riva L, Fagan RP, Fairweather NF. 2011. The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7:e1002024 doi:10.1371/journal.ppat.1002024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Romling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 46. Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross P, et al. 1986. Control of cellulose synthesis Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101–117 [Google Scholar]
- 48. Ryan RP, et al. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49. Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 50. Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sebaihia M, et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 53. Seshasayee AS, Fraser GM, Luscombe NM. 2010. Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res. 38:5970–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shang F, et al. 2009. The Staphylococcus aureus GGDEF domain-containing protein, GdpS, influences protein A gene expression in a cyclic diguanylic acid-independent manner. Infect. Immun. 77:2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimizu T, et al. 2002. Complete genome sequence of Clostridium perfringens, an anerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 57. Simm R, Morr M, Remminghorst U, Andersson M, Romling U. 2009. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 386:53–58 [DOI] [PubMed] [Google Scholar]
- 58. Skarin H, Håfström T, Westerberg J, Segerman B. 2011. Clostridium botulinum group III: a group with dual identity shaped by plasmids, phages and mobile elements. BMC Genomics 12:185 doi:10.1186/1471-2164-12-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith CJ, Markowitz SM, Macrina FL. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sommerfeldt N, et al. 2009. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331 [DOI] [PubMed] [Google Scholar]
- 61. Stabler RA, et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102 doi:10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Starnbach MN, Lory S. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459–469 [DOI] [PubMed] [Google Scholar]
- 63. Sudarsan N, et al. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tamayo R, Schild S, Pratt JT, Camilli A. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect. Immun. 76:1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thormann KM, et al. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tischler ADa AC. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Twine SM, et al. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vial PA, et al. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 158:70–79 [DOI] [PubMed] [Google Scholar]
- 73. Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.