Abstract
The Entner-Doudoroff (ED) pathway has recently been shown to play an important role in sugar catabolism for many organisms although very little information is available on the functionality of this pathway in Vibrio cholerae, the causative agent of cholera. In this study, activation of the genes edd and eda, encoding 6-phosphogluconate dehydratase and 2-keto-3-deoxy-6-phosphogluconate aldolase, was used as a marker of a functional ED pathway in V. cholerae. Transcriptional activation analyses and gene silencing experiments with cells grown in sugar-supplemented M9 medium demonstrated that the ED pathway is functional in V. cholerae and is obligatory for gluconate catabolism. Importantly, selective activation of the ED pathway led to concurrent elevation of transcripts of prime virulence genes (ctxA and tcpA) and their regulator (toxT). Further, lowering of these transcript levels and cholera toxin production in vitro by an ED pathway-defective mutant (strain N16961 with a Δedd mutation [ΔeddN16961 strain]) suggested the importance of this pathway in regulating V. cholerae virulence. The in vivo relevance of these data was established as the mutant failed to colonize in suckling mice intestine or to induce fluid accumulation in ligated rabbit ileal loops. Activation of the ED pathway in V. cholerae was shown to inhibit biofilm formation in vitro that could be reversed in the mutant. As further support for these results, comparative transcriptome analysis with cells grown in the presence of glucose or gluconate revealed that a functional ED pathway led to activation of a subset of previously reported in vivo expressed genes. All of these results suggest the importance of the ED pathway in V. cholerae pathogenesis.
INTRODUCTION
Carbohydrates play an important role in the assimilation of energy by living cells and act as carbon sources for the synthesis of important metabolites. The breakdown of sugars through different catabolic pathways provides energy to the bacterial cells in the form of ATP and other reducing equivalents. The best-characterized pathways for sugar catabolism in bacteria are the Embden-Meyerhof-Parnas (EMP), the pentose phosphate (PP), and the Entner-Doudoroff (ED) pathways. These pathways are widespread in prokaryotes and are crucial for their physiology and nutrition. The overall schemes of the ED and EMP pathways are quite similar: 6-carbon sugars are primed by phosphorylation and subsequently cleaved by the aldolase enzyme into two 3-carbon intermediates (27). The two key enzymes distinctive to the ED pathway are (i) 6-phosphogluconate dehydratase (Edd) (EC 4.2.1.12), which catalyzes dehydration of 6-phosphogluconate to form 2-keto-3-deoxy-6-phosphogluconate (KDPG), and (ii) KDPG aldolase (Eda) (EC 4.1.2.14), which cleaves KDPG to pyruvate and glyceraldehyde 3-phosphate, the latter being further catabolized through the EMP pathway and tricarboxylic acid (TCA) cycle (6). The ED pathway was discovered in 1952 in Pseudomonas saccharophila (9) and several years later was shown to be present in Escherichia coli (8). Initially, the ED pathway was considered to be restricted to Gram-negative bacteria, but current studies indicate that it is widely distributed from Archaea to Eukarya (6). It is now believed that the ED pathway predates the EMP pathway in the evolution of microbes (30).
The enzymes Edd and Eda of the ED pathway play key roles in gluconate (Gnt) catabolism. Gnt, a sugar acid which is available in the intestinal milieu, can be utilized by various microorganisms as an energy source. The organism E. coli utilizes Gnt through the ED pathway (27). This bacterium has high-affinity (GntT) and low-affinity (GntU) Gnt transporters and a thermoresistant gluconokinase enzyme (GntK) (EC 2.7.1.12) involved in phosphorylation of Gnt. Catabolism of Gnt via the ED pathway in the organism is controlled by the repressor protein GntR. The edd, eda, gntT, gntU, and gntK genes are crucial for functioning of the ED pathway, and their expression is negatively regulated by gntR, whereas Gnt acts as a true inducer (26). Recent studies have suggested that the ED pathway may play an important role in the physiology of E. coli bacteria toward their adaptation specific for the intestinal milieu (4). Apart from E. coli, the ED pathway may be important in the survival of pathogenic organisms like Salmonella enterica, Neisseria gonorrhoeae, Klebsiella pneumoniae, Helicobacter pylori, Pseudomonas aeruginosa, Legionella pneumophila, Pasteurella pestis, Xanthomonas campestris, Pectobacterium carotovorum, etc. in their respective host systems (2, 3, 10, 15, 20, 21, 23, 24, 31, 33).
The Gram-negative gammaproteobacterium Vibrio cholerae, the etiological agent of the diarrheal disease cholera, has a life cycle that includes its existence in intestinal (in humans) as well as extraintestinal (marine aquatic) environments, preferably by forming biofilms (12). Clearly, the adaptation of the organism in such distinct environments would require appropriate metabolic pathways to be functional and efficient to derive energy from diverse nutrient sources. To date, our knowledge is quite limited in understanding the physiological changes that V. cholerae adopts during the transition between the two extreme environments and their pathological consequences. An analysis of genomic data of V. cholerae has revealed the presence of a Gnt utilization system including the ED pathway genes with a unique arrangement (29). The present study was undertaken with the following objectives: (i) to determine whether the ED pathway is functional in V. cholerae, (ii) to determine whether activation of the ED pathway has any role in the expression of virulence genes in vitro, (iii) to ascertain the relevance of a functional ED pathway toward the expression of its pathogenicity in vivo, and (iv) to evaluate the role of a functional ED pathway in biofilm formation. The significance of our results was analyzed and discussed using a set of comparative microarray-based gene expression data generated with the organism grown separately in the presence of glucose (Glu) or Gnt as the sole carbon source.
MATERIALS AND METHODS
Bacteriology.
Strains of V. cholerae O1 classical (O395) and El Tor (N16961) biotypes were included in the study. Studies were carried out with V. cholerae strains grown in M9 minimal medium supplemented individually with 0.2% concentrations of different sugars such as Glu, N-acetylglucosamine (GlcNAc), Gnt, and ribose and incubated at 37°C for 12 h.
Real-time PCR assay.
V. cholerae strains were grown in minimal medium supplemented with different sugars. Total RNA was extracted by using the TRIzol method. Reverse transcriptase PCR was performed with 4 μg of total RNA as the template with random hexamers and MultiScribe reverse transcriptase (high-capacity cDNA synthesis reverse transcriptase kit; Applied Biosystems). The cDNA thus synthesized was subsequently used for a SYBR green-based real-time PCR assay performed in an ABI 7900 instrument using gene-specific primers that included genes for metabolic activities, virulence factors, regulatory elements, and certain other genes of interest (see Table S1 in the supplemental material). The transcript levels of recA served as an internal control. All of the real-time PCR assays were performed with total RNA (before cDNA synthesis), which showed no amplification, and this was to exclude the possibility of DNA contamination in the total RNA as extracted from the cells.
GM1 ELISA for CT estimation.
The ability of V. cholerae strains to express cholera toxin (CT) in vitro was assayed by GM1 enzyme-linked immunosorbent assay (ELISA) using cell-free culture supernatants of cells grown for 12 h(14). For each set of experiments, a known amount of purified CT was used to generate a standard curve showing the interrelationship between the optical density at 492 nm (OD492) and the amount of CT. The OD492 average obtained from triplicate wells of the experimental sets was considered to estimate the amount of CT present in the culture supernatant using the standard curve (7).
Development of plasmid constructs for antisense-mediated inhibition.
An antisense strategy was utilized to inhibit functional activity of genes encoding 6-phosphofructokinase (pfk), 6-phosphogluconate dehydratase (edd), and 6-phosphogluconate dehydrogenase (gnd), key enzymes of the EMP pathway, ED pathway, and PP pathway, respectively. PCR amplicons specific for pfk, edd, and gnd were initially cloned into the vector pCR2.1. Cloned pfk, edd, and gnd were subsequently subcloned into the EcoRI site of a modified pBAD vector to generate plasmid constructs pTPApfk, pTPAgnd, and pTPAedd, respectively. Partial deletion was introduced into the catabolite activator protein (CAP) binding site of pBAD using a QuikChange II site-directed mutagenesis kit (Stratagene) to generate a modified pBAD, which was used for generating antisense constructs. The genes pfk, gnd, and edd present in pTPApfk, pTPAgnd, and pTPAedd, respectively, existed in reverse orientation relative to the arabinose-inducible promoter. These constructs were electroporated into the V. cholerae N16961 strain, and the effect of conditional as well as specific gene silencing was evaluated using minimal medium supplemented with or without the inducer arabinose. The enzyme encoded by edd converts 6-phosphogluconate to KDPG, which is subsequently converted to pyruvate and glyceraldehyde 3-phosphate by the action of the KDPG aldolase encoded by eda. Deactivation of eda will therefore lead to accumulation of KDPG within the cells. It has been reported earlier that accumulated KDPG at higher concentrations resulted in bacteriostasis (11). Thus, we selected edd but not eda for antisense suppression assays.
Generation of an in-frame deletion mutant of edd.
Construction of a Δedd V. cholerae mutant was generated by an allelic exchange procedure. For this, an internal deletion was introduced in the edd gene by a PCR fragment-joining method, and the resulting amplicon was cloned into SacI sites of a suicide vector, pDS132 (chloramphenicol resistant [Cmr]) (28), to generate the suicidal construct pTPedd, which can replicate only in the presence of pir protein. The suicidal construct pTPedd was mobilized into recipient V. cholerae strains by conjugal transfer, and the transconjugants were selected on plates containing 7 μg/ml chloramphenicol and 100 U/ml polymyxin B. Selection of these transconjugants on the Cm-containing plates caused integration of pTPedd into the V. cholerae genome by homologous recombination in the edd locus. These integrants were subsequently passaged on plates containing 10% sucrose to generate an in-frame deletion mutant of edd (due to removal of the vector backbone along with a functional copy of edd). Successful removal of the integrated vector backbone led to Cm sensitivity in the Δedd mutant of strain N16961 (ΔeddN16961 strain). In-frame deletion of edd was confirmed by nucleotide sequencing. Deactivation of edd resulted in a nonfunctional ED pathway in this mutant. The ΔeddN16961 mutant was further modified by electroporation with the antisense plasmid construct pTPApfk to create a double mutant with a defective ED pathway and EMP pathway under arabinose-inducible conditions.
Animal experimentations.
The potential of the V. cholerae strains to colonize the intestine was evaluated using the suckling mouse model. Following 18 h of challenge, animals were sacrificed, and their intestines were removed, washed, and homogenized in normal saline. The challenge dose and the intestinal homogenates were serially diluted and plated on Luria agar plates to determine the number of CFU. The colonization potential was determined as the ratio of the number of CFU recovered to the number of CFU used for oral challenge (5). The overall virulence potential of V. cholerae strains was also determined by a rabbit ileal loop (RIL) assay. After 18 h of challenge, rabbits were killed, and the fluid accumulation (FA) ratio was determined by measuring the amount of fluid accumulated per unit length of the loop. Reduction of the FA ratio served as an index of the attenuation of the virulence phenotype.
Biofilm assay.
The biofilm-forming capacities of the V. cholerae strains were determined by crystal violet staining after 18 h of incubating the culture in borosilicate glass tubes at 37°C under static conditions (5) in minimal medium supplemented with either Glu, Gnt, or GlcNAc. Absorbed dye in the attached cells was solubilized in dimethyl sulfoxide (DMSO) and the OD570 was measured spectrophotometrically (18).
Gene expression profiling.
The V. cholerae O1 El Tor strain N16961 was grown in minimal medium supplemented with either 0.2% Glu or 0.2% Gnt for 12 h, and total RNA was isolated using TRIzol reagent (Invitrogen), treated with RNase-free DNase I (Ambion), and finally purified with the use of an RNeasy kit (Qiagen). The microarray experiment was performed by Genotypic Technology, India. Polyadenylation of the bacterial RNA samples using Epicentre's A-plus Poly(A) polymerase tailing kit was followed by T7 oligo(dT)-based cDNA synthesis and subsequent amplification and labeling by in vitro transcription using Cy3-CTP, Agilent's low-input linear amplification kit. The normalization of the signals generated after hybridization was done using GeneSpring GX, version 11.0, software, and final data were obtained after normalization using the signals from cells grown Glu-supplemented medium.
Statistical analysis.
Comparative real-time PCR analysis of the transcript levels of various genes was analyzed by the INSTAT2 program, and P values were calculated using a paired t test.
RESULTS
Catabolic pathway preferences for the utilization of Gnt and GlcNAc by V. cholerae.
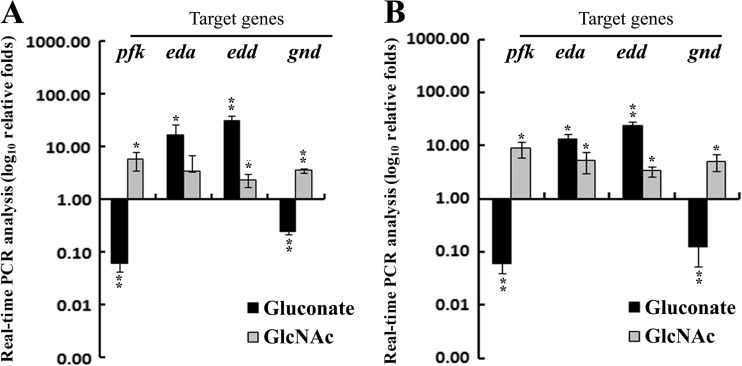
Relative transcript levels of pfk, gnd, eda, and edd were evaluated by real-time PCR assays using cells grown in M9 minimal medium supplemented individually with either Glu or Gnt or GlcNAc, which served as an indicator for involvement of the EMP, PP, or ED pathway, respectively. The real-time PCR results were normalized to the transcript level of recA, and the respective transcript levels in cells grown in the M9 medium with Glu supplementation (M9-Glu) served as a reference (Fig. 1). Data generated with strains belonging to El Tor (N16961) and classical (O395) biotypes showed elevated levels of both the eda and edd marker genes for the ED pathway in cells grown in M9 medium with Gnt supplementation (M9-Gnt) and lower transcript levels of the pfk and gnd genes, markers for the EMP and PP pathways, respectively, compared to cells grown in M9-Glu medium. Supplementation in M9 medium with GlcNAc (M9-GlcNAc) caused elevation of pfk, gnd, eda, and edd transcript levels compared to cells grown in M9-Glu medium.
Fig 1.
Relative transcript levels were measured by real-time PCR assay in V. cholerae strains of El Tor (N16961) (A) and classical (O395) (B) biotypes grown separately in M9 minimal medium supplemented with a 0.2% concentration of either glucose (Glu) or gluconate (Gnt) or N-acetylglucosamine (GlcNAc) as a sole carbon source. Normalization of the transcript levels was made against cells grown with Glu as a reference. Error bars show standard deviations for triplicate samples. pfk, 6-phosphofructokinase; eda, 2-keto-3-deoxy-6-phosphogluconate aldolase; edd, 6-phosphogluconate dehydratase; gnd, 6-phosphogluconate dehydrogenase. *, 0.005 > P < 0.05; **, P ≤ 0.005. Activation of pfk, eda and edd, and gnd has been considered an index for functional activity of the EMP, the ED, and the PP pathways, respectively.
Antisense-mediated suppression of pathway-specific marker genes and its effect on V. cholerae grown in sugar-supplemented medium in vitro.
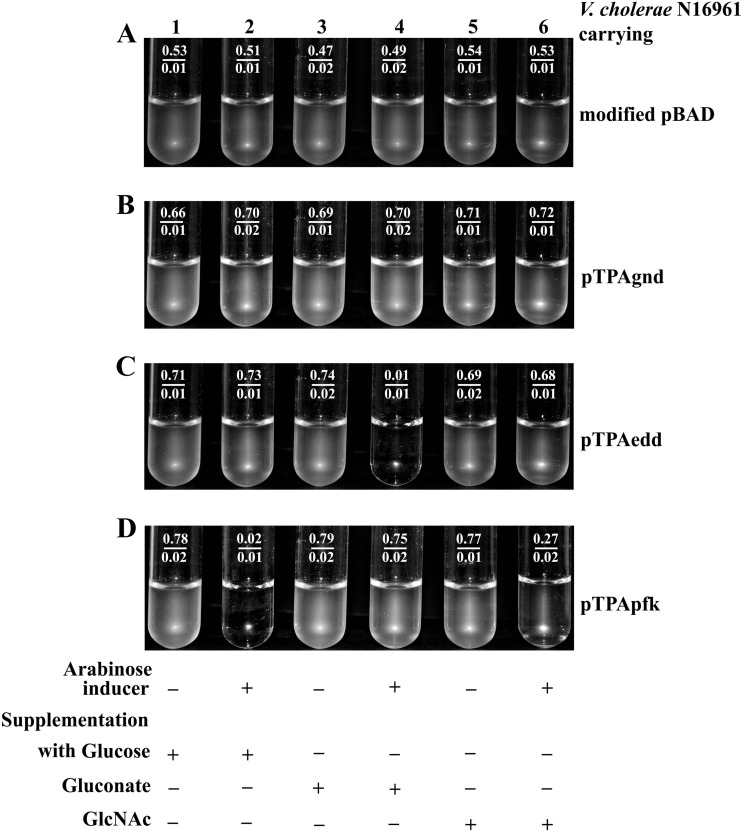
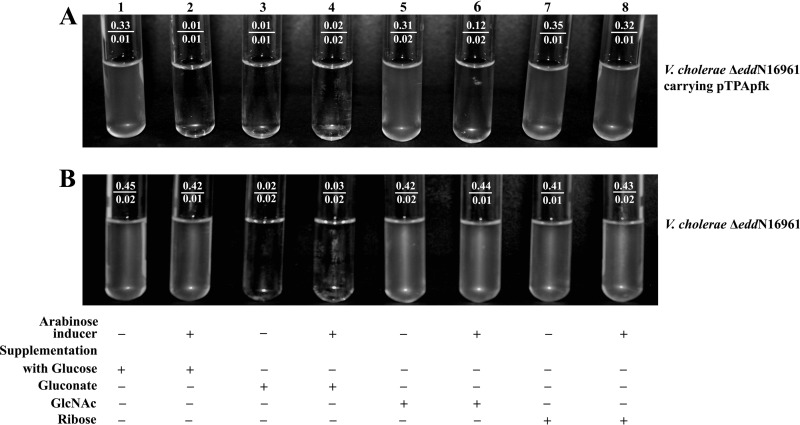
Antisense-mediated suppression of pathway-specific marker genes (pfk, edd, and gnd) was performed by the introduction of the appropriate construct (pTPApfk, pTAAedd, and pTPAgnd) into V. cholerae El Tor strain N16961. Expression of antisense RNA was controlled by an arabinose-inducible promoter for all of these constructs. As a negative control, V. cholerae N16961 carrying only a modified pBAD vector was also included in this assay. Figure 2A shows that vector alone did not confer any effect on the growth characteristics of a V. cholerae strain grown in minimal medium supplemented with different sugars and in the presence or absence of the inducer arabinose. V. cholerae strains carrying an antisense construct were allowed to grow in the M9 minimal medium with or without the inducer arabinose to evaluate functional involvement of different pathways, if any. A V. cholerae strain carrying pTPAedd, antisense to edd, failed to grow in M9-Gnt medium in the presence of the inducer arabinose (Fig. 2C, tube 4) (final OD600 of 0.01 from an OD600 of 0.01 due to the addition of inoculum). Arabinose-induced antisense pTPAedd caused no growth in M9-Gnt medium but failed to show any growth-inhibitory effect in either M9-Glu or M9-GlcNAc medium. Similarly, a strain carrying pTPApfk, antisense to pfk, showed no growth in M9-Glu medium with the inducer arabinose (Fig. 2D, tube 2) (final OD600 of 0.02 from an OD600 of 0.01). Arabinose-induced pTPApfk-mediated growth inhibition was observable in M9-GlcNAc medium (Fig. 2D, tube 6) (final OD600 of 0.27 from an OD600 of 0.02), but no such effect was noted in medium supplemented with Gnt. The construct pTPAgnd failed to show any growth-inhibitory effect in minimal medium supplemented with Glu or Gnt or GlcNAc, even in the presence of the inducer arabinose (Fig. 2B). The above findings established involvement of the ED and EMP pathways in V. cholerae for Gnt and Glu utilization, respectively. Assessment of the involvement of the PP pathway was further addressed through growth experiments using a ΔeddN16961 mutant carrying pTPApfk (a double mutant due to a defective ED pathway and an arabinose-inducible defective EMP pathway) in M9 medium supplemented individually with all of the above sugars as well as with ribose, a pentose sugar. Results are presented in Fig. 3. As expected no growth was observed in M9 medium supplemented with Gnt (Fig. 3A, tubes 3 and 4). A conditional double mutant showed good growth in M9 medium supplemented with either Glu or GlcNAc or ribose in the absence of the arabinose inducer (Fig. 3A, tubes 1, 5, and 7). Induction of pfk antisense caused no growth in M9 medium supplemented with Glu (Fig. 3A, tube 2) (final OD600 of 0.01 from an OD600 of 0.01). The observed absence of growth in either Gnt-supplemented medium or under arabinose-induced conditions for Glu-supplemented medium confirmed nonfunctional ED and EMP pathways under arabinose-induced conditions. This mutant grew very well in M9-GlcNAc medium (Fig. 3A, tube 5) but showed growth inhibition under arabinose-inducible conditions (Fig. 3A, tube 6) (final OD600 of 0.12 from an OD600 of 0.02). These data indicated that a conditional double mutant can utilize GlcNAc through the PP pathway with growth suppression. Studies with ribose as a supplement in M9 medium further confirmed that the PP pathway is operative in V. cholerae as the conditional double mutant showed good growth in M9-ribose medium with or without the inducer arabinose (Fig. 3A, tubes 7 and 8).
Fig 2.
Growth characteristics in medium supplemented with Glu (tubes 1 and 2), Gnt (tubes 3 and 4), and GlcNAc (tubes 5 and 6) for V. cholerae strain N16961 carrying a modified pBAD vector or carrying the antisense construct pTPAgnd, pTPAedd, or pTPApfk to suppress gnd, edd, or pfk, respectively, under arabinose-inducible conditions (even-numbered tubes). Growth of V. cholerae is represented by OD600 values, as shown in the numerator in each of the tubes. OD600 values in the denominator represent baseline values due to the addition of inoculum. The presence (+) and absence (−) of either inducer or sugar supplements are indicated.
Fig 3.
Growth characteristics in medium supplemented with Glu (tubes 1 and 2), Gnt (tubes 3 and 4), GlcNAc (tubes 5 and 6), and ribose (tubes 7 and 8) for an isogenic in-frame edd deletion mutant of N16961 (ΔeddN16961) carrying pTPApfk (A) and the ΔeddN16961 strain (B). Antisense construct pTPApfk suppresses pfk under arabinose-inducible conditions (even-numbered tubes). Growth of V. cholerae is represented by OD600 values shown in the numerator in each of the tubes. OD600 values in denominator represent baseline values due to the addition of inoculum. The presence (+) and absence (−) of either inducer or sugar supplements are indicated.
Expression of virulence factors in V. cholerae cells grown under different culture conditions.
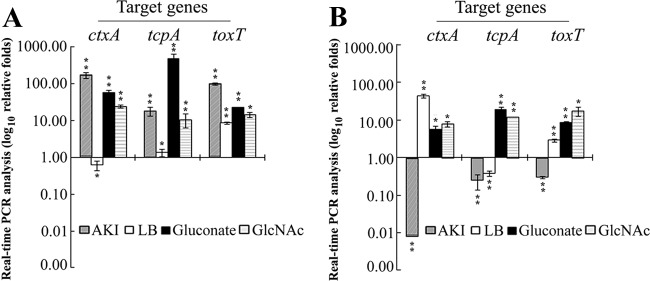
Real-time PCR-based analysis was made on relative expression of transcripts of ctxA, tcpA, and their regulatory element toxT in V. cholerae strains grown for 12 h at 37°C in M9 medium supplemented with either Glu or Gnt or GlcNAc. Elevation in the transcripts of the virulence genes (ctxA and tcpA) and of their regulator toxT was evident in cells grown in M9-Gnt and M9-GlcNAc media compared to cells grown in M9-Glu medium (Fig. 4). Similar data were also generated with V. cholerae cells grown in Luria broth (LB), pH 6.5, at 30°C or in AKI medium (0.5% NaCl, 0.4% yeast extract, 1.5% Bacto peptone, and 0.3% NaHCO3) at 37°C for a comparison. As expected, elevation of ctxA transcripts was observed in AKI medium but not in LB for El Tor strain N16961 (Fig. 4A) while classical strain O395 showed elevation of ctxA transcripts in LB but not in AKI medium (Fig. 4B). The CT production abilities of the V. cholerae strains were evaluated in cell-free culture supernatants by growing the cells for 12 h under various culture conditions. The El Tor biotype strain N16961 produced more CT in AKI medium at 37°C than in LB, pH 6.5, at 30°C, and this correlated well with relative transcript levels of ctxA under the respective culture conditions (Table 1 and Fig. 4A). The classical biotype strain O395 produced the largest amount of CT with growth in LB (pH 6.5) at 30°C, a condition which also produced a significant increase in ctxA transcripts (Table 1 and Fig. 4B). Strains of both of the biotypes produced a good amount of CT in M9 medium supplemented with Gnt but not with Glu or GlcNAc (Table 1). However, detection of comparative levels of ctxA transcripts was evident when cells were grown in M9 medium supplemented with either Gnt or GlcNAc (Fig. 4). Apart from the effect of medium and the supplements, the major reason for discrepancies between ctxA transcript levels and CT estimation may be due to certain inherent basic differences between these two assays. The real-time PCR-based detection of ctxA transcripts reflected relative amounts of the transcripts present at the 12-h growth time point as mRNAs are known to be very unstable in nature. On the other hand, cell-free CT estimation in the spent medium reflected the cumulative effect of CT production over a 12-h growth period.
Fig 4.
Relative transcripts levels of virulence genes were measured by real-time PCR assay in V. cholerae strains of El Tor (N16961) (A) and classical (O395) (B) biotypes grown under different culture conditions for 12 h. Media and conditions used are as follows: AKI medium, pH 7.4 at 37°C; LB, pH 6.5 at 30°C; M9 medium supplemented with a 0.2% concentration of either Glu or Gnt or GlcNAc at 37°C. Transcript levels were normalized to those of cells grown in M9-Glu medium. Error bars show standard deviations for triplicate assays. *, 0.005> P < 0.05; **, P ≤ 0.005.
Table 1.
Ability of V. cholerae O1 strains of classical (O395) and El Tor (N16961) biotypes to produce CT under various cultural conditions and assayed using cell-free culture supernatant by GM1 ELISA method
| V. cholerae strain | CT amt (ng ml−1 OD600 unit−1) by culture conditiona |
||||
|---|---|---|---|---|---|
| LB, pH 6.5, 30°C | AKI medium, pH 7.4, 37°C | M9-Glu medium, 37°C | M9-Gnt medium, 37°C | M9-GlcNAc medium, 37°C | |
| O395 | 835 ± 27** | 93 ± 24** | 32 ± 7 | 150 ± 40* | 39 ± 1* |
| N16961 | 22 ± 3* | 569 ± 26** | 13 ± 1 | 163 ± 19** | 26 ± 3** |
Values are means ± standard deviations.
, 0.005 > P < 0.05;
, P < 0.005.
Effect of edd deletion (defective ED pathway) on V. cholerae pathogenesis.
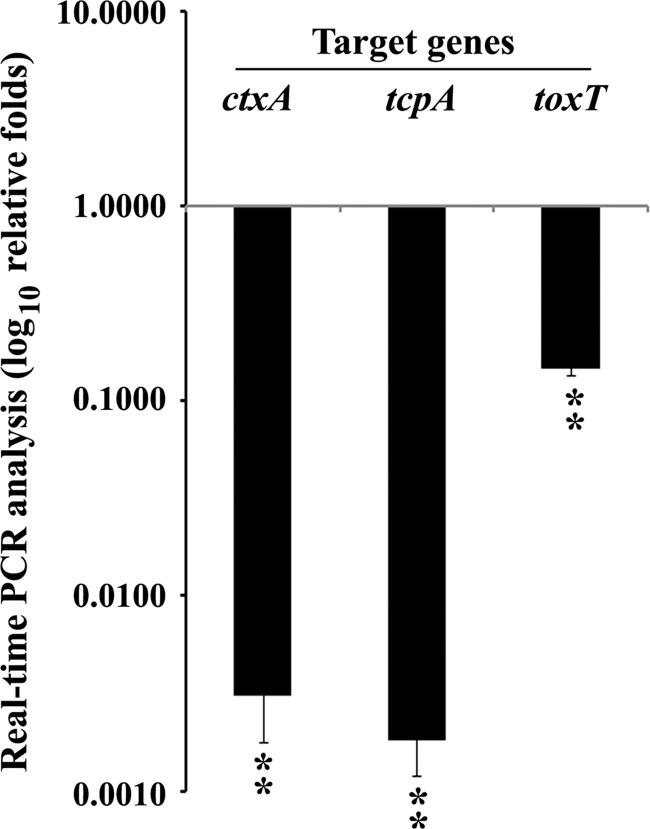
Genetic manipulation studies were carried out to generate an in-frame edd deletion (ED pathway-defective) mutant of V. cholerae El Tor biotype strain N16961. An in-frame edd deletion mutant (ΔeddN16961 strain) failed to grow in M9-Gnt medium but grew well in M9-Glu and M9-GlcNAc media. This evidence confirmed the obligatory involvement of the ED pathway in Gnt utilization by V. cholerae. Transcriptional differences of the virulence genes such as ctxA, tcpA, and their regulatory element toxT were estimated between the wild-type and the mutant strains grown in AKI medium at 37°C. Significantly decreased levels of the transcripts for ctxA, tcpA, and toxT were evident in the Δedd mutant strain compared to wild-type levels (Fig. 5). These data corroborated the observed decrease in CT production by the ΔeddN16961 strain compared to the wild type (Table 2). The in vivo experiments with the ΔeddN16961 mutant showed severe attenuation of fluid accumulation in the ligated RIL compared to its isogenic wild-type strain (Table 2). Animal experiments also showed that the mutant failed to colonize in the suckling mouse intestine.
Fig 5.
Real-Time PCR-based assay for a comparative analysis of the transcript levels between O1 El Tor wild-type V. cholerae strain N16961 and an isogenic in-frame edd deletion mutant (ΔeddN16961 strain) when grown in AKI medium, pH 7.4, at 37°C. Transcript levels were normalized to strain N16961 grown under identical conditions. Error bars show standard deviations for triplicate assays. **, P ≤ 0.005.
Table 2.
Comparison of functional activities between a wild-type (N16961) strain and its isogenic in-frame edd deletion mutant (ΔeddN16961 strain)
| V. cholerae strain | Growth in M9-Gnt mediuma | Phenotypeb |
||
|---|---|---|---|---|
| CT amt (ng ml−1 OD600 unit−1)c | RIL fluid accumulation (ml of fluid cm−1 of IL)d | Biofilm production (OD570)e | ||
| N16961 | + | 569 ± 26 | 1.46 ± 0.12 | 0.51 ± 0.30 |
| ΔeddN16961 strain | − | 118 ± 37** | 0.07 ± 0.02* | 0.96 ± 0.24** |
+, positive growth; −, no growth.
Values are means ± standard deviations. *, 0.005 > P < 0.05; **, P < 0.005.
V. cholerae strains were grown in AKI medium, pH 7.4, at 37°C for 12 h. The amount of CT produced by the strains was measured by GM1 ELISA.
Fluid accumulation was measured in the RIL assay. IL, intestinal loop.
Retention of crystal violet dye by V. cholerae strains that formed biofilm on an abiotic surface was estimated at 570 nm.
Role of the ED pathway in biofilm formation.
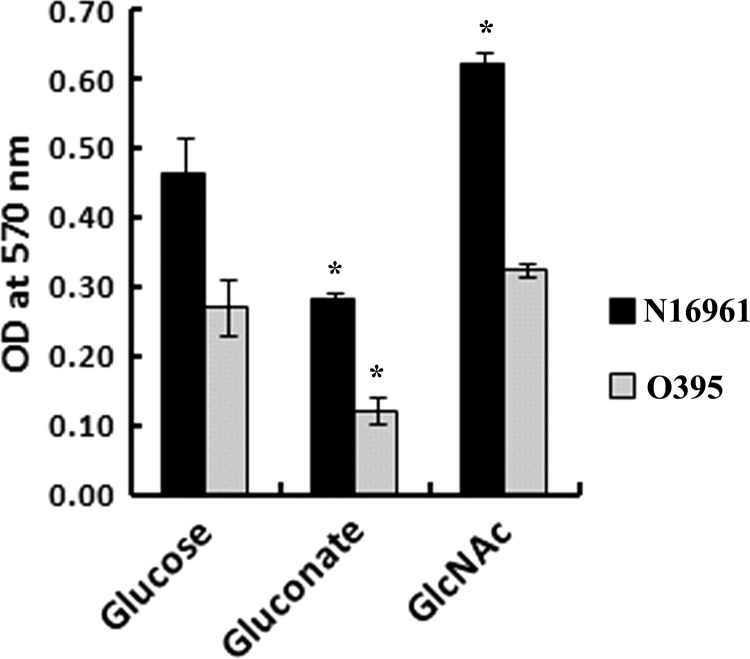
The culture condition-dependent biofilm-forming ability of V. cholerae strains was assayed by growing the cells in minimal medium supplemented with either Glu or Gnt or GlcNAc at 37°C under static conditions. The dye retention capacity of the adhered cells served as an index for the extent of biofilm formation, which was measured in DMSO-solubilized cells at 570 nm. OD570 values obtained under different culture conditions are presented in Fig. 6. V. cholerae strains produced biofilms, but the extent of biofilm formation varied with the culture conditions, as evident by the various OD570 values. The OD570 for El Tor biotype strain N16961 was higher than that of the classical biotype strain O395 (Fig. 6), which is in accordance with the reported high biofilm formation ability of El Tor strains compared to classical strains. Biofilms formed by the strains grown in M9-Gnt and M9-GlcNAc supplemented media were compared with cells grown in M9-Glu supplemented medium a as reference. Comparative analysis showed that GlcNAc supplementation caused significant induction of biofilm formation in the case of strain N16961 (1.33-fold, P < 0.05), while strain O395 showed a 1.18-fold elevation in biofilm formation compared to Glu supplementation. Gnt-supplemented medium, on the other hand, caused a significant (P < 0.05) reduction of biofilm formation (1.67-fold and 2.25-fold lower for N16961 and O395, respectively). Deactivation of the ED pathway by the deletion of edd resulted in an increased ability of the mutant to produce biofilm (Table 2) (P ≤ 0.005), which substantiated the negative impact of a functional ED pathway on biofilm formation by V. cholerae.
Fig 6.
Biofilm formation ability of V. cholerae strains N16961 and O395 when grown in M9 medium supplemented with either Glu or Gnt or GlcNAc at 37°C for 18 h under static conditions. The extent of dye retention capacity (OD570) of the adhered cells served as an index for the extent of biofilm formation by the cells. Error bars show standard deviations for triplicate assays. *, 0.005 > P < 0.05, when estimated against values obtained with cells grown in Glu.
Comparative transcriptome profiles of V. cholerae cells grown in M9-Gnt versus M9-Glu medium.
Genome-wide transcriptional profiles of V. cholerae N16961 were determined by growing the cells in M9-Gnt and M9-Glu media. Data generated through microarray-based hybridization analysis revealed that 2,284 genes were differentially regulated while the transcript levels of 1,625 genes remained unchanged. Among the differentially regulated genes, 1,285 and 999 genes were upregulated and downregulated, respectively, in cells grown in M9-Gnt medium compared to growth in M9-Glu medium. A comparative analysis of selected genes is presented in Table S2 in the supplemental material along with expression profiles adopted from the Gene Expression Omnibus (GEO) database (accession number GSE24405-07) showing V. cholerae comparative in vivo expression profiles. A fold change (increase or decrease) of 0.6 or more (log2 expression ratio) were considered significant. Among 115 selected genes, 69 and 18 genes showed significant upregulation and downregulation, respectively (see Table S2). Real-time PCR assays were also carried out on a subset of 42 genes for validation, which showed that the trend as detected in the microarray experiments remains valid in real-time PCR assays (see Table S2).
Cells grown in M9-Gnt medium showed higher expression levels of the genes involved in Gnt utilization, including gntP, gntK, edd, eda, and gntR. However, the genes involved in the PP pathway, such as tkt, tka, gnd, and zwf, and the genes pgi and pfk involved in the upper part of the EMP pathway were downregulated. Remarkably, the transcription levels of genes like glgA and glgX and debranching enzymes participating in glycogen synthesis for energy storage were also upregulated in cells grown in M9-Gnt medium (see Table S2 in the supplemental material). These trends were further validated through real-time PCR assays carried out using primers specific to pathway-specific key enzymes (see Table S2).
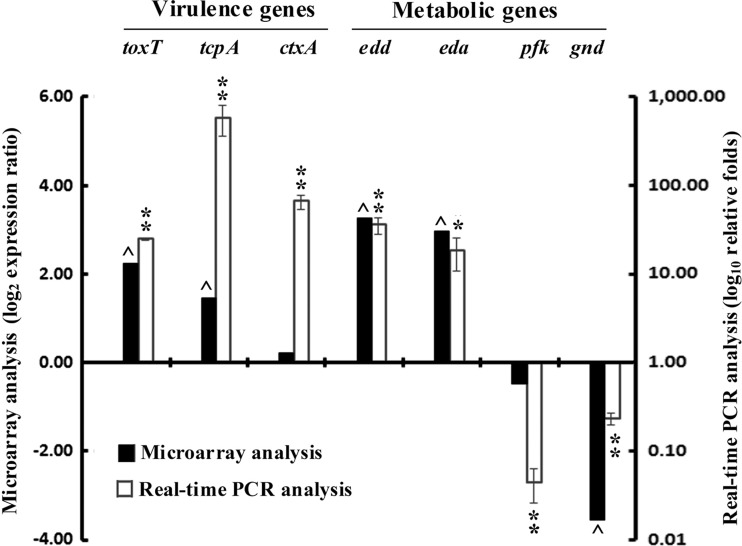
Transcript levels of genes that encode the major toxigenic factors of V. cholerae including ctxB, zot, and rtxA were upregulated in M9-Gnt medium compared to growth in M9-Glu medium. There was a clear trend suggesting that genes responsible for intestinal colonization, like tcpA, mshABCD, acf, and cep, were strongly expressed. Higher transcription levels of the regulatory elements, such as tcpPH, toxR, toxT, tcpT, tcpS, toxS, and aphAB, were evident in cells with a functionally active ED pathway, i.e., in cells grown in M9-Gnt medium compared to those grown in M9-Glu medium. Another virulence factor, ompT, was also transcribed well under M9-Gnt growth conditions. In addition to upregulation of the virulence factors and their regulatory elements, elevated transcript levels were also apparent for genes involved in motility, chemotaxis, and acquisition of iron uptake. These include a set of genes predicted to be involved in chemotaxis, including cheA, cheB, cheW, cheR, and several other methyl-accepting chemotaxis protein-encoding genes. Further, transcript levels of genes such as flgA, fliA, and flaC participating in bacterial motility were upregulated with Gnt supplementation and not with Glu supplementation. The genes participating in iron acquisition, vibriobactin synthesis, iron(II) transport (feoAB), and hemin ABC transporter complex were also upregulated. In addition, other functional genes (cadA, tprG, acnA, gltB1, ppc, thiF, pqiA, sdaC2, Na+ symporter, histidine sensor kinase, etc.) that were reported to be expressed during in vivo growth were also active in vitro under M9-Gnt growth conditions (see Table S2 in the supplemental material). Microarray data on key virulence genes and metabolic genes relative to real-time PCR data are presented in Fig. 7 for comparison as well as for validation.
Fig 7.
Comparative microarray analysis of gene expression (key virulence and metabolic genes) of V. cholerae N16961 grown in M9-Gnt medium compared to growth in M9-Glu medium. Relative expression levels of the genes as determined in microarray experiments are represented by log2 expression ratio. Microarray data have been compared to real-time PCR analysis determined as log10 relative fold. Data were normalized to cells grown with Glu supplementation. Error bars show standard deviations for triplicate real-time assays. *, 0.005 > P < 0.05; **, P ≤ 0.005. A change of gene expression level by a factor of 0.6 or more in microarray analysis is indicated by a caret.
DISCUSSION
Analysis carried out in this study showed that genes for Gnt utilization and the ED pathway were located at different loci in the chromosomes of most of the pathogenic bacteria. The observed ubiquitous presence of the ED pathway in these organisms is an indication of its importance in energy metabolism, along with the involvement of the more efficient EMP pathway. V. cholerae whole-genome analysis revealed that the genes involved in the oxidative phase of the PP pathway are localized in a single operon, but the organization of genes involved in the nonoxidative phase and the EMP pathway is scattered within the two chromosomes. Further analysis revealed variations in the average GC content of the EMP and the PP pathways relative to predicted Gnt utilization systems, including ED pathway genes. The average GC content of most of the genes involves in the EMP and PP pathways is about 50%. On the other hand, the average GC content of the predicted Gnt utilization system including the ED pathway genes is 54%, much higher than that of the genome of V. cholerae, which is 47% (13). The predicted Gnt utilization system in V. cholerae is comprised of two operons, gntP-eda and gntK-edd, which are located in a single cluster adjacent to gntR. The unique organization and monocistronic arrangement of the genes of the ED pathway and Gnt utilization system in the V. cholerae genome indicate that V. cholerae might activate the ED pathway, at least in growth medium with Gnt supplementation.
Results presented in this study establish that the ED pathway is functional in V. cholerae and that utilization of Gnt is mediated through this pathway (Table 2 and Fig. 2). Growth experiments in M9 medium supplemented with sugars also confirmed that Glu is utilized primarily through the EMP pathway (Fig. 2). Growth experiments with a conditional double mutant (ΔeddN16961 strain carrying pTPpfk) further show the existence of a functional PP pathway in V. cholerae, at least with pentose sugar ribose supplementation (Fig. 3A, tubes 7 and 8). Growth experiments with the conditional double mutant showed that deactivation of both the ED and EMP pathways caused growth suppression in GlcNAc-supplemented M9 medium (Fig. 3A, tube 6), but no such effect was apparent in case of deactivation of only the ED pathway. It is evident that GlcNAc utilization can occur through the PP pathway but with less efficiency. All results generated through growth experiments point out the fact that catabolism of Glu and Gnt occurs through the EMP and ED pathways, respectively; V. cholerae utilizes GlcNAc by using all or at least two of the three pathways examined. This is not surprising since this sugar is available in abundance as the breakdown product of the chitinous surface of copepods and other crustacean exoskeletons to which V. cholerae attaches itself for survival in the environment (34). The mucous-rich environment of the intestinal milieu, where pathogenic V. cholerae survives to cause infection in humans (1), is also a rich source of GlcNAc.
Biofilm formation on an abiotic surface is a multistep process and is believed to be essential for the survival of organisms in the environment. Increased levels of biofilm formation were noted in V. cholerae cells grown in the presence of GlcNAc compared to Glu-supplemented medium. A decreased ability to form biofilm was also noted in medium supplemented with Gnt compared to Glu supplementation (Fig. 6). These results suggest that activation of the ED pathway may not be conducive to biofilm formation. Further support for this conclusion is provided by data generated with an edd deletion mutant, where deactivation of the ED pathway led to an increase in the biofilm formation ability of the mutant compared to its wild-type control (Table 2). It may be noted, however, that though biofilm formation aids in the survival of V. cholerae in the environment, this may not be true in the intestine. The biofilm structure may enhance survival of V. cholerae during passage through the gastric acid barrier, but after the bacteria reach the intestine, dispersal of the individual cells from the ingested biofilm clumps is critical to facilitate the colonization process and to establish infection in the gut (36). This may be achieved through the activation of the ED pathway in the intestine, where Gnt is reported to be available from degraded food material, epithelial cells, etc. (27).
A major finding of this study is that the activation of the ED pathway leads to upregulation of the expression of the virulence genes (toxT, tcpA, and ctxA) of V. cholerae that are responsible for its colonization and toxin production (Fig. 4). Interestingly, upregulation of these virulence-associated genes in V. cholerae grown in the presence of Gnt was found to be independent of its biotype (Fig. 4). It may be mentioned here that V. cholerae strains belonging to different biotypes exhibit considerable dependence on their culture conditions in vitro for the expression of CT (16, 32). Although we have not been able to explain the basis of the observed upregulation of CT production by the ED pathway, it would be prudent to assume that the regulation of the activation of virulence genes in V. cholerae may be quite different in complex enriched medium from that in simple defined medium, as used in this study. The above considerations raise an important question regarding the relevance of functional ED pathway data obtained in vitro to the pathogenesis of V. cholerae in vivo. To address this issue, we generated an ED pathway-defective mutant (ΔeddN16961 strain) which resulted in marked attenuation of its ability to express these virulence-associated genes in vitro (Fig. 5). In vivo studies demonstrated that the mutant failed to establish colonization in the suckling mouse model. Additionally, it also failed to cause CT-induced fluid accumulation in the RIL model (Table 2). These results suggest that a functional ED pathway is apparently essential for the expression of the pathogenic potential of V. cholerae strains in vivo.
Further support for the conclusions drawn above was obtained from comparative microarray-based gene expression studies. Whole-genome transcriptome profile analysis showed that most of the virulence genes, their regulatory genes, and the virulence-associated genes (like those involved in flagellar movement, chemotaxis, and iron acquisition, etc.) were upregulated in cells grown in M9-Gnt medium compared to cells grown in M9-Glu medium. Other studies have shown that anaerobic conditions exist within the RIL or in the human intestine and that this might drive V. cholerae to express increased levels of genes involved during anaerobic respiration and in pathogenesis (35). To survive under poorly oxygenated conditions, V. cholerae may drive increased expression levels of the genes that are involved in anaerobic metabolism or respiration. Such an anaerobic condition may give a crucial signal in expressing the virulence factors (17, 22). Studies with V. cholerae grown under in vitro anaerobic conditions showed a reduction in CT production, and this can be attributed to the effect of repression by the H-NS protein (19). But when the pathogenic V. cholerae resides inside the intestine under anaerobic conditions, it produces CT. Our expression profile shows the elevated level of ctxA transcription followed by the downregulation of the gene encoding the regulatory H-NS protein. A subset of genes known to be expressed only under in vivo conditions (25) was transcribed in vitro when cells were grown in M9-Gnt medium. Considering the transcriptional preferences of virulence gene expression, it can be considered that there must be a significant role for the operative ED pathway during the pathogenic cycle of V. cholerae.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the kind support of Dominique Schneider, who provided the pDS132 vector. We acknowledge the expert help of Anuradha Sinha in real-time PCR assays.
This work was supported in part by the Department of Biotechnology (grants BT/PR6918/BRB/10/454/2005) and the Indian Council of Medical Research (TDR/491/2008-ECD-II), Government of India. T.P. is the recipient of Senior Research Fellowships from Indian Council of Medical Research.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1. Almagro-Moreno S, Boyd EF. 2009. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun. 77:3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Britigan BE, Chai Y, Cohen MS. 1985. Effects of human serum on the growth and metabolism of Neisseria gonorrhoeae: an alternative view of serum. Infect. Immun. 50:738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruggemann H, et al. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 8:1228–1240 [DOI] [PubMed] [Google Scholar]
- 4. Chang DE, et al. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chatterjee S, et al. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conway T. 1992. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 9:1–27 [DOI] [PubMed] [Google Scholar]
- 7. De K, et al. 2004. Molecular characterisation of rough strains of Vibrio cholerae isolated from diarrhoeal cases in India and their comparison to smooth strains. FEMS Microbiol. Lett. 232:23–30 [DOI] [PubMed] [Google Scholar]
- 8. Eisenberg RC, Dobrogosz WJ. 1967. Gluconate metabolism in Escherichia coli. J. Bacteriol. 93:941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Entner N, Doudoroff M. 1952. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J. Biol. Chem. 196:853–862 [PubMed] [Google Scholar]
- 10. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 11. Fuhrman LK, Wanken A, Nickerson KW, Conway T. 1998. Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner-Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiol. Lett. 159:261–266 [DOI] [PubMed] [Google Scholar]
- 12. Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]
- 13. Heidelberg JF, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmgren J. 1973. Comparison of the tissue receptors for Vibrio cholerae and Escherichia coli enterotoxins by means of gangliosides and natural cholera toxoid. Infect. Immun. 8:851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hommes RW, Postma PW, Tempest DW, Neijssel OM. 1989. The influence of the culture pH value on the direct glucose oxidative pathway in Klebsiella pneumoniae NCTC 418. Arch. Microbiol. 151:261–267 [DOI] [PubMed] [Google Scholar]
- 16. Iwanaga M, et al. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075–1083 [DOI] [PubMed] [Google Scholar]
- 17. Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J. Bacteriol. 192:4181–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lauriano CM, Ghosh C, Correa NE, Klose KE. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186:4864–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634 [DOI] [PubMed] [Google Scholar]
- 20. Lessie TG, Phibbs PV., Jr 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359–388:359–388 [DOI] [PubMed] [Google Scholar]
- 21. Lu GT, et al. 2009. Glyceraldehyde-3-phosphate dehydrogenase of Xanthomonas campestris pv. campestris is required for extracellular polysaccharide production and full virulence. Microbiology 155:1602–1612 [DOI] [PubMed] [Google Scholar]
- 22. Marrero K, et al. 2009. Anaerobic growth promotes synthesis of colonization factors encoded at the Vibrio pathogenicity island in Vibrio cholerae El Tor. Res. Microbiol. 160:48–56 [DOI] [PubMed] [Google Scholar]
- 23. Mole B, Habibi S, Dangl JL, Grant SR. 2010. Gluconate metabolism is required for virulence of the soft-rot pathogen Pectobacterium carotovorum. Mol. Plant Microbe Interact. 23:1335–1344 [DOI] [PubMed] [Google Scholar]
- 24. Mortlock RP. 1962. Gluconate metabolism of Pasteurellapestis. J. Bacteriol. 84:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Osorio CG, et al. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73:972–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peekhaus N, Conway T. 1998. Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 180:1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peekhaus N, Conway T. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255 [DOI] [PubMed] [Google Scholar]
- 29. Rodionov DA, Mironov AA, Rakhmaninova AB, Gelfand MS. 2000. Transcriptional regulation of transport and utilization systems for hexuronides, hexuronates and hexonates in gamma purple bacteria. Mol. Microbiol. 38:673–683 [DOI] [PubMed] [Google Scholar]
- 30. Romano AH, Conway T. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448–455 [DOI] [PubMed] [Google Scholar]
- 31. Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wanken AE, Conway T, Eaton KA. 2003. The Entner-Doudoroff pathway has little effect on Helicobacter pylori colonization of mice. Infect. Immun. 71:2920–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Q, Dziejman M, Mekalanos JJ. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U. S. A. 100:1286–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.