Abstract
In Escherichia coli, putrescine is metabolized to succinate for use as a carbon and nitrogen source by the putrescine utilization pathway (Puu pathway). One gene in the puu gene cluster encodes a transcription factor, PuuR, which has a helix-turn-helix DNA-binding motif. DNA microarray analysis of an E. coli puuR mutant, in which three amino acid residues in the helix-turn-helix DNA binding motif of PuuR were mutated to alanine to eliminate DNA binding of PuuR, suggested that PuuR is a negative regulator of puu genes. Results of gel shift and DNase I footprint analyses suggested that PuuR binds to the promoter regions of puuA and puuD. The binding of wild-type PuuR to a DNA probe containing PuuR recognition sites was diminished with increasing putrescine concentrations in vitro. These results suggest that PuuR regulates the intracellular putrescine concentration by the transcriptional regulation of genes in the Puu pathway, including puuR itself. The puu gene cluster is found in E. coli and closely related enterobacteria, but this gene cluster is uncommon in other bacterial groups. E. coli and related enterobacteria may have gained the Puu pathway as an adaptation for survival in the mammalian intestine, an environment in which polyamines exist at relatively high concentrations.
INTRODUCTION
Escherichia coli can use various compounds as carbon and nitrogen sources. We previously reported that E. coli can utilize putrescine as a carbon and nitrogen source by the putrescine utilization pathway, in which putrescine is degraded via γ-glutamylated intermediates (7). Putrescine is a polyamine, a class of polycationic biogenic amines that are widely distributed in living organisms, where they are deeply involved in the regulation of cellular functions (15, 23, 24).
The biosynthesis of polyamine has been particularly well studied in E. coli (2). Putrescine is formed either by the decarboxylation of ornithine (catalyzed by SpeC) or by the decarboxylation of arginine into agmatine (catalyzed by SpeA), followed by the hydrolysis of agmatine into putrescine and urea by an agmatine ureohydrolase (SpeB). E. coli can import environmentally available putrescine across the cell membrane, and four putrescine-specific importers have been reported to date. PotE catalyzes both the uptake and excretion of putrescine (4, 5). PotFGHI was reported as an ATP-dependent putrescine transporter that is a member of the ATP-binding cassette (ABC) transporter family (16). PuuP is a member of the Puu pathway and depends on the proton motive force for putrescine import. Of these reported three polyamine importers, only PuuP has been found to be essential for E. coli growth on minimal medium with putrescine as the sole carbon or nitrogen source (11). PlaP (previously named YeeF) was recently discovered and shows a low affinity for putrescine but is essential for the surface motility response induced by extracellular putrescine (10).
Two putrescine utilization pathways, the Puu pathway and the YgjG-YdcW pathway, are known in E. coli. The Puu pathway metabolizes putrescine via γ-glutamylated intermediates and γ-amino butyric acid (GABA) to succinic semialdehyde, a precursor of succinate (7). The YgjG-YdcW pathway metabolizes putrescine to GABA without γ-glutamylation (18, 19). The Puu pathway is thought to be the main putrescine utilization pathway. No influence on E. coli growth with putrescine as a sole carbon or nitrogen source was observed even when ygjG and ydcW were deleted, but the deletion of genes encoding the Puu pathway removed the ability of E. coli to utilize putrescine as a sole carbon or nitrogen source (9).
In the Puu pathway (Fig. 1B), following the import of putrescine by PuuP, PuuA ligates the γ-carboxyl group of glutamic acid to an amino group of putrescine to generate γ-glutamyl-putrescine (9). It is thought that PuuB and PuuC oxidize a second amino group of γ-glutamyl-putrescine to form γ-glutamyl-γ-amino butyric acid via γ-glutamyl-γ-aminobutyraldehyde (7). PuuD hydrolyzes γ-glutamyl-γ-amino butyric acid to glutamic acid and GABA (8). PuuE deaminates GABA to succinic semialdehyde (6). All members of the Puu pathway are encoded by the puu gene cluster, which includes an open reading frame encoding PuuR, a predicted transcriptional regulator (7) (Fig. 1A). GabT, an enzyme 67.4% homologous to PuuE, catalyzes the same reaction as PuuE but does not reside within the puu gene cluster (6, 20). Finally, YneI or GabD, enzymes that are also encoded by genes separate from the puu gene cluster, oxidizes succinic semialdehyde to produce succinic acid (6, 20). In previous studies, the enzymatic activity of PuuD and the transcription of both puuA and puuP were found to be strongly enhanced by the deletion of puuR (7, 9, 11); however, the detailed mechanisms of regulatory functions of PuuR have not been elucidated.
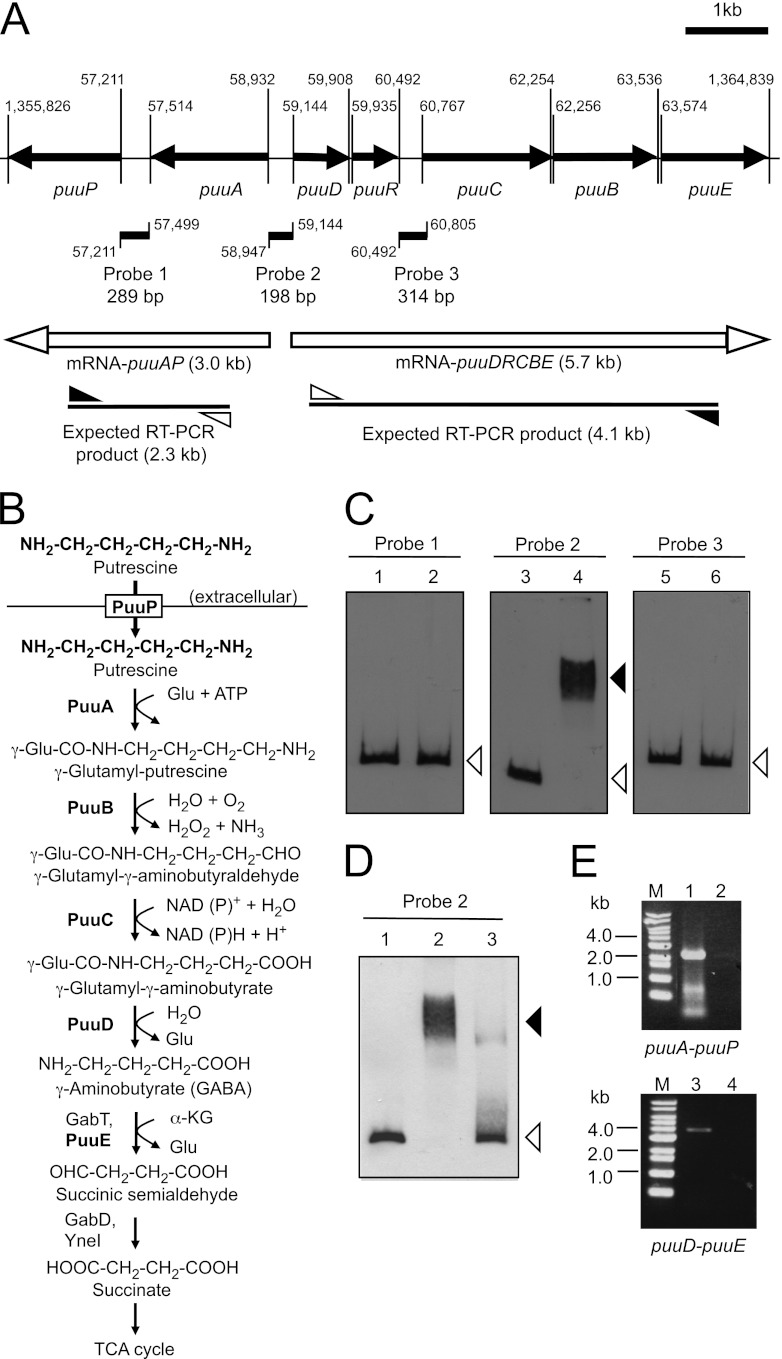
Fig 1.
Gel mobility shift assay to detect PuuR and DNA probe complexes. (A) Depiction of the puu gene cluster and locations of probes, which were designed for gel mobility shift assay at the intergenic regions between puuP and puuA (probe 1), puuA and puuD (probe 2), and puuR and puuC (probe 3). puuR and other puu genes are shown as black arrows with the gene names below. The locations of the three probes are indicated by black lines annotated with the probe names and lengths. Numbers indicate the start and end bases of the probes and open reading frames in the E. coli genome. The numbers omit the first two digits, except for the ends of puuP and puuE. mRNAs expressed from puuA and puuD promoters are labeled with white arrows and the estimated sizes of the transcripts. The expected RT-PCR products from mRNA-puuAP and mRNA-puuDRCBE are labeled with lines and the estimated sizes. White and black triangles indicate forward (puuA-R for mRNA puuAP and puuD-RT1 for mRNA-puuDRCBE) and reverse (puuP-RT1 for mRNA-puuAP and puuE-RT2 for mRNA-puuDRCBE) primers, respectively. (B) The Puu pathway of E. coli. PuuP works as a putrescine importer from extracellular space. PuuA, PuuB, PuuC, and PuuD make up the Puu pathway that degrades putrescine to GABA via γ-glutamylated intermediates. Glu, glutamic acid. α-KG, α-ketoglutarate. (C) Gel mobility shift assay using three probes and His6-PuuR. Assays were conducted using 5% PAGE in 0.5× TBE buffer. Thirty femtomoles (final concentration, 3 nM) of DIG-labeled probe 1, probe 2, and probe 3 was incubated at 4°C for 20 min in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 40 pmol (final concentration, 4 μM) of purified His6-PuuR. Black and white triangles indicate signals of the shifted band and free probe, respectively. (D) Gel mobility shift assay using probe 2 and cell extract from KEI23 (His6-PuuR) and GM13 (His6-PuuR3Ala). Cell extracts were prepared from the cells after 5 h of IPTG induction. For each assay, 3.6 μg cell extract protein was mixed with 30 fmol of DIG-labeled probe 2. Lane 1, absence of cell extract. Lane 2, presence of cell extract with His6-PuuR. Lane 3, presence of cell extract with His6-PuuR3Ala. Black and white triangles indicate signals of the shifted band and free probe, respectively. (E) Transcription unit of the puu genes identified by RT-PCR and agarose gel electrophoresis of RT-PCR products. Total RNA was prepared from strain KEI12, and cDNA was synthesized by an RT reaction using primer puuP-RT1 or puuE-RT2, respectively. PCR was then performed using the primer sets described above with the cDNA as the template. Control reactions without RT enzyme were performed by the same method. Lanes contain the following samples: M, 1-kb DNA ladder (Bioneer); 1 and 2, RT-PCR product and control sample without RT using the puuA-R and puuP-RT1 primer set; 3 and 4, RT-PCR product and control sample without RT using the puuD-RT1 and puuE-RT2 primer set.
In this study, we report the identification of binding sites and recognition motifs for PuuR DNA binding in the puu gene cluster and discuss the mechanisms by which PuuR regulates the Puu pathway and intracellular putrescine concentration.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
All bacterial strains were derivatives of E. coli K-12. The bacterial strains and plasmids used in this study are listed in Table 1. The growth of bacterial cultures was monitored by determining the optical density at 600 nm (OD600) using a UV-1700 spectrophotometer (Shimadzu). M9-tryptone (M9 minimal medium [13], modified in that 1% Bacto tryptone was used instead of 0.2% glucose) (7) was used to grow the E. coli strains from which RNA was extracted. For other experiments, LB medium (Becton, Dickinson and Company) was used. Antibiotics were added at the following concentrations: ampicillin, 50 μg ml−1; kanamycin, 30 μg ml−1; tetracycline, 15 μg ml−1; and chloramphenicol, 30 μg ml−1. P1 transduction, DNA manipulation, and transformation were performed using standard methods (13, 17).
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid name | Genotype | Source |
|---|---|---|
| Strains | ||
| MG1655 | F− prototrophic | C. A. Gross |
| KEI12 | MG1655 except kan+ ymjA+ puuP+A+D+R(3Ala) C+B+E+ pspF+ cat+ | 6 |
| KEI23 | pKEI20/SK212 | This study |
| GM13 | pGM13/SK212 | This study |
| NNE1 | pKJ133/KEI12 | This study |
| NNE3 | pNNB1/KEI12 | This study |
| SH639 | F− Δggt-2 | 22 |
| SK123 | SH639 except ΔpuuDR::kan+ | This study |
| SK167 | SH639 except ΔpuuADR::kan+ | This study |
| SK212 | SH639 except ΔpuuR::kan+ | 6 |
| SK226 | SH639 except ΔpuuA::FRT kan+ FRT | This study |
| Plasmids | ||
| pQE-80L | ColE1 replicon, bla+ lacIq T5p (His)6 | Qiagen |
| pGM05 | pSK103 but puuD+R+ with 266-bp upstream region of puuD | This study |
| pGM06 | pSK103 but puuD+R+ with 214-bp upstream region of puuD | This study |
| pGM07 | pSK103 but puuD+R+ with 193-bp upstream region of puuD | This study |
| pGM08 | pSK103 but puuD+R+ with 156-bp upstream region of puuD | This study |
| pGM09 | pSK103 but puuD+R+ with 100-bp upstream region of puuD | This study |
| pGM13 | pQE-80L but T5p (His)6 puuR(3Ala) | This study |
| pGM14 | pSK103 but puuA+D+R+ with 421-bp upstream region of puuA | This study |
| pGM15 | pSK103 but puuA+D+R+ with 310-bp upstream region of puuA | This study |
| pGM16 | pSK103 but puuA+D+R+ with 229-bp upstream region of puuA | This study |
| pGM17 | pSK103 but puuA+D+R+ with 124-bp upstream region of puuA | This study |
| pGM18 | pSK103 but puuA+D+R+ with 60-bp upstream region of puuA | This study |
| pKEI20 | pQE-80L but T5p (His)6 puuR+ | This study |
| pKJ133 | pUC19 but puuD+R(3Ala) | 6 |
| pKJ134 | pACYC184 but puuP+A+D+R+C+B+E+ | 6 |
| pNNB1 | pUC19 but puuA+D+R(3Ala) with 229-bp upstream region of puuA | This study |
| pNNB2 | pKJ133 but harbors mutation for probe 10 mut1 | This study |
| pNNB3 | pKJ133 but harbors mutation for probe 10 mut2 | This study |
| pNNB4 | pKJ133 but harbors mutation for probe 10 mut3 | This study |
| pNNB5 | pKJ133 but puuA+D+R(3Ala) with 421-bp upstream region of puuA | This study |
| pNNB6 | pSK116 but puuA+D+ΔR with 421-bp upstream region of puuA | This study |
| pSK103 | pUC19 but puuD+R+ | 6 |
| pSK116 | pUC19 but puuD+ΔR | This study |
The plasmid pKEI20, which carries puuR with a His6 tag coding sequence at its 5′ end, under the control of the T5 promoter and two lac operator boxes, was prepared as follows. The puuR gene was amplified by PCR using the primers puuR-1 and puuR-2, which include one BamHI site external to each end of the open reading frame (sequences for the oligonucleotides used in this study are shown in Table 2). The amplified product was digested with BamHI and then cloned into the multiple cloning site of pQE-80L. A 12-amino-acid sequence with His tag of pQE-80L, MRGSHHHHHHGS, was added to the N-terminal end of the PuuR protein to form His6-PuuR protein. The plasmid pKEI20 was transformed into E. coli strain SK212 (ΔpuuR) to obtain strain KEI23.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| antisense1 | TGCTCAATCTCACAAAGTGGA |
| antisense6 | TTTTCTTGCAGAGTCTGGGTC |
| Fc-QC-Fw-CttC3 | GGACTAAATTATCGCCCTTCCTGCGCTTTAAATGACC |
| Fc-QC-Rv-CttC3 | GGTCATTTAAAGCGCAGGAAGGGCGATAATTTAGTCC |
| Fc-QC-Fw-aCa3 | CTCACAAAGTGGACTACATTATCGCCATTACTGCGC |
| Fc-QC-Rv-aCa3 | GCGCAGTAATGGCGATAATGTAGTCCACTTTGTGAG |
| Fc-QC-Fw-tCt | CAAAGTGGACTAAATTCTCGCCATTACTGCGC |
| Fc-QC-Rv-tCt | GCGCAGTAATGGCGAGAATTTAGTCCACTTTG |
| gapA-F | TGAAAGGCGTTCTGGGCTACAC |
| gapA-R | CGAAGTTGTCGTTCAGAGCGATACC |
| GSA-sense2 | CCAGGTAGCGTTTCACTTCC |
| GSA-anti2 | CAAACCAAAGGCGAAGAATC |
| GSA-sense3 | CATGATTCTTCGCCTTTGGT |
| GSA-anti3 | TTGCATACAGATTCGAATGGTGG |
| GSA-sense4 | CCACCATTCGAATCTGTATGC |
| GSA-anti4 | TCAATCTAGCAGTGGGTTTTC |
| GSA-sense5 | GAAAACCCACTGCTAGATTGA |
| GSA-anti5 | TGCTCAATCTCACAAAGTGGA |
| GSA-sense6 | TCCACTTTGTGAGATTGAGCA |
| GSA-anti7 | TTTTCTTGCAGAGTCTGGGTC |
| GSA-probe4.5-sense | AAATGTTAAATTGAGTTTGC |
| GSA-probe4.5-anti | ACTGCGCTTTAAATGACCTT |
| GSA-Probe5.5-sense2 | AAGGTCATTTAAAGCGCAG |
| GSA-Probe5.5-antisense | TCCATGCTCAATCTCACAAA |
| puuA-anti | GCTCTAGATTACGCGTTTTTCAACATCC |
| puuA-sense1 | CGGGATCCAAGATAAATGCCATCGAGTT |
| puuA-sense2 | CGGGATCCATTCAGGTACTTTTCTTGCA |
| puuA-sense3 | CGGGATCCGTTCATTATATTTTCCATGC |
| puuA-sense4 | CGGGATCCGTTTTCATTTTTGCAAACTC |
| puuA-sense5 | CGGGATCCGGTGGTCATTATATTTTACG |
| puuA-F | AAGGAACCGAGAACAGGAACAC |
| puuA-R | GCAATGGATATTCTGGGCAAC |
| puuA-RT2 | GCAATGGATATTCTGGGCAAC |
| puuC-anti-primer | CGCTTTATCCTGCCAGTAAGCC |
| puuC-sense-275 | ATCTTTTTGTTCTGTAAGCCGGGT |
| puuD-RT1 | TGGCTTAGTTGAGGCGGTTAG |
| puuD-RT2 | CGTACTCGCTACTGTTCCATTCC |
| puuD + 60_anti_Afl2 | GGGCTTAAGCCTGTTCCTGCACATTAC |
| puuD-100_sense | CGGGGTACCGCAAAAATGAAAACCCACTG |
| puuD-155_sense | CGGGGTACCCCACCATTCGAATCTGTATGC |
| puuD-190_sense | CGGGGTACCTGTTTTCCGCTCGTTATCAA |
| puuD-220_sense | CGGGGTACCCATGATTCTTVGCCTTTGGT |
| puuD-265_sense | CGGGGTACCGCCTCTCTTCTGACTGCTGAA |
| puuE-RT2 | TGATACGCGGTGTGGGTAAA |
| puuP-anti | TAATTGCTGCCTCCCCTGCG |
| puuP-sense-270-GSA | TTGTTTCATCGGCCTGAGCG |
| puuP-RT1 | ATAAAGATAACGCCACCGTAGACC |
| puuR-1 | GCGGATCCATGAGTGATGAGGGACTGGCGCCAGGAAAA |
| puuR-2 | GCGGATCCTTAAAACGTGGTGGGCGTATGGGCGCTGAT |
| sense1 | GGTTTGTTTTCCGCTCGTTA |
| sense4 | CCACCATTCGAATCTGTATGC |
To obtain pNNB1, a 1.7-kb fragment containing puuA was amplified by PCR using puuA-sense3 and puuA-anti primers and digested with BamHI and XbaI. The 1.7-kb puuA fragment was ligated to the 5.7-kb BamHI and XbaI fragment of pKJ133.
Plasmids harboring different lengths of the upstream regions of puuA and puuD were constructed from pSK103. The upstream region of puuA was amplified with forward primer (puuA-sense1 for pGM14, puuA-sense2 for pGM15, puuA-sense3 for pGM16, puuA-sense4 for pGM17, and puuA-sense5 for pGM18), which was designed at different positions upstream of puuA, adding a BamHI site, and reverse primer (puuA_anti), which was designed from the XbaI site downstream from puuA. The PCR products were digested with XbaI and BamHI and cloned into the 5.7-kb XbaI-BamHI fragment of pSK103. A 1.6-kb EcoRI fragment that includes puuD and partial puuR of pSK103 was subcloned into the EcoRI site of pUC19 to obtain pSK116. pNNB5 and pNNB6 were obtained by cloning a PCR product of puuA-sense1 and puuA-anti into the 5.7-kb XbaI-BamHI fragment of pKJ133 or pSK116. The constructed plasmids were introduced into E. coli strain SK167 (ΔpuuADR). The upstream region of puuD was amplified with forward primer (puuD-265_sense for pGM05, puuD-220_sense for pGM06, puuD-190_sense for pGM07, puuD-155_sense for pGM08, and puuD-100_sense for pGM09), which was designed at different positions upstream of puuD, adding a KpnI site, and reverse primer (puuD + 60_anti_Afl2), which was designed to an AflII site within puuD. The PCR products were digested with KpnI and AflII and cloned into the 4.9-kb KpnI-AflII fragment of pSK103. The constructed plasmids were introduced into strain SK123 (ΔpuuDR).
Overexpression and purification of His6-PuuR.
To overexpress His-tagged PuuR protein, strain KEI23 was grown at 37°C in LB medium containing ampicillin. Cells at an OD600 of 0.4 to 0.8 were treated with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside followed by incubation for 5 h at 37°C. Cells were harvested at 4°C, resuspended in 5 ml of lysis buffer (50 mM HEPES-KOH, pH 7.5, 200 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 1 mM dithiothreitol), and disrupted by sonication. The cell lysate was centrifuged at 20,000 × g at 4°C for 15 min. The supernatant was purified using a HiTrap chelating 1-ml column (GE Healthcare), following the manufacturer's instructions. The purified protein was dialyzed against the lysis buffer and stored at −20°C after the addition of glycerol (final concentration, 30%, vol/vol).
Size exclusion chromatography.
Size exclusion chromatography for purified His6-PuuR was performed using a Superdex 200 HR 10/30 column (GE Healthcare) with gel filtration buffer (50 mM Tris-HCl, pH 7.4, 200 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 1 mM dithiothreitol) at a flow rate of 0.4 ml/min.
DNA gel mobility shift assay.
To visualize binding of PuuR protein to the target DNA region, gel mobility shift assays were performed. Probes were synthesized by PCR using KOD-plus DNA polymerase (Toyobo). We used pKJ134 as the template DNA with the following paired primers: puuP-sense-270-GSA and puuP-anti for probe 1, sense1 and antisense1 for probe 2, puuC-sense-275 and puuC-anti-primer for probe 3, GSA-sense2 and GSA-anti2 for probe 4, GSA-sense3 and GSA-anti3 for probe 5, GSA-sense4 and GSA-anti4 for probe 6, GSA-sense5 and GSA-anti5 for probe 7, GSA-sense6 and GSA-anti7 for probe 8, GSA-probe4.5-sense and GSA-probe4.5-anti for probe 9, and GSA-Probe5.5-sense2 and GSA-Probe5.5-antisense for probe 10. Probes for FC site mutation were obtained using the GSA-Probe5.5-sense2 and GSA-Probe5.5-antisense primer set, with the following template DNA sequences: pNNB2, pNNB3, and pNNB4 for probe 10 mut1, probe 10 mut2, and probe 10 mut3. PCR conditions were as follows: a denaturation cycle at 94°C for 2 min, followed by 30 cycles at 94°C for 15 s, 60°C for 30 s, and 68°C for 1 min. The PCR products were purified by 8% PAGE in 0.5× Tris-acetate-EDTA (TAE) buffer after column purification using the Wizard SV Gel and PCR cleanup system (Promega). The probes were digoxigenin (DIG) labeled by terminal transferase incorporation of DIG-ddUTP (Roche). Gel mobility shift assays followed the protocols of the DIG gel shift kit, 2nd generation (Roche). The DIG-labeled DNA probes were mixed with His6-PuuR protein in 10 μl of binding reaction solution, containing 0.5 μg of poly(dI-dC), 3 μg of bovine serum albumin (BSA), 5 mM MgCl2, and 5× binding buffer, at 4°C for 20 min. The DNA and protein mixtures were subjected to 5% PAGE in 0.5× Tris-borate-EDTA (TBE) buffer (44.5 mM Tris, 44.5 mM boric acid, and 1 mM EDTA). The gel contents were then transferred to an equilibrated positively charged nylon membrane, Immobilon-Ny+ (Millipore), by contact blotting. The luminescence signal was developed in CSPD (disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro) tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate) working solution (Roche) at 37°C for 10 min following the incubation of the nylon membrane with an anti-DIG antibody (Roche). The nylon membrane was exposed to an X-ray film, RX-U (Fuji Film).
Reverse transcriptase PCR (RT-PCR).
Total RNA was isolated using an RNeasy minikit (Qiagen) from strain KEI12, which was grown in M9 with 1% tryptone at an OD600 of 0.5 to 0.8. RNA concentration and quality were evaluated by measuring the OD260 and the 260 nm/280 nm ratio, respectively. cDNA was synthesized using SuperScriptIII reverse transcriptase (Invitrogen) from 1 μg of DNase I (amplification grade; Invitrogen)-treated total RNA using the primers puuP-RT1 for the analysis of mRNA-puuAP and puuE-RT2 for the analysis of mRNA-puuDRCBE. As a control, the same reactions were performed without RT enzyme. PCRs were carried out with KOD-plus DNA polymerase (Toyobo) using puuP-RT1 and puuA-R primers for puuAP and puuE-RT2 and puuD-RT1 primers for puuDRCBE using the following program: 30 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 4 min, executed soon after a denaturation cycle at 94°C for 2 min.
DNase I footprint assay.
DNA segments used for DNase I footprint assay were 32P-labeled with T4 polynucleotide kinase (TaKaRa) with [γ-32P]ATP (3,000 Ci/mmol; PerkinElmer). PCR was performed with 32P-labeled primers antisense1 and sense4 and unlabeled primers sense1 and antisense6 for segments 1 and 2, respectively. Labeled primers were purified using a QIAquick nucleotide removal kit (Qiagen) before the PCR. Labeled PCR products were then purified with the Wizard SV gel and PCR cleanup system (Promega). The PCR program and reactions for binding between DNA segments and purified His6-PuuR were performed in the same way as for the gel mobility shift assays. DNase I (0.25 units) (TaKaRa) was added to each reaction mixture and incubated for 1 min at room temperature prior to the addition of 2.5 μl of 0.5 M EDTA solution to stop the DNase I digestion. Digested DNA fragments were purified twice by phenol-chloroform extraction and ethanol precipitation using Quick-Precip Plus solution (Edge Bio). Dideoxynucleotide sequencing of the region was performed using a Thermo sequenase primer cycle sequencing kit (GE Healthcare) with the corresponding radiolabeled primer. The samples were analyzed on a 6% PAGE gel containing 8 M urea. The gels were dried onto Whatman 3MM paper and exposed to X-ray film RX-U (Fuji Film).
Site-directed mutagenesis of the FC region.
Site-directed mutagenesis of the FC region was performed using the QuikChange method (Stratagene), except that KOD-plus DNA polymerase (Toyobo) was used. Plasmid pKJ134 was used as a template, and the mutagenic primers (Fc-QC-Fw-CttC3 and Fc-QC-Rv-CttC3 for probe 10 mut1, Fc-QC-Fw-aCa3 and Fc-QC-Rv-aCa3 for probe 10 mut2, and Fc-QC-Fw-tCt and Fc-QC-Rv-tCt for probe 10 mut3) were used for the introduction of the mutations. Mutated plasmids were transformed into E. coli strain DH5α, and the presences of the desired mutations were confirmed by DNA sequencing. The plasmid constructs pNNB2, pNNB3, and pNNB4, which harbor mutations for probe 10 mut1, probe 10 mut2, and probe 10 mut3, respectively, were successfully produced.
Primer extension analysis.
RNA was isolated from cells grown at an OD600 of 0.5 to 0.8 using an RNeasy minikit (Qiagen) for strains NNE1 (for puuD) and NNE3 (for puuA), which were grown in M9 with 1% tryptone supplemented with 0.2% putrescine. RNA concentration and quality were evaluated by measuring the OD260 and the 260 nm/280 nm ratio, respectively. The 5′ ends of the transcripts were determined by primer extension analysis. Five micrograms of total RNA was reverse transcribed using SuperScriptIII reverse transcriptase (Invitrogen) with 32P-labeled puuA-RT2 and puuD-RT2 primers for puuA and puuD, respectively. The 32P-labeling method was the same as described above. Products of the primer extension reactions were compared with the products of dideoxynucleotide sequencing reactions, which were prepared and analyzed in a manner similar to that described for DNase I footprint analysis.
Transcription assay.
The transcription of puuA and puuD was quantified by real-time PCR using SYBR green (Applied Biosystems) following a method described previously (9). Total RNA was extracted from the constructed strain, and real-time PCR was performed using puuA-F and puuA-R primers for puuA and puuD-RT1 and puuD-RT2 primers for puuD. The reactions were carried out in a MicroAmp fast optical 48-well plate (Applied Biosystems) with the following program: 40 cycles of 95°C for 15 s, followed by 60°C for 60 s, executed soon after a denaturing cycle at 95°C for 10 min. gapA was used as the internal reference gene and was amplified using gapA-F and gapA-R primers. The relative expression level of puuA and puuD was measured by using a previously described method (9).
RESULTS AND DISCUSSION
Characterization of PuuR.
We previously reported that puuR is located in the middle of the puu gene cluster (7) (Fig. 1A). We predicted that puuR encodes a transcriptional regulator because a secondary structure prediction made from the amino acid sequence of PuuR revealed a helix-turn-helix (HTH) DNA binding motif. The molecular weight of purified His6-PuuR was determined to be 96,000 by size exclusion chromatography (data not shown). Because the molecular weight of the His6-PuuR monomer was found to be 26,000 by SDS-PAGE analysis, it was suggested that native PuuR forms tetramers. Primary sequence analysis revealed that the PuuR protein exhibits two distinct regions. The N-terminal region of PuuR (residues 1 to 70) shows significant similarity to members of the HTH-XRE family of transcriptional regulators (25). The C-terminal region (residues 110 to 180) shows similarity to cupin superfamily proteins, which share a 6-stranded β-barrel core (21).
We also previously reported that the enzymatic activity of PuuD and the transcription of puuA and puuP were strongly enhanced by the deletion of puuR (7, 9, 11). Additionally, transcriptions of puuE, puuR, puuC, and puuB were enhanced in the DNA-binding-deficient puuR(3Ala) mutant. In that study, microarray analysis yielded the following fold increases in transcription: puuE, 18.8; puuR, 14.8; puuC, 15.0; and puuB, 18.6 (6). Three amino acids in the puuR(3Ala) mutants that were predicted to comprise a key portion of the HTH DNA-binding motif of PuuR by a secondary structure prediction program (Genetyx software, Genetyx Corp.) were mutated to alanine residues (Thr33Ala, His34Ala, and Ser35Ala) (6). These residues exist on the turn and helix of the HTH motif. According to the program, this alanine substitution ought to disable the formation of the PuuR HTH motif. This result suggests that PuuR functions as a negative regulator of all puu genes.
Gel mobility shift assay for analysis of the PuuR binding region.
To determine whether PuuR binding sites exist on intergenic regions of the puu gene cluster, the purified His6-PuuR protein was subjected to gel mobility shift assays. Three probes were designed for the three intergenic regions between puuP and puuA (probe 1), puuA and puuD (probe 2), and puuR and puuC (probe 3) (Fig. 1A). These regions were identified as potential regulatory regions for puu gene expression because of the length of the intergenic regions. Purified His6-PuuR bound to probe 2. However, no shifted band was found with probe 1 or 3 (Fig. 1C). These results suggest that PuuR binds to the intergenic region between puuA and puuD for repression of the puu genes, but PuuR does not bind to the intergenic region between puuP and puuA or puuR and puuC. A gel mobility shift assay was performed with cell extracts of His6-PuuR or His6-PuuR3Ala from overexpressed cells using probe 2. The shifted band for His6-PuuR3Ala is much fainter than for His6-PuuR (Fig. 1D). This result suggests that the puuR(3Ala) mutation effectively weakened the interaction between PuuR and probe 2.
Transcripts of puuAP and puuDRCBE were produced from the puuA and puuD promoters.
The gel mobility shift assay results suggested that PuuR binds at the intergenic region between puuA and puuD and regulates the expression of the puu genes as a unit. To confirm this speculation, mRNA transcripts of puuP and puuCBE were examined by RT-PCR. For examination of the puuP transcript, total RNA was prepared from strain KEI12 [puuR(3Ala)] and RT-PCR was performed using a reverse primer (puuP-RT1) designed from an internal region of puuP for the RT reaction and a forward primer (puuA-R) designed from an internal region of puuA for the amplification of a reverse-transcribed product with the reverse primer (Fig. 1A). If all mRNAs for puuP and puuA were transcribed independently, no RT-PCR product would be detected with the primer set. As a result of the RT-PCR, a band of the predicted size (2.3 kb) was detected (Fig. 1E, lane 1). No bands were observed when the RT enzyme was not included in the reaction mixture (Fig. 1E, lane 2). This suggests that at least some transcription of puuP is initiated at the puuA promoter along with puuA, although this result does not exclude the possibility that an independent mRNA transcript of puuP may exist.
Similarly, RT-PCR was performed using a reverse primer (puuE-RT2) designed from an internal region of puuE for the RT reaction and a forward primer (puuD-RT1) designed from an internal region of puuD for the amplification of a reverse-transcribed product with the reverse primer (Fig. 1A). If all mRNAs for puuCBE and puuDR were transcribed independently, no RT-PCR product would be detected with this primer set. As a result of the RT-PCR, a product of the predicted size (4.1 kb) was detected (Fig. 1E, lane 3). No bands were observed when RT enzyme was not included in the reaction (Fig. 1E, lane 4). This result does not imply the lack of an independent promoter and mRNA transcript of puuCBE (14). However, at least some transcription of puuCBE is initiated at the puuD promoter with puuDR. The RT-PCR products were sequenced using the internal primers. The intergenic regions between puuA and puuP and between puuR and puuC were analyzed, and the linkages between puuP and puuA and between puuR and puuC in the RT-PCR products were confirmed (data not shown).
Identification of the PuuR-binding region and predicted recognition motif.
To characterize the PuuR-binding region between puuA and puuD, a DNase I footprint assay was performed with two 32P-labeled DNA segments, corresponding to the coding and the noncoding strands of puuD (Fig. 2A). Four different loci, FA, FB, FC, and FD, were protected from DNase I digestion when preincubated with His6-PuuR, suggesting that PuuR binds to these four loci (Fig. 2A, B, and C).
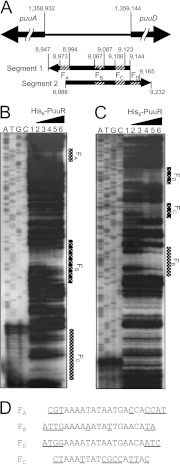
Fig 2.
DNase I footprint analysis of the intergenic region between puuA and puuD. (A) Location of two [32P]ATP-labeled DNA segments between puuA and puuD. Arrows indicate directions from 5′ to 3′ on 32P-labeled DNA strands. Striped areas labeled FA, FB, FC, and FD indicate PuuR binding regions, which were identified by DNase I footprint analysis. Numbers indicate the start and end bases of the segment and the PuuR binding region in the E. coli genome. The numbers omit the first three digits, except for the start of puuA and puuD. (B and C) DNase I footprint assay with His6-PuuR. The 32P-labeled probes (1.35 fmol) were incubated in the absence (lane 1) or presence of increasing concentrations of purified His6-PuuR (lane 2, 1 pmol; lane 3, 5 pmol; lane 4, 10 pmol; lane 5, 20 pmol; lane 6, 40 pmol), and then subjected to DNase I footprinting assays (panel B represents segment 1 from panel A; panel C represents segment 2 from panel A). Lanes A, T, G, and C represent the respective sequence ladders. (D) An alignment of the nucleotide sequences of all four sites protected by PuuR, which were observed in the DNase I footprint assay. Bases in the predicted 15-bp PuuR recognition sequence (AAAATATAATGAACA) are shown, and bases not in the predicted consensus sequence are shown with underlined letters.
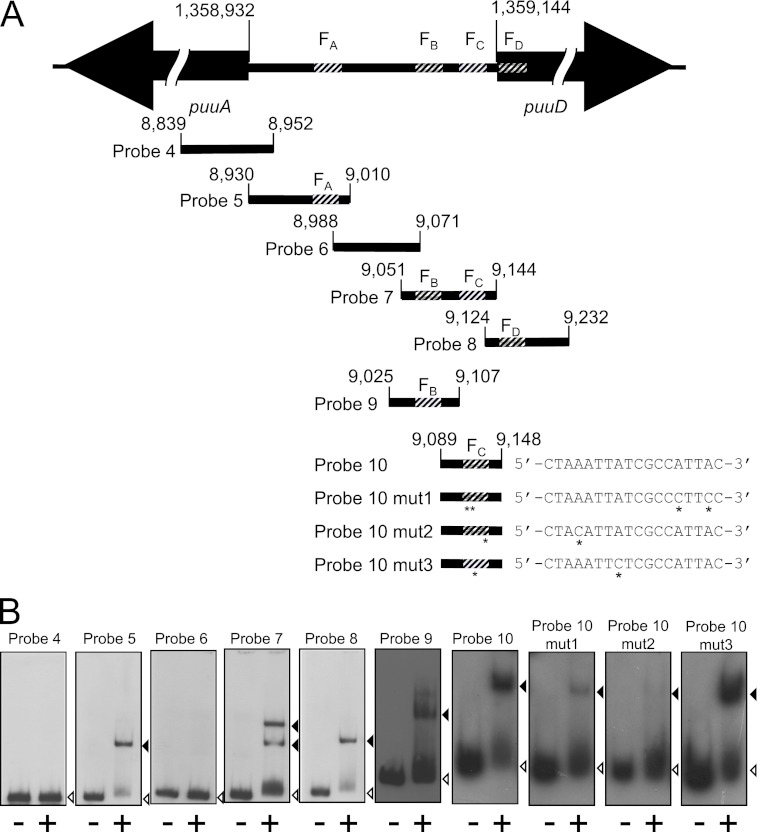
Nucleotide sequence alignments of all four sites protected by PuuR revealed a predicted 15-bp PuuR recognition motif (Fig. 2D). Three of the four binding sites were aligned in the same direction on the DNA, whereas FC was aligned in the opposite direction. DNA probes were prepared between puuA and puuD to confirm the binding of PuuR at these four sites in detail (Fig. 3). These probes each contained either one of the four sites or no site. Probes 5, 8, 9, and 10, containing the FA, FD, FB, and FC sites, respectively, produced shifted bands by gel mobility shift assay (Fig. 3B), although the motif sequence of the FC site is partially matched with the consensus sequence. Probes 4 and 6, which did not include the FA, FB, FC, or FD site, did not produce a shifted band. These results suggest that PuuR interacts with each of these four sites. To further investigate the importance of individual bases in the predicted 15-bp PuuR recognition motif for PuuR binding, point mutations were introduced into the FC site (Fig. 3). Mutations were introduced into the FC site because while the FC site is partially matched to the consensus sequence, PuuR recognizes the sequence motif in the FC site for binding. The important bases for recognition by PuuR must reside in the FC site. Two cytosine bases were substituted in probe 10 mut1 for the 14th and 17th adenine bases from the 5′ end of the FC region (Fig. 3A, bases marked with asterisks). Probe 10 mut1 produced a very slight band shift (Fig. 3B). Probe 10 mut2, in which an adenine-to-cytosine transversion was introduced in the 4th adenine base from the 5′ end of the FC region (Fig. 3A, base marked with asterisk), also produced a faint band shift (Fig. 3B). The 8th adenine base from the 5′ end of the FC region was changed to cytosine to obtain probe 10 mut3 (Fig. 3A, base marked with asterisk). The signal of Probe 10 mut3 was shifted to almost the same degree as probe 10, which harbors a nonmutated FC site (Fig. 3B). These results indicate that the adenine residues in the mutation site on probe 10 mut1 and mut2 are important for binding PuuR, whereas the residues in probe 10 mut3 are not.
Fig 3.
Gel mobility shift assays using probes designed from the intergenic region of puuA and puuD. (A) Intergenic region between puuA and puuD, and location of each probe used in gel mobility shift assays. The base number of the 5′ end of puuA and puuD in the E. coli genome is indicated above each gene. The four PuuR binding sites, FA, FB, FC, and FD, are shown as striped areas. The probe location is indicated with the base numbers in the E. coli genome. The number omits the first three digits except for the start of puuA and puuD. Sequence alignments of each FC site mutation are shown. Asterisks below probes 10 mut1, 10 mut2, and 10 mut3 and sequences indicate mutation sites, which were introduced using the QuikChange method in the FC site. (B) Gel mobility shift assays were performed following the same method as described for Fig. 1 except for the concentration of the probes. Probe 9, probe 10, probe 10 mut1, probe 10 mut2, and probe 10 mut3 were prepared at 1.5 pmol/μl, and 1 μl of each probe was used in the reactions. Lanes on the left (marked with minus symbols below each lane) were incubated in the absence of His6-PuuR, and lanes on the right (marked with a plus symbol below each lane) were incubated in the presence of His6-PuuR. Black and white triangles indicate signals of the shifted band and free probe, respectively.
Determination of the 5′ end of the mRNA transcripts of puuA and puuD.
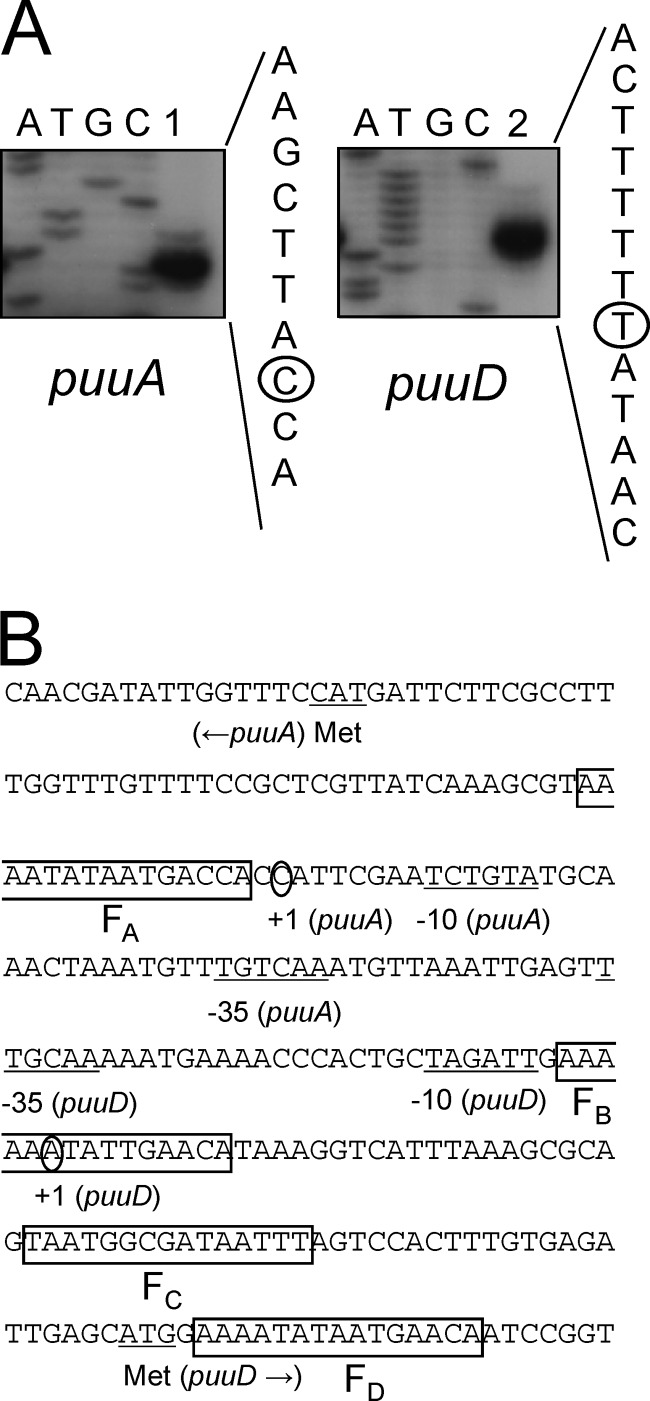
The results shown above suggest that the promoters for puuA and puuD are located in the intergenic regions of puuA and puuD and are negatively regulated by PuuR. The 5′ ends of the mRNA transcripts of puuA and puuD were determined by primer extension analysis using total RNA isolated from the E. coli strains NNE3 and NNE1. The NNE3 and NNE1 strains were designed for the high expression of puuA and puuD mRNA transcripts, respectively, from the original promoters. These strains were designed by increasing puuA and puuD copy numbers using a pUC19 vector in the puuR(3Ala) host cell, in which repression of puu gene expression by PuuR was disrupted. The 5′ ends of mRNAs for puuA and puuD were mapped at 60 and 68 nucleotides upstream from the ATG translation initiation codons of puuA and puuD, respectively (Fig. 4). The putative −35 and −10 hexamers were found upstream from each transcription start site (Fig. 4B).
Fig 4.
Primer extension analysis and promoter region sequences for puuA and puuD. (A) Identification of the transcription start sites of puuA and puuD. The sizes of the extended products, lane 1 for puuA and lane 2 for puuD, were determined by comparison with the DNA sequencing ladder, lanes A, T, G, and C. An expanded view of the nucleotides surrounding the transcription start site (denoted by a black circle) in the coding strand for puuA and puuD is shown. (B) Sequence of the intergenic region between puuA and puuD. The nucleotide sequence from position −210 to +90 for the puuD transcription start site is indicated. The transcription start site (+1) and the inferred −10 and −35 hexamers of the puuA and puuD promoters are indicated. The initiation codons of puuA and puuD are also shown, marked with underlining and the label “Met.” The predicted 15-bp PuuR-binding motifs in FA, FB, FC, and FD, which were determined by DNase I footprint assay, are marked by boxes.
Mapping of the four PuuR binding sites revealed that FA is located 2 to 16 bp downstream of the puuA transcription start site, FB overlaps with the transcription start site of puuD, FC is located between the transcription start site and the initiation codon of puuD, and FD is located downstream from the initiation codon of puuD (Fig. 4B). Within the family of Enterobacteriaceae, Shigella flexneri, Citrobacter rodentium, Klebsiella pneumoniae, and Enterobacter cloacae are phylogenetically related to E. coli. Whole-genome sequences of these microorganisms have already been determined, and we found the puu gene cluster in their genomes by using the KEGG SSDB gene cluster search (http://www.genome.jp/kegg/ssdb/). FA, FB, and FD sites are well conserved between puuA and puuD in these four species. The FC site is conserved among E. coli, S. flexneri, and C. rodentium but not in K. pneumoniae and E. cloacae (see Fig. S1 in the supplemental material). These results indicate that PuuR is likely to regulate the transcription of puu genes by binding to the conserved recognition motifs in not only E. coli but also closely related enterobacteria.
Expression analysis of constructs with different lengths of promoter region sequence.
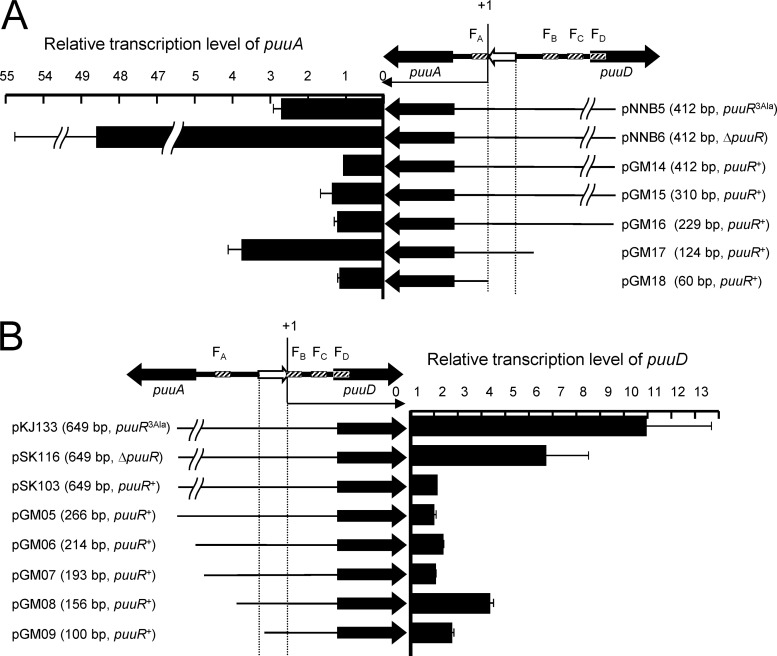
Several constructs with different lengths of the promoter regions for puuA and puuD were designed for expression analysis. The puuA transcript from pGM14, which had a stretch of 412 bp in front of the initiation codon of puuA, was used as a relative standard for the transcription level of puuA. pGM17, which lacked the FB, FC, and FD regions but retained the putative −35 and −10 hexamers, exhibited a 3.7-fold higher transcription of puuA than pGM14 (Fig. 5A). pGM15 and pGM16, which have shorter upstream regions than pGM14 but do not lack any of the PuuR binding regions, exhibited the same level of transcription as pGM14. pGM18, in which the putative promoter region of puuA is deleted, exhibited low puuA transcription. The relative transcription level of puuR(3Ala) (pNNB5) and ΔpuuR (pNNB6) were much higher than that of puuR+ (pGM14), even though they have the same upstream length.
Fig 5.
Expression analysis of the constructs with different lengths of the promoter region sequence. (A) Relative transcription levels of puuA. (B) Relative transcription levels of puuD. puuA, puuD, and the intergenic region between puuA and puuD are shown. The positions of the four PuuR binding sites, FA, FB, FC, and FD, are noted. Promoter regions for puuA and puuD are shown with white arrows and vertical dashed lines. The transcription start sites of puuA (A) and puuD (B) are labeled +1. The name of each construct and the lengths of the promoter regions from the start codon in each plasmid sequence are shown. Constructed plasmids were transformed into strain SK167 (ΔpuuADR) (A) or strain SK123 (ΔpuuDR) (B). Each construct was cultured in M9 containing 1% tryptone and collected at an OD600 of 0.5 to 0.8, followed by extraction of total RNA. Transcription of puuA or puuD in each construct was measured using real-time PCR. pGM14 transcription was used as the control for puuA relative transcription values, and pSK103 was used as the control for puuD calculations. The expression values of pGM14 and pSK103 were set at 1. Error bars indicate standard deviations, and experiments were performed in triplicate.
Constructs with different lengths of the promoter region for puuD were prepared in the same manner. The puuD transcript from pSK103, which has a stretch of 649 bp in front of the initiation codon of puuD, was used as a relative standard for the transcription level of puuD. pGM08, in which the FA region was deleted, exhibited a 3.2-fold-higher transcription of puuD than pSK103. pGM05, pGM06, and pGM07, which deleted the upstream region but did not lack any PuuR binding region, exhibited the same transcription level as pSK103. pGM09, which carries a deletion in the promoter region of the putative −35 hexamer for puuD, exhibited low transcription of puuD. Relative transcription levels in puuR(3Ala) (pKJ133) and ΔpuuR (pSK116) were highly increased, much more than in puuR+ (pSK103), although they have the same upstream length.
These findings indicate that the DNA region containing the PuuR binding motif is of primary importance for the repression of puuA and puuD. Repression by PuuR was reduced when the number of PuuR binding sites was decreased, although deletion of the FA site for the effects on puuA expression and deletion of the FB, FC, and FD sites for the effects on puuD expression could not be tested because of the locations of these promoter regions. These expression analysis results are in accordance with the results of primer extension analysis and PuuR binding region analysis by DNase I footprint assay.
Effects of polyamines on the interaction between DNA and PuuR.
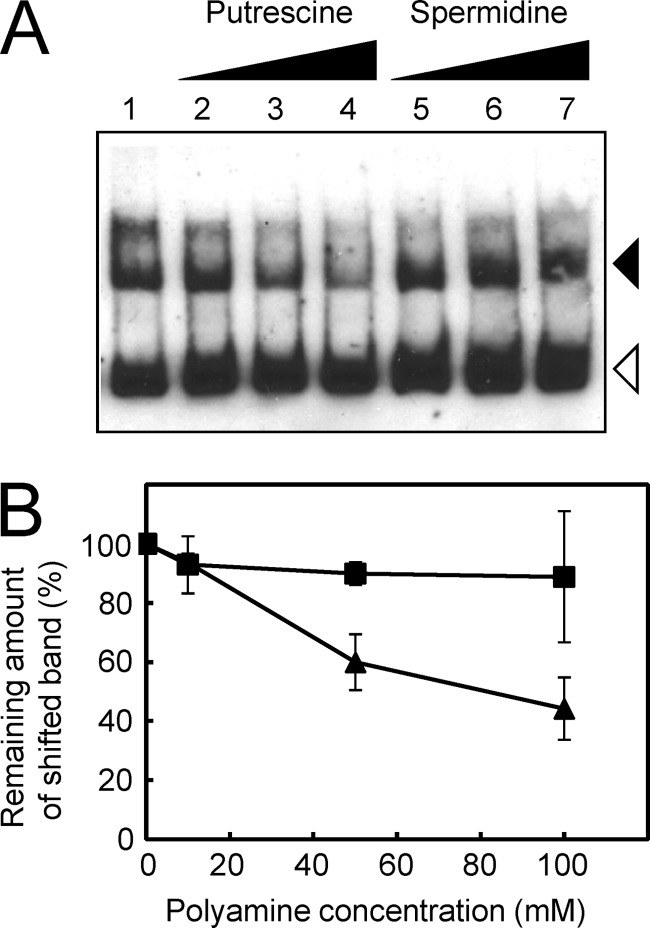
The effect of polyamines on the interaction between DNA and PuuR was analyzed by gel mobility shift assay. One picomole of purified His6-PuuR (final concentration, 100 nM) was mixed with 30 fmol of DIG-labeled probe 2 corresponding to the intergenic region between puuA and puuD (final concentration, 3 nM), and then polyamine solutions were added. The amount of the shifted band decreased as the concentration of putrescine increased, but there was no such change in band shift proportion with increasing spermidine concentrations (Fig. 6). The amino groups of polyamines have charge interaction with the phosphate groups of the nucleic acids in DNA and RNA (3, 24). In each molecule, spermidine contains three amino groups and putrescine contains two amino groups. Because the shifted bands still remained under high concentrations of spermidine, we concluded that the effect of putrescine on the DNA binding activity of PuuR is not the result of a nonspecific charge interaction. The result suggests that putrescine works as an effector for dissociation of PuuR from the binding sites on DNA, leading to transcription of the puu genes, but spermidine does not. The effective putrescine concentration may seem relatively high in this assay; however, the concentration of free putrescine in the E. coli cell is reported to be 12.5 mM, and the total putrescine concentration in the cell is 32.2 mM (3). Since putrescine is involved in the regulation of important cellular functions, E. coli requires a homeostatic mechanism to prevent any large decreases in intracellular putrescine. The Km value for putrescine (44.6 mM) of PuuA is extraordinarily high (9). This low affinity for putrescine of PuuR, the puu repressor, and PuuA, the first enzyme of the Puu pathway, maintains the relatively high putrescine concentration in the cell.
Fig 6.
Effects of putrescine and spermidine on DNA binding by PuuR. (A) A representative gel mobility shift assay with several concentrations of polyamines. Assays were conducted in the same manner as described for Fig. 1C using DIG-labeled DNA probe (30 fmol of probe 2) and 1 pmol of purified His6-PuuR with 0 mM (lane 1), 10 mM (lanes 2 and 5), 50 mM (lanes 3 and 6), or 100 mM (lanes 4 and 7) (final concentration) of putrescine (lanes 2 to 4) or spermidine (lanes 5 to 7). Black and white triangles indicate signals of the shifted band and free probe, respectively. (B) The remaining amount of shifted band was calculated from the results of the gel mobility shift assays depicted in panel A. Densitometry was performed to quantify the shifted bands using ImageJ software (version 1.37). Closed triangles indicate the proportion of shifted bands remaining when putrescine was added, and closed squares indicate the proportion remaining when spermidine was added. Error bars indicate standard deviations, and experiments were performed in triplicate.
A model of the puu gene expression regulation mechanism by PuuR.
If the free putrescine concentration in the cell reaches a sufficiently high level to change the conformation of PuuR, PuuR dissociates from DNA and the transcription of all puu genes begins. After the Puu pathway utilizes putrescine and its concentration in the cell decreases, PuuR may bind to DNA again and represses the transcription of puu genes (Fig. 7). In previous experiments, the expression of PuuD was significantly repressed by cultivation conditions such as low aeration or a high concentration of glucose, succinate, or ammonium chloride in the medium and during lag phase to early log phase, whereas the expression of PuuD was upregulated under high aeration conditions (8). Regulation of the Puu pathway by PuuR seems to be a simple, primary regulatory system that maintains the putrescine concentration in the cell, although there is a possibility of other regulatory mechanisms such as an aeration shift by FNR or ArcA for expression of the Puu pathway (14).
Fig 7.
Model of the mechanisms by which PuuR regulates puu gene expression. At low intracellular putrescine concentrations, PuuR binds to the intergenic region between puuA and puuD and represses the expression of the puu genes. At high intracellular putrescine concentrations, PuuR dissociates from DNA, and transcription of the puu genes occurs. At least some of the transcripts are expressed as mRNA-puuAP and mRNA-puuDRCBE together from the puuA and puuD promoters, respectively. Circled PuuR indicates PuuR protein. The horizontal thick black arrows indicate the puu gene cluster. White arrows between puuA and puuD indicate the puuA and puuD promoters. The expressions of mRNAs from each promoter are shown by fine arrows below the gene names. The vertical black arrows indicate shifts of intracellular putrescine concentration.
Working hypothesis of the Puu pathway.
The results presented above suggest that the Puu pathway will work when the putrescine concentration in the cell is relatively high, because PuuR remains bound to DNA at low concentrations of putrescine. The Puu pathway involves five enzymatic reactions and consumes ATP for the γ-glutamylation step by PuuA. To avoid unnecessary expression of the Puu pathway enzymes, it is profitable for the expression of the Puu pathway to be induced only under high intracellular concentrations of putrescine. Recently, we reported that PlaP (previously named YeeF) was considered to function as a sensor for extracellular putrescine (10). PlaP was characterized as a low-affinity putrescine importer, and the ΔplaP ΔpotABCD ΔspeAB ΔspeC E. coli strain did not exhibit surface motility on a plate containing putrescine, while the plaP+ ΔpotABCD ΔspeAB ΔspeC strain exhibited surface motility on the plate (10). The incorporation of 14C-labeled putrescine was also significantly decreased in the ΔplaP strain even when genes encoding other putrescine importers were not disrupted (10). Although the expressions of the regulatory mechanisms of E. coli putrescine importers have not been investigated under this experimental condition, E. coli has at least four such importers that may be used selectively, depending on growth conditions. Under the conditions used in our experiments, PlaP may function as the main putrescine importer (10). PlaP is a homologue (63% identity) of PuuP, although the affinity of PuuP for putrescine is much higher than that of PlaP. If the extracellular putrescine concentration is sufficiently high for importation of putrescine by PlaP, the intracellular putrescine concentration elevates and PuuR that has interacted with putrescine dissociates from DNA. As a result, transcription of the puu genes occurs and the subsequently expressed PuuP further imports putrescine for utilization as a carbon and nitrogen source through the Puu pathway. PlaP and PuuP might have evolved by the duplication of a single ancestral putrescine transporter. Since a low-affinity gate importer could prevent waste expression of the puu gene, PlaP might have evolved toward a lower affinity for putrescine.
The puu gene cluster is only found in Enterobacteriaceae, but not in Pseudomonas aeruginosa (26), and is not found in other bacteria, including those in the phyla Firmicutes and Bacteroidetes, which constitute an absolute majority in the human intestinal flora (1, 27). In the human intestine, there are 400 to 500 different bacterial species, and there are 1010 to 1011 bacterial cells per gram of feces, but E. coli cells constitute less than 1% of the total cell number (1). The fecal putrescine concentration of healthy adults is approximately 1 mM and decreases in aged adults (12). It seems probable that E. coli and closely related enterobacteria gained the Puu pathway and survive in mammalian intestines during sugar limitation or nitrogen starvation conditions by using putrescine as a carbon or nitrogen source.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), Japan, and by Grant-in-Aid for Scientific Research No. 21380059 and for Challenging Exploratory Research No. 23658079 to H.S., from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print 20 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arumugam M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glansdorff N. 1996. Biosynthesis of arginine and polyamines, p 408–433 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella, cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 3. Igarashi K, Kashiwagi K. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271:559–564 [DOI] [PubMed] [Google Scholar]
- 4. Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K. 1992. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K. 1997. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J. Biol. Chem. 272:6318–6323 [DOI] [PubMed] [Google Scholar]
- 6. Kurihara S, Kato K, Asada K, Kumagai H, Suzuki H. 2010. A putrescine-inducible pathway comprising PuuE-YneI in which γ-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192:4582–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurihara S, et al. 2005. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 280:4602–4608 [DOI] [PubMed] [Google Scholar]
- 8. Kurihara S, Oda S, Kumagai H, Suzuki H. 2006. γ-Glutamyl-γ-aminobutyrate hydrolase in the putrescine utilization pathway of Escherichia coli K-12. FEMS Microbiol. Lett. 256:318–323 [DOI] [PubMed] [Google Scholar]
- 9. Kurihara S, et al. 2008. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 283:19981–19990 [DOI] [PubMed] [Google Scholar]
- 10. Kurihara S, Suzuki H, Oshida M, Benno Y. 2011. A novel putrescine importer required for type 1 pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J. Biol. Chem. 286:10185–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurihara S, et al. 2009. The putrescine importer PuuP of Escherichia coli K-12. J. Bacteriol. 191:2776–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsumoto M, Benno Y. 2007. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol. Immunol. 51:25–35 [DOI] [PubMed] [Google Scholar]
- 13. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and bacteria, p 263–274, 437 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 14. Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 281:27806–27815 [DOI] [PubMed] [Google Scholar]
- 15. Pegg AE. 1986. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 234:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pistocchi R, et al. 1993. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J. Biol. Chem. 268:146–152 [PubMed] [Google Scholar]
- 17. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, p 1.1–2.110 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 18. Samsonova NN, Smirnov SV, Altman IB, Ptitsyn LR. 2003. Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 3:2 doi:10.1186/1471-2180-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samsonova NN, Smirnov SV, Novikova AE, Ptitsyn LR. 2005. Identification of Escherichia coli K12 YdcW protein as a γ-aminobutyraldehyde dehydrogenase. FEBS Lett. 579:4107–4112 [DOI] [PubMed] [Google Scholar]
- 20. Schneider BL, et al. 2002. The Escherichia coli gabDTPC operon: specific γ-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 184:6976–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stipanuk MH, Simmons CR, Karplus PA, Dominy JE., Jr 2011. Thiol dioxygenases: unique families of cupin proteins. Amino Acids 41:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki H, Kumagai H, Echigo T, Tochikura T. 1989. DNA sequence of the Escherichia coli K-12 γ-glutamyltranspeptidase gene, ggt. J. Bacteriol. 171:5169–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas T, Thomas TJ. 2001. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 58:244–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wintjens R, Rooman M. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294–313 [DOI] [PubMed] [Google Scholar]
- 26. Yao X, He W, Lu CD. 2011. Functional characterization of seven γ-glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and β-alanine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 193:3923–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zoetendal EG, Rajilić-Stojanović M, de Vos WM. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605–1615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.