Abstract
The capacity of pathogens to respond to environmental signals, such as iron concentration, is key to bacterial survival and establishment of a successful infection. Bacillus cereus is a widely distributed bacterium with distinct pathogenic properties. Hemolysin II (HlyII) is one of its pore-forming cytotoxins and has been shown to be involved in bacterial pathogenicity in a number of cell and animal models. Unlike many other B. cereus pathogenicity factors, HlyII is not regulated by pleiotropic transcriptional regulator PlcR but is controlled by its own regulator, HlyIIR. Using a combination of in vivo and in vitro techniques, we show that hlyII expression is also negatively regulated by iron by the global regulator Fur via direct interaction with the hlyII promoter. DNase I footprinting and in vitro transcription experiments indicate that Fur prevents RNA polymerase binding to the hlyII promoter. HlyII expression profiles demonstrate that both HlyIIR and Fur regulate HlyII expression in a concerted fashion, with the effect of Fur being maximal in the early stages of bacterial growth. In sum, these results show that Fur serves as a transcriptional repressor for hlyII expression.
INTRODUCTION
Bacillus cereus sensu lato is a group of bacteria with various pathogenic properties (23). Three important species that belong to the B. cereus group are entomopathogenic Bacillus thuringiensis, deadly Bacillus anthracis, and B. cereus sensu stricto, the last of which has widely varying properties that determine its role both as a harmless, spore-forming soil microorganism and as a causative agent of food poisoning and endophthalmitis (19, 38). These three species are closely related genetically and are now classified as a single species (22, 32). B. cereus is considered to be an emerging pathogen (8), warranting a detailed investigation of the mechanisms and regulation of B. cereus toxin production.
B. cereus produces a broad range of secreted cytotoxic factors, including at least four hemolysins, several phospholipases, proteases, an emetic toxin, and a score of pore-forming toxins (38). Hemolysin II (HlyII) of Bacillus cereus was discovered to be one of the secreted factors responsible for causing hemolysis (35). It has widespread expression among B. cereus group members (10, 34) and is found with increased probability in pathogenic strains (11).
Recently, we have succeeded in purifying HlyII and demonstrating its cytotoxicity toward human cell lines, indicating its potential functionality in vivo (4). In concert with these in vitro data, we showed that expression of HlyII in B. subtilis renders this organism virulent for the crustacean Daphnia magna (36) and leads to membrane damage in the alga Chara corallina (25); the role of HlyII in the virulence of B. thuringiensis in mice and insects was demonstrated by others (40). The toxic properties of HlyII rely on its ability to disrupt cellular and artificial membranes by pore formation (3). The prevalence of the hlyII genes among various B. cereus pathogenic strains is a significant indicator of their potential importance in virulence and pathogenesis (11).
The maximal expression of HlyII in bacterial cultures occurs during the late exponential growth phase (36), coinciding with transcription activator PlcR-regulated expression (37) of other major B. cereus secreted cytotoxins, such as enterotoxins (15), cytotoxin K (29), phospholipases, and proteases. However, PlcR was shown to have no effect on HlyII production (17), suggesting the existence of alternative regulatory pathways. Moreover, disruption of the plcR gene does not lead to eradication of B. cereus pathogenicity in a macrophage-based assay, provided that the hlyII and inhA1 protease genes are intact (40). This observation highlights the important role of hemolysin II in pathogenesis and suggests that the action of HlyII occurs in a regulated, concerted manner.
Expression of HlyII is regulated by the specialized transcriptional regulator HlyIIR (9, 36), a member of the TetR family of dimeric transcriptional regulators (26). As was demonstrated in vitro and in vivo, HlyIIR represses hlyII transcription (9) via direct interaction with the operator region of the hlyII gene. Interestingly, unlike most transcriptional repressors, HlyIIR forms a ternary complex with RNA polymerase (RNAP) and may slow transcription and inhibit further steps of transcription initiation after formation of the closed RNAP-DNA complex and potentially after open complex formation (9). The mechanism of the modulation of HlyIIR activity that triggers HlyII production is unknown.
The predicted sequence for the ferric uptake regulator (Fur) box was found in the hlyII promoter region (21). The Fur protein in many bacteria acts as a global transcriptional regulator of iron homeostasis (6). Fur interprets changes of Fe2+ concentration in the environment, repressing a number of the bacterial genes that are responsible for iron uptake and oxidative stress adaptation (28). In many bacteria, Fur regulates various genes that are linked to bacterial pathogenesis (12, 39). Earlier, the B. cereus Fur homologue was identified and characterized (21). The pathogenic properties of the fur-null strain are significantly impaired in the insect infection model. However, no difference in hemolysis between wild-type (wt) and fur mutant B. cereus strains was detected on blood agar (21). Thus, we explored the mechanisms and conditions of Fur-dependent hlyII regulation in bacterial cells and in vitro.
Here, we present the first experimental evidence that Fur is a transcriptional regulator that participates in control of B. cereus HlyII expression by obstructing the binding of RNA polymerase to the hlyII promoter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. Escherichia coli was routinely grown in a liquid medium (20 g/liter tryptic soy broth [Difco], 5 g/liter yeast extract, 10 g/liter NaCl). For cultivation of B. subtilis and B. thuringiensis strains, 1× morpholinepropanesulfonic acid (MOPS) minimal medium (14) or LB medium was used. For simulation of Fe-rich and Fe-depleted conditions, 0.1 to 0.3 mM FeCl3 and 0.2 mM dipyridyl (DPD) were used as indicated below. Antibiotics, when required for culture, were added at the following final concentrations: for E. coli, ampicillin at 100 μg/ml and kanamycin at 50 μg/ml; for Bacillus, erythromycin, chloramphenicol, and kanamycin at 10 μg/ml.
Table 1.
Strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Description | Reference or source |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| CU1065 | trpC2 | 41 |
| HB2501 | CU1065 fur::kan | 20 |
| BD170 | BD168 thr-5 trpC2 | E. U. Poluektova |
| EH2 | CU1065 amyE::hlyII | This work |
| EH2R | CU1065 amyE::hlyII-hlyIIR | This work |
| B. cereus VKM B-771 | 35 | |
| B. thuringiensis VKM B-1555 | VKM | |
| E. coli Z85 | thi Δ(lac-proAB) Δ(srl-recA) hsdR supE Tn10(Tcr) (F+ traD proAB lacI ΔM15) | DH5α derivative |
| Plasmids | ||
| pUJ1 | pUC19 with 2.9-kb EcoRI fragment | 35 |
| pHT304-18Z | Promoterless lacZ plasmid | 2 |
| pPH2Z-B771 | hlyII promoter region in pHT304-18Z | This work |
| pEHB | pDG364 derivative | 36 |
| pEH2 | pEHB with hlyII gene | 36 |
| pEH2R | pEHB with hlyII and hlyIIR genes | 36 |
| pFUR6His | fur in pET29(b) | This work |
| pHT01 | MoBiTec | |
| pFUR | B. cereus fur in pHT01 | This work |
| pFUR-Km | Cmr replaced by Kmr in pFUR | This work |
| Oligonucleotides | ||
| hlyIIp-for | GTATCTGGATCCAGGCTGTAATAAGTAAATG | |
| hlyIIp-rev | ACAATGTAGAAGCTTATTAATCTTTATGCC | |
| pHTFur-for | GTCGCTCTAGAATGGAAGAAAGAATTGAACGAATTAAG | |
| pHTFur-rev | ATCGCCCCGGGTTATTTTTCATCCGTTTCATTTTCTT | |
| H2Z-for | AGGAATTTTAGATTATTATGAATGGAAAGG | |
| H2Z-rev | CATTATAACGGACGCTACGGCAACA | |
| Km-NheI-for | CAATTGGCTAGCTTCAACAAACGGGCCAG | |
| Km-SphI-rev | CTTAAGGCATGCCGCCATGACAGCCATG |
Plasmids and strains construction.
The plasmid pFUR6His, expressing His6-tagged Fur, was obtained by cloning the PCR-amplified B. cereus fur (oligonucleotides pETFur-for and pETFur-rev). After digestion at NdeI and XhoI sites, the PCR product was cloned into pET-29(b) (Novagen) that had been digested with the same enzymes. DNA sequences were confirmed.
The plasmid pFUR for Fur expression in B. subtilis was prepared by cloning the B. cereus fur gene into the vector pHT01 (MoBiTec, Germany) between XbaI and SmaI sites using the primers pHTFur-for and pHTFur-rev. pHT01carries a strong σA-dependent promoter preceding the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible groE operon of B. subtilis, which has been converted into an efficiently controllable (IPTG-inducible) promoter by addition of the lac operator.
pFUR-Km was prepared by replacement of the NheI-SphI fragment of pFUR, containing a chloramphenicol resistance determinant, by kan obtained by PCR amplification of pUB110 using oligonucleotides Km-NheI-for and Km-SphI-rev.
pPH2Z-B771, carrying the hemolysin II gene promoter (positions +228 to −204 relative to the transcription start point) was PCR amplified from chromosomal DNA of the B. cereus VKM B-771 strain using the primers H2Z-for and H2Z-rev, cloned into the EcoRV site of pUC128, digested with BamHI and HindIII, and cloned into the same sites of pHT304:18Z (2).
B. subtilis EH2 and EH2R were constructed as described in reference 36.
Expression and purification of Fur-His6.
Fur-His6 was overexpressed in E. coli BL21(DE3)(pFUR6His) by IPTG induction; 1 mM IPTG was added to cells grown to mid-exponential phase, and cells were harvested 4 h later. Cell pellets were resuspended in 50 mM Tris-HCl (pH 8.0)–200 mM NaCl, and cells were lysed by ultrasonication. Cell extract was prepared by centrifugation at 40,000 × g at 4°C for 1 h, followed by filtration through a DEAE-cellulose column to remove nucleic acids. Fur-His6 was purified by Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography (Sigma), followed by anion-exchange chromatography (MonoQ) and hydrophobic chromatography on phenyl-Sepharose CL-4B. The protein-containing fractions were dialyzed against 10 mM Tris-HCl (pH 8.0)–200 mM NaCl–5 μM ZnCl2–20% glycerol at 4°C and stored at −20°C. Fur was judged to be ∼ 95% pure by SDS-PAGE.
HlyIIR was purified as described before (9). B. subtilis RNA polymerase was purified as described previously (24).
Electrophoretic mobility shift assay (EMSA).
Target promoter DNA corresponding to the hlyII promoter region was PCR amplified using promoter-specific oligonucleotides hlyIIp-for and hlyIIp-rev (Table 1). The fragments were 32P labeled with T4 DNA kinase and purified using the Qiagen gel extraction kit. Binding reaction mixtures (20 μl) contained 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 20 ng/μl chicken erythrocyte DNA, 7.5% glycerol, 1 or 2 nM DNA, Fur-His6 (various concentrations), and 0.1 mg/ml xylene cyanol. Reaction mixtures were incubated for 15 min at 25°C to allow Fur-DNA complex formation to occur. Samples were fractionated through native 6% polyacrylamide or 0.7% agarose gels using 0.5× Tris-acetate (TA) as the gel running buffer. Radioactive bands were visualized by autoradiography and quantified using ImageQuant 5.2 software (Molecular Dynamics). The apparent equilibrium binding constant (KD) of the Fur-PhlyII DNA interaction was calculated as described previously (27).
In vitro transcription and DNA footprinting.
In vitro transcription and DNA footprinting were performed as described elsewhere with some modifications (9). A DNA region containing PhlyII was prepared as described for EMSA. To form open promoter complexes, 300 nM RNAP σA holoenzyme was incubated in transcription buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10 mM MgCl2) with 100 nM PhlyII DNA fragment for 10 min at 37°C. For multiround transcription, purified Fur and HlyIIR proteins were added to the reaction mixture either before or after RNAP addition and open complex formation. The reaction mixture was incubated for 10 min at 37°C. Transcription complexes were used in footprinting or were supplemented with 500 μM GTP, 50 μM ATP and CTP, and [α-32P]UTP to analyze in vitro transcription products. Transcription reactions were allowed to proceed for 10 min at 37°C before termination by addition of formamide buffer. In single-round transcription assays, Fur was added to the 10 nM DNA simultaneously with 125 nM Bacillus RNAP, and the mix was incubated for 3 min at 37°C. The reaction was initiated by addition of 100 μM GTP, 100 μM ATP, 100 μM CTP, 10 μM UTP, ∼0.02 to 0.2 μM [α-32P]UTP, 50 ng/μl heparin for runoff assay, and 100 μM GpA, 10 μM UTP, and ∼0.02 to 0.2 μM [α-32P]UTP for abortive initiation. Reaction mixtures were incubated for 5 min, and reactions were stopped by addition of formamide buffer. Reaction products were separated on polyacrylamide gels with 6 M urea and revealed by autoradiography. Apparent inhibition constants were determined by digitizing the scanned gel pattern, and determination of the fraction of residual transcription products was performed using ImageQuant 5.2 software (Molecular Dynamics). Data were fit to the Hill equation in SigmaPlot. For footprinting analysis, hlyII DNA fragment was 32P end labeled at the nontemplate strand. Samples containing hlyII promoter complexes were footprinted with DNase I (33). The reaction was stopped as described above, and the samples were then analyzed on a 6% sequencing gel.
β-Galactosidase assay.
Bacterial cultures were grown at 28°C in LB or the specified medium. Cells were collected at various times during growth. The amounts of β-galactosidase in recombinant B. subtilis and B. thuringiensis cell extracts were determined using lysozyme cell permeabilization with a quantitative colorimetric assay with o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, as described elsewhere (31). The activity was determined as Miller units, i.e., optical density at 420 nm (OD420)/(OD600 × hydrolysis time × relative volume of cell lysate).
Quantitative hemolysis assay.
The quantitative hemolysis assay has been described earlier (36). The procedure is based on measuring the hemoglobin released upon erythrocyte lysis. It was routinely done using human erythrocytes. Control samples for estimation of spontaneous and complete lysis of erythrocytes were run in parallel. For complete lysis, osmotic shock with water was used. The extent of lysis was determined using the following equation: hemolytic activity (HA) = 2(n − 1), where HA is in hemolytic units (HU) per ml and n is the number of dilutions. To measure cholesterol-independent hemolytic activity in the B. cereus culture medium, the first dilution was supplemented with 0.02% cholesterol and then incubated for 30 min at 25°C.
Statistical analysis.
Statistical analysis was performed using SigmaPlot 9 for Student's t test assuming unequal variances. P values of <0.05 were accepted as statistically significant.
RESULTS
Fur operator PhlyII is found in many B. cereus genomes.
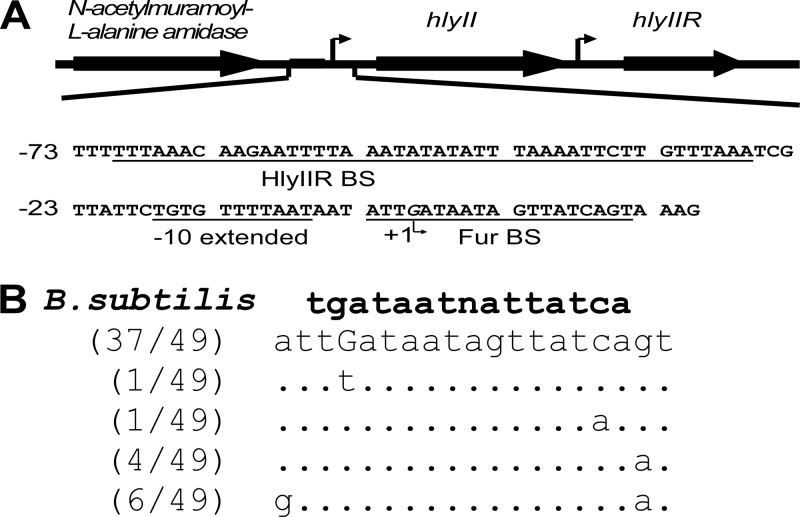
The structural organization of the hlyII transcriptional unit is shown on Fig. 1A. We analyzed the sequences of 49 hlyII promoters of B. cereus sensu lato in the bacterial genome database (NCBI) and identified Fur box-like sequences similar to the canonical B. subtilis Fur binding site (BS) (7). We found that in all these strains the Fur box overlapped the transcription starting point (9). The extracted BS sequences were aligned using FASTA (Fig. 1B). The potential Fur box of B. cereus VKM B-771 (the strain from which hemolysin II was originally isolated) was identified as a widespread variant and is therefore suited to the current study.
Fig 1.
Sequence of the hlyII promoter with corresponding regulatory elements. (A) hlyII and adjacent genes in B. cereus VKM B-771 and B. thuringiensis VKM B-1555. The transcription start site of hlyII is marked by a bent arrow. The HlyIIR binding site and −10 promoter element are underlined. The sites were mapped by Budarina et al. (9). The putative Fur binding site is shown (21). Numbering is with respect to the hlyII transcription start. (B) Single nucleotide polymorphisms (SNPs) in the Fur binding site. Multiple alignments were constructed using BLAST (word size, 16 bp), searching a 60-bp sequence of the Fur box neighborhood of the PhlyII regions in the B. cereus, B. thuringiensis, B. anthracis, B. mycoides, B. pseudomycoides, B. weihenstephanensis, and B. cytotoxis genomic databases (49 entries). The consensus B. subtilis Fur binding site is shown in bold (16).
Expression of hemolysin II is iron sensitive.
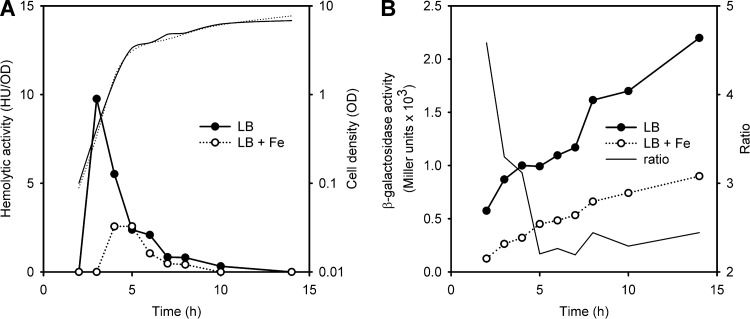
In agreement with previous reports (21), we found that high total multicomponent hemolytic activity of B. cereus strains is not affected by iron supplementation in the medium (data not shown). To test whether iron could specifically affect expression of hlyII, we introduced a PhlyII-lacZ fusion on the plasmid pPH2Z-B771 into B. thuringiensis VKM B-1555, which has authentic hlyII-hlyIIR genes in its chromosomal DNA (34). Next, cholesterol-independent HlyII activity in the culture medium and β-galactosidase activity in the cell were tested at different time points (Fig. 2).
Fig 2.
Expression of hemolysin II is reduced in the presence of FeCl3. Cells were grown in LB at 37°C. Experiments were repeated 2 times, and representative curves are shown. (A) HlyII-specific cholesterol-independent hemolytic activity of B. thuringiensis VKM B-1555 cultures. Corresponding growth curves are shown in by thin lines. (B) Expression pattern of an hlyII-lacZ fusion in B. thuringiensis VKM B-1555. The ratio for β-galactosidase activity with or without added FeCl3 is shown in by the thin line. ●, control; ○, 0.3 mM FeCl3.
At the beginning of stationary phase, B. cereus cells secrete a number of extracellular proteases (18) that may affect the HlyII lifetime in the culture. Thus, it was no surprise that the overall expression profiles of hlyII and hlyII-lacZ were different but both hlyII-lacZ promoter activity and specific HlyII hemolytic activity were repressed 2- to 3-fold by 0.3 mM FeCl3. Variation of Fe concentrations in the range of 0.1 to 0.3 mM FeCl3 did not affect the repression level. Thus, all further experiments were performed with 0.1 mM FeCl3. The effect of iron was more pronounced at the beginning of the growth curve but continued to be significant in the log growth phase (P = 0.05) (Fig. 3A). Specifically, iron delayed onset of HlyII production for about 2 h and decreased maximal hemolytic activity by 3-fold (Fig. 2).
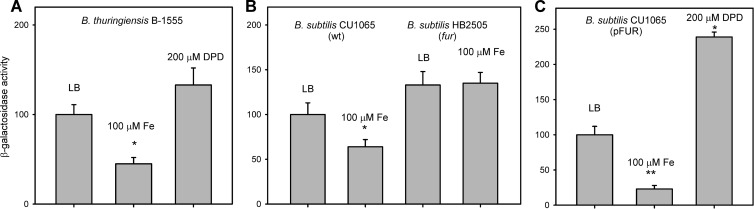
Fig 3.
Fur regulates hlyII-lacZ expression in Bacillus. All strains contain pPH2Z-B771 with PhlyII-lacZ fusion. The results show the means and standard deviations from at least three independent experiments. The significance of results was statistically analyzed using the paired t test (SigmaPlot) (*, P < 0.05; **, P < 0.01). Each experiment was performed at least three times. (A) β-Galactosidase activity in B. thuringiensis VKM B-1555(pPH2Z-B771) under iron-replete (100 μM FeCl3) and iron-depleted (100 μM DPD) conditions (6 h of growth, late exponential phase). (B) Effect of iron supplementation (100 μM FeCl3) on β-galactosidase activity in the wt B. subtilis CU1065 and fur-deficient B. subtilis HB2501. (C) β-Galactosidase activity in the wt B. subtilis CU1065 bearing pFUR-expressed B. cereus fur under iron-replete (100 μM FeCl3) and iron-depleted (100 μM DPD) conditions (2 h of growth, early exponential phase).
To mimic low-iron conditions, bacteria were grown in the presence of the iron scavenger 2,2-dipyridyl (DPD) (200 μM). We found a weak derepression effect on specific hlyII-lacZ activity (P = 0.1) (Fig. 3A). However, it was observed that DPD delays bacterial growth.
Therefore, we have shown that hlyII expression is reduced under iron-rich conditions, but it is unlikely that iron-deficient conditions induce hlyII expression. Next, we conducted experiments to verify that the effect of iron on hlyII expression is governed by Fur.
Fur mediates the effect of iron on hlyII transcription.
Both the amino acid sequences and recognition sites of the B. cereus and the B. subtilis Fur proteins are similar, with 95% amino acid identity in DNA binding region (16, 21), justifying the use of B. subtilis strains in our experiments. Expression of the hlyII-lacZ fusion was examined in wild-type (wt) B. subtilis CU1065(pPH2Z-B771) and its Fur-deficient isogenic mutant HB2501(pPH2Z-B771) (Δfur) (20). Only weak differences in hlyII-lacZ expression were found between these strains, and average β-galactosidase activity was only slightly higher in B. subtilis HB2501 (Fig. 3B, compare bars 1 and 3) (P = 0.1). However, when both strains were challenged with 0.1 mM FeCl3, hlyII-lacZ activity was decreased 2-fold in the wt CU1065(pPH2Z-B771) but was not affected in the fur-deficient strain (Fig. 3B). This observation strongly suggests that the effect of iron on hlyII expression is due to regulation by Fur.
Since little is known about the regulation of the B. cereus fur gene itself, we decided to determine whether overexpression of B. cereus fur will amplify the repression of the B. subtilis Fur. We introduced the fur gene on the plasmid pFUR into B. subtilis BD170(pPH2Z-B771). We found that the β-galactosidase activity of hlyII-lacZ was 4- to 5-fold lower in the presence of pFUR than in the presence of the empty vector pHT01 at 0.1 mM FeCl3 (Fig. 3C).
Derepression by DPD was found to be weak under most conditions that we tested. The most pronounced differences were observed at low bacterial densities. However, addition of 200 μM DPD to the culture medium of B. subtilis BD170(pPH2Z-B771, pFUR) increased detectable β-galactosidase activity about 2-fold (Fig. 3C), demonstrating that B. cereus fur overexpression increases the observed derepression effect of decreased iron-replete conditions. No significant derepression effect of DPD was found in the late exponential or stationary phase of growth. In summary, our in vivo results confirmed that B. cereus Fur is one of the transcriptional regulators of HlyII expression.
Fur repression is pronounced in the early log phase.
B. cereus cells express a number of substances with hemolytic activities; some of them are not characterized. While a mutant that lacks all possible sources of hemolysis besides HlyII is difficult to obtain, we decided to reconstitute the Fur regulation of HlyII expression in B. subtilis.
The hlyII gene is expressed in B. cereus under the control of its own regulator, HlyIIR. To determine where additional Fur-mediated inhibition takes place, we used a pair of B. subtilis strains that express chromosomal copies of either hlyII alone (EH2) or hlyII-hlyIIR (EH2R) in tandem, obtained by integration into B. subtilis CU1065.
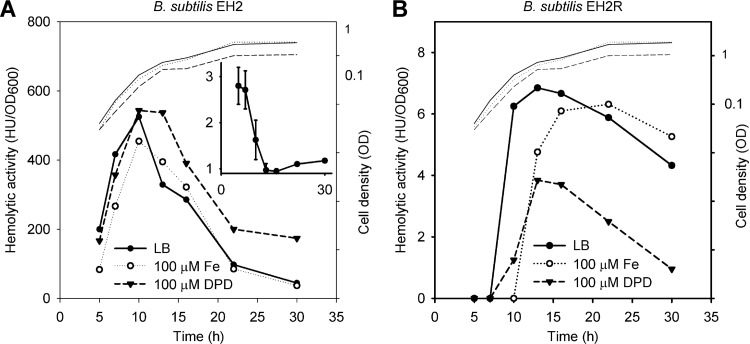
As expected, in the absence of HlyIIR (EH2), the observed HlyII activity was enormous, with the maximum occurring at the end of exponential phase, a fast decline in stationary phase, and minimal hemolytic activity at the end of stationary phase (Fig. 4A). In fact, the resultant strain was found to be the best source of hemolysin II for protein purification. Under the conditions used, the presence of HlyIIR (EH2R) decreased hemolytic activity 100-fold, but the expression followed the same pattern as without HlyIIR (Fig. 4B). It seems that HlyII production depends on cis-acting HlyIIR at all growth phases. Essentially, the expression curves obtained are in agreement with the expression pattern in B. thuringiensis VKM B-1555 (Fig. 2). When the bacteria were challenged with iron, HlyII production decreased in both cases. The maximal effect was found at the beginning of exponential phase for both strains. In B. subtilis EH2, HlyII production was decreased 2.5-fold, but in B. subtilis EH2R, in the presence of HlyIIR, the onset of HlyII activity was delayed by several hours in the presence of iron.
Fig 4.
Fur delays onset of hlyII expression. Cells were grown in LB at 28°C. Corresponding growth curves are shown in each panel (thin lines). Each experiment was performed at least three times. Results from representative experiments are shown. (A) Effects of iron and DPD on HlyII production in B. subtilis EH2. The average ratio of hemolytic activities between curves is shown in the inset. (B) Effects of iron and DPD on HlyII production in B. subtilis EH2R.
Only a weak DPD derepression effect on hemolytic activity was observed in strain EH2 (Fig. 4A). However, in strain EH2R, DPD significantly decreased HlyII expression (Fig. 4B). We verified that DPD itself has no effect on the hemolysis reaction. Due to the negative influence of DPD on bacterial growth, direct comparison of the results is problematic.
Therefore, we conclude that Fur is an auxiliary repressor of hlyII expression with the maximal effect at the beginning of cell growth, when the concentration of HlyIIR may be low. However, experiments under iron-depleted conditions indicate that Fur may be required for full hlyII expression and may play a dual role in this regulation.
Fur and HlyIIR abrogate HlyII-specific activity in B. subtilis.
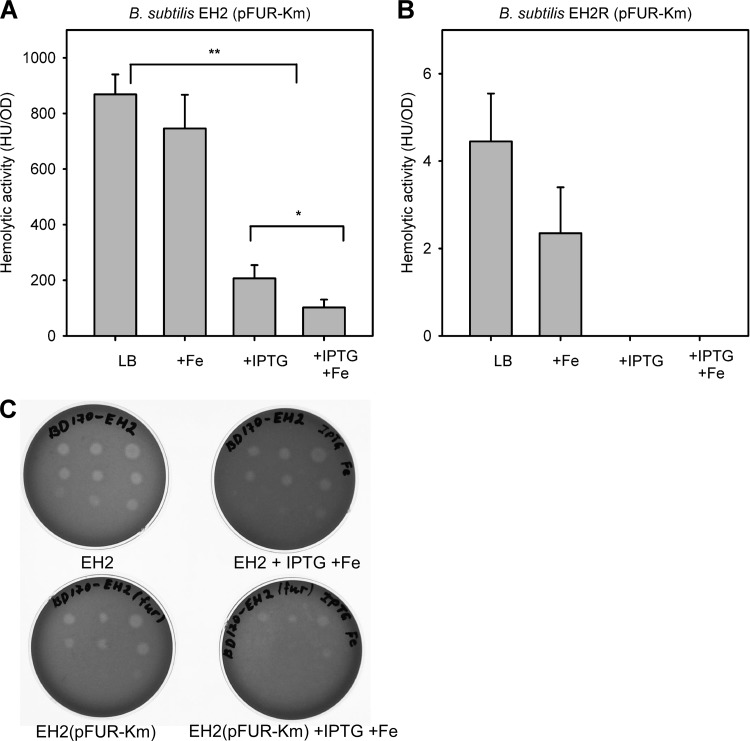
We next introduced the fur gene on the plasmid pFUR-Km into strain EH2 and measured the HlyII activity in various conditions. The results for EH2(pFUR-Km) are presented in Fig. 5. Induction of Fur expression by 0.25 mM IPTG decreased HlyII activity 4-fold (P < 0.01), and supplementation of the culture medium with IPTG and Fe additionally decreased the hemolytic activity 2-fold (P < 0.05) (Fig. 5A). Thus, the maximum repression effect of B. cereus Fur under iron-saturated conditions is of the same order of magnitude as the effect of HlyIIR (Fig. 4, compare maximum hemolytic activities of EH2 and EH2R). We next tested the effect of fur coexpression on ability EH2 to produce clearance zones on blood agar (Fig. 5C). As we showed before, the EH2 hemolysis zones are observed before bacterial growth became visible (36), thus allowing us to monitor very early regulatory events. The results obtained were entirely similar to the results that were obtained in liquid culture experiments, demonstrating a gradual decrease of hemolysis upon Fe supplementation and induction of Fur expression by IPTG. We conclude that, at high intracellular concentrations, Fur inhibits the HlyII activity efficiently even in the absence of HlyIIR.
Fig 5.
Fur-dependent regulation of HlyII activity. (A) Effect of Fur coexpression on hemolytic activity in liquid cultures of B. subtilis EH2(pFUR-Km). (B) Effect of Fur coexpression on hemolytic activity in liquid cultures of B. subtilis EH2R(pFUR-Km). Cells were grown in LB at 28°C. Points were taken in the middle of the exponential stage of growth (OD ∼ 0.5). The results show the means and standard deviations from at least three independent experiments. The significance of results was statistically analyzed using the paired t test (SigmaPlot) (*, P < 0.05; **, P < 0.01). (C) Effect of Fur coexpression on development of hemolysis on blood agar. From left to right are serial dilutions (2 μl) of corresponding overnight cultures of B. subtilis EH2 and EH2(pFUR)in the presence and absence of Fe and IPTG (0.25 mM). Plates were incubated for 10 h at 24°C. No visible bacterial growth was observed. Top rows, no dilution and 2- and 4-fold dilutions; middle rows, 8-, 16-, and 32-fold dilutions; bottom rows, 160-, 800-, and 1,600-fold dilutions. The contrast of the image was uniformly adjusted using Adobe Photoshop CS3.
In agreement with the results shown in Fig. 4, iron-depleted conditions do not affect hlyII expression significantly in EH2(pFUR-Km). However, induction of Fur expression in the presence of 100 μM DPD leads to 3- to 5-fold inhibition of HlyII production (data not shown).
To verify the synergistic action of Fur and HlyIIR, we introduced pFUR-Km in EH2R. In the absence of IPTG, the presence of pFUR-Km does not affect the hemolytic activity of strain EH2R. However, EH2R completely lost its residual hemolytic activity in the presence of either IPTG or Fe or both (Fig. 5B). Thus, we found that only combined action of both transcriptional regulators eliminates the hemolytic activity completely.
Fur binds to the hlyII promoter.
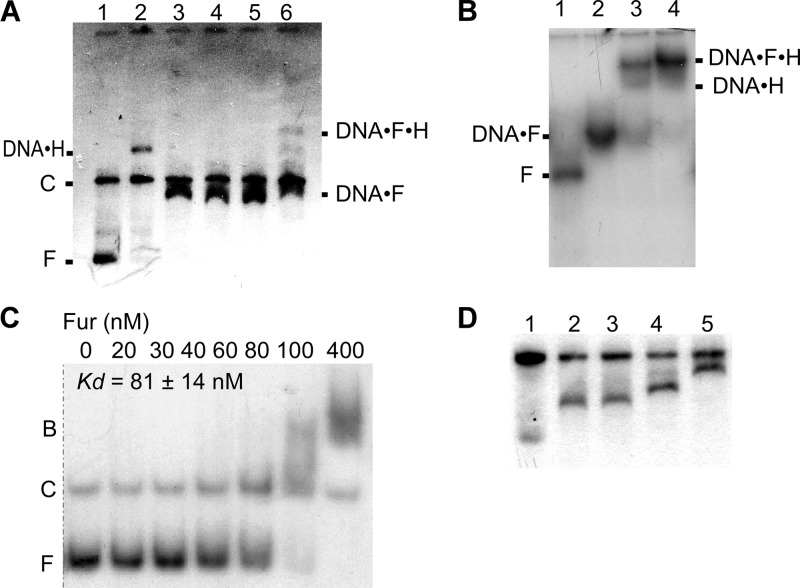
In order to prove that Fur regulation of HlyII expression occurs on PhlyII, we conducted in vitro experiments. First, we showed in gel shift experiments that Fur, after having been expressed in and purified from E. coli, directly binds to PhlyII. The B. cereus Fur protein was used in EMSA. Plasmid pUJ1 (35) was digested with EcoRI and BamHI (Fig. 6A). A small fragment (400 bp; positions +198 to −204 relative to the transcription start) contains the entire sequence upstream of hlyII and is predicted to bind the Fur protein, while the rest of the plasmid has no predicted Fur binding sites. The results (Fig. 6A, lanes 3 to 5) show that the electrophoretic mobility of only the 400-bp vector fragment changed in the presence of Fur, whereas the large DNA fragment did not compete for the Fur binding site, confirming the specificity of Fur binding to the hlyII regulatory region. Individual and mutual binding of both HlyIIR and Fur (Fig. 6A [lanes 2 and 6, respectively] and B) was observed, which is in agreement with the fact that their sites are separated. However, the concentration of HlyIIR that was sufficient to produce a full shift of the PhlyII fragment alone was not sufficient to do so with the hlyII fragment in complex with Fur (Fig. 6A, lane 6). In competition experiments, a 250-fold excess of unlabeled 400-bp fragment diminishes both Fur and HlyIIR binding (data not shown). Thus, we have confirmed that Fur specifically binds to the hlyII promoter.
Fig 6.
Electrophoresis mobility shift analysis of Fur-hlyII DNA interaction. (A) EMSA of Fur and HlyIIR binding to PhlyII. Plasmid pUJ1 was digested with EcoRI and BamHI. Lanes: 1, no protein added; 2, HlyIIR at 0.1 μM; 3 to 5, Fur at 0.4, 0.8, and 1.2 μM, respectively; 6, Fur at 0.8 μM after 5 min incubation (25°C) with 0.1 μM HlyIIR added. Positions of free (F) and competitor (C) DNA and DNA-protein complexes are shown. (B) HlyIIR and Fur bind to a 400-bp DNA fragment of the entire hlyII promoter-operator. Lanes: 1, no protein added; 2, Fur at 0.8 μM; 3, Fur at 0.8 μM and HlyIIR at 0.2 μM; 4, Fur at 0.8 μM and HlyIIR at 0.4 μM. (C) Determination of the apparent equilibrium binding constant (KD) of Fur-PhlyII interactions by EMSA. 5′-end-labeled DNA fragments (20 nM) were incubated in transcription buffer with the concentration of Fur shown below each lane. The samples were separated on a 6% polyacrylamide gel. Positions of bound (B) and free (F) DNA are shown. (D) Resolution of Fur-PhlyII complexes with different stoichiometries. DNA was processed as for Fig. 4A. Lanes: 1, no protein, with doubled DNA to ensure visibility of diffuse small fragments; 2, Fur at 0.3 μM; 3, Fur at 0.6 μM; 4, Fur at 1.2 μM; 5, Fur at 2 μM. Experiments for panels A, B, and D were done using 0.7% agarose.
Next, we characterized the Fur-PhlyII interaction via quantitative EMSA using a DNA fragment that corresponds to the region from position −94 to +140 relative to the start of hlyII transcription (Fig. 6C). The transition between bands occurs at between 60 and 100 nM Fur, indicating that the apparent KD value is around 80 nM. The curve for binding of Fur protein to the DNA was sigmoid, with a Hill coefficient higher than 2, which may indicate that more than one Fur dimer binds to the DNA. Although the stoichiometry of the Fur-DNA complexes is unknown, in some experiments it was clear that as the concentration of Fur was increased, a portion of shifted complexes with higher molecular weight was observed (Fig. 6C and D). At least 3 slower migration complexes were resolved. Thus, slower-migrating DNA species may contain more than one Fur dimer per DNA fragment.
Additionally, we verified that supplementation of purified Fur protein with iron changed neither the binding constant nor the in vitro EMSA pattern (data not shown). These results suggest that the protein preparation was saturated with iron or that iron does not directly affect Fur binding properties in vitro. In summary, these gel shift experiments prove that Fur binds to the hlyII promoter specifically and may interfere with the binding of HlyIIR.
Fur represses hlyII transcription by preclusion of RNA polymerase binding.
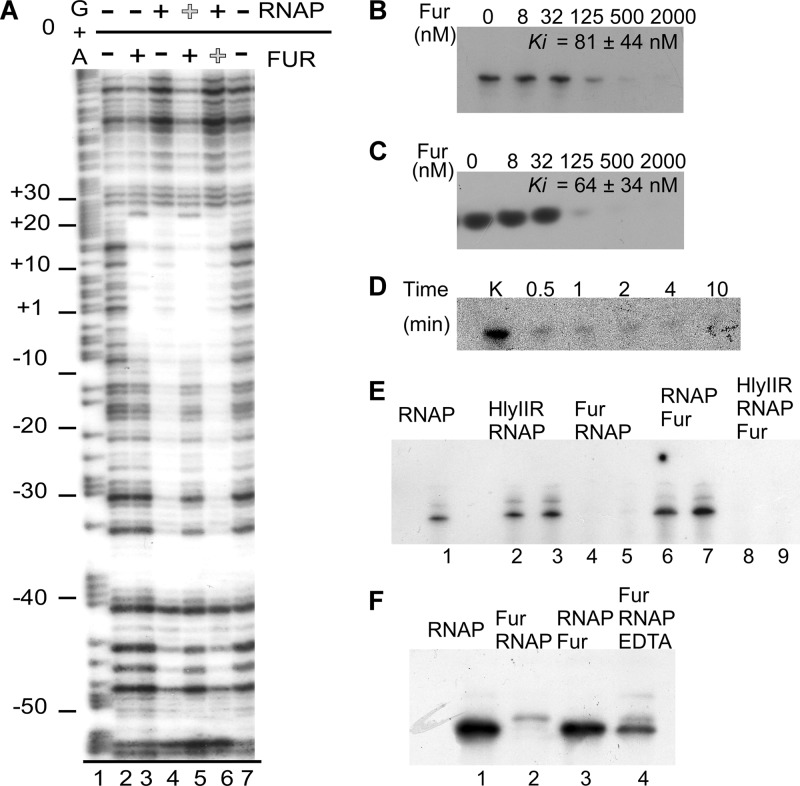
Next, we confirmed that Fur binds to the predicted Fur box within the hlyII promoter (Fig. the 7A). Fur prevented DNase I-specific DNA cleavage of the extended region around transcription site, between positions −10 and +22. The Fur-protected region overlaps well with a footprint obtained in the presence of RNA polymerase (−50 to +25) (lane 4) but not HlyIIR (−60 to −17) (9). To further explore the mechanism of Fur repression, we performed the DNase I footprinting in the presence of both Fur and RNAP (Fig. 7A, lanes 5 and 6). The results indicated that binding of Fur and binding of RNAP were mutually exclusive and strongly dependent on the order of protein binding. Only the footprint of the protein added first was observed. This suggests that Fur binding may obstruct RNAP from binding to the hlyII promoter.
Fig 7.
Fur recognizes a Fur box within the hlyII promoter and inhibits hlyII transcription in vitro by competing with RNA polymerase. (A) DNase I footprint analyses for the noncoding strand (template strand) of the hlyII promoter. Lane 1, product of G/A sequencing reaction. Lanes 2 to 7, results from DNase I footprinting. Lanes 2 and 7, empty DNA; lane 3, no RNAP, 1.4 μM Fur; lane 4, 0.1 μM RNAP, no Fur; lane 5, 1.4 μM Fur added first and then 0.1 μM RNAP; lane 6, 0.1 μM RNAP and then 1.4 μM Fur. (B) Runoff product formation. (C) Abortive initiation. (D) Time-dependent synthesis of runoff product in the presence of 0.25 μM Fur. (E) Fur and HlyIIR may work together in transcription inhibition. Fifty nanograms of 400-bp PhlyII fragments and proteins was combined in the presence of GTP, ATP, CTP, and [α-32P]UTP. Lane 1, 0.1 μM RNAP; lanes 2 and 3, 2 and 4 μM HlyIIR, respectively, and then 0.1 μM RNAP; lanes 4 and 5, 0.7 and 1.4 μM Fur, respectively, and then 0.1 μM RNAP; lanes 6 and 7, 0.1 μM RNAP and then 0.7 and 1.4 μM Fur, respectively; lanes 8 and 9, 2 and 4 μM HlyIIR, respectively, 0.1 μM RNAP, and 0.7 μM Fur added last. (F) EDTA can reduce the effect of Fur inhibition. Protein concentrations are as in Fig. 5E.
Since our footprint results demonstrated that Fur binds to the PhlyII transcription start point, in vitro transcription was performed to determine whether bound Fur inhibits hlyII transcription and how it interacts with HlyIIR in in vitro transcription. First, we studied the effects of different concentrations of purified Fur on RNA transcription in vitro (Fig. 7B and C). In single-round transcription assays, where heparin was used to prevent multiple initiation rounds, Fur was added to the preformed transcription complexes, with an additional 10 min of incubation to achieve equilibrium; this protocol blocked the RNA transcription equally efficiently in both abortive initiation and runoff assays. The inhibition curves were strikingly sigmoid, as was observed in EMSA. In this system, once Fur was added in an inhibiting concentration, prolonged incubation was not able to overcome the Fur inhibition effect (Fig. 7D). We conclude that rather than delaying transcription, Fur prevents transcription initiation.
To explore the potential interactions between two transcriptional regulators, we performed multiround runoff transcription experiments using both HlyIIR and Fur proteins (Fig. 7E). In agreement with the footprinting experiments, Fur completely blocked hlyII transcription when added before RNAP (lanes 4 and 5). The same concentrations of Fur were not sufficient to prevent transcription of preformed hlyII-RNAP complexes (lanes 6 and 7). HlyIIR at chosen suboptimal concentrations, when added first, was not able to prevent hlyII transcription, but at higher concentrations it reduced the yield of transcription (9). However, the same concentrations of HlyIIR (before RNAP) and Fur (after RNAP) regulators added together were able to prevent transcription completely. This observation suggests that Fur and HlyIIR act together in transcription inhibition.
The Fur metallation status seems to be important to hlyII transcription repression. First, EMSA experiments failed when running buffers were supplemented with EDTA (1 mM). Second, the presence of 0.5 mM EDTA in the transcription buffer partially restored the Fur-inhibited RNAP activity in a multiround transcription assay (Fig. 7F).
Overall, our in vitro results demonstrate that Fur competes with RNAP binding to the hlyII promoter and prevents RNAP binding.
DISCUSSION
Bacterial pore-forming toxins, such as hemolysin II, are essential for adaptation of microorganisms to the challenges of the environment (5). The production of cytotoxins should be very well tuned to particular conditions because of the high cost of synthesis, strong disruption potential, and latent danger to membranes of bacterial cells producing them (13). Bacterial cells interpret a broad range of environmental signals through the use of different mechanisms, including transcriptional and posttranscriptional regulation; iron is one of these signals. Inside mammalian hosts, iron is mostly bound to proteins, and free iron concentration could be as low as 10−24 M (30). Iron-rich and iron-deficient conditions are triggers that sharply switch bacterial metabolism, changing protein expression patterns, modifying bacterial cell behavior, and allowing for fast adaptation. In many cases, the effect of iron on bacteria is mediated by the ferric uptake regulator Fur, which is a known regulator of virulence in bacterial pathogens (12). For example, a Staphylococcus aureus Δfur mutant demonstrates impaired expression of immunomodulatory proteins, but expression of cytolysins, which may potentially enhance the immune response, is increased. This suggests that Fur organizes expression of S. aureus virulence factors (39). Accordingly, a Fur-deficient strain of B. cereus has markedly decreased virulence (21), but the identities of the Fur-regulated genes responsible for this effect are unknown. Most probably, fur-deficient B. cereus has decreased fitness in the model organism due to increased sensitivity to oxidative stress, or bacterial cells lose tight control under the expression of iron uptake determinants, such as hemolysin II. This study presents the first experimental evidence that expression of one of the B. cereus pathogenicity factors, hemolysin II, is regulated by iron concentration and that this regulation depends on the ferric uptake regulator.
The regulatory circuitry of the hlyII gene provides for a very delicate fine-tuning. In the most commonly used bacterial medium, LB, the concentration of iron ions is ambient (about 17 μM [1]). Under these conditions, expression of hlyII is downregulated by Fur in a growth phase-specific way: iron-replete conditions suppress HlyII production by 2- to 4-fold during the whole exponential phase of growth (Fig. 2B), while under conditions of iron depletion, expression is derepressed (2- to 3-fold) only in the early exponential phase, at essentially low cell densities (Fig. 3C). Regulation by Fur is coordinated with the HlyIIR-mediated regulation in an additive manner. Maximum levels of expression of hemolysin II require that both regulators are at low concentrations or inactive (Fig. 4A). Moderate expression of HlyII is observed when HlyIIR is absent but Fur is active and loaded with iron (Fig. 5A). When both regulators are present in high concentrations, only negligible amounts of HlyII protein are synthesized (Fig. 5B). Thus, our data indicate that, hierarchically, Fur has a secondary role in hlyII expression compared to HlyIIR. However, in the presence of HlyIIR, the role of Fur is very important during the early exponential phase of growth, where HlyII hemolytic activity is delayed in the presence of iron (Fig. 2A).
However, Fur regulation is never this straightforward. Thus, the effect of iron-replete conditions is much more complex. In an hlyIIR background, low production of HlyII is additionally impaired under iron-deficient conditions. In the absence of HlyIIR, the activating effect of iron-replete conditions is never strong. This suggests that Fur (and iron) may regulate hemolysin II expression indirectly. The mechanisms of this regulation are obscure, may be on a transcriptional or posttranscriptional level, and will be the subject of our future research.
Our complementary in vitro results provide additional evidence of Fur-dependent regulation of hlyII expression. DNase I footprinting experiments (Fig. 7A) mapped the Fur box overlapping the previously identified transcription start point (9). The Fur BS is located downstream of the −10 sequence of the hlyII promoter, suggesting a direct competition of Fur with RNAP. The sites of HlyIIR and Fur are separated; however, in EMSA experiments we observed slightly decreased affinity of HlyIIR (Fig. 6A). This interaction is specific and is not competed out by the addition of excess nonspecific DNA. The sharp cooperativity observed in the EMSA experiments suggests that more than one Fur dimer binds to PhlyII. We reliably detected at least three different Fur binding events (Fig. 6D). Given that our footprinting assays unambiguously show only one mapped Fur BS, the results suggest that Fur may oligomerize on its binding site.
To investigate the kinetic aspects of Fur-mediated hlyII regulation, we performed a series of in vitro transcription experiments in which we varied the order of the addition of components. Fur-mediated inhibition of in vitro transcription was much more efficient when the regulator was added to the naked DNA, with RNAP added last (Fig. 7E). The simple sequestration model can rationalize this observation: Fur BS partially overlaps the −10 sequence of PhlyII, and by priming the reaction with the addition of Fur, we efficiently obstruct the RNAP binding site. Fur inhibition constants for runoff and abortive transcription (Fig. 7B and C) were in agreement with the binding constant that was observed in EMSA experiments (Fig. 6C).
Therefore, we conclude that Fur is a supplementary regulator, allowing for precise control of hemolysin II expression.
ACKNOWLEDGMENTS
This work was supported by grant 1357 from Federal Target Program “Research and Scientific-Pedagogical Personnel of Innovative Russia” for 2010–2012 (Z.I.A.-K.) and partially by the Russian Foundation for Basic Research (12-04-00558) (A.S.S.).
We are grateful to John Helmann (Cornell University) for providing B. subtilis CU1065 and HB2501, to Andrei Pomerantsev (NIH, Bethesda, MD) for help with the plasmid construction, to Marina Zakharova (IBPM) for providing B. subtilis RNAP, and to Vasili Hauryliuk (University of Tartu) for critical reading and valuable suggestions.
Footnotes
Published ahead of print 20 April 2012
REFERENCES
- 1. Abdul-Tehrani H, et al. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agaisse H, Lereclus D. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97–107 [DOI] [PubMed] [Google Scholar]
- 3. Andreeva ZI, et al. 2007. The properties of Bacillus cereus hemolysin II pores depend on environmental conditions. Biochim. Biophys. Acta 1768:253–263 [DOI] [PubMed] [Google Scholar]
- 4. Andreeva ZI, et al. 2006. Purification and cytotoxic properties of Bacillus cereus hemolysin II. Protein Expr. Purif. 47:186–193 [DOI] [PubMed] [Google Scholar]
- 5. Andreeva-Kovalevskaya ZI, Solonin AS, Sineva EV, Ternovsky VI. 2008. Pore-forming proteins and adaptation of living organisms to environmental conditions. Biochemistry (Mosc.) 73:1473–1492 [DOI] [PubMed] [Google Scholar]
- 6. Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 7. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottone EJ. 2010. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23:382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Budarina ZI, et al. 2004. A new Bacillus cereus DNA-binding protein, HlyIIR, negatively regulates expression of B. cereus haemolysin II. Microbiology 150:3691–3701 [DOI] [PubMed] [Google Scholar]
- 10. Budarina ZI, et al. 1994. Hemolysin II is more characteristic of Bacillus thuringiensis than Bacillus cereus. Arch. Microbiol. 161:252–257 [DOI] [PubMed] [Google Scholar]
- 11. Cadot C, et al. 2010. InhA1, NprA, and HlyII as candidates for markers to differentiate pathogenic from nonpathogenic Bacillus cereus strains. J. Clin. Microbiol. 48:1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ceuppens S, et al. 2011. Regulation of toxin production by Bacillus cereus and its food safety implications. Crit. Rev. Microbiol. 37:188–213 [DOI] [PubMed] [Google Scholar]
- 14. Chen L, James LP, Helmann JD. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175:5428–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fagerlund A, Lindbäck T, Storset AK, Granum PE, Hardy SP. 2008. Bacillus cereus Nhe is a pore-forming toxin with structural and functional properties similar to the ClyA (HlyE, SheA) family of haemolysins, able to induce osmotic lysis in epithelia. Microbiology 154:693. [DOI] [PubMed] [Google Scholar]
- 16. Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185:6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gohar M, et al. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793 doi:10.1371/journal.pone.0002793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gohar M, et al. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784–791 [DOI] [PubMed] [Google Scholar]
- 19. Granum PE. 2005. Bacillus cereus, p 409 In Fratamico P, Bhunia A, Smith J. (ed), Foodborne pathogens: microbiology and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 20. Guedon E, Helmann JD. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48:495–506 [DOI] [PubMed] [Google Scholar]
- 21. Harvie DR, Vilchez S, Steggles JR, Ellar DJ. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151:569–577 [DOI] [PubMed] [Google Scholar]
- 22. Helgason E, et al. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen GB, Hansen BM, Eilenberg J, Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631–640 [DOI] [PubMed] [Google Scholar]
- 24. Kashlev M, et al. 1996. Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol. 274:326–334 [DOI] [PubMed] [Google Scholar]
- 25. Kataev AA, Andreeva-Kovalevskaya ZI, Solonin AS, Ternovsky VI. 2012. Bacillus cereus can attack the cell membranes of the alga Chara corallina by means of HlyII. Biochim. Biophys. Acta 1818:1235–1241 [DOI] [PubMed] [Google Scholar]
- 26. Kovalevskiy OV, Lebedev AA, Surin AK, Solonin AS, Antson AA. 2007. Crystal structure of Bacillus cereus HlyIIR, a transcriptional regulator of the gene for pore-forming toxin hemolysin II. J. Mol. Biol. 365:825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lapouge K, et al. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol. Microbiol. 66:341–356 [DOI] [PubMed] [Google Scholar]
- 28. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 29. Lund T, De Buyser ML, Granum PE. 2000. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38:254–261 [DOI] [PubMed] [Google Scholar]
- 30. Raymond KN, Dertz EA, Kim SS. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U. S. A. 100:3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Schmidt T, Scott E, Dyer D. 2011. Whole-genome phylogenies of the family Bacillaceae and expansion of the sigma factor gene family in the Bacillus cereus species-group. BMC Genomics 12:430 doi:10.1186/1471-2164-12-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Severinova E, Severinov K, Darst SA. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA1. J. Mol. Biol. 279:9–18 [DOI] [PubMed] [Google Scholar]
- 34. Shadrin AM, Shapyrina EV, Siunov AV, Severinov KV, Solonin AS. 2007. Bacillus cereus pore-forming toxins hemolysin II and cytotoxin K: polymorphism and distribution of genes among representatives of the cereus group. Mikrobiologiia 76:462–470 [PubMed] [Google Scholar]
- 35. Sinev MA, Budarina ZI, Gavrilenko IV, Tomashevskii A, Kuz'min NP. 1993. Evidence of the existence of hemolysin II from Bacillus cereus: cloning the genetic determinant of hemolysin II. Mol. Biol. (Mosc.) 27:1218–1229 [PubMed] [Google Scholar]
- 36. Sineva EV, et al. 2009. Expression of Bacillus cereus hemolysin II in Bacillus subtilis renders the bacteria pathogenic for the crustacean Daphnia magna. FEMS Microbiol. Lett. 299:110–119 [DOI] [PubMed] [Google Scholar]
- 37. Slamti L, Lereclus D. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579–606 [DOI] [PubMed] [Google Scholar]
- 39. Torres VJ, et al. 2010. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78:1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tran SL, et al. 2011. Haemolysin II is a Bacillus cereus virulence factor that induces apoptosis of macrophages. Cell. Microbiol. 13:92–108 [DOI] [PubMed] [Google Scholar]
- 41. Vander Horn PB, Zahler SA. 1992. Cloning and nucleotide sequence of the leucyl-tRNA synthetase gene of Bacillus subtilis. J. Bacteriol. 174:3928–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]