Abstract
The common colonization factor of Escherichia coli, the Mat (also termed ECP) fimbria, functions to advance biofilm formation on inert surfaces as well as bacterial adherence to epithelial cells and subsequent colonization. We used global mini-Tn5 transposon mutagenesis to identify novel regulators of biofilm formation by the meningitic E. coli isolate IHE 3034. Of the 4,418 transformants, we found 17 that were impaired in biofilm formation. Most of these mutants were affected in lipopolysaccharide synthesis and were reduced in growth but not in Mat fimbria expression. In contrast, two mutants grew well but did not express Mat fimbria. The insertions in these two mutants were located at different sites of the rcsB gene, which encodes a DNA-binding response regulator of the Rcs response regulon. The mutations abrogated temperature-dependent biofilm formation by IHE 3034, and the phenotype correlated with loss of mat expression. The defect in biofilm formation in the rcsB mutant was reversed upon complementation with rcsB as well as by overexpression of structural mat genes but not by overexpression of the fimbria-specific activator gene matA. Monitoring of the mat operon promoter activity with chromosomal reporter fusions showed that the RcsB protein and an RcsAB box in the mat regulatory region, but not RcsC, RcsD, AckA, and Pta, are essential for initiation of mat transcription. Gel retardation assays showed that RcsB specifically binds to the mat promoter DNA, which enables its function in promoting biofilm formation by E. coli.
INTRODUCTION
Bacterial growth in biofilm provides many advantages compared to a planktonic lifestyle. In a biofilm, bacteria gain resistance to antimicrobial agents and defense systems of the host as well as better protection from predation and other environmental assaults, such as hydrodynamic shear forces and desiccation (9, 20). This protective effect probably explains why bacterial biofilm growth is common in many environmental, clinical, and industrial settings and why it complicates the elimination of biofilm-associated bacteria. Following initial attachment to a surface, bacterial cells undergo profound physiological and metabolic adjustments along the transition from a single-cell state into a communal lifestyle. This complex process of biofilm development requires integration of a variety of environmental, cell-to-cell, and intracellular signals into an interplay of regulatory networks (25, 50).
Escherichia coli is a commensal bacterium in the gastrointestinal tract as well as a pathogen of intra- and extraintestinal body sites. E. coli expresses an arsenal of surface appendages that confer tissue-adhesive and biofilm formation capacity upon the bacterium (27, 28). Meningitis-associated and temperature-regulated (Mat) fimbriae (or E. coli common pili [ECP]) promote biofilm growth on inert surfaces (34) and adhesion to human epithelial cell lines in vitro (4, 32, 53, 55). Available data also indicate that the fimbria is expressed in vivo in the human body and in the intestine of infant mouse (32, 53). Gene screening assays covering a total of ca. 600 E. coli isolates have detected the major fimbrillin gene matB (or ecpA) in over 91% of the pathogenic and commensal E. coli isolates (4, 6, 21, 46, 53, 55, 59). A similar prevalence is observed for the entire matABCDEF operon in the 45 available E. coli genomic sequences, where the mat genes also are highly conserved in sequence and chromosomal location (T. A. Lehti, P. Bauchart, M. Kukkonen, U. Dobrindt, T. K. Korhonen, B. Westerlund-Wikström, submitted for publication). The mat operon clearly is a fitness determinant of E. coli; thus, a better understanding of the factors that regulate its expression is of interest. In a previous study we demonstrated that mat expression is subject to silencing by the nucleoid-associated protein H-NS and that the mat operon-encoded regulator MatA is required in order to overcome this repression and initiate transcription from the mat promoter in the newborn meningitis E. coli (NMEC) isolate IHE 3034. The promoter controls the expression of matABCDEF transcript from which an abundant matB transcript is processed. The mature matB transcript is highly stable at 20°C, whereas at 37°C it is unstable and of very low abundance, and MatA is indirectly involved in matB mRNA turnover. Transcription of matB is modulated by temperature, growth phase, low pH, and high acetate; the last two cues lead to expression of Mat fimbria at 37°C in NMEC isolates of the virulence-associated phylogenetic group B2 (Lehti et al., submitted).
In order to better understand mat expression and biofilm formation by E. coli cells, we performed a genetic search in IHE 3034, a well-characterized NMEC isolate of the serotype O18ac:K1:H7 and the phylogroup B2 (1, 29, 34, 41). Our aim was to identify genes that affect biofilm formation by IHE 3034, and we found that RcsB of the Rcs phosphorelay has a role in the control of biofilm formation and Mat fimbria expression by E. coli IHE 3034.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains and plasmids employed in this study are listed in Table 1. All cultivations were performed at 20 or 37°C. For biofilm assays, the cultures were grown in M63 minimal medium (M63 salts [44] supplemented with 0.4% glucose and 1% Casamino Acids) supplemented with the appropriate antibiotics. For other experiments, the bacteria were grown in Luria-Bertani (LB) broth. The effect of osmotic shock, a condition known to activate the Rcs phosphorelay (64), was tested by adding NaCl (final concentration, 500 mM) or 15% (wt/vol) sucrose to logarithmic-phase cultures and then incubating for 1 h (64). The effect of glucose and zinc was tested by preculturing bacteria to logarithmic phase at 37°C and then growing the bacteria at 20°C for 3 h in the presence of 0.4% glucose and 1 mM ZnCl2 (19). The antibiotic added was ampicillin (Ap, 100 μg/ml), chloramphenicol (Cm, 25 μg/ml), kanamycin (Km, 25 μg/ml), rifampin (Rif, 75 μg/ml), or streptomycin (Sm, 100 μg/ml). Gene expression from pSE380-based plasmids in recombinant E. coli strains was induced as described earlier (34, 46) by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 5 μM.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| IHE 3034-Sm | Streptomycin-resistant IHE 3034, O18ac:K1:H7 | 45 |
| IHE 3034-203 | IHE 3034-Sm rrnB T14-pmatA-lacZ [−608 to +81 from the GTG of matA (pA)] | T. A. Lehti |
| IHE 3034-204 | IHE 3034-Sm rrnB T14-pmatA-lacZ [−608 to +745 from the GTG of matA(A536C) (pAB)] | T. A. Lehti |
| IHE 3034-213 | IHE 3034-Sm rcsB::mini-Tn5cat rrnB T14-pmatA-lacZ [−608 to +81 from the GTG of matA (pA)] | This study |
| IHE 3034-214 | IHE 3034-Sm rcsB::mini-Tn5cat rrnB T14-pmatA-lacZ [−608 to +745 from the GTG of matA(A536C) (pAB)] | This study |
| IHE 3034-215 | IHE 3034-Sm rrnB T14-pmatA-lacZ [−608 to +745 with G−340T−337C−336A−331G−330C−327 from the GTG of matA(A536C) (pABRcsAB box mut)] | This study |
| IHE 3034-216 | IHE 3034-204 ΔrcsC (+61 to +2790 from the TTG of rcsC) | This study |
| IHE 3034-217 | IHE 3034-204 ΔrcsD (+61 to +1834 from the ATG of rcsD) | This study |
| IHE 3034-218 | IHE 3034-204 ΔackA Δpta (+61 to +3362 from the ATG of ackA) | This study |
| IHE 3034-Rif | Rifampin-resistant IHE 3034, O18ac:K1:H7 | 46 |
| IHE 3034-102 | IHE 3034-Rif matA(A536C), resulting in the substitution MatA H179P | T. A. Lehti |
| IHE 3034-120 | IHE 3034-Rif rrnB T14-prcsB-lacZ (−803 to +20 from the ATG of rcsB) | This study |
| IHE 3034-121 | IHE 3034-Rif rrnB T14-prcsD-lacZ (−259 to +16 from the ATG of rcsD) | This study |
| IHE 3034-122 | IHE 3034-102 rrnB T14-prcsB-lacZ (−803 to +20 from the ATG of rcsB) | This study |
| IHE 3034-123 | IHE 3034-102 rrnB T14-prcsD-lacZ (−259 to +16 from the ATG of rcsD) | This study |
| IHE3034-96 | IHE 3034-Rif ΔmatBCDEF (+61 from the ATG of matB to +753 from the ATG of matF) | 34 |
| S17-1 λpir | recA thi pro hsdR RP4-2-Tc::Mu Km::Tn7 Smr TpR λpir | 63 |
| Sm10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 63 |
| BL21(DE3) | F− ompT dcm hsdSB (rB− mB−) gal lon λDE3 | 65 |
| BL21(AI) slyD | F− ompT dcm hsdSB(rB− mB−) gal araB::T7RNAP-tetA ΔslyD::cat | 10 |
| Plasmids | ||
| pSE380 | Expression vector, ptrc, lacI, lacO, Apr | Invitrogen |
| pRCSB1 | IHE 3034 rcsB under ptrc in pSE380 | This study |
| pMAT6 | IHE 3034 matBCDEF under ptrc in pSE380 | 46 |
| pMAT19 | IHE 3034 matA under ptrc in pSE380 | T. A. Lehti |
| pUC19-placZ | IHE3034 lacI-pRS551 rrnB T14-partial lacZ in pUC19 | T. A. Lehti |
| pUC19-pAB | IHE3034 pmatA [−608 to +745 from the GTG of matA(A536C)] in pUC19-placZ | T. A. Lehti |
| pCVD442 | Suicide vector, pir-dependent ori R6K, mob RP4, sacB, Apr | 12 |
| pUTmini-Tn5 Cm | Transposon vector, mini-Tn5cat, pir-dependent ori R6K, mob RP4, Apr Cmr | Biomedal |
| pMAL-c2x | MBP fusion vector, ptac, malE, lacI, Apr | New England Biolabs |
| pMAL-RCSA1 | IHE 3034 rcsA in pMAL-c2x | This study |
| pBAU1 | IHE 3034 matA in pMAL-c2x | P. Bauchart |
| pQE30 | N-terminal 6× His tag fusion vector, pT5, lacO, Apr | Qiagen |
| pQE30-RCSB1 | IHE 3034 rcsB in pQE30 | This study |
| pQE30-RCSB2 | IHE 3034 rcsB(T168G) in pQE30, resulting in RcsBD56E | This study |
| pREP4 | Repressor plasmid, lacI, Kmr | Qiagen |
Transposon mutagenesis.
E. coli strain Sm10 λpir was used as the donor to conjugally transfer plasmid pUTmini-Tn5 Cm into IHE 3034-Sm. The transconjugants, with the mini-Tn5 transposon integrated into the chromosome, were selected on LB agar plates containing chloramphenicol and streptomycin. Sodium citrate (0.5%) was added to the agar plates during mating and during transconjugant selection. A total of 4,418 colonies were picked into 96-well NunclonΔ-polystyrene microtiter plates (Nunc) containing 160 μl of M63 medium. After incubation at 20°C for 48 h, the cultures were diluted 1:100 in M63 medium and transferred into 96-well polyvinyl chloride (PVC) microtiter plates (Falcon). Cultures were further incubated at 20°C for 48 h and crystal violet (CV) stained for biofilm formation analysis as described below.
Biofilm formation on PVC microtiter plates.
The ability to form biofilm on PVC microtiter plates (Falcon) was determined as described previously (34). In brief, a culture grown in M63 medium for 48 h at 20°C was diluted 1:200 in M63 medium, and 160 μl of this cell suspension was transferred to each of six parallel wells on a 96-well PVC microtiter plate. After incubation at 20°C for 48 h, the planktonic cells were removed from the wells and the optical density of the cell suspension was measured at 595 nm (OD595) using an enzyme-linked immunosorbent assay (ELISA) recorder (Multiskan EX, Thermo Scientific) as a measure of total growth. Furthermore, the adherent cell layers on the wells were washed twice with phosphate-buffered saline (PBS), pH 7.1, dried, and stained with 0.1% CV for 10 min. After being washed two times with PBS and dried, cells were destained with 180 μl of ethanol-acetone (80:20, vol/vol) and diluted 1:4 in ethanol-acetone (80:20, vol/vol). The mixture (160 μl) was transferred to a new 96-well plate, and the absorbance at 595 nm was measured in an ELISA recorder.
Identification of mini-Tn5 insertion sites.
To identify the localization of the transposon insertion, chromosomal DNA from the selected clones was extracted using Blood & Cell Culture DNA Midi Kit with genomic-tip 100/G (Qiagen) to obtain DNA pure enough for direct genomic sequencing. Chromosomal DNA was subjected to sequencing using primer 013Tn5Seq (Table 2). Sequencing was performed at the Institute of Biotechnology at the University of Helsinki. The DNA sequences obtained were compared against the complete genome of E. coli IHE 3034 (GenBank accession no. CP001969) using the BLASTN algorithm.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) | Purpose or target |
|---|---|---|
| 013Tn5Seq | ATGTTACCCGAGAGCTTG | Sequencing of mini-Tn5 insertion sites |
| RrcsB2F | CGGGAATTCTAAGGAGGTAATACATGAACAATATGAACGTA | pRCSB1 |
| RcsBR | GCCGTCGACTTAGTCTTTATCTGCCGGA | pRCSB1; rcsB DIG-probe |
| RcsBEC-up | GCGGATCCATGAACAATATGAACGTAATTATTGC | pQE30-RCSB1 and pQE30-RCSB2 |
| RcsBEC-low | CCAAGCTTAGTCTTTATCTGCCGGACTTAAGG | pQE30-RCSB1 and pQE30-RCSB2 |
| RcsBD56EF | GATTACCGAGCTCTCCATGCCTGGC | pQE30-RCSB2 |
| RcsBD56ER | ATGGAGAGCTCGGTAATCAACACATGC | pQE30-RCSB2 |
| RcsAEC-up | GGGGATCCATGTCAACGATTATTATGGATTTATG | pMAL-RCSA1 |
| RcsAEC-low | GAAAAGCTTAGCGCATGTTGACAAAAATACC | pMAL-RCSA1 |
| prcsB-803F | CGCGGATCCAGACCCAAAACGATCG | prcsB-lacZ fusion |
| prcsB20R | CGCGGATCCATTACGTTCATATTGTTCATG | prcsB-lacZ fusion |
| prcsD-259F | CGCGGATCCGCTTGTTAACTATTTCAC | prcsD-lacZ fusion |
| prcsD16R | CGCGGATCCTCTCTTTCTGACGCATTC | prcsD-lacZ fusion |
| RcsABmutF | CAACACATGAATCCTATAGCTCACACCTCAGGGCAAAG | RcsAB box mutation |
| RcsABmutR | GAGGTGTGAGCTATAGGATTCATGTGTTGTGTAATGGAC | RcsAB box mutation |
| rcsCoutF | GCAGAGCTCCACGAATAGCTGTTAGCG | rcsC deletion |
| rcsCoutR | GCATCTAGAGCAACCTGTATCACACCC | rcsC deletion |
| rcsCinF | CATGTTCAGAGCAGTGATAAAACAGACGCTGAC | rcsC deletion |
| rcsCinR | GTCTGTTTTATCACTGCTCTGAACATGTAGCGC | rcsC deletion |
| rcsDoutF | GCAGAGCTCGTTTGGAGCTGGAGATCG | rcsD deletion |
| rcsDoutR | GCATCTAGAGCAATATCGTTCTCGAC | rcsD deletion |
| rcsDinF | GGGAGCATTACCGAGAAGGCGTAAGCATTCATG | rcsD deletion |
| rcsDinR | TTACGCCTTCTCGGTAATGCTCCCCGGTAG | rcsD deletion |
| ackAoutF | GCAGAGCTCTTTGCACCGCACGCGG | ackApta deletion |
| ptaoutR | GCATCTAGACCGCTAATGGCATTGG | ackApta deletion |
| ackAptainF | TGAAATTTGCCATCGTTGATGATATCGTCTACACC | ackApta deletion |
| ackAptainR | ACGATATCATCAACGATGGCAAATTTCAGTGAAG | ackApta deletion |
| 035ZaplacZr | GTCGGTTTATGCAGCAAC | Sequencing of promoter-lacZ fusions |
| 190pRS551R | GGCCAGTGAATCCGTAATC | Sequencing of promoter-lacZ fusions |
| rcsCseqF | GCGTTAGTGTCTTATCTGGC | Sequencing of rcsC deletion |
| rcsDseqF | CGCTTGTTAACTATTTCAC | Sequencing of rcsD deletion |
| ackAseqF | CGCAAAATGGCATAGACTC | Sequencing of ackApta deletion |
| MATB-F | ATGAAAAAAAAGGTTCTGGCA | matB DIG-probe |
| RT-MATB | TTAACTGGTCCAGGTCGCGTC | matB DIG-probe |
| RCSB-F | ATGAACAATATGAACGTAATTA | rcsB DIG-probe |
| AF | CATTAAGACTATTCCTAACAC | pmatA fragment |
| AR | GCCATGTCACTACTTTCC | pmatA fragment |
| A2F | CCAATAACCTTATGAAACTG | pmatA2 fragment |
| A3F | TTTGTGCGGCATGTTAATC | pmatA3 fragment |
| A3R | CCTGATGAGTGTCAATATGT | pmatA3 fragment |
| A4F | GCCATCGTTCCTGTGACA | pmatA4 fragment |
| A4R | TGTTGTGTAATGGACTAGTA | pmatA4 fragment |
| BF | GTGTGAAGACAGTGTATAC | pmatB fragment |
| BR | CGTTACCAGAGCTATTGC | pmatB fragment |
Construction of expression plasmids.
The plasmids described below were generated using standard recombinant DNA techniques (56), the Phusion High-Fidelity DNA Polymerase (Finnzymes), and IHE 3034 chromosomal DNA as the template for PCRs. For the construction of pRCSB1, a 0.7-kb EcoRI/SalI DNA fragment containing rcsB coding sequence and a canonical ribosome binding site AGGAGG inserted 7 nucleotides (nt) upstream of the ATG start codon was amplified by using the oligonucleotide primers RcsB2F and RcsBR (Table 2). The amplicon was cloned into the EcoRI-SalI sites of pSE380 under the IPTG-inducible ptrc promoter.
The primers RcsBEC-up and RcsBEC-low described by Kelm et al. (26) (Table 2) were used to generate PCR product containing rcsB together with upstream BamHI and downstream HindIII restriction sites. The amplified DNA was cloned into vector pQE30, and the resulting plasmid was designated pQE30-RCSB1. To express a D56E mutant form of RcsB with a histidine tag, two PCR products were amplified (i) with the upstream primer RcsBEC-up and the mutagenesis primer RcsBD56ER and (ii) with the mutagenesis primer RcsBD56EF and the downstream primer RcsBEC-low (Table 2). The overlapping PCR products were purified and mixed, and a fusion product was amplified with the up- and downstream primers. The fusion DNA fragment was digested with BamHI and HindIII and cloned into vector pQE30, to generate pQE30-RCSB2.
To generate pMAL-RCSBA1, a 0.6-kb BamHI/HindIII DNA fragment containing the rcsA coding sequence was amplified by RcsAEC-up and RcsAEC-low primers (26) (Table 2) and inserted between the BamHI and HindIII restriction sites of the pMAL-c2x vector.
Construction of single-copy lacZ reporter strains.
Specific promoter-lacZ reporters were integrated into the chromosomal lac locus using an allelic-exchange strategy as previously described (Lehti et al., submitted). Briefly, the promoter region of prcsB (bp −803 to +20 from the translational start of rcsB), using primers prcsB-803F and prcsB20R, and the promoter region of prcsD (bp −259 to +16 from the translational start of rcsD), using primers prcsD-259F and prcsD16R (Table 2), were amplified from IHE 3034 genomic DNA and cloned individually into a BamHI site on the pUC19-placZ plasmid. The pUC19-placZ plasmid contains the IHE 3034 lacI gene and its upstream region (bp −420 to +1101 from the translational start of lacI), as well as four rrnB T1 terminators, a promoter cloning site and a part of the promoterless lacZ gene from reporter plasmid pRS551 (bp −756 to +2066 from the EcoRI site). The resulting plasmids were digested with XbaI and SacI, and the restriction fragments containing the prcsB or prcsD promoter with lacI and lacZ flanking regions were subcloned into a pir-dependent suicide vector pCVD442. The pCVD442-based suicide plasmids for integration of mat regulatory regions pA (bp −608 to +81 from the translational start of matA) and pAB (bp −608 to + 745 from the translational start of matA) into the chromosomal lac locus were available from previous work (Lehti et al., submitted). To mutate the RcsAB box in the pAB regulatory region, site-directed mutagenesis was performed using an inverse PCR strategy with mutagenic primers RcsABmutF and RcsABmutR, Phusion High-Fidelity DNA polymerase (Finnzymes), and 5% dimethyl sulfoxide (DMSO) as a PCR additive. The plasmid pUC19-pAB used as the template was available from previous work (Lehti et al., submitted). The purified PCR product was treated with DpnI in order to digest the parental plasmid strands, and the reaction mixture was directly transformed into DH10B. After intracellular circularization of the PCR product in DH10B, the resulting plasmid was digested with XbaI and SacI, and the XbaI-SacI fragment containing the pAB region with the substituted sequence was subcloned into pCVD442.
In-frame deletions of rcsC, rcsD, and ackApta on the chromosome of the pAB-lacZ reporter strain IHE 3034-204 were performed by site-specific mutagenesis using recombinant PCR and pCVD442 essentially as described by Mobley et al. (40). The 5′ and 3′ flanking sequences of each deletion site were amplified from IHE 3034 chromosomal DNA, fused with each other by recombinant PCR, and then cloned into the SacI-XbaI site of pCVD442. Primers for the mutagenesis are listed in Table 2.
The suicide plasmids were mobilized into IHE 3034 derivatives via conjugation from S17-1 λpir or Sm10 λpir. The transconjugants were subjected to a two-step sucrose selection, first in LB broth supplemented with 10% sucrose and then on LB agar plate with 10% sucrose. The correctly integrated single-copy promoter fusions replacing the native lacZ promoter as well as the gene deletions were verified by DNA sequencing using primers listed in Table 2.
β-Galactosidase assay.
For the measurement of β-galactosidase activities, overnight cultures were diluted 1:100 and grown to mid-logarithmic phase (OD600 of 0.5 to 0.6), and the cells from 1-ml aliquots were pelleted and resuspended in 1 ml ice-cold Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) with 50 mM β-mercaptoethanol. β-Galactosidase activities were determined as described by Miller (39) using ο-nitrophenyl-β-d-galactopyranoside as a substrate. Differences between average values were tested for significance by performing an unpaired 2-tailed Student' t test, and P values of less than 0.05 were considered statistically significant.
RNA isolation and Northern blotting.
Overnight cultures of bacteria were diluted 1:100 and grown to mid-logarithmic phase (OD600 of 0.5 to 0.6) and stationary phase (OD600 of about 2.2), and 1-ml aliquots were withdrawn at these phases. After centrifugation at 4°C, the cell pellets were resuspended in 0.1 ml of Tris-EDTA (TE) buffer containing lysozyme (400 μg/ml), and total bacterial RNA was isolated using the RNeasy Minikit (Qiagen) following the manufacturer's protocol (RNeasy Mini handbook). The RNA concentrations were determined by measuring the optical density at 260 nm, and Northern blot analysis was carried out with 2.5 μg of total RNA. The RNA samples were separated next to digoxigenin (DIG)-labeled RNA molecular weight markers (Roche Applied Science) by electrophoresis on 1.2% agarose gel containing 0.2 M formaldehyde and 0.1 μg/ml ethidium bromide, transferred onto a positively charged nylon membrane (Roche Applied Science) by overnight capillary transfer, and cross-linked by UV radiation. Prehybridization and hybridization were performed in a high-SDS hybridization buffer (7% SDS, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 0.1% N-lauroylsarcosine, 2% blocking solution [Roche (as else-where): Applied Science], 50 mM sodium phosphate at pH 7.0) at 50°C according to the DIG application manual for filter hybridization (Roche Applied Science). Membranes were probed with DIG-labeled single-stranded DNA (ssDNA) probes complementary to the coding sequences of matB or rcsB. The probes were generated by linear PCR using PCR-amplified DNA templates (prepared with the primer sets MATB-F/RT-MATB and RCSB-F/RcsBR) (Table 2), the appropriate reverse primer, and PCR DIG labeling mix (Roche Applied Science). Detection was carried out using the DIG luminescent detection kit (Roche Applied Science) according to the manufacturer's instructions, and chemiluminescence was visualized by X-ray films (Agfa).
Protein purification.
His-tagged RcsB and RcsBD56E proteins were purified from BL21(AI) slyD (pREP4) cells harboring expression plasmid pQE30-RCSB1 or pQE30-RCSB2. The strains were grown at 20°C in LB media with appropriate antibiotics to an OD600 of 0.3, followed by IPTG (1 mM) induction for 3 h. The His-tagged recombinant proteins were purified under native conditions using Ni-nitrilotriacetic acid resin according to the QIAexpress System (Qiagen). BL21(DE3) (pMAL-RCSA1), BL21(DE3) (pBAU1), and BL21(DE3) (pMAL-c2x) cells were used for purification of maltose-binding protein (MBP)-RcsA, MBP-MatA, and MBP. These cells were cultivated at 20°C in LB with 0.2% glucose and appropriate antibiotics to an OD600 of 0.6, at which point the culture temperature was reduced to 15°C and IPTG was added to a final concentration of 0.3 mM. After 4 h of incubation, the cells were harvested. MBP-RcsA, MBP-MatA, and MBP were purified using amylose resin according to the manufacturer's instructions (New England BioLabs). Finally, the purified proteins were dialyzed against 15 mM Tris-HCl at pH 7.5, 45 mM NaCl, 7.5 mM MgCl2. The protein concentrations were determined from Coomassie-stained SDS-PAGE gels comparing whole-band intensities with internal bovine serum albumin (BSA) standards of known concentrations using the Tina (v2.0) image analysis program (Raytest Isotopenmessgeräte GmbH).
EMSA.
The DNA fragments pmatA to pmatA4 and pmatB for the electrophoretic mobility shift assay (EMSA) were generated with PCR amplification using the primer sets AF/AR, A2F/AR, A3F/A3R, A4F/A4R, and BF/BR (Table 2) and IHE 3034 genomic DNA as a template. For each DNA fragment, 0.2 pmol was incubated with increasing concentrations of purified recombinant proteins in the presence of 10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 30 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1% Nonidet P-40, 0.05 mg/ml BSA, 5% glycerol. The final reaction volume was 10 μl. After incubation at 20°C for 25 min, samples were run on a 6% nondenaturing PAGE in 0.5× Tris-borate-EDTA buffer at 100 V at 20°C. Following staining with ethidium bromide, the DNA fragments were visualized under UV irradiation.
Detection of Mat fimbria expression by serum agglutination.
Bacterial agglutination in antisera was assessed as described previously (54) by gently mixing equal volumes of a bacterial suspension and a 1:100 dilution of anti-Mat antiserum (34, 46) in PBS on glass slides.
RESULTS AND DISCUSSION
Isolation of biofilm-defective and biofilm-enhanced transposon mutants.
We performed a mini-Tn5-based transposon mutagenesis in IHE 3034-Sm to identify genes involved in biofilm formation. A total of 4,418 individual transposon insertion mutants were examined for their ability to develop biofilm on polyvinyl chloride (PVC) 96-well plates. After 48 h of static incubation in M63 medium (34) at 20°C, biofilm formation was determined by crystal violet staining. Through this analysis, 31 mutants that showed reproducibly altered biofilm formation at 20°C were identified; 29 mutants had impaired ability to form biofilm, and two mutants (19E6 and 3E5) (Table 3) exhibited enhanced biofilm formation. The last two mutants were also tested for their ability to form biofilm at 37°C, but no change compared to the parental wild-type strain in the level of biofilm formation was observed (data not shown).
Table 3.
Mini-Tn5 transposon biofilm mutants of E. coli IHE 3034-Sm
| Mutant | Locus tag | Gene name | Gene product or function | Transposon position in the chromosome | No. of identical insertions in the gene | % Biofilm formationc | % Final cell densityd | Expression of Mat fimbriaee |
|---|---|---|---|---|---|---|---|---|
| 17E7 | ECOK1_0281 | matA | Positive regulator of mat operonb | 310678 | 1 | 6 ± 1 | 107 ± 2 | − |
| 6C4 | ECOK1_0281 | matA | Positive regulator of mat operonb | 310652 | 1 | 7 ± 2 | 108 ± 1 | − |
| 1H5 | ECOK1_1929 | pabB | Aminodeoxychorismate synthase | 1957754 | 1 | 26 ± 7 | 30 ± 1 | (+) |
| 30H2 | ECOK1_2452 | rcsB | Capsular synthesis regulator component B | 2530902 | 2 | 16 ± 5 | 99 ± 1 | − |
| 32C8 | ECOK1_2452 | rcsB | Capsular synthesis regulator component B | 2531153 | 1 | 14 ± 4 | 103 ± 3 | − |
| 4B10 | ECOK1_3592 | metY | tRNA-Met | 3691157 | 1 | 39 ± 13 | 90 ± 1 | (+) |
| 1H3 | ECOK1_4062 | rfaD | ADP-glyceromanno-heptose 6-epimerase | 4167517 | 1 | 6 ± 3 | 37 ± 13 | + |
| 39A11 | ECOK1_4065 | rfaL | O-antigen polymerase | 4170848 | 2 | 49 ± 18 | 58 ± 10 | + |
| 16C10 | ECOK1_4066 | rfaV | Glycosyltransferase, group 2 family protein | 4172535 | 1 | 51 ± 4 | 66 ± 6 | + |
| 49D7 | ECOK1_4067 | rfaJ | LPS 1,2-glucosyltransferase | 4172986 | 2 | 54 ± 16 | 76 ± 5 | + |
| 47G11a | ECOK1_4067 | rfaJ | LPS 1,2-glucosyltransferase | 4173659 | 1 | 46 ± 10 | 74 ± 8 | + |
| and | and | and | and | |||||
| ECOK1_4309 | tatC | Twin arginine-targeting protein translocase TatC | 4421714 | 1 | ||||
| 8E5 | ECOK1_4068 | rfaY | LPS core biosynthesis protein RfaY | 4174260 | 1 | 21 ± 10 | 78 ± 3 | + |
| 19E7 | ECOK1_4068 | rfaY | LPS core biosynthesis protein RfaY | 4173977 | 6 | 7 ± 6 | 67 ± 15 | + |
| 23G5 | ECOK1_4068 | rfaY | LPS core biosynthesis protein RfaY | 4174029 | 1 | 5 ± 2 | 77 ± 4 | + |
| 27H3 | ECOK1_4069 | rfaJ | LPS 1,2-glucosyltransferase | 4175175 | 2 | 5 ± 1 | 69 ± 9 | + |
| 1F12 | ECOK1_4070 | rfaI | LPS 1,3-galactosyltransferase | 4176489 | 1 | 14 ± 18 | 69 ± 7 | + |
| 18B12 | ECOK1_4072 | rfaG | LPS core biosynthesis protein RfaG | 4178099 | 4 | 4 ± 1 | 41 ± 4 | + |
| 19E6 | ECOK1_2267 | rfbA | Glucose-1-phosphate thymidylyltransferase | 2326200 | 1 | 163 ± 8 | 98 ± 5 | + |
| 3E5 | ECOK1_3052 | mprA | Transcriptional repressor MprA | 3121426 | 1 | 122 ± 11 | 100 ± 2 | + |
A mutant with a double transposon insertion at two different locations.
This gene description is provided according to Lehti et al. (submitted).
Percentages of biofilm formation relative to that of the wild-type IHE 3034-Sm. The data are mean values ± standard deviations from at least three independent experiments.
Percentages of final cell density of planktonic cells (optical density at 595 nm) reached at the end of the cultivation for each mutant relative to that of wild-type IHE 3034-Sm. The data represent the means ± standard deviations from three or four independent experiments.
Detected in planktonic cells at the end of cultivation by agglutination in anti-Mat fimbria antiserum. −, no agglutination; (+), weak agglutination; +, strong agglutination.
Characterization of biofilm-altered mutants.
We next purified genomic DNA from the 31 mutant strains, and the precise mini-Tn5 insertion sites were determined by direct DNA sequencing of the transposon/genome junctions using a primer that read out of the transposon. A BLASTN algorithm for homology search against the complete genome sequence of IHE 3034 identified a total of 19 separate insertion sites in 15 distinct genes (Table 3). The majority of the transposons were clustered in the genomic rfa (also known as waa) region involved in lipopolysaccharide (LPS) biosynthesis. This finding is in line with the observations showing that truncation of LPS structure abolishes biofilm formation on abiotic surfaces in E. coli isolates (2, 5, 42, 51). As differential growth might explain the observed differences in biofilm formation, we compared the cell densities of planktonic wild-type and mutant cells after 48 h of static incubation in M63 medium. The 11 mutants with insertions in the rfa LPS biosynthesis locus and the mutant carrying an insertion in pabB, which encodes an enzyme involved in folic acid biosynthesis, reached final optical cell densities that were 78% or less of that of the wild-type strain (Table 3). These mutants apparently had growth defects and were not further investigated. The remaining eight mutants, of which six were confirmed to be biofilm-defective and two biofilm-enhanced variants, were chosen for further analyses.
Mutations affecting biofilm formation.
The mutant 3E5 with the insertion in the coding sequence of mprA (also known as emrR) displayed moderately increased biofilm formation (Table 3). MprA functions as a negative regulator of the emrRAB operon that encodes a multidrug extrusion pump (35), which contributes to E. coli survival at acidic pH (11). The enhanced biofilm phenotype of the mprA mutant is presumably due to increased expression of emrAB. This is in line with previous studies showing that inactivation of efflux pumps, including EmrAB, decreases biofilm formation by E. coli (31, 38). In another biofilm-enhanced mutant, 19E6, the insertion was located in the rfbA gene, which encodes the first enzyme in the biosynthetic pathway of l-rhamnose, a component of the LPS structure (61) including the O18ac antigen of IHE 3034 (17).
One of the biofilm-defective mutants, 4B10 (Table 3), harbored the transposon in the metY gene that encodes tRNAMet. The impact of this mutation on biofilm formation likely is due to polar effects on the multicistronic operon that includes metY and encodes, e.g., the transcription termination factor NusA, the translation initiation factor IF2, the ribosome binding factor RbfA, and polynucleotide phosphorylase PNPase (52, 57).
Five of the biofilm-defective mutants carried, as did the mprA mutant, the transposon insertion in genes encoding transcriptional regulators. In two of these (17E7 and 6C4) (Table 3), the insertion was located at different positions in the first gene of the mat operon, matA. We have recently shown that the gene product MatA is a positive regulator of Mat fimbria expression and a negative regulator of bacterial motility (33; Lehti et al., submitted).
Two isolates harbored two individual transposon insertions in the rcsB gene (30H2 and 32C8) (Table 3). RcsB is a pleiotropic regulator of the Rcs (regulator of colanic acid capsule synthesis) phosphorelay signaling pathway conserved among the members of the Enterobacteriaceae family (22). A phosphorelay is an expanded form of the canonical two-component signal transduction system that involves additional receiver and phosphotransferase domains for multiple phosphotransfer events and provides more steps to fine-tune the pathway (3, 74). The Rcs pathway controls the expression of a number of genes involved in cellular processes such as synthesis of cell surface-associated structures, e.g., downregulation of flagellum biosynthesis and upregulation of synthesis of the exopolysaccharide colonic acid, and biofilm formation (13, 19, 43). Loss of positive Rcs control of the synthesis of cell surface components may be the reason for the impaired biofilm formation by the two rcsB transposon mutants.
We next assessed whether expression of the Mat fimbria is affected in the biofilm mutants. The matA and rcsB mutants 17E7, 6C4, 30H2, and 32C8 showed significant reduction in the surface expression of Mat fimbriae (Table 3). metY mutant cells exhibited a partial decrease in Mat expression level, whereas rfbA and mprA mutants retained the wild-type fimbriation capacity.
Role of the rcsB gene in Mat fimbria-mediated biofilm formation.
E. coli surface appendages reported to be affected by the Rcs system include flagella, curli, and type 1 fimbriae, which are involved in biofilm formation under different growth conditions (42, 48, 70). RcsB, together with its cofactor RcsA, represses the activity of the flagellar master operon flhDC (15) and the two csg operons for curli expression (13, 69). In contrast, RcsB positively controls the expression of type 1 fimbriae by controlling the transcription of the site-specific recombinase genes fimB and fimE, thereby favoring the “phase-on” orientation of the fimA promoter (62). The impaired Mat fimbriation phenotype of the rcsB mutants indicated a novel regulatory target for the RcsB DNA-binding protein. To confirm the involvement of RcsB in Mat-mediated biofilm formation, we introduced the plasmid pRCSB1 into the rcsB mutant 30H2 for further analysis. Complementation of the rcsB mutation completely restored Mat fimbria expression (data not shown) and biofilm formation (Fig. 1). The results thus confirmed that the observed defects in Mat fimbriation and biofilm formation are due to the inactivation of the rcsB gene and not to a polar effect of the Tn5 insertion. Overexpression of rcsB did not enhance surface expression of Mat fimbriae or the capacity to form biofilm in the wild-type IHE 3034 background, nor did it rescue Mat fimbriation in the IHE 3034 matA mutant (data not shown).
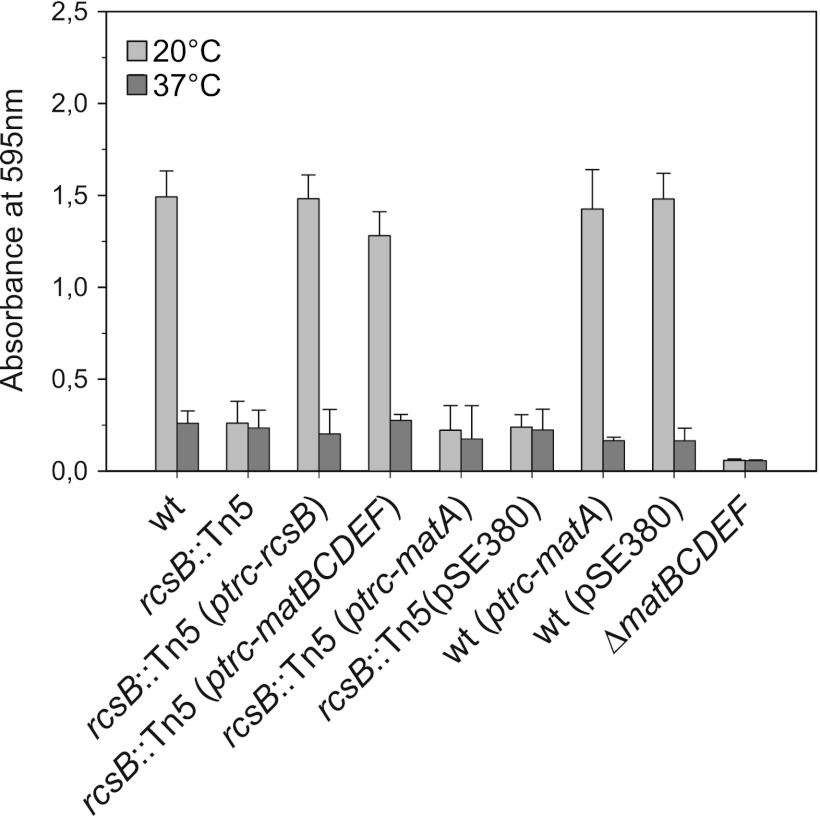
Fig 1.
RcsB is required for temperature-dependent biofilm formation by IHE 3034 via Mat fimbria expression. The wild-type strain IHE 3034-Sm and the isogenic rcsB mutant strains were grown on PVC microtiter plates in M63 medium containing 0.4% glucose at 20°C or 37°C for 48 h without shaking and then washed and stained with crystal violet to measure biofilm formation. For complementation, pSE380 plasmids carrying the rcsB, matA, or matBCDEF genes from IHE 3034 under the IPTG-inducible ptrc promoter were used. The Mat fimbria-deficient matBCDEF mutant was included in the assays for comparison. The data shown were reproduced in at least three independent experiments, and error bars represent standard deviations.
Coexpression of matBCDEF genes from the inducible plasmid pMAT6 complemented the biofilm formation phenotype of the rcsB derivative (Fig. 1), indicating that the effect of rcsB on Mat-dependent biofilm formation mainly is on the level of Mat fimbria expression. The result further supports a major role of Mat fimbriae in biofilm growth at 20°C (34).
MatA is required for activation of mat expression, and its ectopic overexpression bypasses the temperature-dependent control of Mat fimbria expression (Lehti et al., submitted). We therefore assessed whether introduction of the multicopy plasmid pMAT19 carrying inducible matA would override the requirement for rcsB in the transition from planktonic to biofilm growth. We found that the biofilm-defective phenotype remained unaltered in the rcsB strain harboring the MatA-overexpressing plasmid and at an equally low level as in the strain IHE 3034 ΔmatBCDEF (Fig. 1). Moreover, the presence of pMAT19 in the wild-type IHE 3034 background did not enhance the biofilm-promoting property at 20°C or at 37°C. This observation further corroborates that the induced overexpression of Mat fimbriae is insufficient to promote biofilm formation at 37°C (34).
Mutation of rcsB abolishes matB transcription.
A key feature in fimbrial assembly is the regulation of expression of the gene encoding the major structural subunit. Earlier work (Lehti et al., submitted) had shown that matB is transcribed in IHE 3034 as a part of the polycistronic matABCDEF mRNA, which is rapidly processed to a stable transcript of 0.67 kb containing the intact matB mRNA. To investigate the effects of rcsB on matB transcription, a Northern blot analysis was performed, using a probe complementary to matB. As shown in Fig. 2, matB transcript accumulated in logarithmic-phase cells of wild-type IHE 3034 grown at 20°C. Inactivation of rcsB by transposon mutagenesis resulted in undetectable levels of matB mRNA, whereas complementation of the mutation increased the matB transcript level close to that seen in the wild-type cells from 20°C. However, the growth phase dependence and temperature dependence of matB regulation remained unaffected by ectopic overexpression of rcsB. No matB mRNA was observed in total RNA isolated from the rcsB mutant harboring the matA-bearing plasmid pMAT19 or the empty expression vector pSE380. These findings confirmed that RcsB is involved in transcriptional control of the mat operon, in addition to MatA, as an indispensable regulatory protein. Further, the lack of matB transcript in rcsB-deficient cells in the presence of multicopy matA was independent of growth temperature, indicating that RcsB is involved in the regulation of Mat fimbria expression at 20°C and at 37°C. An interesting difference was observed between RcsB- and MatA-mediated activation of matB transcription: overexpression of matA in trans in the wild-type IHE 3034 background (Fig. 2) or in the matA mutant (Lehti et al., submitted) loosened the temperature and growth phase dependency in matB transcription, whereas no accumulation above normal levels of matB transcript was evident after overexpression of rcsB in the rcsB mutant (Fig. 2). Thus, the cellular concentration of MatA but not of RcsB is critical for the temperature control of Mat fimbria expression.
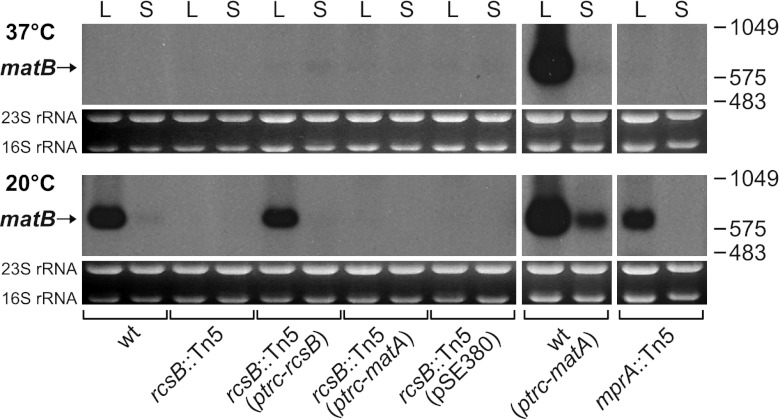
Fig 2.
Disruption of rcsB prevents transcription of matB fimbrillin gene. Northern blot analysis of matB mRNA levels in wild-type IHE 3034-Sm, the rcsB mutant, the mprA mutant, and strains overproducing rcsB or matA from a plasmid-borne IPTG-inducible ptrc promoter. Total RNA (2.5 μg) isolated from cells grown to mid-logarithmic (L) and stationary growth phases (S) in LB at 20 or 37°C was hybridized with a DIG-labeled probe specific to matB. As RNA loading controls, the panels below the Northern blots show the abundance of 23S and 16S rRNA in each sample after denaturing agarose gel electrophoresis and ethidium bromide staining of the gels used for blotting. The migration positions of DIG-labeled RNA molecular weight markers (bases) are indicated on the right.
We also analyzed by Northern hybridization total RNA samples isolated from the biofilm-enhanced mprA mutant 3E5, which carried the transposon in a regulatory gene. The inactivation of mprA did not alter the transcription of matB (Fig. 2), which is in accordance with the observed reactivity of the strain 3E5 with anti-Mat fimbria antibodies (Table 3).
RcsB is required for activation of mat promoter.
We next individually integrated the mat promoter-lacZ fusions pA-lacZ and pAB-lacZ as single copies into the lacZ locus of the rcsB mutant 30H2. The pA-lacZ fusion encompasses 608 bp of the mat upstream region and the first 81 bp of matA. The pAB-lacZ fusion includes the mat upstream region, a missense-substituted matA(A536C) and a 73-bp matA-B intergenic region followed by the first 81 bp of the matB open reading frame. See Fig. 3A for an outline of the promoter fusions. The A536C substitution (H179P in the MatA protein) inactivates DNA binding, and it was here included in the pAB-lacZ fusion to avoid two functional copies of matA in the chromosome. The IHE 3034-derived sequence in the pAB-lacZ fusion contains a potential H-NS-repressed downstream regulatory element and supports lower LacZ activity in the wild-type background (Lehti et al., submitted) (Fig. 3A). In particular, in IHE 3034 cells from 37°C, the pAB-lacZ fusion directs only basal levels of lacZ expression. This expression profile correlates with the matB mRNA levels detected by Northern hybridization (Fig. 2).
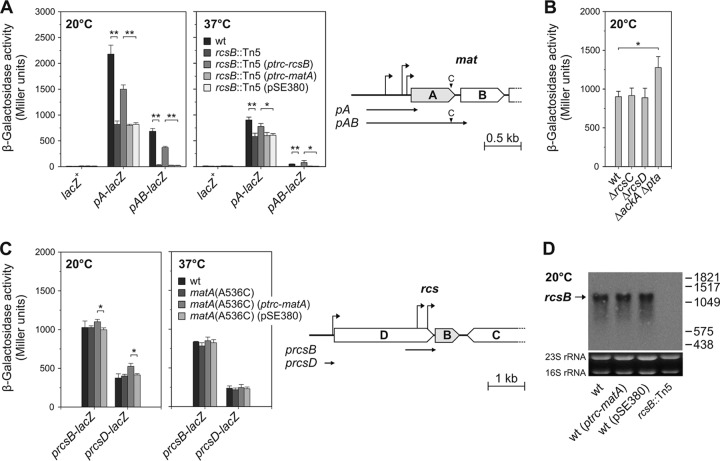
Fig 3.
Activation of mat promoter is dependent on RcsB. (A) The graphs on the left present β-galactosidase expression directed by the chromosomal pA-lacZ and pAB-lacZ fusions inserted at the lac locus in the wild-type IHE 3034-Sm and the rcsB mutant in the presence or absence of rcsB or matA expression plasmid. An outline of the IHE 3034 DNA fragments used to construct chromosomal single-copy transcriptional pA-lacZ and pAB-lacZ fusions is shown on the right. The matA gene encoding the MatA regulatory protein is shown in gray, and right-angled arrows indicate the three transcription start sites identified by 5′ RACE (Lehti et al., submitted). The pA DNA fragment used for the reporter fusion construct contains a matA upstream region and a short downstream region, and the pAB fragment contains the same upstream sequences and a more extended downstream region with an uncharacterized putative H-NS repression site. The location of an introduced A536C missense mutation in chromosomal matA and matA in the pAB fragment is indicated by the letter C and a black triangle in the scheme. (B) β-Galactosidase expression directed by the chromosomal pAB-lacZ fusion inserted at the lac locus in wild-type IHE 3034-Sm and the rcsC, rcsD, and ackA-pta mutants. (C) The graphs on the left show the effects of matA on prcsB-lacZ and prcsD-lacZ expression. An outline of the IHE 3034 DNA fragments used to construct chromosomal single-copy transcriptional prcsB-lacZ and prcsD-lacZ fusions is shown on the right. The rcsB gene encoding the RcsB regulatory protein is shown in gray, and right-angled arrows indicate transcription start sites (30). The prcsB and prcsD DNA fragments used for reporter fusion constructs contain an upstream region and a short downstream region. (D) Northern blot showing steady-state levels of rcsB mRNAs at 20°C in IHE 3034-Sm and its derivatives carrying a matA expression plasmid or vector pSE380. The isogenic rcsB transposon mutant served as a negative control. The position of the primary rcsB transcript is indicated with an arrow, and the lengths of DIG-labeled RNA markers (bases) are shown on the right. Ethidium bromide-stained 23S and 16S rRNAs are shown below the Northern blots as RNA loading controls. In panels A, B, and C, reporter strains were grown to mid-logarithmic phase in LB at 20°C or 37°C and assayed for β-galactosidase activity. The results are the averages and standard deviations of three independent experiments. Significant differences, as calculated by Student's t test, are indicated by asterisks (**, P < 0.005; *, P < 0.05).
Measurements of β-galactosidase expression at exponential growth phase showed that the disruption of the rcsB gene significantly decreased (P < 0.005) the activities of both reporter fusions (Fig. 3A). In the absence of RcsB, the expression directed by pA-lacZ fusion was reduced to 41% of that of IHE 3034 at 20°C and to 64% at 37°C. The RcsB dependence for mat promoter activation was especially pronounced when the pAB-lacZ fusion was utilized; no induction of transcription occurred at 20°C and 37°C in the rcsB background. Introduction of the RcsB expression plasmid into the rcsB::Tn5 reporter strains led to a distinct stimulation of expression, although the promoter activities were not completely restored to the levels of the wild-type strain. Consistent with the biofilm and Northern blot results, providing MatA in trans had no effect on the expression of the mat promoter-lacZ fusions in the rcsB mutant (Fig. 3A).
The core of the Rcs pathway (67) is composed of the two inner membrane-bound proteins, the sensor kinase RcsC and the histidine phosphotransferase RcsD, and the response regulator RcsB harboring a phosphoreceiver domain and an effector domain for DNA binding. Upon stimulation by reduced temperature, acid stress, osmotic shock, cell envelope disturbance, or a solid surface (8), RcsC becomes autophosphorylated. The phosphoryl group is transferred to RcsD, which in turn modulates the phosphorylation state of RcsB by interacting with its phosphoreceiver and effector domains (60). There is also evidence that RcsB directly can accept the phosphoryl group from acetyl phosphate, linking the Rcs system to central metabolism (16). Phosphorylated RcsB binds as a homodimer to a subset of target promoters, including the promoter of the biofilm-dependent modulation gene bdm (14) and the small RNA RprA (37), whereas other Rcs-regulated promoters that contain an RcsAB box are recognized by RcsB in association with the accessory protein RcsA (15, 72). In some cases RcsB specifically recognizes the RcsAB box in the absence of RcsA (49). To analyze whether additional genes of the Rcs phosphorelay system affect mat regulation, we deleted the rcsC and rcsD genes as well as the ackA-pta genes, encoding enzymes involved in the maintenance of intracellular acetyl phosphate concentrations, from the IHE 3034 pAB-lacZ reporter strain. As shown in Fig. 3B, deletion of rcsC and rcsD did not affect the β-galactosidase activity of the pAB-lacZ fusion, whereas the ackA and pta double deletion had a minor increasing effect on the reporter activity. We also attempted to activate the Rcs phosphorelay of IHE 3034 using published Rcs-activating conditions (19, 24, 64). No effect on transcription from the mat promoter was observed when the logarithmic-phase IHE 3034 pAB-lacZ reporter cells grown at 20°C and 37°C in Luria broth were treated with 500 mM NaCl, 15% sucrose, or 1 mM ZnCl2 and 0.4% glucose, or when the rcsC and rcsD derivatives of the IHE 3034 pAB-lacZ reporter strain were grown in the presence of 500 mM NaCl (data not shown). The results thus showed that RcsB, but not the entire Rcs phosphorelay, is required for activation of the mat promoter and suggested that a phosphoryl group is not provided for RcsB phosphorylation by RcsC or RcsD. Since (i) published conditions for activation of the Rcs phosphorelay did not affect transcription from the mat promoter and (ii) deletion of ackA and pta did not decrease transcription, the results thus indicated that the phosphoryl group comes from an alternative unknown donor or that RcsB activates mat transcription in an unphosphorylated state.
We also tested whether RcsB and MatA regulate each other. Given that rcsB is the second gene in a rcsDB operon and is transcribed either individually or as a bicistronic mRNA covering rcsDB (30), the effect of MatA was tested on the activities of both prcsB and prcsD promoters installed as single individual copies into the chromosome of wild-type IHE 3034 and the matA(A536C) mutant (see Fig. 3C for an outline of the promoter fusions). No changes in promoter activities were apparent in the absence of functional MatA, and during plasmid-derived MatA overexpression only a minor increase in promoter activities was observed at 20°C (Fig. 3C). Ectopic expression of MatA had no effect on the steady-state level of rcsB transcripts, of approximately 1,200 and 900 nucleotides as reported by Krin et al. (30), in wild-type IHE 3034 at 20°C (Fig. 3D). These results indicate that the promoters directing expression of rcsB are controlled independently of MatA.
RcsB binds directly to the mat regulatory region.
Analysis of the sequences at the mat promoter regulatory region revealed the presence of a potential RcsAB box, a nonpalindromic consensus sequence TaAGaatatTCctA (where the uppercase letters indicate highly conserved residues) defined as a site for RcsB-RcsA heterodimer DNA binding (72). The putative RcsAB box was centered 178 bp upstream of the presumed −35 region of the primary transcriptional start site T2 of the mat operon (Lehti et al., submitted). In IHE 3034, 13 of 14 bases in this DNA sequence fit the consensus sequence (Fig. 4A), suggesting that the mat promoter might be directly recognized by RcsB and RcsA. The RcsAB box is fully conserved and present in all 41 complete E. coli genome sequences that contain the mat regulatory region. Interestingly, the four available Klebsiella genomes harbor the mat operon together with a perfect RcsAB box, although the promoter sequences (bp −608 to −1 relative to the translational start of matA) in E. coli and Klebsiella strains overall are only 64 to 68% identical, indicating a conserved regulatory function of the RcsAB box in the mat promoter.
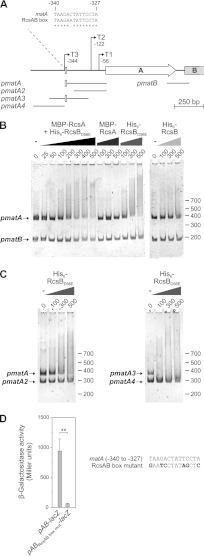
Fig 4.
Binding of RcsB to the mat promoter region and the effect of the RcsAB box on mat expression. (A) Schematic presentation of the matA region of IHE 3034 and DNA fragments used in EMSAs. Names of the DNA fragments are shown on the left. The three transcriptional start sites upstream of matA (T1 to T3; Lehti et al., submitted) are indicated with right-angled arrows. The positions of putative RcsAB box are shown as white boxes, and the alignment result of the matA upstream region with the consensus sequence of the RcsAB box (72) is indicated. Numbering denotes the distances from the translational start site of matA. (B) Competitive EMSA performed in the presence of pmatA and pmatB fragments with increasing concentrations (nM) of MBP-RcsA and His6-RcsBD56E proteins at an equimolar RcsA/RcsB ratio, or with either protein alone. Binding of His6-RcsB is also shown. (C) Competitive EMSA with increasing concentrations (nM) of His6-RcsBD56E together with pmatA and pmatA2 fragments, or with pmatA3 and pmatA4 fragments. The lengths of DNA standards (bp) are shown on the right for reference. (D) β-Galactosidase expression directed by the chromosomal pAB-lacZ fusion and the pAB-lacZRcsAB box mutant fusion inserted individually at the lac locus in wild-type IHE 3034-Sm. Significant difference, as calculated by Student's t test, is indicated by two asterisks (**, P < 0.005). A comparison between the IHE 3034 RcsAB box and the mutant RcsAB box used is shown on the right.
The capacity of RcsB and RcsA to bind to a pmatA fragment derived from upstream of IHE 3034 matA and, as a control, to a pmatB fragment containing the matA-matB intergenic region (see Fig. 4A for an outline of the fragments) was tested in an electrophoretic mobility shift assay (EMSA). The assay was based on the RcsBD56E substitution variant, in which the conserved aspartic acid residue of the phosphoreceiver domain is replaced by a glutamic acid residue, thus mimicking the phosphorylated, active form of the protein (18). The pmatA fragment is a target for binding by the activator MatA and the repressor H-NS (Lehti et al., submitted) and contains the putative RcsAB box at the 5′ end (Fig. 4A). The pmatA fragment was retarded by increasing amounts of RcsBD56E, while only a weak shifting was seen with the fragment pmatB in the same assay (Fig. 4B). The wild-type RcsB protein displayed a weaker interaction with pmatA than did RcsBD56E, as two to three times more protein was required for an equally efficient retardation, which is consistent with the fact that phosphorylated RcsB binds DNA more efficiently (15, 66). RcsA did not bind to pmatA, and the presence of equimolar amounts of RcsA and RcsBD56E resulted in a shifting profile similar to that formed by RcsBD56E alone, indicating that activated RcsB binds stably to the mat regulatory region without a stabilizing effect of RcsA. When DNA regions located on both sides of the box (fragments pmat2 and pmat4 [Fig. 4A]), and thus lacking the putative 14-bp RcsAB box, were tested as target sequences, RcsBD56E showed only a weak DNA binding activity in the EMSA (Fig. 4C). In contrast, combining RcsBD56E with a 330-bp pmat3 that harbors the rcsAB box at its center led to complete shifting of the free DNA at the low protein concentration of 100 nM, which is a considerably lower concentration than that required for the pmatA PCR product. Since RcsB can form a regulatory complex with different coregulators, GadE, TviA, and BglJ (7, 30, 68, 73), in addition to the ability to heterodimerize with RcsA, we combined RcsBD56E with MBP-MatA in the EMSA. However, the binding affinity of RcsBD56E for the mat promoter was not enhanced (data not shown).
The effect on mat expression of the RcsAB box located upstream of the mat operon was next assessed. A chromosomal pAB-lacZ fusion carrying a nonfunctional RcsAB box mutated according to Venkatesh et al. (68) was constructed and inserted at the lac locus in wild-type IHE 3034-Sm. The results showed that no β-galactosidase expression occurred from the reporter carrying a mutated RcsAB box (Fig. 4D), indicating that in the bacterial cell, RcsB binds to the RcsAB box and thereby affects mat expression.
Conclusion.
E. coli is a minor species in the human microbiota as well as a human pathogen that also is able to persist and proliferate in water, sediment, and soil (23, 47, 58, 71). Alternation between host and environmental reservoirs exposes the bacterium to changing stress factors, and survival in different niches demands adaptation of gene expression. The involvement of the Rcs phosphorelay system in the proper development of biofilms is well established (8, 36). Here, we describe a novel member of the Rcs regulon, the common colonization factor Mat fimbria, and demonstrate that the RcsB regulator, but not RcsC and RcsD, is directly required for activation of mat expression, thus playing a pivotal role in biofilm formation by the meningitis-associated E. coli strain IHE 3034. The regulation of the conserved mat operon is apparently complex, and integration of RcsB in this network provides an additional control point and a possible route for incorporation of environmental cues. The RcsB and MatA regulators act jointly at the mat promoter to promote the biofilm mode of growth also via another regulatory network, namely, the flagellar cascade, by repressing the flhDC operon (15; 33). Thus, RcsB directly and indirectly contributes to flagellum-mediated motility, utilizing the RcsB-MatA-flhDC circuit. Cumulatively, the reciprocal regulation of adhesive Mat fimbriae and motility strengthens the role of the Rcs system in controlling sessile, adherent behavior in E. coli.
ACKNOWLEDGMENTS
We thank Michael S. Donnenberg for providing the strain BL21 (AI) slyD and Philippe Bauchart for providing the plasmid pBAU1. We also thank Raili Lameranta and Elina Nummenmaa for technical assistance.
This work was supported by University of Helsinki, Viikki Doctoral Programme in Molecular Biosciences, Academy of Finland (ERA-NET PathoGenoMics grants 118982 and 130202 and general research grant 123900) and the European Network of Excellence in EuroPathoGenomics EPG (number CEE LSHB-CT-2005-512061).
Footnotes
Published ahead of print 20 April 2012
REFERENCES
- 1. Achtman M, et al. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amini S, Goodarzi H, Tavazoie S. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5:e1000432 doi:10.1371/journal.ppat.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Appleby JL, Parkinson JS, Bourret RB. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845–848 [DOI] [PubMed] [Google Scholar]
- 4. Avelino F, et al. 2010. The majority of enteroaggregative Escherichia coli strains produce the E. coli common pilus when adhering to cultured epithelial cells. Int. J. Med. Microbiol. 300:440–448 [DOI] [PubMed] [Google Scholar]
- 5. Beloin C, et al. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackburn D, et al. 2009. Distribution of the Escherichia coli common pilus among diverse strains of human enterotoxigenic E. coli. J. Clin. Microbiol. 47:1781–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castanié-Cornet MP, et al. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 38:3546–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 5:1173–1184 [DOI] [PubMed] [Google Scholar]
- 9. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 10. Daniel A, et al. 2006. Interaction and localization studies of enteropathogenic Escherichia coli type IV bundle-forming pilus outer membrane components. Microbiology 152:2405–2420 [DOI] [PubMed] [Google Scholar]
- 11. Deininger KN, et al. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960 doi:10.1371/journal.pone.0018960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrières L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665–1682 [DOI] [PubMed] [Google Scholar]
- 14. Francez-Charlot A, Castanié-Cornet MP, Gutierrez C, Cam K. 2005. Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3873–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francez-Charlot A, et al. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823–832 [DOI] [PubMed] [Google Scholar]
- 16. Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734–747 [DOI] [PubMed] [Google Scholar]
- 17. Gupta DS, Jann B, Jann K. 1984. Escherichia coli O18ac antigen: structure of the O-specific polysaccharide moiety. Infect. Immun. 45:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupte G, Woodward C, Stout V. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagiwara D, et al. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 21. Hernandes RT, et al. 2011. Fimbrial adhesins produced by atypical enteropathogenic Escherichia coli strains. Appl. Environ. Microbiol. 77:8391–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang YH, Ferrières L, Clarke DJ. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157:206–212 [DOI] [PubMed] [Google Scholar]
- 23. Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kannan G, et al. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8:37 doi:10.1186/1471-2180-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelm O, Kiecker C, Geider K, Bernhard F. 1997. Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol. Gen. Genet. 256:72–83 [DOI] [PubMed] [Google Scholar]
- 27. Klemm P, Hancock V, Schembri MA. 2010. Fimbrial adhesins from extraintestinal Escherichia coli. Environ. Microbiol. Rep. 2:628–640 [DOI] [PubMed] [Google Scholar]
- 28. Korea CG, Ghigo JM, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 33:300–311 [DOI] [PubMed] [Google Scholar]
- 29. Korhonen TK, et al. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krin E, Danchin A, Soutourina O. 2010. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res. Microbiol. 161:363–371 [DOI] [PubMed] [Google Scholar]
- 31. Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74:7376–7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lasaro MA, et al. 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lehti TA, Bauchart P, Dobrindt U, Korhonen TK, Westerlund-Wikström B. 2012. The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC. Microbiology doi:10.1099/mic.0.056499-0 [DOI] [PubMed] [Google Scholar]
- 34. Lehti TA, et al. 2010. Mat fimbriae promote biofilm formation by meningitis-associated Escherichia coli. Microbiology 156:2408–2417 [DOI] [PubMed] [Google Scholar]
- 35. Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405 [DOI] [PubMed] [Google Scholar]
- 37. Majdalani N, Hernandez D, Gottesman S. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813–826 [DOI] [PubMed] [Google Scholar]
- 38. Matsumura K, Furukawa S, Ogihara H, Morinaga Y. 2011. Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci. 16:69–72 [DOI] [PubMed] [Google Scholar]
- 39. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 40. Mobley HL, et al. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of αGal(1-4)βGal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143–155 [DOI] [PubMed] [Google Scholar]
- 41. Moriel DG, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Niba ET, Naka Y, Nagase M, Mori H, Kitakawa M. 2007. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 14:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oshima T, et al. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281–291 [DOI] [PubMed] [Google Scholar]
- 44. Pardee AB, Jacob F, Monod J. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase by E. coli. J. Mol. Biol. 1:165–178 [Google Scholar]
- 45. Pouttu R, et al. 1999. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol. Microbiol. 31:1747–1757 [DOI] [PubMed] [Google Scholar]
- 46. Pouttu R, et al. 2001. matB, a common fimbrillin gene of Escherichia coli, expressed in a genetically conserved, virulent clonal group. J. Bacteriol. 183:4727–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631–640 [DOI] [PubMed] [Google Scholar]
- 48. Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 49. Pristovsek P, et al. 2003. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 278:17752–17759 [DOI] [PubMed] [Google Scholar]
- 50. Prüß BM, Besemann C, Denton A, Wolfe AJ. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Puttamreddy S, Cornick NA, Minion FC. 2010. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect. Immun. 78:2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Régnier P, Grunberg-Manago M. 1989. Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 210:293–302 [DOI] [PubMed] [Google Scholar]
- 53. Rendón MA, et al. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rhen M, Knowles J, Penttilä ME, Sarvas M, Korhonen TK. 1983. P fimbriae of Escherichia coli: molecular cloning of DNA fragments containing the structural genes. FEMS Microbiol. Lett. 19:119–123 [Google Scholar]
- 55. Saldaña Z, et al. 2009. The Escherichia coli common pilus and the bundle-forming pilus act in concert during the formation of localized adherence by enteropathogenic E. coli. J. Bacteriol. 191:3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 57. Sands JF, Régnier P, Cummings HS, Grunberg-Manago M, Hershey JW. 1988. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 16:10803–10816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Savageau MA. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732–744 [Google Scholar]
- 59. Scaletsky IC, Aranda KR, Souza TB, Silva NP. 2010. Adherence factors in atypical enteropathogenic Escherichia coli strains expressing the localized adherence-like pattern in HEp-2 cells. J. Clin. Microbiol. 48:302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmöe K, et al. 2011. Structural insights into Rcs phosphotransfer: the newly identified RcsD-ABL domain enhances interaction with the response regulator RcsB. Structure 19:577–587 [DOI] [PubMed] [Google Scholar]
- 61. Schnaitman CA, Klena JD. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schwan WR, Shibata S, Aizawa S, Wolfe AJ. 2007. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J. Bacteriol. 189:7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 64. Sledjeski DD, Gottesman S. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 66. Sturny R, Cam K, Gutierrez C, Conter A. 2003. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. J. Bacteriol. 185:4298–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440–450 [DOI] [PubMed] [Google Scholar]
- 68. Venkatesh GR, et al. 2010. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 192:6456–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vianney A, et al. 2005. Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151:2487–2497 [DOI] [PubMed] [Google Scholar]
- 70. Vidal O, et al. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274–2288 [DOI] [PubMed] [Google Scholar]
- 72. Wehland M, Bernhard F. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275:7013–7020 [DOI] [PubMed] [Google Scholar]
- 73. Winter SE, et al. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74:175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang W, Shi L. 2005. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology 151:2159–2173 [DOI] [PubMed] [Google Scholar]