Abstract
Colorectal cancer is the third most common cancer worldwide with an annual incidence of ∼1 million cases and an annual mortality rate of ∼655,000 individuals. There is an urgent need for identifying novel targets to develop more sensitive, reliable, and specific tests for early stage detection of colon cancer. Post-translational modifications are known to play an important role in cancer progression and immune surveillance of tumors. In the present study, we compared the N-glycan profiles from 13 colorectal cancer tumor tissues and corresponding control colon tissues. The N-glycans were enzymatically released, purified, and labeled with 2-aminobenzoic acid. Aliquots were profiled by hydrophilic interaction liquid chromatography (HILIC-HPLC) with fluorescence detection and by negative mode MALDI-TOF-MS. Using partial least squares discriminant analysis to investigate the N-glycosylation changes in colorectal cancer, an excellent separation and prediction ability were observed for both HILIC-HPLC and MALDI-TOF-MS data. For structure elucidation, information from positive mode ESI-ion trap-MS/MS and negative mode MALDI-TOF/TOF-MS was combined. Among the features with a high separation power, structures containing a bisecting GlcNAc were found to be decreased in the tumor, whereas sulfated glycans, paucimannosidic glycans, and glycans containing a sialylated Lewis type epitope were shown to be increased in tumor tissues. In addition, core-fucosylated high mannose N-glycans were detected in tumor samples. In conclusion, the combination of HILIC and MALDI-TOF-MS profiling of N-glycans with multivariate statistical analysis demonstrated its potential for identifying N-glycosylation changes in colorectal cancer tissues and provided new leads that might be used as candidate biomarkers.
Colorectal cancer is the third most common cancer worldwide with an annual incidence of ∼1 million cases and an annual mortality of ∼655,000 (1). The most widely used screening procedures for colorectal cancer include flexible sigmoidoscopy, double-contrast barium enema, colonoscopy, and fecal occult blood testing (2–4). Some of the screening methods are invasive and uncomfortable, and several of the methods, such as fecal occult blood testing, are characterized by a low sensitivity and specificity of 53 and 87%, respectively (5). Surgery is the cornerstone of the therapy and the tumor, node, and metastasis staging system remains the gold standard for prognostication of the disease relying entirely on morphological and histopathological appearance of the tumor. However, tumors with similar histopathological characteristics may present a different clinical outcome and responsiveness to therapy. Evidently, a better understanding of the biology of colorectal cancer could accelerate the search for improved methods for noninvasive early detection and characterization of the tumor. The exceptional amount of data coming from high throughput experiments in genomics and proteomics has advanced our understanding of colorectal cancer but has, so far, resulted in a surprising scarcity of well validated, clinically useful markers.
In addition to genomics and proteomics, the emerging field of glycomics is gaining importance in cancer research. Glycosylation is one of the most common and most complex post-translational modifications of proteins and plays a critical role in key cellular processes such as cell differentiation, adhesion, and proliferation. It is therefore not unexpected that the glycosylation of proteins in tumor cells differs from the glycosylation of nonmalignant cells, which already many years ago has been demonstrated by the differential binding of lectins to healthy and cancerous tissues (6, 7). Aberrant glycosylation has been observed in many types of diseases, including different cancers (8–10), and may be caused by changes in the expression levels of glycosyltransferases in the Golgi compartment (11) and the lysosomal exoglycosidases (12), resulting in a change in the levels of regular glycans and expression of new species.
Recently, Arnold et al. (13) analyzed fluorescently tagged N-glycans of serum samples of a cohort of lung cancer patients and controls using hydrophilic interaction liquid chromatography (HILIC)1 and weak anion exchange high performance liquid chromatography (HPLC) resulting in the identification of lung cancer-related glycan alterations. In another study, Abd Hamid et al. (14) analyzed fluorescently tagged serum N-glycans of advanced breast cancer patients using HPLC with MALDI-TOF-MS and found that specific glycans were changing during breast cancer progression. De Loez et al. (15) showed the alteration of the low abundant high mannose glycans in breast cancer using mouse serum samples and patients sera. In addition, several studies have indicated the high expression of some glycan epitopes such as β1–6-linked GlcNAc, sialyl-LeX, sialyl-LeA, and LeY, in cancer (16–19). The branching of N-linked structures promoting invasion and metastasis has been associated with the shorter 5–10-year survival rates of cancer patients. Expression of some other glycan epitopes such as bisecting GlcNAc, which is competing with the expression of β1–6-linked antennae and hence the occurrence of triantennary and tetraantennary structures, has been implicated in the suppression of the tumor progression, leading to higher post-operative survival rates (20, 21).
Most of the biomarker discovery studies based on glycomics have focused on mapping changes in serum and cell cultures (22–24). The biggest challenge to the discovery of biomarkers in body fluids is the fact that a heterogeneous mix of glycoproteins is secreted from a variety of tissues in the body, making it difficult to attribute a candidate marker to a tissue-specific disease. Cell cultures have limitations with regard to the type of research questions that can be addressed. The isolated cell cultures do not mimic the in vivo tumor biology in which the interplay between tumor cells and the neighboring cells is of critical importance (25). In this respect, glycosylation profiling of tumor and the corresponding control tissues to reveal changes in glycosylation is advantageous, because the ambiguity of source tissue is greatly reduced.
Paired proteome analysis of tumor and neighboring normal colorectal mucosa is a common methodology employed to identify those changes in protein expression that can be associated with tumorigenesis (26, 27). Post-translational modifications are playing an important role in oncogenesis, and therefore the search for cancer biomarkers should not only focus on the protein backbone but also on its post-translational modifications. One of the prominent determinants of protein structure and function, N-linked glycosylation, is the result of a complex biosynthetic process that regulates maturation of proteins through secretory pathways (28, 29). Because protein glycosylation is known to change during malignant transformation, altered levels of oligosaccharides present on tumor glycoproteins might represent a new source of biomarkers and might contribute to a better understanding of cancer biology.
In the present study, we investigated N-glycosylation changes occurring in colorectal cancer tumor tissues, and to that end we determined the N-glycan profiles from 13 colorectal cancer tumor tissue samples and the corresponding control colon tissues. We characterized the N-linked glycan compositions of 245 structures present in colorectal cancer tissues based on their m/z value in combination with tandem mass spectrometry analysis and common knowledge of the glycobiology. The combination of HILIC analysis of fluorescently tagged N-glycans and MALDI-TOF(/TOF)-MS analysis with multivariate statistical analysis proved to be a valuable approach for identifying candidate biomarkers and deepened our understanding about glycosylation changes of colorectal tumor tissues. Our study revealed an increase in the level of paucimannosidic structures and sulfated N-glycans and a decrease of N-glycans containing a bisecting GlcNAc in the tumor glycome of patients with colorectal cancer in comparison with surrounding colon control tissue. Furthermore, thorough characterization of N-glycans released from tumor glycoproteins showed the presence of core-fucosylated high mannose N-glycans.
MATERIALS AND METHODS
Chemicals and Solvents
Ammonium formate, 2-aminobenzoic acid (AA), 2-picoline borane, 2-mercaptoethanol, DMSO, Nonidet P-40, and TFA were obtained from Sigma-Aldrich. The 500-mg C18 reverse phase cartridges and methanol were obtained from J. T. Baker (Deventer, The Netherlands), and the 150-mg Carbograph graphitized carbon SPE cartridges were from Alltech (Breda, The Netherlands). ACN was obtained from Biosolve (Valkenswaard, The Netherlands). Chloroform, glacial acetic acid, sodium azide, and SDS were from Merck. N-Glycosidase F was purchased from Roche Applied Science, and 2,5-dihydroxybenzoic acid was from Bruker Daltonics (Bremen, Germany). All of the buffers were prepared using Milli-Q water.
Tissue Samples
Thirteen tissue samples of primary colorectal carcinoma and the corresponding, paired control tissue were collected at the Department of Surgery of Leiden University Medical Center (Leiden, The Netherlands). All of the samples were handled in a coded fashion, according to National Ethical Guidelines (Code for Proper Secondary Use of Human Tissue, Dutch Federation of Medical Scientific Societies).
Clinical data related to the samples used in this study, including age; gender; blood group; Dukes stage; tumor, node, and metastasis stage; CEA preoperative value; tumor stroma ratio; localization of tumor tissue (proximal and distal); and the colon section used for the release of the N-glycans are summarized in Table I. Twenty cryosections of ∼10-μm thickness and ∼1-cm2 size were obtained per sample and used for glycosylation analysis.
Table I. Description of the sample cohort.
| Tumor type | Gendera | Age (years)b | Blood group | TNM stagec | CEA (ng/ml)d | Primary tumor location | Tumor localization | TSRe |
|---|---|---|---|---|---|---|---|---|
| MSIf | M | 63 | O+ | T3N1M0 | 1.0 | Rectosigmoid | Distal | High |
| F | 72 | A+ | T3N0M0 | 12.8 | Coecum | Proximal | High | |
| M | 69 | O+ | T3N2M1 | 1.2 | Coecum | Proximal | Low | |
| F | 78 | O+ | T3N0M0 | 2.1 | Coecum | Proximal | High | |
| Non-MSI | M | 59 | A+ | T3N0M0 | 12.0 | Colon ascendens | Proximal | Low |
| F | 41 | A+ | T3N1M1 | 93.5 | Colon sigmoideum | Distal | High | |
| M | 75 | B+ | T3N1M0 | 23.4 | Coecum | Proximal | High | |
| M | 60 | O+ | T3N2M0 | 1.5 | Colon ascendens | Proximal | High | |
| F | 68 | A+ | T3N1M0 | 4.2 | Colon sigmoideum | Distal | High | |
| Rectum | M | 72 | A+ | T3N0M0 | 1.9 | Rectum | Distal | |
| F | 40 | B+ | T2N1M0 | 2.5 | Rectum | Distal | High | |
| F | 29 | A+ | T3N2M0 | 13.6 | Rectum | Distal | High | |
| F | 55 | O+ | T4N0M0 | Rectum | Distal | High |
a M, male; F, female.
b Age at diagnosis.
c TNM, tumor node metastasis; T, size or direct extent of the primary tumor; N0, absence of lymph node metastasis; N1, invasion of regional lymph nodes; N2, tumor spread to a certain extent; M0, absence of distant metastasis; M1, presence of distant metastasis.
d CEA, preoperative carcinoembryonic antigen levels.
e TSR, tumor stroma ration estimated on the basis of morphological characteristics.
f MSI, microsatellite instability.
Histopathological Protocol
Pathological examination entailed routine microscopic analysis of 5-μm hematoxylin- and eosin-stained sections of the primary tumor. For the identification of microsatellite instability high patients, 5-μm slides were immunohistochemically stained for MLH1 and PMS2 markers (30).
Tissue Homogenization and Isolation of Proteins
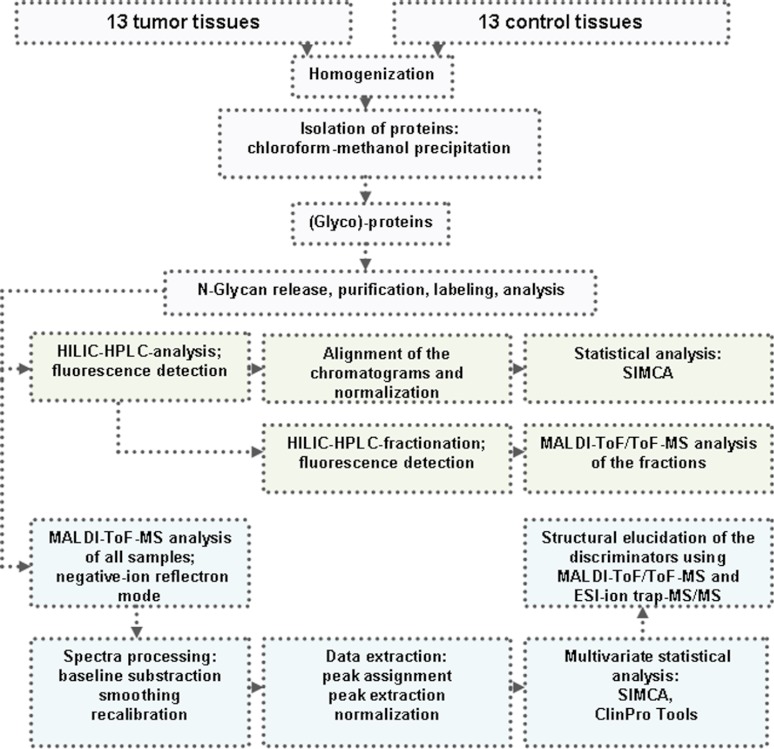
The N-glycans were prepared and analyzed as schematically illustrated in Fig. 1. The isolation of proteins was performed after lipids extraction (31) as described by Wuhrer et al. (32). In short, 1 ml of water was added to the tissue sections followed by homogenization with a sonicator stick (Branson Sonifier 250; Gemini BV, Haaksbergen, The Netherlands). Subsequently, 1.75 ml of methanol were added to the samples, followed by vortexing and sonication in a water bath for ∼15 min. Next, 3.25 ml of chloroform were added, resulting in a phase separation. Vortexing and sonication were repeated once, and the samples were centrifuged at 15,000 × g for 15 min. The upper phase was removed and replaced by methanol/water (50/50, v/v). The samples were vortexed, sonicated, and centrifuged followed by the removal of the upper phase. Methanol/water (50/50, v/v) was added for a second time, followed by homogenization (vortexing), sonication, and centrifugation of the samples and removal of the upper phase. One volume of methanol was added to the lower phase, and the interphase containing most of the proteins. The samples were homogenized (vortexing) and centrifuged at 15,000 × g for 15 min, resulting in the pelleting of the protein fraction. The supernatant was discarded, and the pellet was resuspended in methanol, followed by centrifugation and removal of the supernatant. This step was repeated three times, and subsequently the pellets were dried under a gentle stream of nitrogen.
Fig. 1.
Schematic presentation of comparative N-glycosylation analysis in cancer versus normal colorectal tissue.
Release of N-glycans from Glycoproteins
Next, 1 ml of PBS was added to the pellets followed by homogenization (by vortexing). SDS and 2-mercaptoethanol were added to the samples to final concentrations of 1.3% (w/v) and 0.5% (v/v), respectively. The samples were heated for 10 min at 95 °C and then put on ice. Nonidet P-40 and sodium azide were added to the samples to final concentrations of 1.5 and 0.1%, respectively. Two milliunits of N-glycosidase F were added to each sample followed by 24 h of incubation at 37 °C. Subsequently, another 2 milliunits of N-glycosidase F were added to each sample, followed by overnight incubation at 37 °C. The N-glycosidase F released glycans were purified by C18 reverse phase SPE (flow through) and graphitized carbon SPE (retentate). The C18 reverse phase cartridges were preconditioned with 5 ml of ACN and 5 ml of water/ACN (40/60, v/v) and equilibrated with 5 ml of water. The samples were applied to the cartridges followed by washes with 3 ml of water/ACN (90/10, v/v) and 3 ml of water. The combined flow through and the wash fractions containing the released glycans were further purified by graphitized carbon SPE. For this purpose, the cartridges were preconditioned with 5 ml of ACN and 5 ml of water/ACN (50/50, containing 0.1% TFA) and equilibrated with 10 ml of water. After applying the samples, the cartridges were washed with 10 ml of water, and the glycans were eluted with 5 ml of water/ACN (50/50, containing 0.1% TFA). The eluate containing the purified glycans was dried by vacuum centrifugation.
Glycan Labeling and Purification
For AA labeling, the dried, purified glycan samples were dissolved in 50 μl of water and mixed with 25 μl of a freshly prepared label solution (48 mg/ml AA in DMSO containing 15% glacial acetic acid) (33). Twenty-five μl aliquots of freshly prepared reducing agent solution (1 m 2-picoline borane in DMSO) were added, followed by 10 min of shaking and incubation at 65 °C for 2 h. The reaction mixture was allowed to cool down to room temperature.
After labeling, the glycans were purified using graphitized carbon SPE. To this purpose, the cartridges were preconditioned with 5 ml of ACN and 5 ml of water/ACN (50/50, containing 0.1% TFA) and equilibrated with 10 ml of water. The samples were applied, followed by a wash with 10 ml of water. Glycans were eluted with 5 ml of water/ACN (50/50, containing 0.1% TFA). ACN was removed by vacuum centrifugation, and the samples were freeze-dried and redissolved in 100 μl of water for further analysis by MALDI-TOF-MS and HPLC with fluorescence detection.
HPLC Analysis and Fractionation of Glycans
AA-labeled glycans were profiled and fractionated by hydrophilic interaction liquid chromatography (HILIC)-HPLC (TSK amide 80, 3 μm, 150-mm × 4.6-mm inner diameter column; Tosoh Bioscience, Stuttgart, Germany) at a 1 ml/min flow rate with fluorescence detection (360 nm/420 nm). A binary gradient was applied using 80% ACN, 20% 50 mm ammonium formate, pH 4.4 (solvent A), and 50 mm ammonium formate, pH 4.4 (solvent B), from 3% solvent B (0 min) to 43% solvent B (50 min). For analytical runs, N-glycan samples (2 μl) were brought to 77% ACN by the addition of 2.6 μl of H2O and 15.4 μl of ACN. The samples were analyzed randomly to avoid possible batch effects. For preparative runs, two pools of AA-labeled glycans were prepared (control and cancer samples separately). The pools were freeze-dried and dissolved in 500 μl of 77% ACN, followed by HILIC-HPLC. Fractions of 30 s were collected, dried by vacuum centrifugation, and reconstituted in 30 μl of water.
MALDI-TOF(/TOF)-MS
Prior to mass spectrometric analysis, unfractionated AA-labeled N-glycan samples were purified by HILIC micro-SPE (34). HILIC microtips were prepared by putting ∼0.5 mg of cotton wool obtained from cotton wool pads into a pipette tip. HILIC microtips were washed with water and equilibrated with 90% ACN containing 0.1% TFA. Part of the sample (20 μl) was brought to 77% ACN and applied to the HILIC microtips by pipetting the sample 10 times up and down. The microtips were washed four times using 10 μl of 90% ACN containing 0.1% TFA, and AA-labeled glycans were eluted with 2 μl of water onto a stainless steel MALDI target plate (Bruker Daltonics). Subsequently 1.3 μl of 2,5-dihydroxybenzoic acid (10 mg/ml in water/ACN, 50/50, containing 0.1% TFA) was applied on top of each sample and allowed to dry at room temperature.
Five μl of dried and reconstituted HILIC-HPLC fractions were desalted using a C18 ZipTipTM (Millipore, Billerica, MA) following the manufacturer's instructions. AA-labeled glycans were eluted with 1.5 μl of 2,5-dihydroxybenzoic acid (10 mg/ml in 50/50, ACN/water containing 0.1% TFA) directly onto a stainless steel MALDI target plate and allowed to dry.
MALDI-TOF-MS was performed on an UltrafleXtremeTM mass spectrometer controlled by FlexControl 3.1 software package (Bruker Daltonics). The instrument was externally calibrated using the Bruker peptides calibration kit. The spectra were acquired in the negative ion reflectron mode over the m/z range from 700 to 5000 for a total of 5000 shots. For the MALDI-TOF/TOF-MS analysis, precursors were accelerated and selected in a time ion gate, after which fragments arising from metastable decay were further accelerated in the LIFT cell and detected after passing the ion reflector. MS and MS/MS data were searched using Glyco-Peakfinder (http://www.glyco-peakfinder.org) using search windows of 400 ppm and 0.5 Da, respectively.
LC-ESI-Ion Trap-MS/MS
Nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed using an Ultimate 3000 LC system (Dionex, Amsterdam, The Netherlands). Aliquots of the HILIC fractions (5 μl) were applied to a C18 PepMapTM 0.3 mm × 5 mm trapping column (Dionex) and washed with 100% A (0.1% formic acid in water and 0.4% ACN) at 25 μl/min for 10 min. Following valve switching, AA-labeled glycans were separated on a reverse phase column (C18 PepMap 100Å, 3 μm, 75 μm × 150 mm; Dionex) at a flow rate of 300 nl/min. The gradient applied to perform sample separation was as follows: 0–25% eluent B (95% ACN, 5% water) in 15 min and 25–70% eluent B in the next 10 min, followed by an isocratic elution with 70% eluent B for 5 min. The LC system was coupled via an online nanospray source to an Esquire HCT ultra ESI-ion trap-MS (Bruker Daltonics) and was operated in the positive ion mode. For electrospray (1100–1250 V), stainless steel capillaries with an inner diameter of 30 μm (Proxeon, Odense, Denmark) were used. The dry gas temperature was set to 165 °C, and the nitrogen stream was set to 7 liters/min. Using sialylated glycoconjugates, the mass spectrometer was carefully tuned to minimize glycan decay in the ion transfer region, resulting in the following settings: skimmer, 40 V; capillary exit, 106 V; octopole 1 DC, 6 V. The eluting AA-labeled glycans were analyzed using the data-dependent MS/MS mode over a 300–1500 m/z range. Five of the most abundant ions in an MS spectrum were selected for MS/MS analysis by collision-induced dissociation using helium as the collision gas.
Multivariate Data Analysis of HPLC Chromatograms and MALDI-TOF-MS Spectra
LC chromatograms were base line-corrected using a Simplex-type fit of the experimental data by an integral of a combination of logistic functions and aligned using the correlation optimized warping (cow, http://www.cs.kvl.dk/dtw_cow). Prior to multivariate statistics performed using SIMCA P+ software (version 12.0; Umetrics AB, Umea, Sweden), each sample was normalized to unit size by dividing each area value by “total area” under the LC chromatograms.
The peak picking algorithm (adopted from MATLAB) was applied to the internal recalibrated, smoothed, and base line-corrected MS spectra (FlexAnalysis 3.1; Bruker Daltonics). Prior to multivariate statistics performed using SIMCA P+ software (version 12.0; Umetrics AB) each sample was normalized to set the “total area” of all the glycan peaks (1000–2700 Da) to 1000.
Principal component analysis (PCA) was performed to provide an overview of the data set and to detect outliers. Potential outliers were verified based on scores (observations lying outside the 0.95 Hotelling's T2 ellipse), distance to model X values, and subsequent visual inspection of the individual MS spectra and LC chromatograms. No outliers were identified. Whereas PCA works to maximize the variation in the data set, partial least squares discriminant analysis (PLS-DA) seeks the maximum separation between predefined class samples in the data (X) using a matrix that represents an orthogonal unit vector for each class. Therefore, PLS-DA was performed to maximize separation of the samples between the group of N-glycans released from the control tissues and the group of N-glycans released from the tumor tissues. The data were mean-centered to provide all the spectral variables with the same weight. As a result, the loadings only show the variables, which have an impact on the discrimination between classes. Finally, orthogonal signal correction (OSC) was applied to the data set followed by a second PLS-DA analysis to eliminate the effect of intersubject variability among the participants, which was unrelated to colorectal cancer and to extract potential glycans that are correlated with the disease. Because the same chromatogram regions and the same glycans were responsible for the group separation in both PLS-DA analyses before and after OSC correction and because the model obtained after applying a two components OSC showed a better predictive capability, the orthogonal corrected data were used for the detection of differentially expressed glycans. The features represented by at least two consecutive isotopes with a variable importance value (VIP) higher than 1 were considered as potential discriminators (the variables with VIP value larger than 1 are the most relevant for explaining the regression model) (35). Apart from the internal cross-validation, classification efficiency was evaluated for the MS data using ClinPro Tools software (version 2.2; Bruker Daltonics).
ClinPro Tools automatically normalizes all spectra to their total ion current. Spectra classes were loaded into the software, and base-line correction was performed using a convex hull base-line algorithm with 0.6 base-line flatness. Recalibration was performed using at least 30% match to the calibrant peaks allowing a maximum shift of 1000 ppm in the range of 1–5 kDa. The Savitsky-Golay algorithm was used for smoothing using an m/z width of 0.1 with 20 cycles. Peak detection was done on a total average spectrum with the signal to noise threshold ≥5. After applying the t test of difference, a support vector machine algorithm was used to classify the two groups (cancer and control). The Leave-One-Out cross-validation was used to calculate the recognition rate. Moreover, receiver operating characteristic curve analysis and area under the curve (AUC) calculations were performed to determine the sensitivity and specificity of the different markers.
A two-tailed paired sample t test analysis was performed using GraphPad Prism (version 4) to evaluate the statistical significance of differences between the glycan classes (tumor versus control). Gaussian distribution of data was confirmed by the application of the D'Agostino-Pearson normality test. The relative intensities of the glycan classes were used to conduct the analysis.
RESULTS
Identification and Structural Characterization of N-glycans from Colorectal Cancer and Control Tissues
N-Glycans from protein extracts of 13 colorectal cancer tumor tissues (six males and seven females; Table I) and 13 control tissues from the same resection material were enzymatically released, purified, and AA-labeled. The control specimens were collected from accompanying colon tissues that were ∼6 cm apart from the colorectal tumor.
Next, two pools of N-glycans were generated: one containing AA-labeled N-glycans from the tumor tissue and one from the corresponding control tissues. These pooled samples were fractionated by HILIC-HPLC, and subsequently all of the fractions were analyzed by MALDI-TOF-MS. In addition, multiple HILIC fractions were analyzed by ESI-ion trap-MS/MS and MALDI-TOF-MS/MS. These tandem mass spectrometric data are provided in supplemental Fig. 1. Structure elucidation was based on the fragmentation of the discriminative glycans and common knowledge of glycobiology. Structural schemes are not intended to specify linkage positions. For example, no distinction is made between six-branch and three-branch of biantennary glycans, and linkage angles are not intended to imply specific linkage positions.
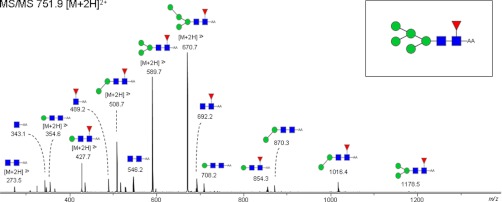
This resulted in the annotation of 245 glycans from the cancer tissue as summarized in supplemental Table 1. Most of the compositions were found to be in accordance with known mammalian N-glycan structures, including oligomannosidic structures as well as hybrid type and complex type glycans that may carry sulfate and sialic acid residues. In addition, several unusual N-glycans such as paucimannosidic structures with and without core fucose were identified. Interestingly, we also observed novel human structures consisting of core-fucosylated high mannose N-glycans (Man5–Man9). A representative example of an ion trap MS/MS spectrum of such a species is shown in Fig. 2.
Fig. 2.
Ion trap-MS/MS spectrum of a core-fucosylated high mannose structure. The MS/MS of m/z 751.9 [M + 2H]2+ clearly indicates the presence of a core fucose (fragment ion at m/z 692.2, 854.3). The doubly charged fragment ions at m/z 670.7, 589.7, 508.7, and 427.7 are indicative for the subsequent loss of the mannoses during the fragmentation. Glycan schemes were prepared using GlycoWorkbench. Red triangle, fucose; green circle, mannose; blue square, N-acetylglucosamine.
MALDI-TOF-MS Profiling of N-glycans from Colorectal and Control Tissues
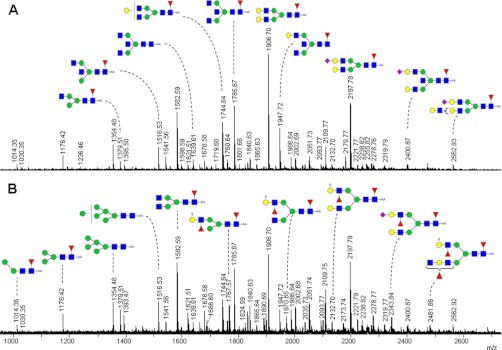
Aliquots of the enzymatically released, AA-labeled, and unfractionated N-glycans from all cancer and control tissues were preconcentrated on HILIC micro-SPE tips and analyzed by negative mode MALDI-TOF-MS. Fig. 3 shows the MALDI-TOF-MS spectra of the AA-labeled glycans enzymatically released from a cancer and its paired control tissue. Although some small differences in the relative abundance of N-glycans of the cancer and control tissue are visible, both spectra contain high mannose type oligosaccharides, complex type and hybrid type glycans, sulfated, and sialylated glycans. Prior to multivariate statistical analysis, mass spectra of tumor and control N-glycan profiles were preprocessed (smoothing and base-line correction), and peak picking and normalization were performed as described under “Material and Methods.”
Fig. 3.
Negative ion mode MALDI-TOF-MS spectra of AA-labeled N-glycans released from a tumor (B) and its paired control tissue (A). Proposed composition of the N-glycan structures released from the colon tissues and deduced from the MALDI-TOF(/TOF)-MS are listed in supplemental Table I. Only major structures are depicted. The m/z values present in both A and B are annotated either in A or in B. Glycan schemes were prepared using GlycoWorkbench. Red triangle, fucose; yellow circle, galactose; green circle, mannose; blue square, N-acetylglucosamine; purple diamond, sialic acid.
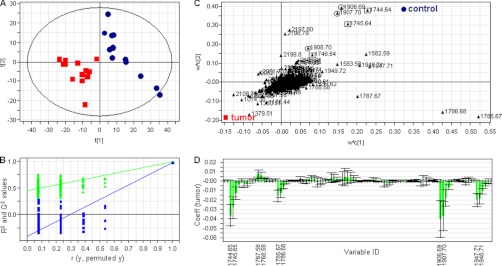
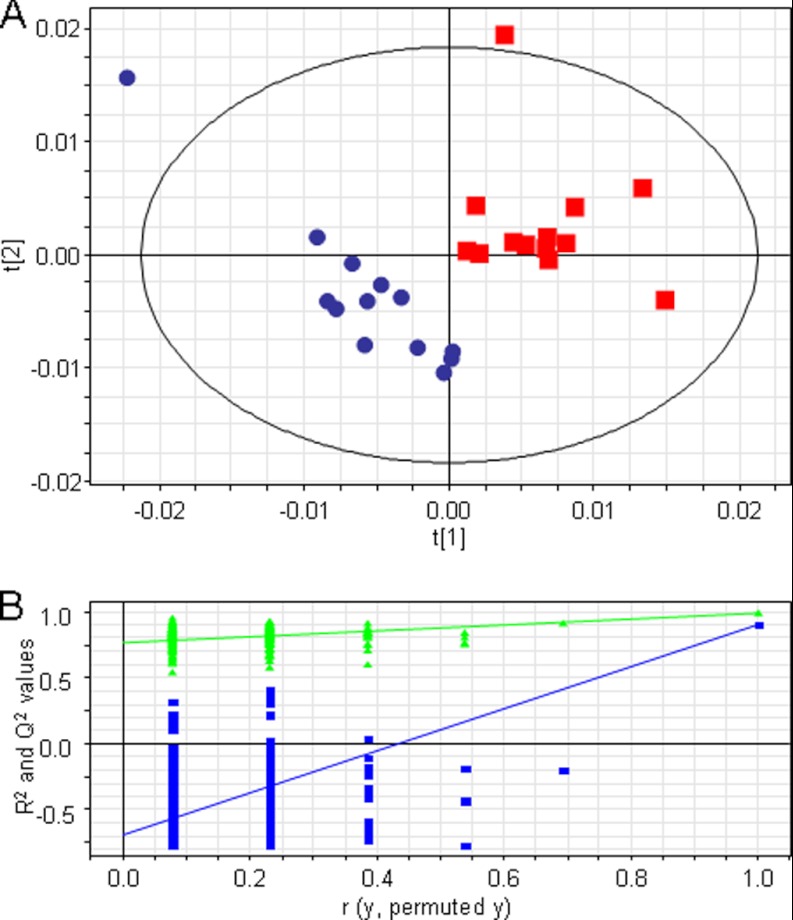
Initial PCA analysis of the MS spectra of the 26 samples showed a homogeneous distribution in the scores, and the score plot of the first two components showed a separation trend between tumor and control samples (supplemental Fig. 2). This separation tendency became more evident in the PCA score plot of the components 3 and 5 (supplemental Fig. 3A). In addition, the score plot of components 3 and 5 showed no clustering according to sex (supplemental Fig. 3B), tumor type (supplemental Fig. 3C), or blood group (supplemental Fig. 3D), indicating that the first components of the PCA analysis are describing the variation between tumor and control samples. To monitor the changes in N-glycosylation caused by colorectal cancer, a PLS-DA two-class model was built using cancer versus control classification as response variable. The quality and the separation power of the model increased after a two-component OSC, and the score plot of the first two components of the PLS-DA model built using the MALDI-TOF-MS spectra of the AA-labeled N-glycans showed a clear discrimination between cancer and control tissues (Fig. 4A). The use of the residual data from this orthogonal model effectively filters obscuring variation in the data set. Following the application of the OSC, the PLS-DA model was constructed with three significant PLS components resulting in an excellent separation R2Y(cum) of 99% and a good prediction ability Q2(cum) of 97%. The validity of the model was evaluated based on a 200-permutation test (intercepts: R2 = 0.43 and Q2 = −0.44) (Fig. 4B).
Fig. 4.
Overview of the PLS-DA model of AA-labeled N-glycans cancer versus control following an OSC approach with outliers removed. A, score plot showing the separation of tumor (red squares) and control (blue circles) tissues based on the information contained in the MS profiles. B, the validity of the model proved based on a 200-permutation test of the three components. Intercepts: R2 = 0.43 (green triangles) and Q2 = −0.44 (blue squares). C, overview of the loading plot corresponding to the score plot. X represents the m/z values that correlate with the Y variables (tumor and control), displaying the individual isotopes of the N-glycans contributing to the differentiation between cancer and control. Squares and circles mark the consecutive isotopes corresponding to two of the N-glycans structures decreased in cancer. D, representative part of the PLS-DA coefficient plot illustrating how strongly Y (samples) is correlated to the systematic part of each of the X variables (individual isotopes of the AA-labeled N-glycans).
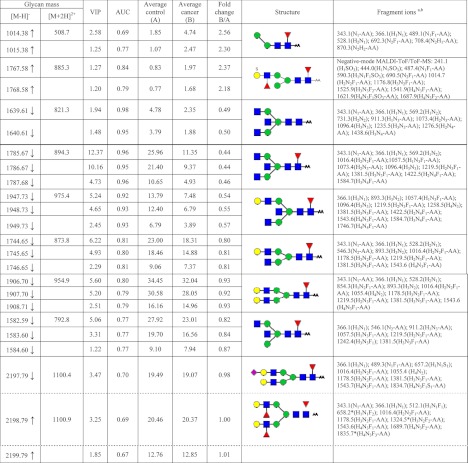
The N-glycans differentially expressed in cancer tissue were identified using the information of the coefficient and loading plots (Fig. 4, C and D) in combination with the VIP value. The diverging peak masses were successive, describing the isotopic pattern of the glycan structures (e.g., m/z 1906.69, 1907.70, and 1908.70). Therefore, based on their VIP value, only the discriminating masses with at least two consecutive isotopes were considered for further analysis. The discriminative glycans and their corresponding VIP values are listed in Table II. Because the small number of samples did not allow evaluation of the model by using a validation sample set, we decided to evaluate the sensitivity and specificity of the differentially expressed structures by calculating their AUC value. Therefore, the MS spectra were imported into ClinPro Tools. The cross-validation recognition rate for the two-class model (cancer versus control) was 93% with the support vector machine algorithm. The t test of difference applied to line spectra of the two groups revealed several peaks discriminating cancer from control. The separation efficiency was evaluated for each individual marker with receiver operating characteristic curve analysis. The AUC value calculated for the individual isotopes of the first three discriminators was higher than 0.9, indicating a highly accurate separation power (36). Based on this model, we calculated the AUC values of the N-glycan structures that were differentially expressed in cancer tissue in comparison with the control tissue (discriminative structures in the PLS-DA analysis) (Table II). The fold increase and decrease was calculated using the average area under the peak and is represented in Table II (fifth and sixth columns).
Table II. Differentially expressed N-glycans in cancer tissue versus control tissue according to multivariate statistics.
N-Glycan masses were determined by negative-mode MALDI-TOF/TOF-MS ([M − H]−) and positive mode LC-ESI-ion trap-MS ([M + 2H]2+). The molecular discriminators were identified based on coefficients and loadings from the two-class model that was build using cancer versus control classification as response variable. The features represented by at list two consecutive isotopes with a variable importance value (VIP) higher than 1 are listed in the third column. The sensitivity and specificity (AUC) calculated for all structures are shown in the fourth column. The average area was calculated for each isotope of the two classes (fifth and sixth columns), and subsequently the fold increase and decrease in cancer samples were calculated. Structure elucidation was based on the fragmentation of the discriminative glycans, and common knowledge of glycobiology and the presence of structural isomers cannot be excluded. Structural schemes are not intended to specify linkage positions. For example, no distinction is made between six-branch and three-branch of biantennary glycans, and linkage angles are not intended to imply specific linkage positions. The glycans marked with a upward arrow are increased in cancer, and the glycans marked with a downward arrow are decreased in cancer. *, glycan rearrangement products caused by internal residue losses (loss of a GlcNAc residue) or migration of terminal monosaccharides such as fucoses. Mass spectrometric glycan rearrangements are dependent on the presence of a mobile proton (71). The fragments indicated in the ninth column are visualized in supplemental Fig. 1.
a AA-labeled glycan species ([M + 2H]2+) were analyzed by ion trap-MS/MS if not indicated differently.
b Next to signal intensity, other relevant criteria used for peak picking were the observation of a proper isotope distribution and mass accuracy.
Structural Elucidation of Differentially Detected N-glycans
The structures of four of the discriminators that were shown to be decreased in cancer tissues contained a bisecting GlcNAc. Representative fragmentation spectra of m/z 1639.61 [M-H]− and of m/z 821.4 [M + 2H]2+ (H3N5-AA) are shown in supplemental Fig. 4. Fragment ions at m/z 911.4 [M+H]+ (H1N3-AA), m/z 1276.6 [M+H]+ (H2N4-AA), and m/z 1274.6 [M-H]− are indicative for the presence of a bisecting GlcNAc. In addition, the fragmentation spectra of the core-fucosylated structures comprise the corresponding fucosylated fragments at m/z 1057.4 [M+H]+ (H1N3F1-AA) and m/z 1422.5 [M+H]+ (H2N4F1-AA) (data not shown). The AUC value calculated for three of the four discriminating structures (m/z 1639.61, 1785.67, and 1947.73 (Table II) containing a bisecting GlcNAc was higher than 0.90, indicating a high sensitivity and specificity (36). Supplemental Fig. 5 shows the receiver operating characteristic curves of the first isotopes of these three structures. The fold change calculated for these structures was ∼0.50, showing a decrease of the glycans containing a bisecting GlcNAc in cancer tissues. The fourth structure (m/z 1582.59) may represent a mixture of a bianntenary N-glycan and an N-glycan containing a bisecting GlcNAc, which could explain the moderate AUC value of 0.77.
In addition to the structures containing a bisecting GlcNAc, core-fucosylated, biantennary N-glycans were found to be slightly decreased in cancer tissues. The AUC value calculated for m/z 1744.65, as well as m/z 1906.69, was ∼0.80. Although the biantennary glycan with m/z 2197.79 was characterized by a relative high VIP value (3.5), the AUC value was moderate (0.7), and the decrease was only minor (Table II). In this region of the MALDI spectrum, there was an overlap of another differential antenna-fucosylated N-glycan (m/z 2198.79; Table II). The second isotope of this glycan showed a slight increase in the cancer tissue. Our data clearly revealed the presence of both the sialylated species (m/z 2197.79 (M+H]+) and the double antenna-fucosylated species (m/z 2198.79 [M+H]+), which were nicely separated during LC-ion trap-MS/MS of the corresponding HILIC fraction (supplemental Fig. 6). The MS/MS of m/z 1100.4 [M + 2H]2+ clearly indicates the presence of a sialic acid (fragment ion at m/z 657.2), whereas the MS/MS of m/z 1100.9 [M + 2H]2+ clearly indicates the presence of a core fucose (fragment ions at m/z 1016.4 and 1178.5) and the presence of a fucose on the antennae (fragment ion at m/z 512.1).
Among the structures shown to be differentially expressed in cancer tissues, we identified a paucimannosidic glycan with m/z 1014.38 [M-H]-(H2N2F1). This structure was found to be elevated in cancer tissues by a factor of 2.5. The sensitivity and specificity calculated by the receiver operating characteristic analysis were good (0.69 for the first isotope and 0.77 for the second isotope).
In addition, the N-glycan with m/z 1767.57 containing a sulfated Lewis X determinant was shown to be increased in cancer (more than 2-fold). This structure proved to have a good sensitivity and specificity for the individual isotopes, with AUC values of 0.84 for the first isotope and 0.79 for the second isotope. Negative mode MALDI-TOF/TOF-MS fragmentation was used for structure elucidation (supplemental Fig. 7). The fragment at m/z 241.1 (H1[S]) indicates the presence of a sulfate group on the terminal hexose. This observation is confirmed by the presence of the fragment ions m/z 444.4 (H1N1[S]) and m/z 590.4 (H1N1F1[S]), indicating a sulfated Lewis X type structure. The presence of a core fucose was confirmed by the fragment ions at m/z 487.4 (N1F1-AA), m/z 690.5(N2F1-AA), m/z 1014.7 (H2N2F1-AA), and m/z 1176.8 (H3N2F1-AA). The fragment at m/z 1525.9 (H3N3F2-AA) shows the presence of a fucose on a N-acetylhexosamine in the antenna. The strong signal at m/z 1687.7 (H4N3F2-AA) indicates the loss of the more labile sulfate group during the negative ion mode fragmentation.
Analysis of N-glycan Classes
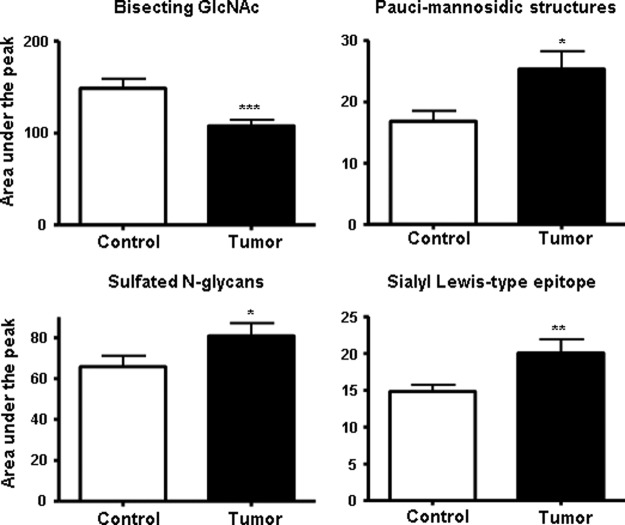
Next, we investigated whether the decrease in related N-glycan structures such as structures containing a bisecting GlcNAc and the increase in paucimannosidic structures and sulfated N-glycans is limited to the set of structures identified with the multivariate statistics or that a general alteration in these clusters of glycans can be observed. For this purpose, the normalized intensities of at least the first two consecutive isotopes of each structure were summed up sample-wise for the different classes of glycans: N-glycans containing a bisecting GlcNAc, sulfated structures, and paucimannosidic structures. As a positive control, sialylated Lewis-type structures were included in this analysis because it has been shown previously that these structures were elevated in colorectal cancer (37, 38), although no individual species belonging to this class was observed to be differential in our MALDI-TOF-MS profiling study. A statistical comparison of the glycan clusters between cancer and control was made by using two-tailed paired t test analysis (Fig. 5).
Fig. 5.
Box plots illustrating the N-glycosylation changes in colon cancer tissues when compared with the accompanying control tissues. The enzymatically released and AA-labeled N-glycans were measured by MALDI-TOF-MS. The structures corresponding to the same class were grouped together to perform a two-tailed paired t test analysis. The N-glycans carrying a sulfate group (lower left panel), a paucimannosidic structure (upper right panel), a bisecting GlcNAc (upper left panel), and a sialyl Lewis-type epitope derived from cancer tissues (lower right panel) were pair-wise compared with the N-glycans derived from the control tissues. The N-glycans containing a sulfate group, the paucimannosidic structure and structures containing a sialyl Lewis-type epitope are shown to be significantly increased in cancer in contrast to structures containing a bisecting GlcNAc that were significantly decreased. The values are reported as the means ± S.E., as indicated in the figure legends. *, p < 0.05; **, p < 0.01; and ***, p < 0.001: statistically significant compared with control.
This analysis showed that the sialylated Lewis-type structures were indeed significantly increased in cancer tissues (p < 0.01). Furthermore, a significant increase (p < 0.05) of sulfated structures and paucimannosidic structures was observed in the cancer tissue. Moreover, the decrease of bisecting GlcNAc structures in cancer cells was found to be highly significant (p < 0.001), which is in line with the fact that these types of structures are the main discriminators in the multivariate statistical analysis.
HILIC-HPLC Profiling of N-glycans from Colorectal and Control Tissues
Next, aliquots of the AA-labeled N-glycans from the individual tumor and control tissues were profiled by HILIC-HPLC with fluorescence detection. The LC chromatograms were base line-corrected and aligned using correlation optimized warping (supplemental Fig. 8). Prior to multivariate statistics, performed using SIMCA P+ software, the aligned data were normalized to unit size. PCA was performed to provide an overview of the data set and to detect possible outliers. The score plot of the components 2 and 5 showed a separation trend between the tumor and control samples (supplemental Fig. 9). Using partial least squares discriminant analysis (PLS-DA) to optimize visualization of the N-glycosylation changes in colorectal cancer, a clear separation of the cancer and control samples was observed. However, the class separation in the PLS-DA model was improved by using a two-component OSC filtering that enhanced the performance of multivariate pattern recognition analysis and the predictive power of the model. Following the application of the OSC, the PLS-DA model was constructed with six significant PLS components, resulting in an excellent separation R2Y(cum) of 99% and a good prediction ability Q2(cum) of 90% (Fig. 6A). The validity of the model was evaluated based on a 200-permutation test (intercepts: R2 = 0.77 and Q2 = −0.69) as shown in Fig. 6B.
Fig. 6.
PLS-DA analysis of tumor versus control using the HILIC-HPLC data. A, PLS-DA score plot of tumor samples (red squares) and control samples (blue dots) after OSC. B, demonstration of the validity of the model using a 200-permutation test of the six components. Intercepts: R2 = 0.77 (green triangles) and Q2 = −0.69 (blue squares).
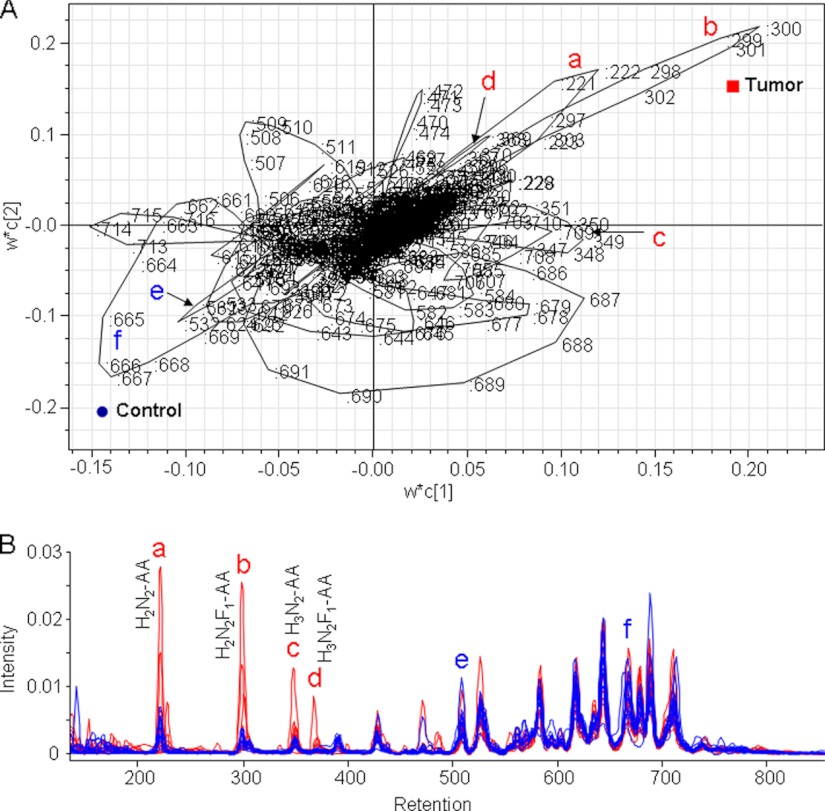
The loading plot of the X- and Y-weights (w*c) of the first PLS component against the second shows how the X variables (chromatogram regions) correlate with the Y variables (tumor and control), displaying the regions corresponding to the chromatogram peaks contributing to the differentiation between tumor and control. As shown in Fig. 7A, these successive discriminating data points are forming loops representing specific areas of the chromatograms. Particularly, the regions 296–303 (increased in cancer) and 660–670 (decreased in cancer) are shown to have a high discriminating power. In addition, several loops corresponding to specific elution ranges and peaks of the chromatograms are extending to intermediate regions of the plot (such as 642–648 and 684–694) and therefore mainly describe the variation within the classes.
Fig. 7.
Demonstration of the major discriminators from the PLS-DA analysis of HILIC-HPLC data. A, loading plot of the PLS-DA model classifying tumor and control tissues. The data point numbers are indicated. B, aligned HILIC chromatograms of tumor (red) and control tissues (blue). The chromatographic regions that contribute to the differentiation between tumor and control are indicated in A and B with a–d (elevated in tumor) and e and f (elevated in control). Compositions of the major glycans are given for the chromatographic peaks that discriminate between tumor and control.
Using the information from the preparative HILIC-HPLC and subsequent structural elucidation described above, we annotated the glycans comprised by the differential chromatographic peaks. The early eluting fractions were of low complexity, containing relatively small N-glycans (paucimannosidic structures) such as H2N2-AA (chromatogram region 220–225; Fig. 7B, peak a), H2N2F1-AA (chromatogram region 296–303; Fig. 7B, peak b), H3N2-AA (chromatogram region 345–352; Fig. 7B, peak c), and H3N2F1-AA (chromatogram region 367–374; Fig. 7B, peak d). The paucimannosidic type N-glycan at m/z 1014.38 (N2H2F1-AA) that was shown to be elevated in cancer tissues based on our MALDI-TOF-MS profiling analysis was also one of the main discriminators in the HILIC-HPLC analysis.
In addition, MALDI-TOF-MS profiling experiments indicated that the biantennary N-glycan containing a bisecting GlcNAc at m/z 1785.67 was decreased in the cancer tissues. This compound eluted in the chromatogram region 524–571. The same region of the chromatogram contains other major compounds that were shown to be increased or decreased in cancer, possibly averaging the effect of individual compounds in this elution range. Nevertheless, part of this region, namely 530–534 (Fig. 7B, peak e) where m/z 1582.59 (a structure also shown to be decreased in cancer) co-elutes with m/z 1785.67, has a high discriminative power in the multivariate statistics analysis of the LC chromatograms.
The sialylated and core-fucosylated bi-antennary glycan at m/z 2197.79, which was shown to be slightly decreased in cancer, elutes in the LC chromatogram region 660–693. The first part of this region, 660–670, (Fig. 7B, peak f) is a discriminator in the multivariate statistics of the LC chromatograms. The co-elution of the N-glycan at m/z 2198.79, which is slightly elevated in cancer, in the second part of the peak, is probably related to the poor discriminative power of this region.
DISCUSSION
In this study we used a combination of liquid chromatography with fluorescent detection, MALDI-TOF-MS profiling, and multivariate statistical analysis to monitor N-glycosylation changes in colorectal cancer tumors as compared with colorectal control tissues. In addition, we analyzed multiple HILIC-HPLC fractions with negative mode MALDI-TOF/TOF-MS and positive mode ESI-ion trap-MS/MS for structural elucidation.
Because of the known lability of sialylated glycans in mass spectrometric analysis, the LC-MS system was carefully tuned to minimize this effect. This involved the adjustment of voltages in the ion transfer region to avoid in-source decay while keeping the sensitivity of the mass spectrometer high. The tuned system allowed the analysis of sialylated, AA-labeled N-glycans without the loss of sialic acids or other labile portions of the molecule (supplemental Fig. 10).
Detection of sialylated, AA-labeled glycans by negative mode MALDI-TOF-MS is influenced by two effects: 1) the more efficient negative mode ionization of sialylated glycans as compared with their nonsialylated counterparts and 2) the in-source desialylation of part of these glycans. Notably, both effects equally apply to tumor and control samples and will therefore not compromise the comparison of these samples.
Usually, the analysis of the LC profiles of complex samples provides information complementary to the analysis of MS spectra. Although MS is characterized by a higher resolution power, incomplete separation of glycan structures is often observed for both techniques. Structurally unrelated N-glycans are co-eluting during HPLC, and therefore the discriminative power of some of the structures is averaged. On the other hand one m/z value may contain several structural isomers. Nevertheless, N-glycosylation changes detected in our study by MALDI-TOF-MS profiling correlated well with the results from the HILIC-HPLC analysis. The N-glycans shown to be differentially expressed in our mass spectrometry analysis were identified as eluting in parts of the LC chromatograms responsible for the separation between tumor and control samples.
One of the observations of our study was that in tumor tissues, N-glycans containing a bisecting GlcNAc were decreased. The AUC value calculated for three of the four discriminating structures (m/z 1639.61, 1785.67, and 1947.73; Table II) containing a bisecting GlcNAc was higher than 0.90, indicating a prominent change of the N-glycans containing a bisecting GlcNAc. Furthermore the decrease of bisecting GlcNAc structures in colorectal cancer tumors was found to be highly significant (p < 0.001). This finding is in agreement with a previous study: the action of GnT-III transferase on the β1,4-mannose results in the formation of a bisecting GlcNAc, which suppresses the β1,6-GlcNAc branch formation (strongly associated with cancer metastasis) catalyzed by GnT-V (39). Bisected complex N-glycans are not substrates for GnT-V and thereby have reduced branching (40, 41). Recently, Song et al. (42) have demonstrated that the bisecting GlcNAc on N-glycans inhibited growth factor signaling and retarded mammary tumor progression. The implication of bisecting GlcNAc for biological functions has been reviewed by Gu et al. (20).
In the current study we observed an increase of sialylated Lewis type epitopes in tumor tissues that is in line with previous research on this topic. Increased levels of sialylated Lewis A and Lewis X epitopes have been earlier described in colorectal cancer (37, 38, 43, 44).
In addition, we observed increased levels of sulfated N-glycans. The expression levels of sulfomucin in colon cancer have been shown to be lower than those in the adjacent normal mucosa (45, 46). On the other hand, the 3-sulfo-Lewis X determinant has been found to be a major carbohydrate motif in mouse xenograft tumors produced by LS174T-HM7 cells, a subline of the human colon carcinoma cell line LS174T with higher metastatic tendency (47). Moreover, an up-regulation of sulfotransferase activities has been described to be associated with human gastric tumorigenesis (48). Recently, Chandrasekaran et al. (11) examined the patterns of glycan-sulfotransferase activities in several cancer cell lines and showed that the 3-sulfo Lewis X and the 3-O-sulfo-Globo unit were uniquely associated with colon cancer cell lines. The sulfation of N-glycans may play both structural and functional roles. The sulfation of Gal as well as GlcNAc residues would provide the N-glycans with a strong negative charge, which provides resistance to degradation by sialidases, galactosidases, and hexosaminidases, potentially increasing their life span by reducing receptor-mediated glycoprotein clearance (49). Moreover, the negative charge of the sulfated group is thought to play an important role in interaction with functional molecules such as growth factors, cellular adhesion molecules, and extracellular matrix proteins (49). Using a galactoside microarray, Horlacher et al. (50) screened the galectin sugar-binding preferences and demonstrated that sulfated glycans are preferentially bound by galectin-1 and galectin-2. Galectin-1 was up-regulated in colorectal cancer and correlated with the degree of dysplasia, suggesting that galectin-1 is related to malignant progression in colorectal cancer (51). Galectins are playing an important role in cancer micrometastasis, and it seems that sulfated glycans may facilitate interaction of colorectal cancer tumors with galectins, resulting in intercellular homotypic and heterotypic adhesion of cancer cells.
One of the glycans shown by both LC and MS analysis to be up-regulated in tumor tissues was a paucimannosidic structure (Man3GlcNAc(Fuc)GlcNAc). Paucimannosidic structures, which are rarely found in vertebrates, have been noticed before in the kidney of a systemic lupus erythematosus mouse model (52). Joosten et al. (53) did report that the human colorectal cancer A33 antigen produced in the SW1222 human colon cancer cell line was decorated with Man3GlcNAc(Fuc)GlcNAc, but the authors did not discuss further the presence of this glycan. A comparison of the early elution range of the HILIC-HPLC profile, published by Joosten et al., with our results (Fig. 5 and supplemental materials) would suggest that the two undetermined structures in their chromatogram may correspond to Man2GlcNAc2 and Man2GlcNAc(Fuc)GlcNAc. The trimmed structures are most probably the result of lysosomal exoglycosidases. Activities of β-galactosidase, α-mannosidase, and neuraminidase (54), as well as β-hexosaminidase (55, 56), have been shown to be significantly increased in colorectal cancer tissue as compared with normal tissue. Moreover, increased levels of lysosomal exoglycosidases have been reported for serum and urine of colorectal cancer patients (12, 57, 58). Although we are not aware of a particular role of paucimannosidic structures in cancer progression, further studies would be needed to investigate whether these structures could be used as a marker for the detection of colorectal carcinoma and the prediction for the response to therapy.
The analysis of the fractionated N-glycans enabled us to characterize the composition of 245 N-glycans present in colorectal tumors. Among the N-glycans released from the tumor proteins, core-fucosylated high mannose structures (Man5–9) were shown to be present. Fucosylated Man5 and Man6 have hitherto only been described for arylsulfatase A purified from human placenta (59), whereas fucosylated Man7–9 have, to the best of our knowledge, not been described on glycoproteins of mammalian tissues or body fluids. It has been established that fucosylation of the innermost GlcNAc occurs almost exclusively in complex and hybrid-type structures because the addition of the core fucose requires at least the presence of a β1,2-GlcNAc unit on the core (α1–3)Man (60, 61). Hence, the occurrence of fucosylated Man5 and Man6 may be explained by core fucosylation of both classical Man5-based hybrids and Man6-based hybrids (62), whereas the Man7–9 described here point to another biosynthetic route involving the direct fucosylation of high mannose N-glycans lacking the β1,2-GlcNAc unit. Although α1,6-fucosyltransferase (FUT8) is supposed to be localized in the medial Golgi, several researchers have demonstrated the cell type-specific Golgi subcompartmentalization of some glycosylation enzymes (63, 64). Magner et al. (65) demonstrated that active mouse thyrotrophs appeared to shift the subcellular site of fucosylation partially from Golgi to rough endoplasmic reticulum. The co-localization of α1,2-mannosidase with α1,6-fucosyltransferase competing for the same substrate may explain the presence of high mannose core-fucosylated structures. Core fucosylation has been shown to be altered in cancer (66), and Kyselova et al. (67) and Saldova et al. (68) describe in two independent studies a significant increase in core fucosylation in prostate cancer patient sera. Furthermore, α1,6-fucosyltransferase was found to be overexpressed in thyroid carcinoma tissue and linked directly to tumor size and lymph node metastasis (69). In contrast to these studies, Tabarés et al. (70) showed a decrease in core fucosylation on the PSA protein isolated from cancer patient sera in comparison with PSA isolated from sera of healthy individuals. Moreover, the expression of FUT8 has been shown to be decreased in anaplastic carcinoma (69). One might conclude that overexpression of α1,6-fucosyltransferase together with the cell type-specific Golgi subcompartmentalization may be specific for a given cancer. The detection of core-fucosylated high mannose structures clearly illustrates the pronounced alterations in the glycosylation machinery of tumor cells.
Analysis of tissue released N-glycans by the combination of mass spectrometry and liquid chromatography represents a new approach in cancer biomarker studies aiming at post-translational modifications rather than the expression of proteins. Changes in glycosylation during cancer progression appear to be characteristic for the disease. To our knowledge an increase in the level of paucimannosidic structures has, up till now, not been associated with colorectal cancer or any other type of cancer, and only few studies have correlated the increase in sulfated glycans and sulfotransferases with cancer in general. Although the decrease of bisecting GlcNAc may not serve as a specific and sensitive marker for colorectal cancer, this might still represent an important factor that affects tumor progression and might open new ways to search for colorectal cancer therapeutics. The fucosylated high mannose structures have been identified only after in-depth analysis of the N-glycan pool released from the tumor samples. So far Man7–9GlcNAc(Fuc)GlcNAc oligosaccharides have not been described on human glycoproteins.
The present data indicate that it might be worthwhile to further study the detection of cancer-associated glycosylation features of specific proteins and aim at the analysis of these glycoproteins in the circulation. These efforts will hopefully result in cancer biomarkers of high sensitivity and specificity.
Supplementary Material
Acknowledgments
We thank Dr. Paul J. Hensbergen for critically reading the manuscript.
Footnotes
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HILIC
- hydrophilic interaction liquid chromatography
- MS/MS
- tandem mass spectrometry
- Fuc
- fucose
- SPE
- solid phase extraction
- PCA
- principal component analysis
- PLS-DA
- partial least squares discriminant analysis
- AA
- 2-aminobenzoic acid
- OSC
- orthogonal signal correction
- VIP
- variable importance in the projection
- AUC
- area under the curve.
REFERENCES
- 1. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) Cancer statistics. CA-Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2. Davies R. J., Miller R., Coleman N. (2005) Colorectal cancer screening: Prospects for molecular stool analysis. Nat. Rev. Cancer 5, 199–209 [DOI] [PubMed] [Google Scholar]
- 3. Levin B., Brooks D., Smith R. A., Stone A. (2003) Emerging technologies in screening for colorectal cancer: CT colonography, immunochemical fecal occult blood tests, and stool screening using molecular markers. CA-Cancer J. Clin. 53, 44–55 [DOI] [PubMed] [Google Scholar]
- 4. Terdiman J. P. (2005) Colonoscopy is superior to flexible sigmoidoscopy for colorectal cancer screening: Now beyond a reasonable doubt? Gastroenterology 129, 1793–1794 [DOI] [PubMed] [Google Scholar]
- 5. Nakazato M., Yamano H., Matsushita H., Sato K., Fujita K., Yamanaka Y., Imai Y. (2006) Immunologic fecal occult blood test for colorectal cancer screening. Jpn. Med. Assoc. J. 49, 203–207 [Google Scholar]
- 6. Saussez S., Marchant H., Nagy N., Decaestecker C., Hassid S., Jortay A., Schüring M. P., Gabius H. J., Danguy A., Salmon I., Kiss R. (1998) Quantitative glycohistochemistry defines new prognostic markers for cancers of the oral cavity. Cancer 82, 252–260 [PubMed] [Google Scholar]
- 7. Turner G. A. (1992) N-Glycosylation of serum proteins in disease and its investigation using lectins. Clin. Chim. Acta 208, 149–171 [DOI] [PubMed] [Google Scholar]
- 8. Hakomori S. I. (1989) Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv. Cancer Res. 52, 257–331 [DOI] [PubMed] [Google Scholar]
- 9. Hakomori S. (2001) Tumor-associated carbohydrate antigens defining tumor malignancy: Basis for development of anti-cancer vaccines. Adv. Exp. Med. Biol. 491, 369–402 [DOI] [PubMed] [Google Scholar]
- 10. An H. J., Kronewitter S. R., de Leoz M. L., Lebrilla C. B. (2009) Glycomics and disease markers. Curr. Opin. Chem. Biol. 13, 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chandrasekaran E. V., Xue J., Neelamegham S., Matta K. L. (2006) The pattern of glycosyl- and sulfotransferase activities in cancer cell lines: A predictor of individual cancer-associated distinct carbohydrate structures for the structural identification of signature glycans. Carbohydr. Res. 341, 983–994 [DOI] [PubMed] [Google Scholar]
- 12. Szajda S. D., Snarska J., Puchalski Z., Zwierz K. (2008) Lysosomal exoglycosidases in serum and urine of patients with colon adenocarcinoma. Hepatogastroenterology 55, 921–925 [PubMed] [Google Scholar]
- 13. Arnold J. N., Saldova R., Galligan M. C., Murphy T. B., Mimura-Kimura Y., Telford J. E., Godwin A. K., Rudd P. M. (2011) Novel glycan biomarkers for the detection of lung cancer. J. Proteome Res. 10, 1755–1764 [DOI] [PubMed] [Google Scholar]
- 14. Abd Hamid U. M., Royle L., Saldova R., Radcliffe C. M., Harvey D. J., Storr S. J., Pardo M., Antrobus R., Chapman C. J., Zitzmann N., Robertson J. F., Dwek R. A., Rudd P. M. (2008) A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology 18, 1105–1118 [DOI] [PubMed] [Google Scholar]
- 15. de Leoz M. L., Young L. J., An H. J., Kronewitter S. R., Kim J., Miyamoto S., Borowsky A. D., Chew H. K., Lebrilla C. B. (2011) High-mannose glycans are elevated during breast cancer progression. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arnold J. N., Saldova R., Hamid U. M., Rudd P. M. (2008) Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics 8, 3284–3293 [DOI] [PubMed] [Google Scholar]
- 17. Hashimoto S., Asao T., Takahashi J., Yagihashi Y., Nishimura T., Saniabadi A. R., Poland D. C., van Dijk W., Kuwano H., Kochibe N., Yazawa S. (2004) α1-Acid glycoprotein fucosylation as a marker of carcinoma progression and prognosis. Cancer 101, 2825–2836 [DOI] [PubMed] [Google Scholar]
- 18. Mizuguchi S., Inoue K., Iwata T., Nishida T., Izumi N., Tsukioka T., Nishiyama N., Uenishi T., Suehiro S. (2006) High serum concentrations of sialyl Lewisx predict multilevel N2 disease in non-small-cell lung cancer. Ann. Surg. Oncol. 13, 1010–1018 [DOI] [PubMed] [Google Scholar]
- 19. Mizuguchi S., Nishiyama N., Iwata T., Nishida T., Izumi N., Tsukioka T., Inoue K., Uenishi T., Wakasa K., Suehiro S. (2007) Serum Sialyl Lewis 19x and cytokeratin 19 fragment as predictive factors for recurrence in patients with stage I non-small cell lung cancer. Lung Cancer 58, 369–375 [DOI] [PubMed] [Google Scholar]
- 20. Gu J., Sato Y., Kariya Y., Isaji T., Taniguchi N., Fukuda T. (2009) A mutual regulation between cell-cell adhesion and N-glycosylation: Implication of the bisecting GlcNAc for biological functions. J. Proteome Res. 8, 431–435 [DOI] [PubMed] [Google Scholar]
- 21. Li M., Song L., Qin X. (2010) Glycan changes: Cancer metastasis and anti-cancer vaccines. J. Biosci. 35, 665–673 [DOI] [PubMed] [Google Scholar]
- 22. Vercoutter-Edouart A. S., Slomianny M. C., Dekeyzer-Beseme O., Haeuw J. F., Michalski J. C. (2008) Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics 8, 3236–3256 [DOI] [PubMed] [Google Scholar]
- 23. Qiu Y., Patwa T. H., Xu L., Shedden K., Misek D. E., Tuck M., Jin G., Ruffin M. T., Turgeon D. K., Synal S., Bresalier R., Marcon N., Brenner D. E., Lubman D. M. (2008) Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J. Proteome Res. 7, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahn Y. H., Kim Y. S., Ji E. S., Lee J. Y., Jung J. A., Ko J. H., Yoo J. S. (2010) Comparative quantitation of aberrant glycoforms by lectin-based glycoprotein enrichment coupled with multiple-reaction monitoring mass spectrometry. Anal. Chem. 82, 4441–4447 [DOI] [PubMed] [Google Scholar]
- 25. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 26. Friedman D. B., Hill S., Keller J. W., Merchant N. B., Levy S. E., Coffey R. J., Caprioli R. M. (2004) Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 4, 793–811 [DOI] [PubMed] [Google Scholar]
- 27. Ma Y., Peng J., Liu W., Zhang P., Huang L., Gao B., Shen T., Zhou Y., Chen H., Chu Z., Zhang M., Qin H. (2009) Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Mol. Cell. Proteomics 8, 1878–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Contessa J. N., Bhojani M. S., Freeze H. H., Ross B. D., Rehemtulla A., Lawrence T. S. (2010) Molecular imaging of N-linked glycosylation suggests glycan biosynthesis is a novel target for cancer therapy. Clin. Cancer Res. 16, 3205–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varki A., Kannagi R., Toole B. P. (2009) Glycosylation changes in cancer, in Essentials of Glycobiology, 2nd Ed., pp. 580–670, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 30. Dierssen J. W., de Miranda N. F., Ferrone S., van P. M., Cornelisse C. J., Fleuren G. J., van W. T., Morreau H. (2007) HNPCC versus sporadic microsatellite-unstable colon cancers follow different routes toward loss of HLA class I expression. BMC Cancer 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 32. Wuhrer M., Koeleman C. A., Deelder A. M., Hokke C. H. (2006) Repeats of LacdiNAc and fucosylated LacdiNAc on N-glycans of the human parasite Schistosoma mansoni. FEBS J. 273, 347–361 [DOI] [PubMed] [Google Scholar]
- 33. Ruhaak L. R., Steenvoorden E., Koeleman C. A., Deelder A. M., Wuhrer M. (2010) 2-picoline-borane: A non-toxic reducing agent for oligosaccharide labeling by reductive amination. Proteomics. 10, 2330–2336 [DOI] [PubMed] [Google Scholar]
- 34. Selman M. H., Hemayatkar M., Deelder A. M., Wuhrer M. (2011) Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83, 2492–2499 [DOI] [PubMed] [Google Scholar]
- 35. Eriksson L., Johansson E., Kettaneh-Wold N., Trygg J., Wikstrom C., Wold S. (2006) Multi- and Megavariate Data Analysis: Part I. Basic Principles and Applications, 2nd Ed., Umetrics Academy, Umea, Sweden [Google Scholar]
- 36. Swets J. A. (1988) Measuring the accuracy of diagnostic systems. Science 240, 1285–1293 [DOI] [PubMed] [Google Scholar]
- 37. Nakagoe T., Sawai T., Tsuji T., Jibiki M., Nanashima A., Yamaguchi H., Kurosaki N., Yasutake T., Ayabe H. (2001) Circulating sialyl LewisX, sialyl LewisA, and sialyl Tn antigens in colorectal cancer patients: Multivariate analysis of predictive factors for serum antigen levels. J. Gastroenterol. 36, 166–172 [DOI] [PubMed] [Google Scholar]
- 38. Nakayama T., Watanabe M., Katsumata T., Teramoto T., Kitajima M. (1995) Expression of sialyl LewisA as a new prognostic factor for patients with advanced colorectal-carcinoma. Cancer 75, 2051–2056 [DOI] [PubMed] [Google Scholar]
- 39. Yoshimura M., Nishikawa A., Ihara Y., Taniguchi S., Taniguchi N. (1995) Suppression of lung metastasis of B16 mouse melanoma by N-acetylglucosaminyltransferase-III gene transfection. Proc. Natl. Acad. Sci. U.S.A. 92, 8754–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schachter H. (1986) Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol. 64, 163–181 [DOI] [PubMed] [Google Scholar]
- 41. Yoshimura M., Ihara Y., Ohnishi A., Ijuhin N., Nishiura T., Kanakura Y., Matsuzawa Y., Taniguchi N. (1996) Bisecting N-acetylglucosamine on K562 cells suppresses natural killer cytotoxicity and promotes spleen colonization. Cancer Res. 56, 412–418 [PubMed] [Google Scholar]
- 42. Song Y., Aglipay J. A., Bernstein J. D., Goswami S., Stanley P. (2010) The bisecting GlcNAc on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Res. 70, 3361–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shimono R., Mori M., Akazawa K., Adachi Y., Sgimachi K. (1994) Immunohistochemical expression of carbohydrate antigen 19-9 in colorectal-carcinoma. Am. J. Gastroenterol. 89, 101–105 [PubMed] [Google Scholar]
- 44. Ugorski M., Laskowska A. (2002) Sialyl LewisA: A tumor-associated carbohydrate antigen involved in adhesion and metastatic potential of cancer cells. Acta Biochim. Pol. 49, 303–311 [PubMed] [Google Scholar]
- 45. Yamori T., Kimura H., Stewart K., Ota D. M., Cleary K. R., Irimura T. (1987) Differential production of high molecular weight sulfated glycoproteins in normal colonic mucosa, primary colon carcinoma, and metastases. Cancer Res. 47, 2741–2747 [PubMed] [Google Scholar]
- 46. Yamori T., Ota D. M., Cleary K. R., Hoff S., Hager L. G., Irimura T. (1989) Monoclonal antibody against human colonic sulfomucin: Immunochemical detection of its binding sites in colonic mucosa, colorectal primary carcinoma, and metastases. Cancer Res. 49, 887–894 [PubMed] [Google Scholar]
- 47. Capon C., Wieruszeski J. M., Lemoine J., Byrd J. C., Leffler H., Kim Y. S. (1997) Sulfated Lewis X determinants as a major structural motif in glycans from LS174T-HM7 human colon carcinoma mucin. J. Biol. Chem. 272, 31957–31968 [DOI] [PubMed] [Google Scholar]
- 48. Chandrasekaran E. V., Xue J., Piskorz C., Locke R. D., Tóth K., Slocum H. K., Matta K. L. (2007) Potential tumor markers for human gastric cancer: An elevation of glycan:sulfotransferases and a concomitant loss of α1,2-fucosyltransferase activities. J. Cancer Res. Clin. Oncol. 133, 599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brockhausen I., Kuhns W. (1997) Role and metabolism of glycoconjugate sulfation. Trends Glycosci. Glycotechnol. 9, 379–398 [Google Scholar]
- 50. Horlacher T., Oberli M. A., Werz D. B., Kröck L., Bufali S., Mishra R., Sobek J., Simons K., Hirashima M., Niki T., Seeberger P. H. (2010) Determination of carbohydrate-binding preferences of human galectins with carbohydrate microarrays. Chembiochem. 11, 1563–1573 [DOI] [PubMed] [Google Scholar]
- 51. Hittelet A., Legendre H., Nagy N., Bronckart Y., Pector J. C., Salmon I., Yeaton P., Gabius H. J., Kiss R., Camby I. (2003) Upregulation of galectins-1 and-3 in human colon cancer and their role in regulating cell migration. Int. J. Cancer 103, 370–379 [DOI] [PubMed] [Google Scholar]
- 52. Hashii N., Kawasaki N., Itoh S., Nakajima Y., Kawanishi T., Yamaguchi T. (2009) Alteration of N-glycosylation in the kidney in a mouse model of systemic lupus erythematosus: Relative quantification of N-glycans using an isotope-tagging method. Immunology 126, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Joosten C. E., Cohen L. S., Ritter G., Batt C. A., Shuler M. L. (2004) Glycosylation profiles of the human colorectal cancer A33 antigen naturally expressed in the human colorectal cancer cell line SW1222 and expressed as recombinant protein in different insect cell lines. Biotechnol. Prog. 20, 1273–1279 [DOI] [PubMed] [Google Scholar]
- 54. Bosmann H. B., Hall T. C. (1974) Enzyme activity in invasive tumors of human breast and colon. Proc. Natl. Acad. Sci. U.S.A. 71, 1833–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gil-Martín E., Rodríguez-Berrocal J., Páez de la Cadena M., Fernández-Briera A. (1997) Alterations of glycosidases in human colonic adenocarcinoma. Clin. Biochem. 30, 17–25 [DOI] [PubMed] [Google Scholar]
- 56. Plucinsky M. C., Prorok J. J., Alhadeff J. A. (1986) β-Hexosaminidase from colon and sera of dukes-classified colorectal cancer patients: Activity levels, isozyme patterns, and kinetic properties. J. Natl. Cancer Inst. 77, 57–62 [PubMed] [Google Scholar]
- 57. Szajda S. D., Snarska J., Jankowska A., Puchalski Z., Zwierz K. (2008) Isoenzymes A and B of N-acetyl-β-d-hexosaminidase in serum and urine of patients with pancreatic cancer. Hepatogastroenterology 55, 695–698 [PubMed] [Google Scholar]
- 58. Szajda S. D., Borzym-Kluczyk M., Snarska J., Puchalski Z., Zwierz K. (2009) N-Acetyl-β-d-hexosaminidase and its isoenzymes A and B in blood serum and urine, as a potential colon cancer markers. Hepatogastroenterology 56, 1287–1298 [PubMed] [Google Scholar]
- 59. Hoja-Lukowicz D., Ciolczyk D., Bergquist J., Lityńska A., Laidler P. (2000) High-mannose-type oligosaccharides from human placental arylsulfatase A are core fucosylated as confirmed by MALDI MS. Glycobiology 10, 551–557 [DOI] [PubMed] [Google Scholar]
- 60. Schwarz P. M., Elbein A. D. (1985) The effect of glycoprotein-processing inhibitors on fucosylation of glycoproteins. J. Biol. Chem. 260, 14452–14458 [PubMed] [Google Scholar]
- 61. Voynow J. A., Kaiser R. S., Scanlin T. F., Glick M. C. (1991) Purification and characterization of GDP-l-fucose-N-acetyl β-d-glucosaminide α1–6-fucosyltransferase from cultured human skin fibroblasts: Requirement of a specific biantennary oligosaccharide as substrate. J. Biol. Chem. 266, 21572–21577 [PubMed] [Google Scholar]
- 62. Crispin M., Aricescu A. R., Chang V. T., Jones E. Y., Stuart D. I., Dwek R. A., Davis S. J., Harvey D. J. (2007) Disruption of α-mannosidase processing induces non-canonical hybrid-type glycosylation. FEBS Lett. 581, 1963–1968 [DOI] [PubMed] [Google Scholar]
- 63. Rabouille C., Hui N., Hunte F., Kieckbusch R., Berger E. G., Warren G., Nilsson T. (1995) Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J. Cell Sci. 108, 1617–1627 [DOI] [PubMed] [Google Scholar]
- 64. Velasco A., Hendricks L., Moremen K. W., Tulsiani D. R., Touster O., Farquhar M. G. (1993) Cell type-dependent variations in the subcellular distribution of α-mannosidase I and II. J. Cell Biol. 122, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Magner J. A., Novak W., Papagiannes E. (1986) Subcellular localization of fucose incorporation into mouse thyrotropin and free α-subunits: Studies employing subcellular fractionation and inhibitors of the intracellular translocation of proteins. Endocrinology 119, 1315–1328 [DOI] [PubMed] [Google Scholar]
- 66. Peracaula R., Barrabés S., Sarrats A., Rudd P. M., de Llorens R. (2008) Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kyselova Z., Mechref Y., Al Bataineh M. M., Dobrolecki L. E., Hickey R. J., Vinson J., Sweeney C. J., Novotny M. V. (2007) Alterations in the serum glycome due to metastatic prostate cancer. J. Proteome Res. 6, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saldova R., Fan Y., Fitzpatrick J. M., Watson R. W., Rudd P. M. (2011) Core fucosylation and α2–3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology 21, 195–205 [DOI] [PubMed] [Google Scholar]
- 69. Ito Y., Miyauchi A., Yoshida H., Uruno T., Nakano K., Takamura Y., Miya A., Kobayashi K., Yokozawa T., Matsuzuka F., Taniguchi N., Matsuura N., Kuma K., Miyoshi E. (2003) Expression of α1,6-fucosyltransferase (FUT8) in papillary carcinoma of the thyroid: Its linkage to biological aggressiveness and anaplastic transformation. Cancer Lett. 200, 167–172 [DOI] [PubMed] [Google Scholar]
- 70. Tabarés G., Radcliffe C. M., Barrabés S., Ramírez M., Aleixandre R. N., Hoesel W., Dwek R. A., Rudd P. M., Peracaula R., de Llorens R. (2006) Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology 16, 132–145 [DOI] [PubMed] [Google Scholar]
- 71. Wuhrer M., Koeleman C. A., Hokke C. H., Deelder A. M. (2006) Mass spectrometry of proton adducts of fucosylated N-glycans: Fucose transfer between antennae gives rise to misleading fragments. Rapid Commun. Mass Spectrom. 20, 1747–1754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.