Abstract
Type III secretion systems are central to the pathogenesis and virulence of many important Gram-negative bacterial pathogens, and elucidation of the secretion mechanism and identification of the secreted substrates are critical to our understanding of their pathogenic mechanisms and developing potential therapeutics. Stable isotope labeling with amino acids in cell culture-based mass spectrometry is a quantitative and highly sensitive proteomics tool that we have previously used to successfully analyze the type III secretomes of Citrobacter rodentium and Salmonella enterica serovar Typhimurium. In this report, stable isotope labeling with amino acids in cell culture was used to analyze the type III secretome of enteropathogenic Escherichia coli (EPEC), an important human pathogen, which, together with enterohemorrhagic E. coli and C. rodentium, represents the family of attaching and effacing bacterial pathogens. We not only confirmed all 25 known EPEC type III-secreted proteins and effectors previously identified by conventional molecular and bioinformatical techniques but also identified several new type III-secreted proteins, including two novel effectors, C_0814/NleJ and LifA, that were shown to be translocated into host cells. LifA is a known virulence factor believed to act as a toxin as well as an adhesin, but its mechanism of secretion and function is not understood. With a predicted molecular mass of 366 kDa, LifA is the largest type III effector identified thus far in any pathogen. We further demonstrated that Efa1, ToxB, and Z4332 (homologs of LifA in enterohemorrhagic E. coli) are also type III effectors. This study has comprehensively characterized the type III secretome of EPEC, expanded the repertoire of type III-secreted effectors for the attaching and effacing pathogens, and provided new insights into the mode of function for LifA/Efa1/ToxB/Z4332, an important family of virulence factors.
Bacterial pathogens utilize a variety of complex protein secretion systems to deliver toxins and effector proteins into host cells. These toxins and effectors impair or hijack host cellular functions and immune response pathways, enabling the pathogens to colonize, invade, and propagate within the hosts and causing various diseases (1, 2). One of the best studied bacterial secretion systems is the type III secretion system (T3SS),1 a macromolecular machinery consisting of a series of multicomponent, ring-shaped protein structures spanning the bacterial envelope, the extracellular space, and the host cellular membrane (3, 4). T3SSs are widely found in many medically and agriculturally significant Gram-negative bacterial pathogens, including Yersinia, Salmonella, Pseudomonas, Shigella, and pathogenic Escherichia spp. (2–5).
T3SSs sequentially secrete a large number of proteins and enable the direct delivery of bacterial effectors into the interior of the host cell. These secreted proteins can be divided into several categories based on their functions and secretion hierarchy, including the T3SS needle and inner rod components (the early substrates), the translocators (the intermediate substrates), and the effectors (the late substrates), as well as some regulatory proteins in certain T3SSs (3, 6). The needle and inner rod proteins are secreted first via a basal body of the T3SS to complete the assembly of the needle complex and are required for secretion of the translocators and effectors. The translocators are presumably secreted next before the effectors, because they are needed for translocating the effectors into host cells.
Enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and Citrobacter rodentium are members of the attaching/effacing (A/E) bacterial pathogens (3, 7). EPEC and EHEC are diarrheagenic in humans and cause significant morbidity and mortality worldwide. EPEC mainly colonizes the small bowel, whereas EHEC primarily inhabits the large bowel. EHEC can also infect cattle, and this serves as a reservoir for subsequent food and water contaminations, resulting in significant economic loss. There are numerous EPEC and EHEC serotypes that infect many other mammals (8). C. rodentium is a mouse-specific pathogen that colonizes murine cecum and large intestines (7). Despite their different hosts and colonization niches, all of the A/E pathogens require a highly conserved T3SS encoded by a chromosomally located pathogenicity island, the locus of enterocyte effacement (LEE), for their virulence and pathogenesis (3, 9). As many as 41 effectors are secreted by certain EHEC serotypes and 30 are secreted by C. rodentium, but there are considerable variations in the number of effector genes present in the different A/E pathogens based on bioinformatic analyses of their genomes (10–16), suggesting strain specificity and plasticity in their effector repertoire. The differences in the effector repertoire may influence the pathogenicity as well as the tissue tropism and host specificity of these different A/E pathogens.
The genome of the prototypical human EPEC strain E2348/69 has been sequenced, and based on bioinformatic analyses of the genome and its comparison with other A/E pathogens, it was estimated that this EPEC strain encodes 21 functional effectors (10). This is a relatively small effector repertoire compared with that of EHEC and C. rodentium (10–16). Because a comprehensive proteomic analysis of type III effectors secreted by EPEC has not been performed, we postulated that this pathogen could secrete additional type III proteins and effectors needed for its colonization of the small bowel rather than the large bowel populated by EHEC and C. rodentium.
An array of genetic, biochemical, bioinformatic, and proteomic approaches has been used extensively to identify and/or predict secretion substrates of T3SSs, because of their critical roles in bacterial virulence and pathogenesis (17–19), and tremendous progress has been made recently in this field with the implementation of modern genomic and proteomic techniques (18). The secreted T3SS structure components (needle and inner rod) and the translocators are evolutionarily and functionally conserved among the T3SSs encoded by different pathogens (3, 4) and can be predicted by bioinformatic methods. However, their effectors are often highly divergent, both in number and in variety. Their discovery therefore requires systematic analyses of individual pathogens by combining bioinformatic, genetic, and proteomic approaches.
We have previously adapted stable isotope labeling with amino acids in cell culture (SILAC), a quantitative proteomic technique (20), to analyze bacterial type III secretome (14, 21). SILAC offers several advantages over conventional proteomic tools, including its simplicity to use by bypassing gel electrophoresis, high throughput and sensitivity, and significant reduction of false positives and nonspecific background by employing an internal standard for relative quantification. This is achieved by differentially labeling the proteomes of a type III secretion-competent or hypersecreting bacterial strain and its T3SS-defective mutant strain with nonradioactive, stable isotope-containing “heavy” amino acids and normal, “light” amino acids, respectively. The culture supernatant containing the secreted proteins from the two strains is then mixed and processed as a single sample for protein precipitation and digestion, peptide fractionation, and mass spectrometry analyses. Peak intensity of the same peptides labeled with either heavy or light amino acids is used to quantitate relative peptide abundance and ratio. Type III-secreted proteins are identified by higher ratios of peptides derived primarily from the type III secretion-competent bacterial strain. The simplicity, sensitivity, and specificity of SILAC are exemplified by our recent successful application of this technique in analyzing the type III secretomes of C. rodentium and SPI-2 (Salmonella enterica serovar Typhimurium pathogenicity island 2)-encoded T3SS (14, 21).
In this report, we used SILAC to systematically analyze the type III secretome of EPEC. Our analysis not only confirmed all 25 EPEC proteins previously identified or predicted to be secreted by the LEE-encoded T3SS (5, 9, 10) but also discovered at least two novel EPEC effectors, C_0814/NleJ and LifA, as well as a number of type III-secreted, but not translocated, proteins. We further showed that Efa1, ToxB, and Z4332, the EHEC homologs of LifA, are also type III effectors. Our results expanded the type III secretome and effector repertoire of EPEC and provided significant insights into the mode of function for the important family of virulence factors LifA/Efa1/ToxB/Z4332.
EXPERIMENTAL PROCEDURES
Oligonucleotide Primers, Bacterial Strains, and Growth Conditions
The oligonucleotide primers used in this study are listed in supplemental Table S1. The prototypical EPEC O127:H6 strain E2348/69, an isolate from a human patient, and EHEC O157:H7 strain EDL933, and C. rodentium strain DBS100, all with sequenced genomes (10, 15, 22, 23), were used in this study. The bacteria were grown in LB broth or agar plates. Dulbecco's modified Eagle's medium (DMEM) from HyClone (Logan, UT) was used as the medium for inducing type III secretion by these bacteria. However, custom-synthesized l-lysine- and l-arginine-deficient DMEM from Caisson Labs (Logan, UT) was used for analyzing the type III secretome of EPEC using SILAC.
Generation of lysA and argH Deletion Mutants in EPEC
Using SILAC to analyze the secreted proteins requires differential labeling of bacterial proteins with isotopic amino acids (Lys and Arg in our study). To ensure efficient labeling of the analyzed proteins, we generated EPEC auxotrophic strains that carry mutations in the de novo biosynthesis of Lys and Arg. The sacB gene-based allelic exchange method was used to generate lysA and argH in-frame deletion mutants in EPEC using the suicide vector pRE112 (24). Two suicide deletion constructs, pRE-ΔlysA and pRE-ΔargH for lysA and argH, respectively, were generated. Two fragments containing the upstream and downstream flanking regions of lysA were amplified by PCR using primer pairs lysA5′-F/lysA5′-R and lysA3′-F/lysA3′-R (supplemental Table S1) and cloned into pRE112 digested with KpnI and SacI to generate pRE-ΔlysA. Similarly, two fragments containing the upstream and downstream flanking regions of argH were amplified by PCR using primer pairs argH5′-F/argH5′-R and argH3′-F/argH3′-R and cloned into pRE112 digested with KpnI and SacI to generate pRE-ΔargH. The constructs pRE-ΔlysA and pRE-ΔargH were created for EHEC strain EDL933 using EHEC genomic DNA as PCR template. Because the lysA and argH genomic regions are nearly identical between EHEC and EPEC, these same constructs were used to generate deletion mutants in EPEC. The suicide vector pRE-ΔlysA was introduced into EPEC by conjugation to generate EPEC ΔlysA using the allelic exchange procedure described before (14, 24), and pRE-ΔargH was subsequently introduced into EPEC ΔlysA to generate EPEC double mutant ΔlysAΔargH. The primer pairs argH-F/argH-R and lysA-F/lysA-R were used to screen by PCR for the ΔargH and ΔlysA mutants, respectively. The pRE-ΔlysA and pRE-ΔargH deletion constructs were sequentially introduced into an EPEC ΔsepDΔescN double mutant (25) to generate EPEC ΔlysAΔargHΔsepDΔescN. Deletion constructs for EPEC escN (26) and sepD (25) in pRE112 were introduced into EPEC ΔlysAΔargH to generate EPEC ΔlysAΔargHΔescN and ΔlysAΔargHΔsepD, respectively. The four EPEC mutant strains, ΔlysAΔargH, ΔlysAΔargHΔescN, ΔlysAΔargHΔsepD, and ΔlysAΔargHΔsepDΔescN, were then used for SILAC analysis of their secreted proteins.
SILAC Analysis
A schematic diagram of the SILAC procedure is shown in Fig. 1A. A detailed protocol was described recently for C. rodentium (14), and only some minor modifications were made in our analysis of EPEC type III secretome. Briefly, the secreted proteins from four EPEC strains (ΔlysAΔargH, ΔlysAΔargHΔescN, ΔlysAΔargHΔsepD, and ΔlysAΔargHΔsepDΔescN) were analyzed using SILAC (20), by pairing strains ΔlysAΔargH and ΔlysAΔargHΔescN to identify type III-secreted proteins by wild type (WT) EPEC and strains ΔlysAΔargHΔsepD and ΔlysAΔargHΔsepDΔescN to identify type III-secreted proteins by the EPEC ΔsepD mutant. The bacteria were grown in l-lysine- and l-arginine-deficient DMEM supplemented with either natural isotopic abundance (light) l-lysine-2HCl (175 mg/liter) and l-arginine-HCl (84 mg/liter) for strains ΔlysAΔargHΔescN and ΔlysAΔargHΔsepDΔescN or heavy [2H4]-lysine (150 mg/liter) and [13C6]-arginine (87 mg/liter) (Cambridge Isotope Laboratories, Cambridge, MA) for strains ΔlysAΔargH and ΔlysAΔargHΔsepD. The strains were grown in 3 ml of the supplemented DMEM overnight at 37 °C in a shaker at 225 rpm and then subcultured 1:20 into 12 ml (four replicates each, 3 ml/well) of the same, prewarmed DMEM in a 6-well cell culture plate (Corning Inc.). The plates were incubated standing in a tissue culture incubator with 5% (v/v) CO2 at 37 °C for 6 h. The cultures for each strain were pooled together from the four wells, and their A600 nm was measured. The cultures were centrifuged at 16,100 × g for 10 min, and the supernatant from ΔlysAΔargH was combined with that from ΔlysAΔargHΔescN in a 1:1 ratio normalized based on their A600 values, whereas the supernatant from ΔlysAΔargHΔsepD and ΔlysAΔargHΔsepDΔescN was pooled similarly. The combined supernatant was filtered through a low protein-binding Millex-GV13 filter unit (0.22 μm; Millipore) to remove any whole bacteria, and trichloroacetic acid was added to a final concentration of 10% (v/v) to precipitate the proteins overnight at 4 °C. The proteins were pelleted by centrifugation at 16,100 × g for 30 min in 2-ml Eppendorf SafeLock tubes, washed with cold acetone, and dried. The protein pellet was solubilized in 6 m urea, 2 m thiourea in 10 mm HEPES (pH 8.0), reduced, alkylated, treated with LysC, and trypsinized exactly as previously described (14, 27). Afterward, the samples were diluted 2-fold with a solution of 1% (v/v) trifluoroacetic acid, 3% (v/v) acetonitrile, and 0.5% (v/v) acetic acid. They were desalted, concentrated, filtered on C18 STop And Go Extraction tips (28), and then eluted directly into a 96-well plate. Peptide mixtures were analyzed on an 1100 Series nanoflow high performance liquid chromatograph (Agilent) on-line coupled via a nanoelectrospray ion source (Proxeon, Odense, Denmark) to a linear trapping quadrupole-Orbitrap (LTQ-OrbitrapXL; ThermoFisher Scientific, Bremen, Germany) tandem mass spectrometer (LC-MS/MS) (29). Peptides were injected directly onto a reversed phase (3-μm-diameter ReproSil-Pur C18; Dr. Maisch, Ammerbuch-Entringen, Germany) column manually packed into a 15-cm-long, 75-μm-inner diameter fused silica emitter. The peptides were eluted directly into the LTQ-OrbitrapXL using a linear gradient from 4.8 to 24% (v/v) acetonitrile (in 0.5% acetic acid) over 60 min at a flow rate of 200 nl/min. The LTQ-OrbitrapXL was set to acquire a full range scan (350–1500 Th, 60,000 resolution, automatic gain control configured to allow 1E+06 ions, and lock mass enabled) in the Orbitrap, from which the five most abundant multiply charged ions were selected for fragmentation in the LTQ (29).
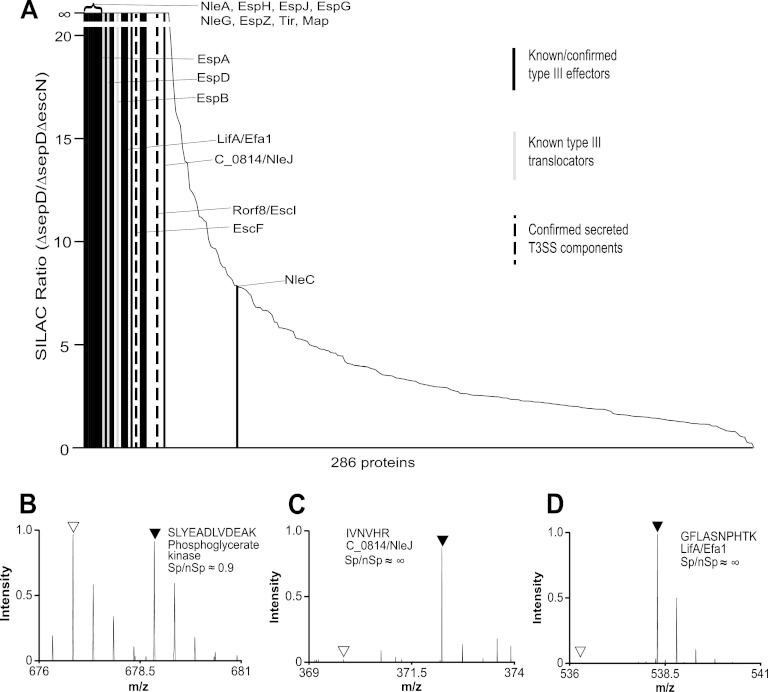
Fig. 1.
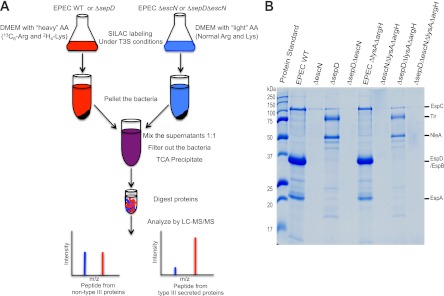
SILAC methodology and protein secretion profiles of strains used for analyzing EPEC type III secretome. A, schematic flow chart of the SILAC protocol. The ΔlysAΔargH derivatives of EPEC WT, ΔescN, ΔsepD, and ΔsepDΔescN strains were grown in DMEM to induce type III secretion. The proteome of ΔescN and ΔsepDΔescN strains was labeled with normal, light amino acids (AA) l-Arg and l-Lys (shown in blue), whereas the proteome of EPEC WT and ΔsepD strains was labeled with heavy isotope-labeled [2H4]Lys and [13C6]Arg (shown in red). The culture supernatant of EPEC WT and ΔescN was combined in a 1:1 ratio, and that of the ΔsepD and ΔsepDΔescN strains was similarly pooled. The mixed supernatant was processed as a single sample and filter-sterilized to remove any remaining bacteria, proteins in the supernatant were precipitated by TCA and digested with trypsin, and the peptides were analyzed by LC-MS/MS. The origin of the peptides was distinguishable by whether they contained heavy or light Lys and Arg, and peak intensity of the peptides was used to quantitate relative peptide abundance. Although heavy (shown in red) and light (shown in blue) peptides from non-type III-secreted background proteins have an equal ratio, peptides from type III-secreted proteins are identified primarily by heavy [2H4]Lys and/or [13C6]Arg-labeled peptides. B, type III secretion profiles of EPEC WT, ΔescN, ΔsepD, and ΔsepDΔescN strains and their ΔlysAΔargH mutants used for SILAC analysis of EPEC type III secretome. The EPEC strains were grown in DMEM to induce type III secretion. Secreted proteins in the supernatant were analyzed in SDS-13% PAGE and stained by Coomassie G-250. Secreted proteins from an equal amount of cultures for each strain (normalized by A600) were loaded in each lane. Protein standards with their molecular masses in kDa are indicated on the left, and the names of the known type III-secreted proteins, translocators EspA, EspB, and EspD and effectors Tir and NleA, are marked on the right. Note that EspC is not a type III-secreted protein.
Mass Spectrometry Data Analysis
Peak lists of fragment ions were generated by Extract_MSN (v3.2, ThermoFisher) using the default parameters. Monoisotopic peak and charge state assignments were checked by DTA Supercharge of the MSQuant suite of software (30). Fragment spectra were searched against a database of typical contaminants (such as keratins and albumin), experimental additives (such as trypsin and immunoglobulins), and the annotated EPEC genome sequences (20,658 sequences with all reversed sequences appended, released January 2010 (10)), using Mascot (version 2.2; Matrix Science) with the following parameters; trypsin specificity allowing up to one missed cleavage, cysteine carbamidomethylation as a fixed modification, [13C6]arginine and [2H4]lysine as variable modifications, electrospray ionization trap fragmentation characteristics, 5-ppm mass tolerance for precursor ion masses, and 0.6 Da tolerance for fragment ion masses. Acceptance criteria for protein identifications were set so that only proteins identified by at least two unique peptides of seven or more amino acids with Mascot ion scores of >25 were accepted, criteria resulting in an estimated false discovery rate of less than 1% based on the number of reversed hits meeting the same criteria (FDR = hits versus reversed sequences/(forward hits + reverse hits)). Quantitative ratios were extracted from the raw data using MSQuant (version 1.4.3), which calculates an intensity-weighted average of within-spectra ratios from all spectra across the chromatographic peak of each peptide ion. For automatic quantification, only those proteins with a coefficient of variation less than 30% were accepted with no further verification. For proteins with high coefficients of variation or with only one quantified peptide in a particular experiment, the chromatographic peak assignment was manually verified or rejected. Quantitation and identification were based only on unique peptides, although shared peptides were retained in the peptide lists. In the overall study, the expression ratios for almost all proteins reported here were based on quantification of at least two peptides. Analytical variability of SILAC data in our experiments was typically <20%, and biological variability was addressed by performing at least three independent replicates of each experiment. All of the peptide and protein information acquired in this study is provided in supplemental Table S2.
Construction of β-lactamase TEM-1 Fusions
N-terminal translational fusions to TEM-1 β-lactamase were generated in the vector pCX341 (provided by I. Rosenshine, Hebrew University, Jerusalem, Israel) (31, 32). The coding regions, without the stop codons, of genes encoding the following EPEC proteins, BolA, C_2495, C_0814, FhuF, Ivy, YdeN, YahO, YgiW, C_4114, YebF, C_2649, C_1078, Spy, YkgM, ModA, YdgH, YbgF, FepB, ChuT, YcdO, YcgK, C_1098, and HdeB, were amplified by PCR with an NdeI site added to the forward primers and an EcoRI site incorporated into the reverse primers (supplemental Table S1). The PCR fragments were then cloned into NdeI/EcoRI-digested pCX341 in front of the coding region of TEM-1. Similarly, the coding regions of genes encoding YodA, YehE, HelD, ZnuA, and FkpA were cloned into NdeI/KpnI-digested pCX341. The coding regions of some proteins, such as YhhA and DeoB, have internal NdeI, KpnI, or EcoRI sites, the preferred cloning sites for the vector pCX341, whereas others (LifA, C_1079, and ToxB) are encoded by some of the largest genes present in the EPEC, EHEC, and C. rodentium genomes, making it technically challenging to clone their full-length genes. In these cases, the coding regions for the first 50 (of YhhA, EPEC LifA, C_1079, EHEC EDL933 ToxB, or C. rodentium LifA) or 100 (of DeoB) amino acid residues were cloned into NdeI/EcoRI-restricted pCX341. For Z4332, the EHEC EDL933 homolog of the N-terminal 433 amino acid residues of EPEC LifA, the entire coding region without its stop codon, plus a 17-bp 5′-flanking region, was amplified by PCR and cloned into KpnI/EcoRI-digested pCX341. These constructs were introduced into EPEC strain E2348/69 and its isogenic ΔescN and ΔsepD mutants by electroporation for type III secretion and translocation assays. The mouse antibody against His-tagged TEM-1 β-lactamase from QED Bioscience (San Diego, CA) was used for detection of the TEM-1 fusion proteins by Western blotting using ECL reagents from GE Healthcare.
Chromosomal Fusions of lifA::blaM in EPEC WT and ΔescN and ΔsepD Mutants
To create lifA::blaM in the EPEC chromosome, we first generated a suicide construct by inserting the TEM-1-encoding blaM in front of the stop codon of lifA in the allelic exchange vector pRE112 (24). This was achieved by amplifying by PCR a 1.2-kb fragment immediately upstream (with KpnI and EcoRI sites incorporated into the forward (EPlifA3′-F) and reverse (EPlifA3′-R) primers, respectively) and a 0.99-kb fragment immediately downstream (with XbaI and SacI sites in the forward (EPlifAd-F) and reverse (EPlifAd-R) primers, respectively) of the stop codon of the lifA gene. After KpnI/EcoRI or XbaI/SacI digestion, these two fragments were ligated into KpnI/SacI-cut pRE112 together with an EcoRI/XbaI fragment of pCX341 containing blaM (32) in a four-way ligation. The ligation mixture was transformed into E. coli strain SY327λpir, which allows the replication of the suicide plasmid pRE112. The resulting plasmid, pRE-EPlifA-TEM, created an in-frame fusion of lifA::blaM, and this plasmid was introduced into E. coli strain SM10λpir by electroporation and then transferred into EPEC WT and ΔescN and ΔsepD mutants by conjugation. The transconjugants were subjected to sucrose selection and the candidates with chromosomal lifA::blaM fusions were screened by PCR using the primers EPlifA3′-F and EPlifAd-R (supplemental Table S1).
Type III Secretion and Translocation Assays
Type III secretion assays for EPEC proteins were performed using essentially the same protocol as described before for EPEC (14, 33). Candidate proteins fused to TEM-1 were expressed in WT EPEC strain E2348/69 and its ΔescN and ΔsepD mutants (25, 26) to assess whether their secretion depended on a functional T3SS. Translocation of these TEM-1 fusions into host cells was assayed using the TEM-1 β-lactamase translocation assay (31, 32). Briefly, EPEC wild type and its isogenic ΔescN strains carrying N-terminal translational fusions to TEM-1 β-lactamase were used to infect HeLa cells (CCL2; American Type Culture Collection), and translocation was detected by an Infinite M200 microplate reader (TECAN) using the fluorescent substrate CCF2-AM from Invitrogen as previously described (14).
RESULTS
Experimental Strategy for SILAC Analysis of EPEC Type III Secretome
A schematic flow chart of the SILAC procedure is shown in Fig. 1A, a strategy similar to that used for our SILAC analysis of the C. rodentium type III secretome (14). To identify proteins specifically secreted by the LEE-encoded T3SS (the type III secretome) of EPEC, we analyzed the secreted proteomes of WT EPEC as well as its sepD mutant by taking advantage of our previous observations (16, 25) that, similar to C. rodentium, WT EPEC preferentially secretes the translocators EspA, EspB, and EspD, with only very small amounts of effectors secreted, when grown statically in DMEM at 37 °C in a 5% (v/v) CO2-containing environment, whereas its sepD mutant predominately secretes effectors, with the translocators hardly detectable (Fig. 1B).
To facilitate proteome labeling by SILAC, we first generated deletion mutants of lysA and argH in EPEC WT, ΔescN, ΔsepD, and ΔsepDΔescN strains to create EPEC ΔlysAΔargH, ΔlysAΔargHΔescN, ΔlysAΔargHΔsepD, and ΔlysAΔargHΔsepDΔescN strains. The escN gene encodes an ATPase essential for type III secretion, whereas sepD is critical for the type III secretion of the translocators, with its mutation resulting in unregulated hypersecretion of effectors such as Tir and NleA (Fig. 1B) (16, 25). These lysine and arginine auxotrophic EPEC strains were required for efficient labeling of the bacterial proteome with isotopic amino acids Lys and Arg when grown in medium supplemented with these amino acids during SILAC. As shown in Fig. 1B, the type III secretion profiles of the lysA and argH mutants grown in DMEM were the same as those of EPEC strains without the lysA and argH mutations.
We used SILAC to specifically distinguish type III-secreted proteins from background proteins secreted by other protein secretion pathways of EPEC, as well as cytosolic proteins present in the culture supernatant because of cell lysis. This was achieved by pairing EPEC strain ΔlysAΔargH with ΔlysAΔargHΔescN and pairing ΔlysAΔargHΔsepD with ΔlysAΔargHΔsepDΔescN and comparatively analyzing the secreted proteins of the paired strains. The proteomes of ΔlysAΔargH and ΔlysAΔargHΔsepD were labeled by Lys and Arg with heavy isotopes, whereas those of ΔlysAΔargHΔescN and ΔlysAΔargHΔsepDΔescN were labeled by Lys and Arg with natural, light isotopes. The culture supernatants of the paired strains were combined into one sample and then filter-sterilized before the proteins were precipitated together, digested, and analyzed by LC-MS/MS. Those proteins showing a high differential SILAC ratio (heavy isotope-labeled versus natural, light isotope-labeled) were deemed to represent proteins specifically secreted through the T3SS (therefore EscN-dependent), whereas general background proteins not secreted by the T3SS should have a SILAC ratio of ∼1 (Fig. 1A). Examples of peptide mass spectra from a background, non-type III-secreted protein, phosphoglycerate kinase, and two putative type III-secreted proteins, C_0814/NleJ and LifA/Efa1, are shown in Fig. 2 (B–D), respectively.
Fig. 2.
SILAC analysis of type III secretome of EPEC strain E2348/69. A, plot of the SILAC ratios for all the proteins identified in EPEC ΔsepD (when compared with ΔsepDΔescN), ranked from the highest on the left to the lowest on the right. Each vertical line represents a protein, as depicted in the key. The black bars represent previously known or confirmed type III-secreted effectors, the gray bars show type III-secreted translocators (EspA, EspB, and EspD), and the dotted bars show secreted T3SS components (EscF and rOrf8/EscI). Also indicated are the two novel effectors, LifA/Efa1 and C_0814/NleJ, found in this study. B–D, the MS spectra of sample peptides of phosphoglycerate kinase, a nonspecific background protein (B), C_0814/NleJ (C), and LifA/Efa1 (D) were shown. Natural Lys- and Arg-labeled peptides are indicated by open triangles, and [2H4]Lys- and/or [13C6]Arg-labeled peptides are indicated by filled triangles. Proteins specifically secreted by the T3SS, such as LifA and C_0814, are identified primarily by [2H4]Lys- and/or [13C6]Arg-labeled peptides, whereas background proteins, such as phosphoglycerate kinase, have a close to equal ratio (1:1) of natural, unlabeled peptides (nSp) and labeled peptides (Sp).
Type III-secreted Proteins by EPEC with a Wild Type T3SS
We first analyzed the secreted proteins from EPEC ΔlysAΔargH (with a functional, wild type T3SS) and ΔlysAΔargHΔescN (T3SS deficient) by SILAC. Proteins with high SILAC ratios, probably secreted by the T3SS of WT EPEC, are listed in Table I. The SILAC ratios, as well as protein and peptide information, for all the proteins from four replicates of EPEC ΔlysAΔargH versus ΔlysAΔargHΔescN (i.e., WT versus ΔescN) secreted protein samples are listed in supplemental Table S2. As expected, the three translocators EspB, EspA, and EspD had the highest SILAC ratios in this SILAC analysis (Table I), confirming that they are preferentially secreted under our experimental conditions. Although the translocators are the most abundant type III-secreted substrates of WT EPEC, it is known that WT EPEC can secrete small amounts of some effectors (32, 34). Indeed, several effectors, including both the LEE-encoded (Map, EspG, EspH, Tir, EspF, and EspZ) and the non-LEE-encoded (NleA, NleE, NleD, and EspJ) effectors displayed high SILAC ratios, although the ratios were much lower than those of the translocators (Table I). EscF and rOrf8/EscI were also found to have a relatively high SILAC ratio, consistent with our analysis of C. rodentium type III secretome (14). Another non-LEE-encoded effector, NleF, had a SILAC ratio of 3.79, ranked lower than ribosomal protein S3 and several extracellular and outer membrane proteins (Table I).
Table I. Proteins with high SILAC ratios secreted via the LEE-encoded T3SS by wild type EPEC identified in the EPEC WT versus ΔescN samples.
| Protein name | Average ratioa | CVb | Accession number/category of secreted proteins |
|---|---|---|---|
| EspB | 4618.90 | 0.35 | Q05129/translocator and effector |
| EspA | 2654.72 | 0.12 | CAS11484/translocator |
| EspD | 2133.96 | 0.26 | CAS11483/translocator |
| Map | 625.56 | 1.14 | CAS11490/LEE-encoded effector |
| NleA | 402.87 | 0.97 | YP_002328981/non-LEE-encoded effector |
| EspG | 270.02 | 1.33 | CAS11518/LEE-encoded effector |
| NleE | 180.25 | 1.34 | CAS10780/non-LEE-encoded effector |
| EspH | 137.52 | NA | CAS11492/LEE-encoded effector |
| Tir | 127.17 | 0.31 | YP_002331403/LEE-encoded effector |
| NleD | 102.00 | 0.92 | YP_002328604/non-LEE-encoded effector |
| EscF | 85.49 | NA | YP_002331394/T3SS needle component |
| EspF | 81.02 | 0.35 | YP_002331392/LEE-encoded effector |
| EspZ | 54.99 | 0.73 | YP_002331413/LEE encoded effector |
| EspJ | 29.96 | 0.39 | CAS08271/non-LEE encoded effector |
| rOrf8/EscI | 26.37 | 0.18 | YP_002331414/T3SS inner rod component |
| Pta | 22.82 | 0.97 | YP_002329941/phosphate acetyltransferase |
| YhhA | 14.64 | NA | CAS11236/hypothetical protein |
| EspC | 14.03 | 0.80 | CAS10463/extracellular serine protease |
| RpsC | 11.03 | 1.84 | CAS11125/30S ribosomal protein S3 |
| FepB | 7.59 | NA | YP_002328066/iron-enterobactin transporter |
| Ivy | 6.95 | 0.92 | YP_002327804/inhibitor of vertebrate C-lysozyme |
| NleF | 3.79 | NA | CAS08993/non-LEE encoded effector |
a Average SILAC ratio (heavy isotope/light isotope) measured.
b CV, coefficient of variation as determined from quantification of several peptides when available. NA, not applicable because there was only one peptide quantified.
Type III-secreted Proteins by the Effector Hypersecreting sepD Mutant of EPEC
Several known EPEC effectors were absent from the list of proteins secreted by WT EPEC (Table I and supplemental Table S2) (10), suggesting that WT EPEC is not an ideal source for cataloguing the effector inventory by proteomics under our experimental conditions because of its strong bias toward secretion of the translocators. To identify additional EPEC effectors, we next analyzed the secreted proteins from EPEC ΔlysAΔargHΔsepD and ΔlysAΔargHΔsepDΔescN by SILAC, because EPEC sepD and sepL mutants have been shown to hypersecrete effectors in DMEM (25). Proteins with the top SILAC ratios and their accession numbers are listed in Table II. The SILAC ratios, as well as protein and peptide information, for all of the proteins from five replicates of EPEC ΔlysAΔargHΔsepD and ΔlysAΔargHΔsepDΔescN (i.e., ΔsepD versus ΔsepDΔescN) secreted protein samples are listed in supplemental Table S2.
Table II. Proteins with high SILAC ratios secreted via the LEE-encoded T3SS by the EPEC ΔsepD mutant identified in the EPEC ΔsepD versus ΔsepDΔescN samples.
| Protein name | Average ratioa | CVb | Accession number/category of secreted proteins |
|---|---|---|---|
| NleA | 4642.27 | 1.12 | YP_002328981/non-LEE-encoded effector |
| EspH | 3045.06 | 0.97 | CAS11492/LEE-encoded effector |
| EspJ | 2801.58 | 1.19 | CAS08271/non-LEE encoded effector |
| EspG | 2290.29 | 0.78 | CAS11518/LEE-encoded effector |
| NleG | 2235.06 | 1.54 | YP_002328601/non-LEE encoded effector |
| EspZ | 1753.09 | 0.16 | YP_002331413/LEE-encoded effector |
| Tir | 1288.60 | 0.33 | YP_002331403/LEE-encoded effector |
| Map | 919.71 | 0.58 | CAS11490/LEE-encoded effector |
| EspA | 838.73 | 0.74 | CAS11484/translocator |
| NleF | 791.77 | 1.21 | CAS08993/non-LEE encoded effector |
| EspD | 671.50 | 0.74 | CAS11483/translocator |
| NleH1 | 491.39 | 1.42 | CAS08992/non-LEE-encoded effector |
| NleB1 | 448.94 | 0.95 | YP_002330703/non-LEE-encoded effector |
| LifA/Efa1 | 386.06 | 0.82 | CAS10782/non-LEE-encoded effector |
| EspB | 312.97 | 0.46 | Q05129/translocator and effector |
| C_1079 | 158.65 | 0.88 | YP_002328637/LifA-like protein |
| EspG2 | 156.67 | 1.16 | YP_002330404/non-LEE-encoded effector |
| BolA | 150.14 | 0.41 | CAS07918/regulator of penicillin-binding proteins and β-lactamase transcription |
| NleH2 | 122.20 | 1.22 | YP_002328287/non-LEE-encoded effector |
| C_4114 | 110.00 | NA | CAS11662/hypothetical protein |
| EspL | 99.67 | 0.74 | CAS10778/non-LEE-encoded effector |
| YehE | 97.07 | NA | CAS09806/hypothetical protein |
| EscF | 74.89 | 1.11 | YP_002331394/T3SS needle component |
| C_0814 | 65.54 | 0.11 | YP_002328380/non-LEE-encoded effector |
| NleB2 | 54.42 | NA | YP_002328602/non-LEE-encoded effector |
| NleE | 36.67 | 0.64 | CAS10780/non-LEE-encoded effector |
| EspF | 33.67 | 0.39 | YP_002331392/LEE-encoded effector |
| C_2495 | 32.34 | NA | CAS10043/hypothetical protein |
| ManX | 30.54 | 0.80 | CAS09489/fused mannose-specific PTS enzymes: IIA and IIB component |
| FhuF | 30.41 | 0.77 | CAS12214/ferric iron reductase |
| GlgC | 30.32 | NA | CAS11224/glucose-1-phosphate adenylyltransferase |
| Ivy | 27.38 | NA | YP_002327804/inhibitor of vertebrate C-lysozyme |
| rOrf8 | 26.17 | 0.28 | YP_002331414/T3SS inner rod component |
| HelD | 25.42 | NA | YP_002328511/DNA helicase IV |
| YgiW | 24.11 | 0.39 | YP_002330783/hypothetical protein |
| NleD | 22.50 | 1.28 | YP_002328604/non-LEE-encoded effector |
| YdeN | 22.46 | NA | YP_002329148/hypothetical protein |
| YebF | 22.02 | 0.32 | CAS09520/hypothetical protein |
| AnsB | 19.55 | 1.18 | CAS10758/periplasmic l-asparaginase II |
| Pal | 17.67 | 1.00 | YP_002328194/OM lipoprotein |
| UshA | 16.27 | 0.93 | YP_002327991/bifunctional UDP-sugarhydrolase/5′-nucleotidase |
| DegP | 15.88 | 0.67 | CAS07716/serine endoprotease |
| C_1078 | 15.60 | NA | CAS08626/hypothetical protein |
| C_2649 | 14.47 | NA | CAS10197/hypothetical protein |
| DeoB | 13.86 | 2.04 | YP_002332126/phosphopentomutase |
| DsbA | 13.79 | NA | P0A4L6/thiol-disulfide interchange protein |
| ChuT | 12.53 | NA | YP_002331210/putative heme-binding protein |
| YhhA | 12.38 | NA | CAS11236/hypothetical protein |
| C_1098 | 12.18 | 0.25 | YP_002328652/hypothetical protein |
| OppA | 11.83 | 0.75 | YP_002328908/oligopeptide transporter subunit |
| C_4406 | 11.17 | 0.24 | CAS11954/dipeptide transport system |
| ZnuA | 11.03 | 1.02 | YP_002329501/zinc transporter subunit |
| FepB | 10.94 | 0.43 | YP_002328066/iron-enterobactin transporter |
| TpiA | 9.66 | 1.39 | CAS11771/triosephosphate isomerase |
| YcdO | 9.39 | 0.45 | YP_002328628/hypothetical protein |
| YahO | 8.97 | 0.65 | CAS07837/hypothetical protein |
| SurA | 8.79 | 0.58 | CAS07604/PPIase |
| YodA | 8.75 | 1.06 | YP_002329600/conserved metal-binding protein |
| YdgH | 8.41 | NA | YP_002329211/hypothetical protein |
| YkgM | 8.30 | 0.93 | CAS07802/RpmJ (L36) paralog |
| ModA | 8.18 | 0.54 | CAS08188/molybdate transporter subunit |
| NleC | 7.89 | NA | CAS08590/non-LEE-encoded effector |
| FkpA | 7.42 | 0.87 | YP_002331064/FKBP-type peptidyl-prolyl cis-trans-isomerase |
| HdeB | 6.84 | 0.41 | CAS11299/acid resistance protein |
| Spy | 5.68 | 0.94 | YP_002329391/periplasmic protein |
| YcgK | 5.28 | 0.81 | CAS08845/hypothetical protein |
| YbgF | 4.67 | NA | CAS08174/hypothetical protein |
a Average SILAC ratio (heavy isotope/light isotope) measured.
b CV, coefficient of variation as determined from quantification of several peptides when available. NA, not applicable because there was only one peptide quantified.
As shown in Fig. 2A, Table II, and supplemental Table S2, the top eight proteins (NleA, EspH, EspJ, EspG, NleG, EspZ, Tir, and Map) with the highest SILAC ratios are all known effectors. Furthermore, 12 additional known or predicted effectors (NleF, NleH, NleB, EspB, EspG2, NleH2, EspL, NleB2, NleE, EspF, NleD, and NleC) were also shown to have high SILAC ratios, bringing the total number of confirmed EPEC effectors to 20, just one (NleE2) less than the effector inventory of 21 predicted by bioinformatics (10). Our analyses also showed that EscF and rOrf8/EscI, two T3SS structural components, as well as the translocators EspA, EspD and EspB, displayed high SILAC ratios and were type III-secreted by the EPEC sepD mutant (Table II), consistent with our findings in C. rodentium (14). EspB is considered to be both a translocator and an effector (9, 10). The detection of the translocators having high SILAC ratios verified the high sensitivity of SILAC, because the trace amount of translocators secreted by the EPEC sepD mutant was barely detectable by Western blot analysis (25). A number of proteins listed in Table II and Fig. 2A showed moderate to high SILAC ratios (ranging from ∼8 to 100), similar to those of the known type III-secreted proteins EscF and rOrf8/EscI and some less abundant effectors such as EspF, NleD, and NleC, and these proteins may represent new type III-secreted proteins and effectors.
Novel Type III-secreted Proteins and an Expanded Type III Secretome of EPEC
By combining the SILAC results of both WT EPEC and its ΔsepD mutant, a total of 306 proteins were identified in the culture supernatant of EPEC, and at least 286 of them were quantified (supplemental Table S2). Based on the SILAC ratios, our analyses confirmed that the EPEC type III secretome includes 2 T3SS components (the needle protein EscF and the inner rod protein EscI), 3 translocators (EspA, EspD, and EspB), 6 LEE-encoded effectors (Tir, Map, EspF, EspG, EspH, and EspZ), and 13 non-LEE-encoded effectors (EspG2, EspJ, EspL, NleA, NleB, NleB2, NleC, NleD, NleE, NleF, NleG, NleH, and NleH2) that have previously been identified or predicted in EPEC using conventional proteomic and bioinformatic methods (5, 9, 14). The only previously predicted effector not confirmed by our SILAC screen was NleE2 (10), but this was because NleE2 is nearly identical to NleE except for an internal deletion of 56 amino acid residues, and it would share almost all the peptides with NleE, making the two proteins virtually indistinguishable by LC-MS/MS. It has been shown that NleE2 is type III-secreted, but not translocated, and may represent a cryptic effector (35). Therefore, our SILAC analyses of EPEC type III secretome confirmed all known or predicted EPEC effectors identified thus far by other methods (10).
Although the three translocators showed the highest SILAC ratios in the WT EPEC secretome and the sepD mutant displayed biased secretion of most known effectors, there were many proteins exhibiting SILAC ratios that are comparable with some of the 25 known, confirmed type III-secreted proteins mentioned above (Fig. 2A and Tables I and II). These proteins include a known EPEC/EHEC virulence factor (LifA/Efa1) previously thought to act as an adhesin and colonization factor (2), some putative periplasmic and outer membrane proteins, and many uncharacterized hypothetical proteins. This suggests that EPEC may have a type III secretome considerably larger than the 25 proteins currently estimated (10), and that some of these uncharacterized proteins may represent novel EPEC type III-secreted proteins and effectors.
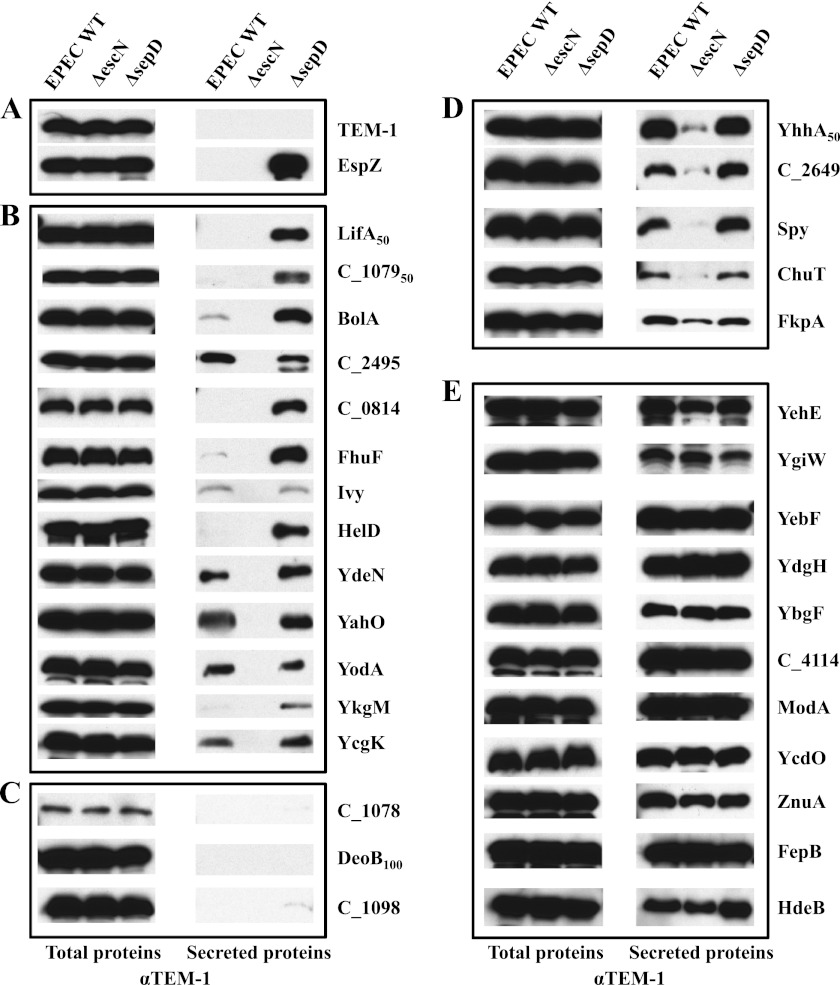
We next attempted to verify the type III secretion status of some of these proteins with relatively high SILAC ratios. We used NleF and NleC, two known effectors that showed the lowest SILAC ratios, as reference points for choosing the candidates from the WT (Table I) and the sepD mutant secretomes (Table II), respectively. A total of 32 candidate proteins were selected for further analysis, including LifA/Efa1, C_1079, BolA, C_4114, C_2495, YehE, C_0814, FhuF, Ivy, HelD, YgiW, YdeN, YebF, C_1078, C_2649, DeoB, ChuT, YhhA, C_1098, ZnuA, FepB, YcdO, YahO, YodA, YdgH, YkgM, ModA, FkpA, HdeB, Spy, YcgK, and YbgF. The coding regions for the selected proteins were cloned into pCX341 to generate constructs expressing TEM-1 fusions at the C terminus, a method commonly used in the A/E pathogens to tag proteins for both assessing type III secretion and monitoring effector translocation into host cells (31, 32). The genes encoding some candidate proteins, such as LifA/Efa1 and C_1079, are large and contain too many internal restriction sites for convenient cloning into the pCX341 vector. In these cases, the coding regions of only the N-terminal 50–100 amino acids were cloned, because as few as 20 amino acid residues of the N terminus of the effectors are usually sufficient for type III secretion and translocation in EPEC (31). These constructs were transformed into EPEC WT, ΔescN, and ΔsepD strains and assayed for type III secretion by Western blot analysis using antibodies against TEM-1.
All 32 TEM-1 fusions, except C_1078, were expressed well in the EPEC strains (Fig. 3). As expected, TEM-1 was not secreted, whereas EspZ-TEM, the TEM-1 fusion to the known effector EspZ, was hypersecreted by the sepD mutant (Fig. 3A). We were able to verify T3SS-dependent secretion for at least 13 proteins, including LifA, C_0814, FhuF, YodA, BolA, C_2495, YkgM, YahO, Ivy, YdeN, YcgK, HelD, and C_1079. The TEM-1 fusions of these proteins were secreted by EPEC WT and/or the sepD mutant, but not the escN mutant (Fig. 3B). C_1078 was not expressed well (Fig. 3C), probably because of its instability, and its type III secretion could not be conclusively verified. The C_1098 and DeoB TEM-1 fusions inhibited general type III secretion of EPEC (data not shown), and their type III secretion could not be conclusively evaluated either (Fig. 3C). Five proteins, YhhA, C_2649, Spy, ChuT, and FkpA, showed enhanced secretion in EPEC WT and the sepD mutant, but they were also secreted at low levels by the escN mutant (Fig. 3D), suggesting that these proteins can be secreted by both the LEE-encoded T3SS and other unknown secretion pathway(s). When overexpressed as TEM-1 fusions on a plasmid in EPEC, the remaining 10 constructs for YehE, YgiW, YebF, YdgH, YbgF, C_4114, ModA, YcdO, ZnuA, and FepB caused significant cell lysis and/or membrane leakage, resulting in high background in secreted proteins (Fig. 3E and data not shown). Their type III secretion status could not be reliably assessed and needs to be verified by an alternate method. One protein, HdeB, appeared to be secreted independently of EscN and the LEE-encoded T3SS (Fig. 3E), because HdeB-TEM did not cause detectable cell lysis (data not shown). Nevertheless, we were able to confirm that the majority of the selected protein candidates that showed moderate to high SILAC ratios in our proteomic screen were indeed secreted in a T3SS-dependent manner. These results demonstrate that EPEC has a type III secretome much larger than previously estimated.
Fig. 3.
Type III secretion assays for the new secreted protein candidates with moderate to high SILAC ratios identified in the SILAC analysis of EPEC type III secretome. The coding regions of EPEC LifA50, C_107950, BolA, C_2495, C_0814, FhuF, Ivy, HelD, YdeN, YahO, YodA, YkgM, YcgK, C_1078, DeoB100, C_1098, YhhA50, C_2649, Spy, ChuT, FkpA, YehE, YgiW, YebF, YdgH, YbgF, C_4114, ModA, YcdO, ZnuA, FepB, and HdeB were cloned into the vector pCX341 to generate N-terminal fusions to TEM-1. These constructs were introduced into EPEC WT, ΔescN, and ΔsepD strains and assayed for type III secretion of the TEM-1 fusion proteins. Proteins from bacterial pellets (total proteins) and secreted proteins in DMEM were separated in SDS-13% PAGE, blotted onto nitrocellulose membranes, and probed with mouse antibodies against TEM-1 by Western blot. The secretion patterns of these fusion proteins can be divided into the following groups: A, empty vector-encoded TEM-1 and the known effector EspZ that were used as negative and positive controls, respectively; B, proteins that were secreted by EPEC WT and/or ΔsepD, but not by ΔescN; C, proteins that were either poorly expressed (C_1078) or inhibited type III secretion when overexpressed as TEM-1 fusions; their type III secretion status could not be assessed; D, proteins that were secreted at low levels by ΔescN but showed enhanced secretion by EPEC WT and ΔsepD; and E, proteins that caused cell lysis when overexpressed in EPEC. Their type III secretion status could not be conclusively determined by this assay. The only exception in this group was HdeB, which did not cause any detectable cell lysis when expressed in EPEC and was therefore secreted by a protein secretion mechanism other than the T3SS.
LifA/Efa1 and C_0814 Are Two Novel Type III Effectors in EPEC
To find out whether any of these new protein candidates are effectors, which should not only be secreted via the T3SS but also translocated into host cells by the T3SS, we took advantage of the fact that the TEM-1 fusions can be used to monitor both type III secretion by Western blot and type III translocation via a fluorescence-based cell infection assay (31). EPEC WT and ΔescN strains expressing the TEM-1 fusion constructs for the 32 protein candidates mentioned above were used to infect cultured HeLa cells to assess whether any of the proteins are translocated into host cells. When loaded with the fluorescent β-lactamase substrate CCF2/AM with excitation at 409 nm, uninfected HeLa cells or cells infected with EPEC strains expressing untranslocated TEM-1 fusions emit green fluorescence, whereas TEM-1 fusions translocated into the cells cleave the CCF2 β-lactam ring generating blue fluorescence (31). An elevated emission ratio (460/530 nm) between blue and green fluorescence indicates translocation of the TEM-1 fusion protein.
As shown in Fig. 4, the TEM-1 vector control was not translocated as expected, whereas the positive control, the TEM-1 fusion to the known effector EspZ, was translocated efficiently into HeLa cells depending on EscN (T3SS). Of the 32 constructs tested, TEM-1 fusions to C_0814 and the N-terminal 50 amino acid residues of LifA were found to be translocated into HeLa cells (Fig. 4). These results demonstrate that C_0814 and LifA are likely novel EPEC type III effectors. This was consistent with our results that LifA/Efa1 and C_0814 showed type III secretion patterns typical for the majority of known effectors (25), because the LifA50- and C_0814-TEM-1 fusions displayed no secretion by the escN mutant, very low levels of secretion by WT EPEC, and hypersecretion by the sepD mutant (Fig. 3B). Interestingly, the TEM-1 fusions of two other candidate proteins, FhuF and HelD (annotated as ferric iron reductase and DNA helicase IV, respectively; Table II), showed a secretion pattern very similar to that of LifA50 and C_0814, but for unknown reasons, the FhuF-TEM-1 and HelD-TEM-1 fusion proteins were not translocated into HeLa cells (Fig. 4).
Fig. 4.
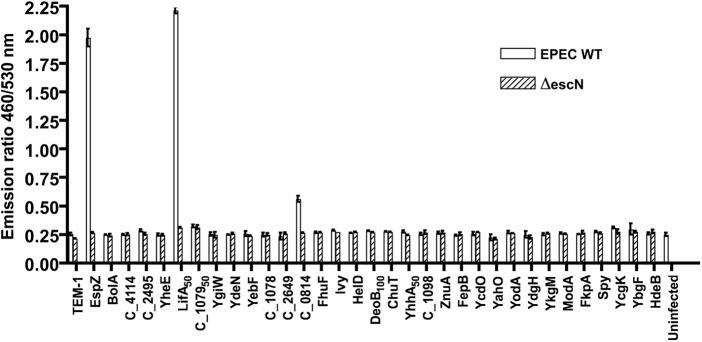
Type III translocation assays for the new secreted protein candidates with moderate to high SILAC ratios identified in the SILAC analysis of EPEC type III secretome. The β-lactamase TEM-1 fusion constructs in pCX341 for the same set of EPEC secreted candidate substrates described in Fig. 3 are used for the type III translocation assays. HeLa cells were infected with EPEC WT and its ΔescN mutant carrying the constructs expressing the TEM-1 fusions. The infected cells were loaded with CCF2/AM and assessed for protein translocation using a fluorescence microplate reader. The fluorescence quantification data shown are averages with standard deviations of triplicate values of the results from one representative experiment of three experiments and are presented as the emission ratio between blue fluorescence (460 nm) and green fluorescence (530 nm). Higher blue fluorescence to green fluorescence ratios by EPEC WT, but not by the ΔescN mutant, indicate positive type III translocation. EPEC EspZ (an effector) and the empty vector encoding TEM-1 were used as positive and negative controls, respectively.
C_0814 Is Encoded by a P2-like Prophage and Contains a Phosphoribosyl Transferase Domain
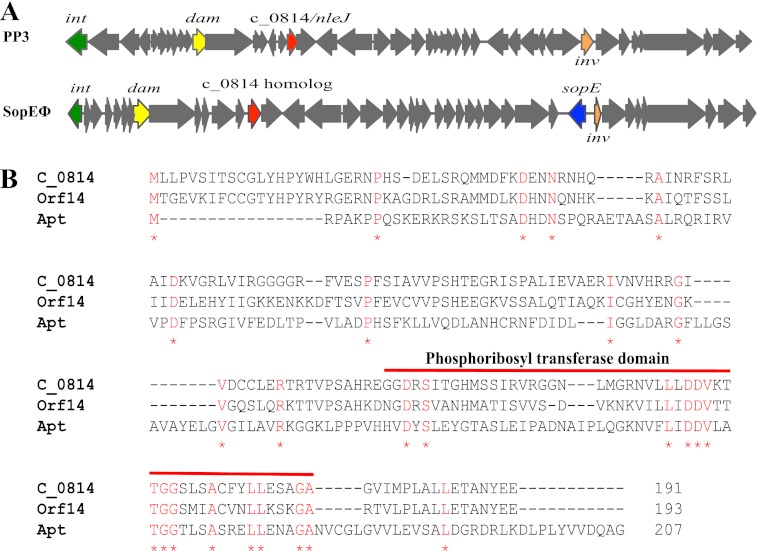
C_0814 is an uncharacterized, hypothetical protein. Its gene is located in the P2-like prophage PP3 in the EPEC chromosome (10). C_0814 does not have a homolog in EHEC or C. rodentium but is present in the pathogenic E. coli strains H591, MS 84-1, 85-81, and 124-1 based on a search of sequenced E. coli genomes (data not shown). C_0814 has 50% identity to a protein (Orf14; GenBankTM accession number AAQ65011) encoded by the SopEΦ phage in S. enterica serovar Typhimurium, the same prophage that carries the gene for SopE, an important type III effector for Salmonella invasion (36). This gene (orf14) is present in many Salmonella strains, with locus tags SL1344_2696 (GenBankTM accession number CBW18798) and STM474_2848 (GenBankTM accession number ADX18498) in Salmonella Typhimurium strains SL1344 and ST4/74, respectively. As shown in Fig. 5A, PP3 of EPEC and SopEΦ of Salmonella are both P2-like phages, and the location and sequence context of c_0814 in PP3 is the same as that of its Salmonella homolog orf14 in SopEΦ. However, the effector gene sopE was noticeably absent from PP3 in EPEC. C_0814 is annotated as a hypothetical protein (10) and predicted to have a phosphoribosyl transferase domain at its C terminus (Fig. 5B). Because we provided evidence that C_0814 is an effector secreted by the LEE-encoded T3SS, we hereby rename C_0814 to NleJ for non-LEE-encoded effector J, to be consistent with our previously proposed nomenclature scheme for effectors encoded outside the LEE (16).
Fig. 5.
EPEC C_0814 is a novel type III effector with a conserved C-terminal phosphoribosyl transferase domain. A, schematic diagram of the P2-like prophage PP3 in EPEC encoding the novel type III effector C_0814/NleJ. The Salmonella SopEφ phage encoding the effector SopE and a homolog of C_0814 is shown for comparison. The nucleotide sequences of the EPEC and Salmonella genomic regions containing these genes were retrieved from GenBankTM accession numbers FM180568 and AY319521 and analyzed using Vector NTI (Invitrogen). The genes encoding the EPEC effector C_0814/NleJ and its homolog (Orf14) from S. enterica Typhimurium strain SL1344, as well as SopE, are highlighted with red and blue arrows, respectively. The phage genes are indicated by gray arrows, whereas the DNA adenine methylase (dam), invertase (inv), and integrase (int) genes are shown with yellow, orange, and green arrows, respectively. B, protein sequence alignment of EPEC C_0814 (GenBankTM accession number YP_002328380), Salmonella phage SopEΦ-encoded Orf14 (GenBankTM AAQ65011), and the adenine phosphoribosyl transferase (Apt) of Corynebacterium jeikeium K411 (GenBankTM YP_250831) using the Cobalt multiple alignment tool at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/tools/cobalt/). The conserved amino acid residues are indicated in red letters and denoted by red asterisks. The conserved phosphoribosyl transferase domain is located near the C terminus and is delimited by a red line. The numbers at the ends of the protein sequences indicate the total numbers of amino acid residues of the proteins.
LifA/Efa1 Is a Member of a New Family of Type III Effectors in the A/E Pathogens and the Largest Type III Effector Identified
Our SILAC proteomic data showed that LifA/Efa1 is secreted by the EPEC sepD mutant but not detectable in the secreted proteins of WT EPEC (Table II and supplemental Table S2). This suggested that LifA is only detectable under effector hypersecreting conditions. The mechanism by which this large protein is secreted was not known previously (2). Indeed, full-length, native LifA has never been detected before in EPEC or EHEC, probably because of its extremely large size, very low abundance, and possible lability, and its existence was only substantiated by genetic evidence (37, 38). Its low abundance may also explain why we did not detect the LifA homolog in the type III secretome of C. rodentium using the same SILAC technology (14). As shown in Table II and Figs. 3 and 4, we demonstrated that LifA is secreted into the culture medium and that its N-terminal 50 amino acid residues contain a signal that is sufficient for T3SS-dependent secretion and translocation.
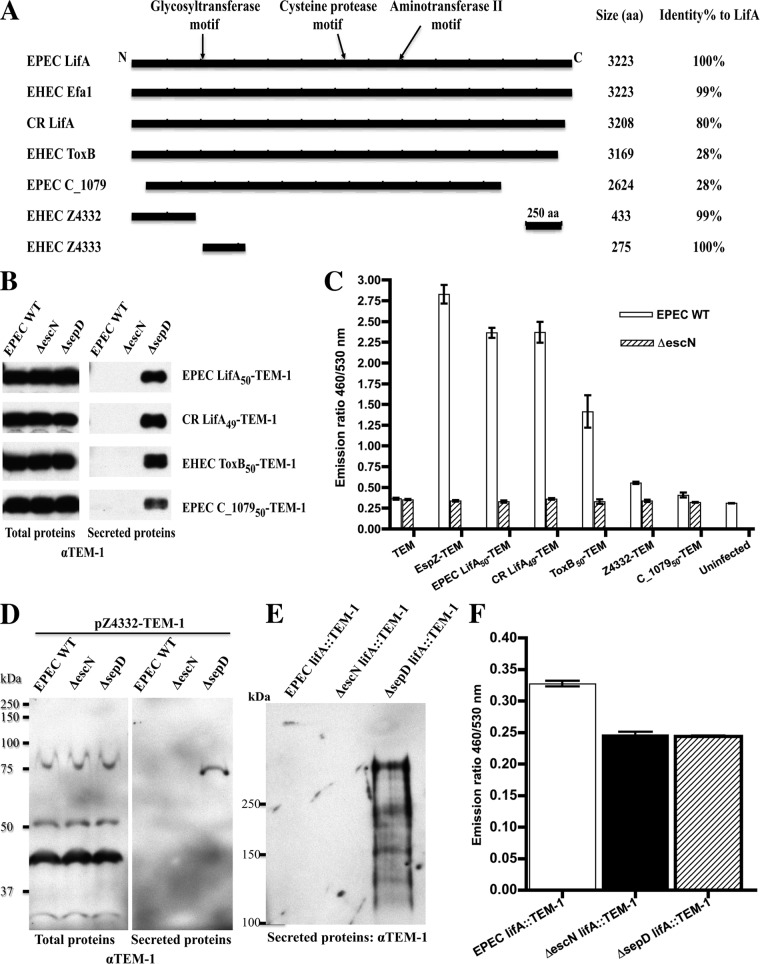
LifA has several closely related homologs in the A/E pathogens, including LifA in C. rodentium, Efa1 in rabbit EPEC and non-O157 EHEC strains, and the less homologous ToxB (L7095) encoded by the pO157 plasmid in EHEC (37–41) (Fig. 6A and supplemental Fig. S1). EPEC LifA and EHEC Efa1 have the same length of 3223 amino acid residues and are 99.9% identical (38). In EHEC O157 strains EDL933 and Sakai (22, 42), the lifA/efa1 gene is split into two open reading frames, Z4332 and Z4333 in EDL933 (Fig. 6A). Z4332 is 99.9% identical to the N-terminal 433 amino acid residues of LifA, and Z4333 contains 275 amino acid residues that match the middle portion of LifA, whereas the rest of LifA is truncated. The EPEC gene encoding Efa1/LifA is located in the integrative element IE6 in the chromosome, downstream of the genes coding for several EPEC non-LEE-encoded effectors, including EspL, NleB, and NleE (10). Another integrative element, IE2, appears to be a duplicate of IE6, but it has diverged considerably from IE6 in sequences. IE2 also contains genes coding for EspL, NleB, and NleE, as well as an Efa1/LifA-like protein C_1079. However, espL and nleB genes in IE2 are pseudogenes because of accumulation of many mutations, and the nleE gene (nleE2) contains mutations and an internal deletion that renders its product, NleE2, not type III-translocated (although it is type III-secreted) (10, 35). The Efa1/LifA-like protein, C_1079, shows less than 30% identity to Efa1/LifA (Fig. 6A and supplemental Fig. S1), and it was produced by EPEC and detected in the culture supernatant of the ΔsepD mutant by SILAC in this study (supplemental Table S2).
Fig. 6.
The LifA/Efa1/ToxB homologs are a new family of type III effectors in the A/E bacterial pathogens. The type III secretion assays and translocation assays are carried out using the same procedure as described in the legends to Figs. 3 and 4, respectively. A, schematic diagrams of the different LifA homologs, their sizes, and their percentages of identity to EPEC LifA in amino acid residues. The locations of the three predicted motifs (glycosyltransferase, cysteine protease, and aminotransferase) of EPEC LifA are indicated at the top. Shown are LifA and C_1079 of EPEC strain E2348/69, Efa1 from EHEC O111:H−, LifA from C. rodentium (CR), and ToxB, Z4332, and Z4333 from EHEC O157:H7 strain EDL933. aa, amino acid residues. B, type III secretion assay and Western blot against TEM-1 for the first 49–50 amino acids of EPEC LifA, C. rodentium LifA, EHEC ToxB, and EPEC C_1079 fused to the N terminus of TEM-1 in pCX341. Total proteins from whole cell lysates are shown in the left panel, and secreted proteins are shown in the right panel. C, type III translocation assay for the TEM-1 fusions to the N-terminal 49–50 amino acids of EPEC LifA, C. rodentium LifA, EHEC ToxB, and EPEC C_1079, as well as full-length Z4332. D, full-length Z4332 from EHEC O157:H7 strain EDL933 is type III-secreted when fused to TEM-1. Shown is a Western blot against TEM-1 using total proteins from whole cell lysates (left panel) and secreted proteins (right panel). Molecular mass standards in kDa are indicated on the left. Note the breakdown products of Z4332-TEM-1 in the total proteins that are not type III-secreted. E, full-length LifA-TEM-1 fusion expressed from EPEC chromosome is type III-secreted. The coding region of TEM-1 was inserted downstream of the lifA coding region before the stop codon in EPEC WT, ΔescN, and ΔsepD strains, and these strains were assayed for type III secretion of the LifA-TEM-1 fusion protein. The proteins were separated in SDS-8% PAGE and immunoblotted against TEM-1. Molecular mass standards in kDa are indicated on the left. F, full-length LifA-TEM-1 fusion expressed from EPEC chromosome is translocated in a T3SS-dependent fashion. EPEC WT, ΔescN, and ΔsepD strains containing chromosomal lifA::blaM fusions were assayed for type III translocation.
To determine whether these LifA homologs are type III-secreted and -translocated, we constructed translational fusions to TEM-1 using the coding regions for the first 50 amino acid residues of EPEC C_1079 and EHEC ToxB, as well as the N-terminal 49 amino acid residues of C. rodentium LifA, which has a 1-amino acid deletion in the N terminus compared with EPEC LifA (supplemental Fig. S1). Because the N-terminal part of EHEC Efa1 and Z4332 is identical to that of EPEC LifA, we did not make an N-terminal 50-amino acid residue fusion to TEM-1 for Efa1 and Z4332. We next checked these constructs in EPEC WT, ΔescN, and ΔsepD strains for type III secretion and translocation. As shown in the right panel of Fig. 6B, ToxB50-TEM-1 and C. rodentium LifA49-TEM-1 fusions showed similar secretion pattern (low levels of secretion by EPEC WT, hypersecretion by ΔsepD, and no secretion by ΔescN) to that of EPEC LifA50-TEM-1 and typical effectors. Indeed, both ToxB50-TEM-1 and C. rodentium LifA49-TEM-1 fusions were translocated efficiently into HeLa cells in the translocation assay (Fig. 6C). This indicated that, like EPEC LifA, C. rodentium LifA, EHEC ToxB, Efa1, and Z4332 also contain an N-terminal type III secretion and translocation signal and are likely type III effectors. However, the C_107950-TEM-1 fusion was not translocated, although it was type III-secreted by the ΔsepD strain, although not as efficiently as the LifA- and ToxB-TEM-1 constructs (Fig. 6, B and C). The N-terminal 75 amino acid residues of C_1079 do not align with Efa1/LifA and ToxB (supplemental Fig. S1). Because the type III secretion and translocation signal is usually contained in the N terminus of the secreted proteins, this may explain why C_107950-TEM-1 was not efficiently type III-secreted and -translocated.
To further demonstrate that these LifA/Efa1 homologs are effectors, we generated a full-length Z4332 fusion to TEM-1 and checked the construct for type III secretion and translocation. As shown in the right panel of Fig. 6D, Z4332-TEM1, with a predicted molecular mass of 79 kDa, was secreted in the effector-hypersecreting mutant ΔsepD, but not by ΔescN. The full-length fusion protein was highly unstable inside the bacteria, and most of the protein was degraded into smaller products (Fig. 6D, left panel). However, only the full-length Z4332-TEM-1 fusion was type III-secreted by ΔsepD (Fig. 6D, right panel). More importantly, although Z4332-TEM-1 was secreted in low abundance compared with the other effector-TEM fusions, it was translocated into HeLa cells in a type III-dependent fashion (Fig. 6C).
Because of its sheer size, it was not feasible to clone full-length EPEC lifA into the TEM-1 fusion vector. To further prove that full-length LifA of EPEC is a type III effector, we generated chromosomally expressed LifA-TEM-1 fusions in EPEC WT, ΔescN, and ΔsepD strains. Although the full-length TEM-1 fusion product was not observed in the total bacterial lysate by Western blot analysis because of either low abundance or protein degradation (data not shown), a large protein approximately the size of LifA-TEM-1 fusion (with a predicted molecular mass of 390 kDa) was seen in the secreted proteins of the effector-hypersecreting ΔsepD strain, but not in the ΔescN mutant (Fig. 6E). Furthermore, LifA-TEM-1 was translocated into HeLa cells and required a functional T3SS (Fig. 6F), although the level of translocation was understandably lower than plasmid-expressed effector-TEM-1 fusions. Collectively, our data demonstrated that except C_1079, the other LifA-related proteins, LifA from EPEC and C. rodentium, Efa1 from EHEC, and Z4332, and ToxB from EHEC O157 strains, are all type III-secreted and -translocated effector proteins.
DISCUSSION
T3SSs are essential virulence factors for many Gram-negative bacterial pathogens, and characterization of their type III secretome and effector repertoire is crucial for our effort in elucidating the molecular pathogenesis mechanisms, as well as developing new therapeutics for these pathogens. In this report, we carried out the first comprehensive proteomic analysis of the type III secretome of the prototypical EPEC strain E2348/69 using SILAC. In addition to verifying all the predicted type III-secreted proteins and effectors in EPEC (10), our SILAC analysis of the EPEC type III secretome led to several new and interesting findings. We showed that the effector repertoire of EPEC was previously underestimated (10). We identified at least two novel EPEC effectors, C_0814/NleJ and LifA, and further demonstrated that Efa1, ToxB, and Z4332, the EHEC homologs of LifA, are also type III effectors. LifA is one of the largest proteins encoded by the A/E pathogens and an important virulence factor (2, 10). LifA/Efa1/ToxB are the largest type III effectors identified thus far for any pathogen, demonstrating the tremendous cargo delivery capacity of the T3SSs and the potential of using the T3SSs as vectors to deliver large proteins or antigens. Furthermore, we discovered a number of type III-secreted, but not translocated, proteins previously unidentified in EPEC (Tables I and II). This suggests that, in addition to the effectors injected into the host cells, EPEC also uses the T3SS to secrete other proteins to the extracellular milieu to facilitate its replication and pathogenesis. Our results indicate that the type III secretome of EPEC is considerably larger than previously predicted.
The comprehensive coverage and confirmation of all previously known EPEC effectors by our SILAC screen further validated the sensitivity of SILAC and the efficacy of our method of coupling proteomics with regulatory mutants of the T3SS. WT EPEC secreted predominantly the three translocators, with only a few effectors, mostly LEE-encoded, detectable by SILAC (Table I). Although the LEE-encoded effectors were found in the culture supernatants of both WT EPEC and its sepD mutant, many non-LEE-encoded effectors were secreted exclusively by the sepD mutant (Table II and supplemental Table S2). Consistent with our previous observations (25), these data indicated that type III secretion of the majority of the effectors is highly regulated and severely suppressed in WT EPEC under our experimental conditions and that identification of the full complement of effectors by proteomics necessitates the utilization of regulatory T3SS mutants that have enhanced effector secretion. Indeed, the combination of proteomic tools and regulatory T3SS mutants, such as the sepD and sepL mutants of the A/E pathogens and the ssaL mutant of Salmonella, has proven powerful for identifying new effectors for C. rodentium, EHEC, Salmonella SPI-2, and now EPEC (Refs. 13, 14, 16, 43, and 44 and this study). Similar schemes should be applicable and beneficial to the identification of secretion substrates in other pathogens with T3SSs or other protein secretory systems.
Although the secreted proteins with top SILAC ratios for both WT EPEC and its sepD mutant were all established type III secretion substrates, there were a few known, not type III-secreted proteins that were enriched in the culture supernatant of WT EPEC and its sepD mutant and displayed moderate SILAC ratios. For example, EspC exhibited a SILAC ratio of 14.03 by WT EPEC when compared with the escN mutant, although EspC is secreted by a type V secretion system, not the LEE-encoded T3SS (45, 46). Intimin, a known outer membrane adhesin, is not a type III-secreted protein but showed a SILAC ratio of 7.67 by the sepD mutant when compared with the sepDescN double mutant. Because EspC and Intimin are either secreted or located in the outer membrane, and their expression is co-regulated with the LEE-encoded T3SS (47), it is possible that their secretion or targeting to the outer membrane is coupled to a functional T3SS, and they are consequently enriched by our sample preparation. Many ribosomal proteins and proteins from various cellular machineries also showed SILAC ratios higher than 2, indicating that there was some cell lysis and/or bacterial membrane leakage during the growth of EPEC under our culture conditions. Ribosomal and intracellular proteins were also found in our SILAC analysis of the C. rodentium type III secretome, but their SILAC ratios were close to 1 (14). This suggests that cell lysis seems to be more biased toward the T3SS-competent WT strain and the sepD mutant in EPEC than in C. rodentium.
We elected to further characterize 32 novel candidate type III-secreted proteins with moderate to high SILAC ratios in our SILAC screen (Tables I and II and Figs. 3 and 4). We were able to verify T3SS-dependent secretion of at least 13 proteins, including two new effectors LifA and C_0814/NleJ. These results suggest that many of the proteins displaying moderate to high SILAC ratios in our proteomic screen are indeed type III-secreted proteins in EPEC. Interestingly, whereas some of them are hypothetical proteins, a number of the new candidate proteins appear to be associated with, or localized to, the bacterial periplasm or membranes. Many of them are annotated as periplasmic or membrane transporters and metabolic enzymes (10). Enrichment of periplasmic proteins and proteins derived from outer membrane vesicles in the supernatant of bacterial cultures has been documented before in studies designed to identify secretion substrates using proteomics (18, 44, 48). It is plausible that some of these proteins have their own mechanisms to be targeted and secreted to the bacterial envelope where they perform nonpathogenic functions, but they can also be secreted by the T3SS when the T3SS is active. Their secretion and presence in the bacterial membranes or at the extracellular milieu may counteract host defense and benefit the invading pathogens, similar to some multitasking, moonlighting bacterial proteins (49). The biological significance of type III secretion of these proteins in EPEC pathogenesis needs to be further investigated.
Our study has now added at least two new members, C_0814/NleJ and LifA/Efa1/ToxB, to the effector repertoire of EPEC. The effectors of A/E pathogens are divided into two groups, the core and accessory effectors (9, 10). The core effectors are shared by all A/E pathogens analyzed to date, whereas the accessory effectors are only found in some A/E pathogens and therefore seem to be strain-specific. C_0814/NleJ appears to be an accessory effector, whereas LifA/Efa1/ToxB seem to be core effectors.
NleJ is encoded by the P2-like prophage PP3 in the EPEC chromosome (Fig. 5A). No close homologs of NleJ are found in C. rodentium and EHEC strains. However, its homolog (Orf14) can be found in Salmonella Typhimurium. The gene organization and context around c_0814/nleJ in EPEC prophage PP3 and SL1344_2696/orf14 in the Salmonella SopEΦ phage are remarkably similar (Fig. 5A), indicating that these two phages share the same evolutionary origin. Our results suggest that, in addition to sopE (36), the SopEΦ phage may carry yet another effector gene. It will be interesting to see whether the Salmonella NleJ homologs (SL1344_2696 and STM474_2848) are secreted by the SPI-1 and/or SPI-2 encoded T3SSs and whether they play any role in Salmonella pathogenesis. Lambdoid phages have been known to play crucial roles in the acquisition and dissemination of effector genes in A/E pathogens, because the vast majority of effectors is encoded by the exchangeable effector loci located within lambdoid prophages in the genomes of these pathogens (10, 13, 15, 22, 42). Our results here indicate for the first time that P2-like phages also disseminate effector genes in A/E pathogens, similar to Salmonella where many temperate phages, including lambdoid, P2-, and P22-like phages, are important vehicles for horizontal transfer of virulence and effector genes.
The nearly identical LifA and Efa1 are encoded only by the eae (therefore LEE)-positive A/E pathogens, and their homologs, including the truncated Z4332 and Z4333 in EHEC O157:H7 strains, are present in almost all of the A/E pathogens analyzed (37, 38, 50). ToxB is encoded on the pO157 plasmid in EHEC O157:H7 strains (41). This suggests that lifA/efa1/toxB and the LEE (and its T3SS) are closely associated and that LifA/Efa1/ToxB are probably core effectors. Indeed, the pathogenicity island bearing the genes for LifA/Efa1 and effectors NleB and NleE is located in the chromosome immediately next to the LEE in some A/E pathogens (51). LifA/Efa1/ToxB have three distinct motifs: a glycosyltransferase motif in the clostridial toxin-homologous region in the N-terminal region and a cysteine protease motif, as well as an aminotransferase II motif in the middle region of the proteins (Fig. 6A) (39, 52, 53). Several proteins with significant homology to LifA/Efa1 are encoded by the genome of Chlamydia spp., and they have been shown to be associated with cytotoxicity (54). It will be interesting to see whether these proteins are effectors of the T3SS in Chlamydia.
The LifA/Efa1/ToxB family of proteins has been shown to be important virulence factors by genetic studies (38–41, 50, 53), but their precise mode of action remains an enigma. Their proposed functions in different A/E pathogens range from being a lymphotoxin and immune modulator (37, 55), an adhesion/colonization factor (38, 40, 41), and a regulator of type III secretion (40, 41, 56), to being a modulator of Rho GTPases and intestinal epithelial-barrier function (52). These observations suggest that LifA/Efa1/ToxB have host cell binding activities and likely exert some of their functions from the bacterial side. The N terminus of LifA/Efa1/ToxB shares significant homology with the N-terminal 470 amino acid residues of Clostridium difficile toxins A and B that contain the glucosyltransferase domain. The clostridial toxins exert their cytotoxicity by glucosylating and inactivating small GTPases Rho, Rac, and Cdc42 and modulate the actin cytoskeleton in targeted eukaryotic cells after gaining entry via receptor-mediated endocytosis using their C-terminal translocation and receptor-binding domains (57). However, LifA/Efa1 proteins do not possess these C-terminal translocation and receptor-binding domains of the clostridial toxins. Efa1 was originally identified as an exported protein, at least to the bacterial periplasm, by TnphoA mutagenesis (38), and there was some evidence by immunofluorescence that Efa1 is associated with the membranes (50). However, LifA/Efa1/ToxB do not have a typical signal sequence for exported proteins at their N termini, and how they gain entry and act within target cells is still unresolved. Our demonstration that LifA/Efa1/ToxB can be secreted and translocated into host cells via the LEE-encoded T3SS offers an instant solution to this paradox and provides possible explanations for many seemingly contradictory functions of LifA/Efa1/ToxB, because type III effectors are known to contribute to bacterial colonization, modulate Rho GTPases and tight junctions, and even affect type III secretion (3, 9, 34).
ToxB represents the first plasmid-encoded effector identified in the A/E pathogens, because all the effectors identified thus far in this family of pathogens are encoded either within integrated elements or in lambdoid prophages in the chromosomes (10, 13, 15). Our results that ToxB is a type III effector may also explain the paradox that LifA, an important virulence factor in C. rodentium and EHEC O5 and O111 strains (38–40), was truncated into a much shorter form (Z4332) in the highly virulent, outbreak-causing EHEC O157:H7 strains (22, 42), whereas the entire ORF of LifA/Efa1 is present in all other A/E pathogens surveyed (50). We have shown that Z4332 is an effector, but Z4332 does not contain either the glycosyltransferase motif DXD or the other functional domains of LifA (Fig. 6A). It is probable that ToxB and LifA/Efa1 have redundant functions. Because ToxB is encoded by plasmid pO157 present in the EHEC O157:H7 strains, toxB may compensate for the mutation or loss of lifA without compromising the virulence of these strains. This would be similar to what has happened to the effector genes in the integrated element IE2 in EPEC (10). Part of IE2 appears to be a duplication of IE6 and contains the important effector genes lifA/efa1, espL, nleB, and nleE. However, although these genes are fully functional in IE6, they have degenerated in IE2 into either pseudogenes (nleB and espL) or genes (nleE2 and C_1079) encoding nonfunctional effectors (9).
In conclusion, our study has further validated SILAC as a simple, robust, highly sensitive, and quantitative proteomic tool for identifying secreted protein substrates. We have reported here an expanded repertoire of type III-secreted proteins and effectors in EPEC. Identification of NleJ/C_0814 and LifA/Efa1/ToxB/Z4332 as type III effectors reveals that, in addition to lambdoid phages and integrative elements, plasmids and P2-like phages have also played an important role in shaping the effector repertoire of the A/E pathogens. Furthermore, our analyses have provided fresh insights into the mechanism of function for effectors LifA/Efa1/ToxB/Z4332, an important family of virulence factors, and demonstrated that the LEE-encoded T3SS has the capacity to deliver very large proteins or antigens.
Supplementary Material
Footnotes
* This work was supported by grants from Genome Canada/Genome British Columbia through the PRoteomics for Emerging PAthogen REsponse project (to L. J. F. and B. B. F.) and operating grants from the Canadian Institutes of Health Research (to L. J. F. and B. B. F.) and the Howard Hughes Medical Institute (B. B. F.). This work was also supported by the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, and the British Columbia Proteomics Network.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- T3SS
- type III secretion system
- A/E
- attaching/effacing
- EPEC
- enteropathogenic E. coli
- EHEC
- enterohemorrhagic E. coli
- LEE
- locus of enterocyte effacement
- Nle
- non-LEE-encoded effector
- SILAC
- stable isotope labeling with amino acids in cell culture
- DMEM
- Dulbecco's modified Eagle's medium
- WT
- wild type.
REFERENCES
- 1. Finlay B. B., Falkow S. (1997) Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61, 136–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaper J. B., Nataro J. P., Mobley H. L. (2004) Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 [DOI] [PubMed] [Google Scholar]
- 3. Cornelis G. R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 4. Schraidt O., Marlovits T. C. (2011) Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331, 1192–1195 [DOI] [PubMed] [Google Scholar]
- 5. Croxen M. A., Finlay B. B. (2010) Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38 [DOI] [PubMed] [Google Scholar]
- 6. Stamm L. M., Goldberg M. B. (2011) Establishing the secretion hierarchy. Science 331, 1147–1148 [DOI] [PubMed] [Google Scholar]
- 7. Mundy R., MacDonald T. T., Dougan G., Frankel G., Wiles S. (2005) Citrobacter rodentium of mice and man. Cell. Microbiol. 7, 1697–1706 [DOI] [PubMed] [Google Scholar]
- 8. Wales A. D., Woodward M. J., Pearson G. R. (2005) Attaching-effacing bacteria in animals. J. Comp. Path. 132, 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong A. R., Pearson J. S., Bright M. D., Munera D., Robinson K. S., Lee S. F., Frankel G., Hartland E. L. (2011) Enteropathogenic and enterohaemorrhagic Escherichia coli: Even more subversive elements. Mol. Microbiol. 80, 1420–1438 [DOI] [PubMed] [Google Scholar]
- 10. Iguchi A, Thomson N. R., Ogura Y., Saunders D., Ooka T., Henderson I. R., Harris D., Asadulghani M., Kurokawa K., Dean P., Kenny B., Quail M. A., Thurston S., Dougan G., Hayashi T., Parkhill J., Frankel G. (2009) Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogura Y., Abe H., Katsura K., Kurokawa K., Asadulghani M., Iguchi A., Ooka T., Nakayama K., Yamashita A., Hattori M., Tobe T., Hayashi T. (2008) Systematic identification and sequence analysis of the genomic islands of the enteropathogenic Escherichia coli strain B171–8 by the combined use of whole-genome PCR scanning and fosmid mapping. J. Bacteriol. 190, 6948–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogura Y., Ooka T., Iguchi A., Toh H., Asadulghani M., Oshima K., Kodama T., Abe H., Nakayama K., Kurokawa K., Tobe T., Hattori M., Hayashi T. (2009) Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tobe T., Beatson S. A., Taniguchi H., Abe H., Bailey C. M., Fivian A., Younis R., Matthews S., Marches O., Frankel G., Hayashi T., Pallen M. J. (2006) An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U.S.A. 103, 14941–14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng W., de Hoog C. L., Yu H. B., Li Y., Croxen M. A., Thomas N. A., Puente J. L., Foster L. J., Finlay B. B. (2010) A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J. Biol. Chem. 285, 6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petty N. K., Bulgin R., Crepin V. F., Cerdeño-Tárraga A. M., Schroeder G. N., Quail M. A., Lennard N., Corton C., Barron A., Clark L., Toribio A. L., Parkhill J., Dougan G., Frankel G., Thomson N. R. (2010) The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192, 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng W., Puente J. L., Gruenheid S., Li Y., Vallance B. A., Vázquez A., Barba J., Ibarra J. A., O'Donnell P., Metalnikov P., Ashman K., Lee S., Goode D., Pawson T., Finlay B. B. (2004) Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lindeberg M., Cartinhour S., Myers C. R., Schechter L. M., Schneider D. J., Collmer A. (2006) Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 18. Bhavsar A. P., Auweter S. D., Finlay B. B. (2010) Proteomics as a probe of microbial pathogenesis and its molecular boundaries. Future Microbiol. 5, 253–265 [DOI] [PubMed] [Google Scholar]
- 19. McDermott J. E., Corrigan A., Peterson E., Oehmen C., Niemann G., Cambronne E. D., Sharp D., Adkins J. N., Samudrala R., Heffron F. (2011) Computational prediction of type III and IV secreted effectors in Gram-negative bacteria. Infect. Immun. 79, 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 21. Auweter S. D., Bhavsar A. P., de Hoog C. L., Li Y., Chan Y. A., van der Heijden J., Lowden M. J., Coombes B. K., Rogers L. D., Stoynov N., Foster L. J., Finlay B. B. (2011) Quantitative mass spectrometry catalogues salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J. Biol. Chem. 286, 24023–24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perna N. T., Plunkett G., 3rd, Burland V., Mau B., Glasner J. D., Rose D. J., Mayhew G. F., Evans P. S., Gregor J., Kirkpatrick H. A., Pósfai G., Hackett J., Klink S., Boutin A., Shao Y., Miller L., Grotbeck E. J., Davis N. W., Lim A., Dimalanta E. T., Potamousis K. D., Apodaca J., Anantharaman T. S., Lin J., Yen G., Schwartz D. C., Welch R. A., Blattner F. R. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533 [DOI] [PubMed] [Google Scholar]
- 23. Schauer D. B., Zabel B. A., Pedraza I. F., O'Hara C. M., Steigerwalt A. G., Brenner D. J. (1995) Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J. Clin. Microbiol. 33, 2064–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edwards R. A., Keller L. H., Schifferli D. M. (1998) Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 [DOI] [PubMed] [Google Scholar]
- 25. Deng W., Li Y., Hardwidge P. R., Frey E. A., Pfuetzner R. A., Lee S., Gruenheid S., Strynakda N. C., Puente J. L., Finlay B. B. (2005) Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gauthier A., Puente J. L., Finlay B. B. (2003) Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71, 3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 29. Chan Q. W., Howes C. G., Foster L. J. (2006) Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5, 2252–2262 [DOI] [PubMed] [Google Scholar]
- 30. Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 31. Charpentier X., Oswald E. (2004) Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based vector. J. Bacteriol. 186, 5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills E., Baruch K., Charpentier X., Kobi S., Rosenshine I. (2008) Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3, 104–113 [DOI] [PubMed] [Google Scholar]
- 33. Zarivach R., Vuckovic M., Deng W., Finlay B. B., Strynadka N. C. (2007) Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria. Nat. Struct. Mol. Biol. 14, 131–137 [DOI] [PubMed] [Google Scholar]
- 34. Thomas N. A., Deng W., Baker N., Puente J., Finlay B. B. (2007) Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J. Biol. Chem. 282, 29634–29645 [DOI] [PubMed] [Google Scholar]
- 35. Nadler C., Baruch K., Kobi S., Mills E., Haviv G., Farago M., Alkalay I., Bartfeld S., Meyer T. F., Ben-Neriah Y., Rosenshine I. (2010) The type III secretion effector NleE inhibits NF-κB activation. PLoS Pathog. 6, e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pelludat C., Mirold S., Hardt W. D. (2003) The SopEφ phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 185, 5182–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klapproth J. M., Scaletsky I. C., McNamara B. P., Lai L. C., Malstrom C., James S. P., Donnenberg M. S. (2000) A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68, 2148–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicholls L., Grant T. H., Robins-Browne R. M. (2000) Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35, 275–288 [DOI] [PubMed] [Google Scholar]
- 39. Klapproth J. M., Sasaki M., Sherman M., Babbin B., Donnenberg M. S., Fernandes P. J., Scaletsky I. C., Kalman D., Nusrat A., Williams I. R. (2005) Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect. Immun. 73, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stevens M. P., van Diemen P. M., Frankel G., Phillips A. D., Wallis T. S. (2002) Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70, 5158–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tatsuno I., Horie M., Abe H., Miki T., Makino K., Shinagawa H., Taguchi H., Kamiya S., Hayashi T., Sasakawa C. (2001) toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., Han C. G., Ohtsubo E., Nakayama K., Murata T., Tanaka M., Tobe T., Iida T., Takami H., Honda T., Sasakawa C., Ogasawara N., Yasunaga T., Kuhara S., Shiba T., Hattori M., Shinagawa H. (2001) Complete genome sequences of enterohaemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22 [DOI] [PubMed] [Google Scholar]
- 43. Li M., Rosenshine I., Yu H. B., Nadler C., Mills E., Hew C. L., Leung K. Y. (2006) Identification and characterization of NleI, a new non-LEE-encoded effector of enteropathogenic Escherichia coli (EPEC). Microbes Infect. 8, 2890–2898 [DOI] [PubMed] [Google Scholar]
- 44. Niemann G. S., Brown R. N., Gustin J. K., Stufkens A., Shaikh-Kidwai A. S., Li J., McDermott J. E., Brewer H. M., Schepmoes A., Smith R. D., Adkins J. N., Heffron F. (2011) Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stein M., Kenny B., Stein M. A., Finlay B. B. (1996) Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178, 6546–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vidal J. E., Navarro-García F. (2008) EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell. Microbiol. 10, 1975–1986 [DOI] [PubMed] [Google Scholar]
- 47. Mellies J. L., Navarro-Garcia F., Okeke I., Frederickson J., Nataro J. P., Kaper J. B. (2001) espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoon H., Ansong C., Adkins J. N., Heffron F. (2011) Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79, 2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henderson B., Martin A. (2011) Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Badea L., Doughty S., Nicholls L., Sloan J., Robins-Browne R. M., Hartland E. L. (2003) Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34, 205–215 [DOI] [PubMed] [Google Scholar]
- 51. Konczy P., Ziebell K., Mascarenhas M., Choi A., Michaud C., Kropinski A. M., Whittam T. S., Wickham M., Finlay B., Karmali M. A. (2008) Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent Verocytotoxin-producing Escherichia coli. J. Bacteriol. 190, 5832–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Babbin B. A., Sasaki M., Gerner-Schmidt K. W., Nusrat A., Klapproth J. M. (2009) The bacterial virulence factor lymphostatin compromises intestinal epithelial barrier function by modulating Rho GTPases. Am. J. Pathol. 174, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deacon V., Dziva F., van Diemen P. M., Frankel G., Stevens M. P. (2010) Efa-1/LifA mediates intestinal colonization of calves by enterohaemorrhagic Escherichia coli O26:H− in a manner independent of glycosyltransferase and cysteine protease motifs or effects on type III secretion. Microbiology 156, 2527–2536 [DOI] [PubMed] [Google Scholar]
- 54. Belland R. J., Scidmore M. A., Crane D. D., Hogan D. M., Whitmire W., McClarty G., Caldwell H. D. (2001) Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U.S.A. 98, 13984–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klapproth J. M., Donnenberg M. S., Abraham J. M., Mobley H. L., James S. P. (1995) Products of enteropathogenic Escherichia coli inhibit lymphocyte activation and lymphokine production. Infect. Immun. 63, 2248–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stevens M. P., Roe A. J., Vlisidou I., van Diemen P. M., La Ragione R. M., Best A., Woodward M. J., Gally D. L., Wallis T. S. (2004) Mutation in toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 72, 5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Voth D. E., Ballard J. D. (2005) Clostridium difficile toxins: Mechanisms of action and role in disease. Clin. Microbiol. Rev. 18, 247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.