Abstract
Symbiotic associations between legumes and rhizobia usually commence with the perception of bacterial lipochitooligosaccharides, known as Nod factors (NF), which triggers rapid cellular and molecular responses in host plants. We report here deep untargeted tandem mass spectrometry-based measurements of rapid NF-induced changes in the phosphorylation status of 13,506 phosphosites in 7739 proteins from the model legume Medicago truncatula. To place these phosphorylation changes within a biological context, quantitative phosphoproteomic and RNA measurements in wild-type plants were compared with those observed in mutants, one defective in NF perception (nfp) and one defective in downstream signal transduction events (dmi3). Our study quantified the early phosphorylation and transcription dynamics that are specifically associated with NF-signaling, confirmed a dmi3-mediated feedback loop in the pathway, and suggested “cryptic” NF-signaling pathways, some of them being also involved in the response to symbiotic arbuscular mycorrhizal fungi.
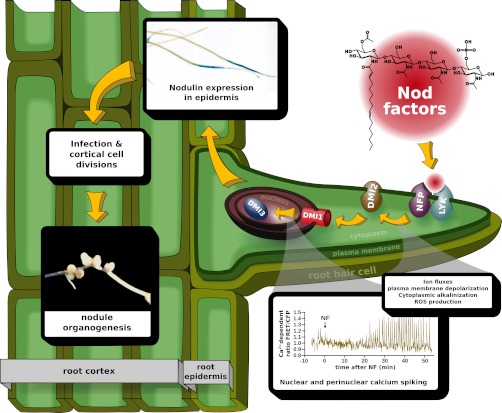
Legumes have the ability to form a very efficient symbiotic association with rhizobia to meet their nitrogen demand. This results in root nodule formation, inside which the rhizobia can fix atmospheric nitrogen efficiently and transfer it to the plants, in exchange for a carbon source. This interaction is often characterized by a high level of host specificity and generally requires an exchange of diffusible signals between legumes and rhizobia. Flavonoids and isoflavonoids present in the legume root exudates induce the expression of nod genes in rhizobia, and are responsible for the production and secretion of bacterial lipochitooligosaccharides (LCOs), known as Nod factors (NF)1 (1, 2). NF are generally required for rhizobial infection and nodule development. Mere application of purified NF at low concentrations (10−8 to 10−12 m) is sufficient to trigger responses in host plants similar to those elicited by the rhizobia themselves (2, 3). Responses are elicited in root cells within a few seconds to minutes after NF application and include changes in ion fluxes across the plasma membrane, as well as accumulation of reactive oxygen species (4, 5). After 15–20 min, oscillations of the nuclear and perinuclear calcium concentration (calcium spiking) are initiated and trigger the expression of early nodulin genes (3). Within the first hour, legume root hairs undergo a cytoplasmic disorganization, which leads to a transient swelling (6), followed by root hair deformations (7).
Forward and reverse genetic approaches, in the model legumes Medicago truncatula (Medicago) and Lotus japonicus (Lotus), have identified several components controlling these events (Fig. 1). NF are perceived with high specificity by LysM receptor-like kinases residing on the plasma membrane. These kinases include nod factor perception (NFP), an essential component of a signaling receptor necessary for early responses to NF and rhizobial infection (8, 9). Mutants in NFP are affected in all known cellular responses to NF reported so far, including calcium spiking, early nodulin genes expression and root hair deformations (9). A leucine-rich repeats (LRR)-receptor kinase, Does not Make Infections 2 (DMI2/NORK), is also localized on the plasma membrane, and acts downstream of NFP. Dmi2 mutants exhibit NF-induced root hair deformations, such as root hair swelling and branching, but are defective in NF-induced calcium spiking and rhizobial infection (10, 11). Signals perceived at the plasma membrane level are transduced downstream to activate signaling components residing at the nuclear level. DMI1, an ion channel, and MCA8, a calcium pump, are localized on the nuclear envelope and control NF-induced calcium spiking (11–14). These complex calcium signatures are decoded by a calcium/calmodulin-dependent protein kinase (CCaMK), called DMI3, which is localized inside the nucleus (15, 16). In response to NF, dmi3 mutants are able to elicit calcium spiking and root hair deformations, but are unable to induce the expression of early nodulin genes and cortical cell divisions (11, 15). Mutations in the auto-inhibitory domain of DMI3 result in constitutive activation of the protein and the spontaneous formation of root nodules even in the absence of rhizobia (17). This indicates that DMI3 is a central regulator of the NF signaling pathway and coordinates downstream nodulation-specific transcriptional events. It has also been shown that dmi3 mutants have increased sensitivity to low concentrations of NF, suggesting a negative feedback in the NF signal transduction pathway (18). Downstream of DMI3, two transcription regulators of the GRAS family, Nodulation Signaling Pathway 1 and 2 (NSP1 and NSP2), control nodulin gene expression in NF-dependent manner (19, 20). NSP1 interacts with NSP2 and binds to the promoter region of Early NODulin 11 (ENOD11) and Nodule INception (NIN), which are popular markers for the study of this signaling pathway (21).
Fig. 1.
Schematic illustration of genetic components involved in legume-rhizobia symbiotic signaling. NF are perceived by a receptor complex containing NFP. Perception of NF triggers early responses, like ion fluxes, depolarization of plasma membrane, production of reactive oxygen species (ROS), and cytoplasmic alkalinization. The signals are transduced downstream to activate LRR-receptor kinase, DMI2 and DMI1. The nuclear ion channel DMI1 is required for NF-induced nuclear calcium spiking. Later these calcium signals are perceived and decoded by calcium/calmodulin-dependent protein kinase, DMI3, which acts as a central player in the NF signaling and coordinates the expression of symbiotic genes, including early nodulin genes. NF signaling culminates with the coordinated and synchronous progression of infection and cortical cell division, resulting in the formation of mature, infected and nitrogen-fixing nodules.
Several genes controlling NF signaling are also required for the establishment of arbuscular mycorrhization (AM), indicating the existence of a “common symbiotic pathway” between these two endosymbioses (22). Similar to rhizobia, AM fungi release diffusible signals, known as Myc factors, which are also LCOs and can be easily collected in germinating spore exudates (GSE; 23, 24). Myc factors trigger responses in host plants similar to those elicited by NF (23). Dmi1, dmi2, and dmi3 mutants are unable to establish AM associations whereas nfp, nsp1, and nsp2 display a wild-type AM phenotype (9, 10, 12, 15). However, some role for NFP and NSP2 in Myc factor signaling was recently identified, suggesting that an overlap between the two LCO pathways may be more significant than previously thought (23). Although Myc factor receptors have not yet been identified, LYR1, a close homolog of NFP, is a potential candidate because its expression is strongly up-regulated during AM symbiosis (25, 26).
Over the last decade, various large-scale approaches such as transcriptomics, proteomics, and metabolomics, have been used to dissect the legume-rhizobia symbiosis (27–29). However, most of these studies looked at relatively late responses to rhizobia or NF and unfortunately were not integrated. The techniques used in the past for analyzing changes in the transcriptome during nodulation include serial analysis of gene expression (SAGE), custom-made macro- and micro-arrays, and Affymetrix GeneChip Genome Arrays (25, 28, 30–33). Except for a few examples (28, 33), most of these studies explored transcriptome changes at late stages of the legume-rhizobia symbiosis (from 1 to 6 days post inoculation, dpi). Moreover, because, many of these studies were performed before the completion of the Medicago genome sequencing project (26), the data are not readily comparable, as they use different probes and nomenclature. Several proteomic studies have been undertaken to explore changes in protein levels during the legume-rhizobia symbiosis, but they all analyzed late stages of the interaction and none of them were quantitative. Many protein kinases have been identified by genetic approaches in the NF signaling pathway and yet, only one phosphoproteomic study has been published so far (34). This very interesting study used two-dimensional gel electrophoresis and radiolabeled phosphoryl groups, allowing the identification of differential phosphorylation events, but unfortunately the identification of the corresponding proteins was not possible.
Although still at the draft stage, the recently released M. truncatula genome assembly (Mt3.5) span a physical distance of 375 milliion base pairs (Mb) of which 246 Mb are nonredundant sequences obtained from 2536 bacterial artificial chromosome library (26). Alignment with expressed sequence tags (ESTs) suggests that about 94% of the expressed genes are represented in the combined data that are newly released. About 47,845 genes have been annotated with experimental and database support with average length of 2211 bp (26). With these salient features, the Medicago genome sequence enables us to utilize next generation RNA sequencing strategy, as well as MS-based methods to explore in depth the early phosphoproteomic, proteomic, and transcriptional responses to NF. In this study, we report an integrated large-scale approach to investigate changes in the phosphoproteome, proteome, and transcriptome that occur one hour after NF treatment. To help place these results within a biological context, we compared results obtained with wild-type Medicago plants to those with two key mutants (nfp and dmi3). Our quantitative study sheds light on the NF-induced transcriptional, translational, and post-translational responses and reveals signaling pathways that previously remained undetected using conventional approaches.
EXPERIMENTAL PROCEDURES
Plant Materials, Growth, and Treatment with NF and GSE
Medicago truncatula Jemalong A17 (wild-type), C31 (nfp-1) (9), and TRV25 (dmi3–1) (15) plants were used for transcriptomics, proteomics, and phosphoproteomics analyses. Plants were grown under three different growth conditions (aeroponic, hydroponic, and plates), which facilitate collection of large quantities of NF-responsive root tissues for RNA/protein extraction. The different growth conditions used in the current study were optimized for efficient response to NF with the help of pENOD11:GUS plants (35). Seeds of Medicago were harvested, acid-scarified, surface sterilized, and germinated (36) before growing in different growth conditions. For growth in the aeroponic system, 3-day old seedlings were placed in the aeroponic system (37) and grown in nitrogen-free modified Fahraeus medium (36) for 14 to 15 d at 22 °C and 24 h of 130 to 200 umol m−2 s−1 light. The roots, however, grew in the dark. For uniform treatment with NF, the modified Fahraeus medium without NF was replaced by the one with 10−8 m NF obtained from Sinorhizobium meliloti strain Rm1021 pRmE43 (pTE3:nodD1) as indicated in (36). One hour after treatment, the root tissues were harvested and flash frozen for phosphoprotein extraction. For growth in the hydroponic system, 2-day old Medicago seedlings were transferred to conical flasks containing liquid modified Fahraeus medium supplemented with 1% sucrose and incubated in the dark at room temperature with constant shaking at 50 rotations per minute. The seedlings were treated with 10−8 m NF, incubated in the medium for one hour and the root tissues were flash frozen for phosphoprotein extraction. For growth in the plate system, 2-day old germinated seedlings were grown on 23 cm × 23 cm square plates containing Fahraeus medium overlaid with moist sterile germination paper. The seedlings were grown in the dark at room temperature for 5 days and treated with 10−8 m NF by flood inoculation to ensure that roots of all the seedlings grown on the plates are treated. One hour after the NF treatment, the root tissues were rapidly harvested and flash-frozen in liquid nitrogen. Proteins were isolated from whole-cell lysate of root tissue or membrane-enriched fractions for phosphoproteomics with the addition of a variety of phosphatase inhibitors as described previously (37). For transcriptomics studies seedlings grown in the plate system were used.
GSE from Glomus intraradices was obtained as described previously (24). Each Medicago seedling (wild-type plant, nfp and dmi3) was treated with GSE obtained from 250 spores dissolved in 1 ml of distilled-sterile water. As a mock inoculation, control seedlings were treated with 1 ml of sterile distilled water. One hour after the treatment, the root tissues were excised and flash-frozen for RNA extraction.
RNA Extraction and TruSeq RNA Sample Preparation
Total RNA was isolated from the root tissues using Qiagen RNeasy Plant mini kit. RNA samples were run on a Bioanalyzer 2100 RNA Pico chip to test the quality of the preparations. RNA from nine samples showing signs of degradation was re-isolated from the same plant material and used in place of the first samples. The final eighteen samples submitted for sequencing had RIN values ranging from 6.2–10.0 (mean 7.7, median 7.7). Sequencing libraries were prepared at the University of Wisconsin Gene Expression Center using the Illumina TruSeq RNA Sample Preparation Kit (mRNA protocol rev. A). Libraries were sequenced at the University of Wisconsin DNA Sequencing Facility on an Illumina HiSeq 2000 system. Samples were multiplexed on three lanes of the flow cell with one replicate of each genotype/treatment on each lane to minimize the effects of technical variation. Data was collected for 100 bp single-end reads.
Sequencing reads were checked for quality using FastQC v0.9.4. Reads were mapped to the publicly available Mt3.5 genome sequence using TopHat v1.3.2 (26, 38). Minimum intron size, maximum intron size, and microexon search parameters were set to 10, 20,000, and true, respectively, and default settings were used for all other parameters. Reads mapping to multiple locations on the genome were filtered out using samtools v0.1.18 (39). The remaining data were used to generate gene-level exon coverage counts based on the published Mt3.5v4 gene annotations using htseq-count from the HTSeq package v0.5.1p2 in non-stranded union mode. Gene-level read counts for the eighteen samples were normalized and analyzed for differential expression using the R package DESeq v1.6.0 with pooled dispersion estimates (40). Test statistics were adjusted for multiple testing using the method of Benjamini and Hochberg and the adjusted p values were used to select differentially expressed genes at a false discovery rate of 5% (41).
Silencing Medicago LYR1
To silence Medicago LYR1, RNAi fragment of about 252 base pairs from LYR1 coding sequences was amplified using “ATGTCGACCAAAGGACATAATCACAGC” and “TTGATATCCTCACAAGCCTTTCTCTTCC” primers and cloned into pENTR1A entry vector. The LYR1-RNAi fragment was cloned into pK7GWIWG2-R hairpin RNAi destination vector carrying DsRed1 as visible reporter by LR-recombination. Medicago Jemalong A17 seedlings were transformed with LYR1-RNAi cassette by Agrobacterium rhizogenes (strain MSU440) -mediated hairy root transformation (42). Transgenic roots expressing LYR1-RNAi cassette were identified by DsRED1 expression (supplemental Fig. S11D-E), and the validation of reduction in LYR1 expression was confirmed by semi-quantitative RT-PCR (supplemental Fig. S11C). The primers used were furnished in the supplemental Table S7.
Validation of RNA Sequencing Data by qRT-PCR
The validation of the expression level of candidate genes by qRT-PCR was performed with RNA samples prepared from three biological replicates each with two technical replicates. The RNA samples were treated with DNase, DNA-free™ (Ambion) to remove any residual DNA contamination present in the samples. RNA samples were quantified using NanoDrop 1000 for precise quantification. For cDNA synthesis, 0.5 μg of RNA was used from all the samples to standardize the RNA concentration. cDNA was synthesized using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas). Quantitative RT-PCR was performed using SYBR® Advantage® qPCR Premix (Clontech, Palo Alto, CA) with respective primer set (given in a separate section) optimized to have an efficiency of 100 ± 5%. The expression levels of candidate genes were normalized using two references genes, MtActin and MtEF1a. The qRT-PCR data were analyzed using GenEx software from Bio-Rad. Primers used for quantitative RT-PCR validation of RNA sequencing data are listed in the supplemental Table S7.
Microsome and Membrane Preparation
Plants were grown as described for RNA extraction with the exception that the tissue was harvested immediately after one hour of NF treatment with no freeze step. All subsequent stages of membrane isolation were carried out in a 4 °C cold room with chilled equipment. Plant tissue was ground in 2× v/w (e.g. 20 ml buffer per 10 g fresh weight tissue) ice-cold Homogenization Buffer (230 mm sorbitol, 50 mm TrisHCl, 10 mm KCl, 3 mm EGTA, pH 7.5) containing freshly added protease inhibitors (1 mm PMSF, 0.7 μg/ml leupeptin, 1.0 μg/ml pepstatin, 1 mm potassium metabisulfite), and phosphatase inhibitors (10 mm NaF, 2 mm Na pyrophosphate, 1 mm ammonium molybdate). Homogenization was performed in a Waring blender using two 20-s pulses. Homogenate was filtered through four layers of Miracloth and centrifuged at 6000 × g for 10 min at 4 °C. The supernatant containing the microsome fraction was transferred to a cold ultracentrifuge tube and spun at 65,000 × g for 30 min at 4 °C. The supernatant was discarded and the microsomal pellet was resuspended in ice-cold Resuspension Buffer (250 mm sucrose, 10 mm KCl, 10 mm HEPES, pH to 7.0 with KOH) with the aid of a Potter teflon homogenizer. The microsomal fraction was either stored at −80 °C for subsequent analysis or further processed as follows to enrich for plasma membrane proteins.
One gram (approximated using 1.0 ml volume) of microsomal sample was added to a four gram (3 g premade + 1 g microsomal) 6% PEG/Dextran two-phase system prepared the previous day in disposable glass culture tubes and stored in the cold room overnight (final concentrations: 6% w/w Dextran T500, 6% w/w PEG-3350, 333 mm sucrose, 5 mm potassium phosphate, 5 mm potassium chloride, 0.1 mm EDTA). Dithiothreitol was added fresh immediately before use to a concentration of 1 mm. After addition of the microsomal sample, the two-phase system was mixed thoroughly by inversion and incubated at 4 °C for 15 min followed by a 15 min spin at 900 × g in a 4 °C centrifuge. The upper phase (enriched for plasma membrane) was removed slowly using a Pasteur pipette, leaving the interface with the lower phase, and transferred to an ultracentrifuge tube. The lower phase was transferred to a separate ultracentrifuge tube. Samples were diluted to 24 ml using resuspension buffer and centrifuged at 100,000 × g for 35 min. at 4 °C. The supernatant was discarded and the pellets were suspended in Resuspension Buffer with the aid of a Potter teflon homogenizer. Protein concentrations in microsomal, upper, and lower phase fractions were quantified using a Bradford assay with a BSA standard curve.
Protein Extraction
For experiments analyzing whole plant extract, protein was precipitated from plant extract via chloroform/methanol extraction or acetone precipitation. Chloroform/methanol extraction was performed by adding four volumes of methanol to one volume of plant extract. The mixture was vortexed and one volume of chloroform was added before additional vortexing. Three volumes of water was added, the solution was vortexed, and subsequently centrifuged for 5 min (4696 × g, 4 °C). The top layer was removed and discarded via serological pipette. Then, three volumes of methanol were added and the solution was vortexed before centrifugation for 15 min (4696 × g, 4 °C). The resulting pellet of protein was washed three times by vortexing with ice cold 80% acetone and centrifuging at 10,000 × g for 5 min. Once washed, the pellet was dried on ice for 30 min and stored at −80 °C.
Acetone precipitation protein extraction consisted of adding four volumes of ice cold acetone to one volume of plant extract. The solution was then vortexed and stored at −20 °C overnight. The sample was then centrifuged for 20 min (4696 × g, 4 °C). The resulting protein pellet was washed three times with ice cold 80% acetone. Once washed, the pellet was dried on ice for 30 min and stored at −80 °C.
Protein Digestion and Isobaric Labeling
For whole plant extract samples, protein pellets were resuspended in ice-cold 8 m urea, 30 mm NaCl, 40 mm tris (pH 8), 2 mm MgCl2, 50 mm b-glycero phosphate, 1 mm sodium orthovanadate, 10 mm sodium pyrophosphate, 1× mini EDTA-free protease inhibitor (Roche Diagnostics), and 1× phosSTOP phosphatase inhibitor (Roche Diagnostics). Total protein was then quantified using a BCA protein assay kit (Thermo Scientific Pierce). For analysis, 1 mg of protein from each sample was reduced by adding DTT to a final concentration of 5 mm, and alkylated with 15 mm iodoacetamide before final capping with 5 mm dithiothreitol. Digestion was carried out by adding LysC (Wako Chemicals) at a 1:100 enzyme-to-protein ratio and incubating at 37 °C for 2 h. At this time, the lysate was diluted with 25 mm tris (pH 8) to a final urea concentration of 1.5 m and further digested for 12 h at 37 °C with trypsin (Promega, Madison, WI) at a 1:100 enzyme-to-protein ratio.
For membrane enriched samples, seven volumes of re-suspension buffer (from above) were added to one volume of membrane-enriched solution (membrane samples contained either 0.5 mg or 1 mg protein). Samples were then reduced as described above. Digestion was carried out by adding LysC (Wako Chemicals) at a 1:120 enzyme-to-protein ratio and incubating at 37 °C for 2 h. At this time, an additional aliquot of LysC was added at a 1:240 enzyme-to-protein ratio before incubation for another 1.5 h at 37 °C. Finally, the lysate was diluted with 25 mm tris (pH 8) to a final urea concentration of 1.5 m and further digested for 12 h at 37 °C with trypsin (Promega) at a 1:100 enzyme-to-protein ratio.
For all experiments, peptides were then acidified with trifluoroacetic acid (TFA) to quench the reaction and de-salted using C-18 solid phase extraction (SPE) columns (Waters). TMT (43) or iTRAQ (44) labeling was carried out per manufacturer's directions (Thermo Scientific Pierce, TMT; Applied Biosystems, iTRAQ). To confirm BCA measurements, aliquots of total protein were taken from each sample, combined in a 1:1:1:1, 1:1:1:1:1:1, or 1:1:1:1:1:1:1:1 ratio (4-Plex, 6-Plex, and 8-Plex experiments, respectively), and analyzed via mass spectrometry. Summed reporter ion ratios from this experiment were used to adjust mixing ratios of the remaining labeled digests, after which the mixed samples were again de-salted using SPE.
Strong Cation Exchange Fractionation
Strong cation exchange (SCX) was carried out using polysulfoethylaspartamide column (9.4 × 200 mm; PolyLC) on a Surveyor LC quaternary pump (Thermo Scientific; flow rate of 3.0 ml/min.). The mixed and dried isobaric label sample was resuspended in buffer A before separation. The following gradient was used for separation: 0–2 min, 100% buffer A, 2–5 min, 0–15% buffer B, 5–35 min, 15–100% buffer B. Buffer B was held at 100% for 10 min. The column was washed extensively with buffer C and water prior to recalibration. Buffer compositions were as follows: buffer A [5 mm KH2PO4, 30% acetonitrile (pH 2.65)], buffer B [5 mm KH2PO4, 30% acetonitrile, 350 mm KCl (pH 2.65)], buffer C [50 mm KH2PO4, 500 mm KCl (pH 7.5)]. Typically 8–14 fractions were collected over a 50 min elution period. Samples were collected by hand, frozen, lyophilized, and desalted by SPE.
Phosphopeptide Sample Preparation
Following SCX fractionation, phosphopeptides were enriched via immobilized metal affinity chromatography using magnetic beads (Qiagen, Valencia, CA). Following equilibration with water, the beads were treated with 40 mm EDTA (pH 8.0) for 30 min with shaking, and washed three times with water again. The beads were then incubated with 100 mm FeCl3 for 30 min with shaking and finally were washed three times with 80% acetonitrile/0.1% TFA. Samples were likewise resuspended in 80% acetonitrile/0.15% TFA and incubated with beads for 45 min with shaking. The resultant mixture was washed three times with 1 ml 80% acetonitrile/0.1% TFA, and eluted using 1:1 acetonitrile/0.7% NH4OH in water. Eluted phosphopeptides were acidified immediately with 4% formic acid and lyophilized to ∼5 μl.
Nano-High Performance Liquid Chromatography
For all samples, online reverse-phase chromatography was performed using a NanoAcquity UPLC system (Waters, Milford, MA). Peptides were loaded onto a precolumn (75 μm ID, packed with 7 cm Magic C18 particles, Bruker-Michrom) for 10 min at a flow rate of 1 μm/min. Samples were then eluted over an analytical column (50 μm ID, packed with 15 cm Magic C18 particles, Bruker-Michrom) using either a 90 or 120 min linear gradient from 2% to 35% acetonitrile with 0.2% formic acid and a flow rate of 300 nL/min.
Mass Spectrometry
All experiments were performed on an ETD-enabled LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). High resolution MS1 scans in the orbitrap were used to guide data-dependent MS/MS scans that used electron transfer dissociation (ETD; (45)), collisionally activated dissociation (CAD), or high-energy CAD (HCD; (46)) to produce sequence informative ions analyzed in the orbitrap. In five of the 11 experiments, HCD alone was used to produce reporter tags, the remaining experiments utilized QuantMode (QM) MS/MS scans. QM is a recently described method that uses gas-phase purification to improve quantitative accuracy and dynamic range (47).
The LTQ Orbitrap Velos used firmware version 2.6.0.1065 SP3. MS1 scans were performed in the Orbitrap at 30,000 or 60,000 resolution at a max injection time of 500 ms and a target value of 1,000,000. Dynamic exclusion duration was set to 30 or 60 s, with a max exclusion list of 500, and an exclusion width of 0.55 Th below and 2.55 Th above the selected average mass. HCD MS2 scans were also acquired in the Orbitrap at a resolution of 7500 and with HCD normalize collision energy (NCE) of 45 or 50%, a max inject time of 200 or 500 ms, and an AGC target of 50000. ETD MS2 scans were acquired in the Orbitrap at a resolution of 7500 and with a charge independent reaction time of 70 ms, a max inject time of 200 ms, an AGC target of 50,000, and a reagent ion target of 400,000. QuantMode (QM) scans were acquired in the Orbitrap at a resolution of 7500 and with a charge dependent PTR reaction time to enable gas-phase purification, HCD NCE of 65–80% to produce isobaric reporter tags, and CAD NCE of 35% to produce sequence informative ions (47). For QM scans an AGC target of 400,000, a reagent ion AGC target of 400,000, and max injection time of 500 ms was used. All Thermo .RAW files are deposited in PeptideAtlas and have been assigned the following entry numbers PASS00056 (iTRAQ 4-Plex Data), PASS00057 (TMT 6-Plex Data), PASS00058 (iTRAQ 8-Plex Data).
Database Searching and FDR Estimation
MS/MS data were analyzed using the Coon OMSSA Proteomics Software Suite (COMPASS; (48)). The Open Mass Spectrometry Search Algorithm (OMSSA; version 2.1.8; (49)) was used to search spectra against a concatenated target-decoy database consisting of Medicago protein sequences from the following databases: MT3.5, Uniprot, NCBI, and an internal Wisconsin Medicago Group database. This composite database contained 51,426 entries and is available for download at http://more.biotech.wisc.edu/. We have previously demonstrated a combined database returns a greater number of identifications (37). For all searches, tryptic peptides were created in silico allowing up to three missed cleavages. The precursor mass tolerance was set to ±4.5 Da and monoisotopic mass tolerance was set to ±0.015 Da for fragments ions. It has previously been demonstrated that using a large precursor mass tolerance coupled with post-search filtering based on precursor mass, such as the algorithm employed in FDROptimizer, increases the number of target identifications (50). Carbamidomethylation of cysteines, isobaric label (TMT or iTRAQ) on the N terminus, and isobaric label on lysines were included as fixed modifications, whereas oxidation of methionines, phosphorylation of s, phosphorylation of t, phosphorylation of y, and isobaric label on tyrosines was added as a variable modification. Results from each experiment were then filtered to a 1% FDR using high resolution batch FDROptimizer. The FDROptimzer conducts false discovery analysis at the peptide level using a two dimensional analysis. One of these dimensions is OMSSA e-value, whereas the other is the precursor mass error.
Protein and Peptide Quantification and Phosphosite Localization
For each experiment, isobaric label quantification was performed using TagQuant. Peptides from all experiments were then combined into protein groups and quantified at the protein level using ProteinHoarder. The ProteinHoarder performs protein grouping of identifications from all experiments, a p-score for both target and decoy proteins is then calculated by multiplying the p values of all peptides contained within the protein group and is used to conduct false discovery rate analysis at the protein level. For phosphopeptides, the Phosphinator software was used to localize phosphorylation sites and combine quantitative data for phosphoisoforms within experiments (51). Following phosphosite localization, any instance in which a peptide failed to have all phosphosites localized, was discarded and not used in subsequent quantitative analysis. Isobaric reporter tag data from all 11 experiments were consolidated in Microsoft Excel. Experimental ratios and p values (Student's t test assuming equal variance) were then determined using Microsoft Excel.
Protein and Phosphoisoform Functional Analysis
To determine functional analysis enrichment, we developed a program, MedicaGO, which links gene ontology (GO) terms available from interPro to corresponding MT3.5 accessions. The GO terms are linked to unique gene identifies, however protein grouping can result in many gene identifiers being combined into a larger compilation of proteins. To account for this, we assigned GO terms from any gene identifier within a protein group to the group as a whole. Any GO term that was significantly represented in a subgroup as compared with the entire proteome/phosphoproteome was considered enriched (p < 0.05, Fisher's exact test with Benjimini-Hochberg correction for multiple comparisons).
Building Medicago-Omics Repository (MORE)
All data presented here (transcriptomic, proteomic, phosphoproteomic) will be freely available on a newly created Medicago-Omics Repository (MORE; http://more.biotech.wisc.edu/). By entering Medicago gene identification number or protein name (e.g. Medtr4g030140 or MtDRP2B) users will obtain RNA, protein, and phosphorylation quantitative data. Overall wild-type, nfp, and dmi3 ratios (+NF/-NF) are displayed for each protein entry. Expandable displays will show the measurements for each individual experiment for each protein and phosphorylation site, allowing users to compare the relative response between different experimental conditions. MORE also houses raw nHPLC-MS/MS data (Thermo .RAW files), and downloadable spreadsheets including all RNA, protein, and phosphorylation data collected during the course of this experiment.
RESULTS
Large-scale Detection and Quantification of the Medicago Proteome and Phosphoproteome
NF-induced dynamics in protein and phosphorylation levels were analyzed in Medicago by high mass accuracy tandem mass spectrometry. Data were collected from 11 quantitative isobaric tag-based (43, 44) nano-HPLC-MS/MS experiments comprising various plant genotypes (wild-type, nfp and dmi3), time points (0, 10, 30, and 60 min), cellular extracts (whole cell, microsome, and upper or lower membrane fractions) and growth conditions (aeroponic, hydroponic, and plates). supplemental Fig. S1 depicts the proteomic workflow used to generate the quantitative data. Proteins extracted from Medicago were subjected to tryptic digestion. To enable large-scale multiplexed quantification, peptides were labeled with isobaric tags (TMT or iTRAQ) and fractionated via strong cation exchange (SCX) to reduce sample complexity. All SCX fractions were enriched for phosphopeptides using immobilized metal affinity chromatography (IMAC). Both unmodified and phosphopeptide fractions were analyzed using an ETD-enabled Velos hybrid linear ion trap-orbitrap mass spectrometer (Thermo-Fisher Scientific). A flow diagram demonstrating MS/MS analysis beginning with MS1 acquisition and resulting in quantitation of significantly altered phosphopeptides is depicted in supplemental Fig. S2.
Tandem mass spectra were searched against a concatenated target-decoy database comprising Medicago protein sequences. Across all experiments, more than 2400 h of instrument time generated ∼10,000,000 high resolution MS/MS spectra of which 2,033,877 were confidently (1% FDR) assigned to 78,635 unique peptide sequences mapping to 7739 proteins (Table I). This includes 15,335 unique phosphopeptides, containing 13,506 nonredundant sites of phosphorylation localized to specific amino acids within 3926 phosphoproteins, resulting in an average of ∼3 phosphosites per protein. A complete list of all phosphoproteins and respective phosphosites, along with peptide spectrum matching (PSM) counts, fold change and p values, are furnished in supplemental Table S1. Protein levels for respective phosphoproteins 1 hour post-treatment are listed in supplemental Table S2. Additional meta-analyses of experiments described here are available in the supplementary information. This discussion includes analysis of protein and phosphoprotein identification overlap between experiments (supplemental Fig. S3), global ratio distributions in each experiment containing a wild-type plant (supplemental Fig. S4), comparison to data collected in Grimsrud et al. (37) (supplemental Fig. S5), as well as analysis of protein and phosphoprotein identification increases because of cellular fractionation.
Table I. Summary of proteomic and phosphproteomic analyses. NF = Nodulation Factor.
| Experiment | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Overall |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | ±NF | |
| Growth | Aeroponic | Hydroponic | Hydroponic | Hydroponic | Plate | Plate | Plate | Plate | Plate | Plate | Plate | – |
| Cellular Fraction | Whole Cell | Whole Cell | Whole Cell | Whole Cell | Upper Membrane | Lower Membrane | Microsome | Whole Cell | Whole Cell | Upper Membrane | Lower Membrane | – |
| Genotype | WT, dmi3 | WT, dmi3 | WT, nfp | WT, nfp | nfp | nfp | WT, nfp, dmi3 | WT | WT | WT | WT | – |
| Biological Replicates | 1 | 1 | 1 | 1 | 3 | 3 | 1 | 3 | 3 | 3 | 3 | – |
| Time Points | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 10/30 min | N/A | N/A | – |
| Unique Peptidesa | 34195 | 37436 | 28976 | 27723 | 11129 | 9152 | 13073 | 12218 | 8581 | 7430 | 10309 | 63290 |
| Unique Phosphopeptidesb | 5975 | 9694 | 8231 | 5471 | 2976 | 4069 | 2055 | 4129 | 2782 | 1032 | 1898 | 15335 |
| Proteins | 6315 | 6983 | 6290 | 6521 | 3996 | 4071 | 3956 | 4256 | 3603 | 2865 | 3604 | 7739 |
| Phosphoproteins | 2392 | 3076 | 2800 | 2382 | 1351 | 1706 | 1000 | 1807 | 1320 | 565 | 977 | 3926 |
| Localized Phosphosites | 5588 | 7988 | 6982 | 4936 | 2656 | 3643 | 1242 | 2627 | 1879 | 829 | 1582 | 13506 |
a Unique based on amino acid sequence, does not include phosphopeptides.
b Unique based on amino acid sequence and number of phosphosites.
The Medicago Proteome is Largely Unaltered During the First Hour of Response to NF
Quantitative proteome analysis revealed few changes in protein expression in response to NF (0.16%). Specifically, only 13 proteins are significantly altered (p < 0.05, Student's t test, assuming equal variances) more than 1.35-fold in wild-type plants (supplemental Table S2). In contrast, transcriptomic data revealed 136 genes significantly altered in wild-type plants (see below). The disparity between proteomic and transcriptomic changes is expected, as changes in RNA expression require time to affect protein level changes. The largely unchanged proteome suggests rapid cellular responses (i.e. calcium spiking, membrane depolarization, etc.) result from NF-induced post-translational modifications (i.e. phosphorylation), which are independent of protein translations.
NF Induce Rapid Changes in Medicago Phosphoproteome
The phosphoproteomic response of Medicago plants to NF treatment was quantified for 13,185 unique phosphoisoforms. The 454 phosphoisoforms exhibited a significant change in phosphorylation upon NF treatment (p < 0.05, Student's t test, assuming equal variances; supplemental Table S3). Considering only phosphorylation changes greater than 1.35-fold results in 98 candidate phosphoisoforms (supplemental Table S3), several of these targets are described in detail in the discussion. Proteins showing significant differential phosphorylation one hour after NF treatment are grouped into functional categories in supplemental Fig. S6.
To elucidate the phosphorylation dynamics within the first hour of NF treatment we designed an eight-plex time course experiment, including three biological replicates for the 0 and 60 min time points and one biological replicate for the 10 and 30 min time points. This experiment, listed as experiment 9 in Table I, produced 1468 localized and quantified phosphoisoforms (supplemental Table S1). The log2 ratio of each time point compared with the 0 min time point was calculated (using the average for both the 0 and 60 min time points) and grouped into 10 nodes using K-means clustering (Fig. 2A). Functional analysis determined node 1 was enriched for the gene ontology (GO) term calmodulin binding (Fig. 2B), whereas node 2 was enriched for the GO terms phosphorylation, water transport, and integral to membrane (Fig. 2C). No significant enrichment of GO terms was determined for the remaining nodes of the time course (supplemental Fig. S7). Calmodulin binding proteins are an interesting group of candidates as calcium signatures both at the plasma membrane and nuclear levels play an essential role in NF signaling. To further investigate the phosphorylation dynamics within the time course, the number of phosphorylation events altered greater than 1.5 fold was counted for each time point (Fig. 2D). The greatest number of 1.5-fold changes occurs at 30 min, corresponding to the amount of time before calcium spiking is initiated within the cell.
Fig. 2.
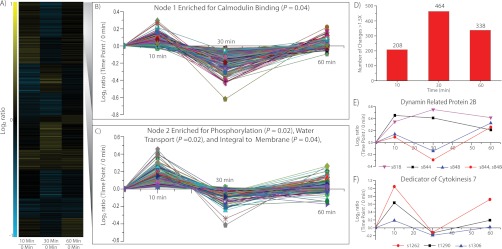
Time course analysis of NF-induced phosphorylation dynamics. A, Heat map representation of phosphorylation changes at 10, 30, and 60 min post NF treatment. Quantitative information for 1468 localized phosphoisoforms was grouped using hierarchical clustering and then pooled into 10 expression patterns using K-means clustering. B, Node 1 of time course expression analysis. Functional analysis of the phosphoisoforms comprising node 1 identified the gene ontology (GO) term calmodulin binding as significantly enriched (p < 0.05, Fisher exact test with Benjimini-Hochberg correction for multiple comparisons). C, Node 2 of time course expression analysis. Functional analysis of the phosphoisoforms comprising node 2 identified the GO terms phosphorylation, water transport, and integral to membrane as significantly enriched (p < 0.05, Fisher exact test with Benjimini-Hochberg correction for multiple comparisons). D, Global phosphorylation changes for each time point. The number of 1.5-fold phosphorylation changes are plotted for each time point. Phosphorylation changes occur rapidly, 10 min post NF treatment and continue to intensify until 30 min. At 60 min, the number of changes is still large with 338 phosphoisoforms displaying a 1.5-fold change. E, Dynamin 2B phosphorylation dynamics within the first hour post NF treatment. Time course phosphorylation dynamics of four dynamin 2B (DRP2B) phosphoisoforms (s818, s844, s848, and s844/s848). The s848 and s844/s848 isoforms are changing in a similar manner suggesting s848 is being regulated, whereas the decrease in s844 from 30 to 60 min coincides with an increase in s844/s848 suggesting some s844 is being converted into s844/s848. F, DOCK7 phosphoryation dynamics within the first hour post NF treatment. Time course dynamics of three DOCK7 phosphosites (t1290, s1262, and s1306) demonstrates time dependent phosphorylation regulation. All three phosphoisoforms display similar time course changes, suggesting global protein phosphorylation regulation in response to NF.
Two proteins highlighted the importance of time course data. Dynamin-related protein 2B (DRP2B, Medtr4g030140) belongs to a GTPase family of proteins responsible for membrane dynamics, vesicle trafficking, and is likely a key player in the NF signaling cascade (Fig. 2E). Two separate phosphosites (s818 and s848) demonstrate a significant increase (p < 0.05) in phosphorylation in the time course experiment (Fig. 2E). Two additional DRP2B phosphoisoforms, s844 and the doubly phosphorylated s844/s848, are also plotted in Fig. 2E. For DRP2B, s848 and the doubly phosphorylated s844/s848 share a distinct expression pattern, suggesting the removal and addition of s848 from these isoforms. Although the s844 and s818 phosphoisoform exhibit a rapid and sustained increase in phosphorylation over the course of the experiment, further implicating DRP2B in the NF signaling cascade. Similarly, time course data reveals three phosphosites at s1263, t1290, and s1306 for Dedicator of Cytokinesis 7 (DOCK7, Medtr8g056900). The s1306 phosphosite belongs to the group of 98 proteins significantly altered in the overall analysis, vide supra, whereas s1263 and t1290 fall short of statistical significance, but follow a similar phosphorylation pattern over the time course (Fig. 2F). These data provide evidence of global phosphorylation changes on DOCK7 in response to NF treatment. Although it escapes the scope of this article, the cases of DRP2B and DOCK7 demonstrate the power of analyzing time course data for each individual protein.
Symbiosis-defective Mutants Display Altered Phosphoproteome Upon NF Treatment
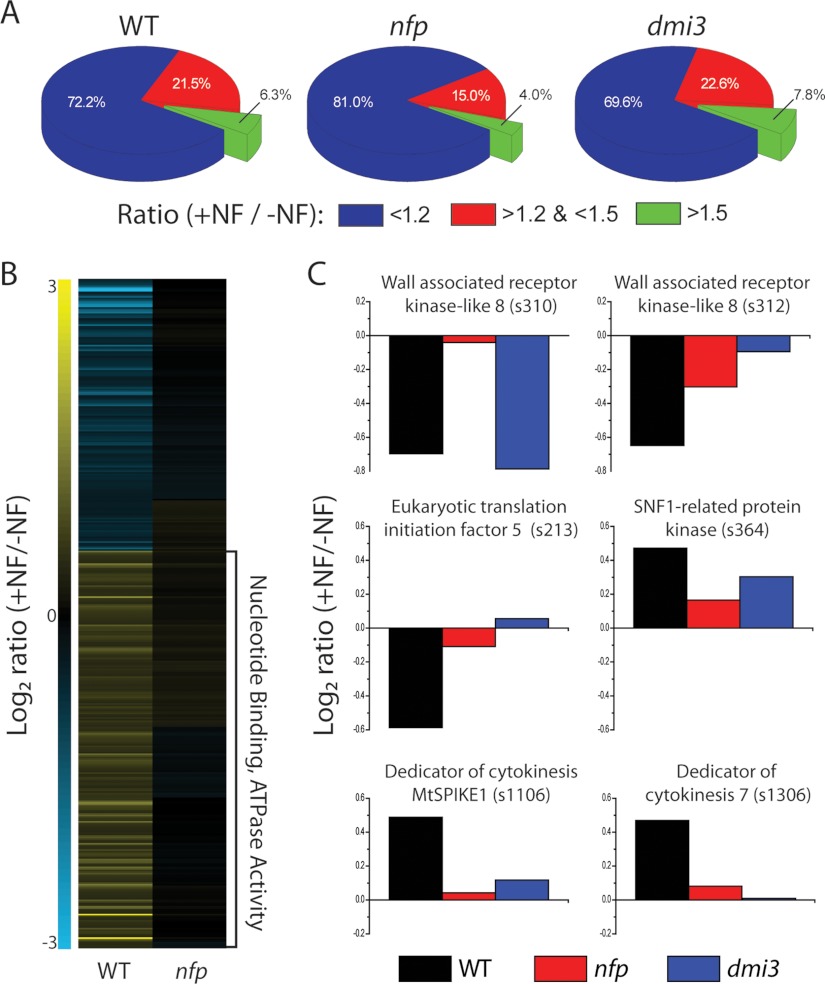
To investigate the dependence of NF-induced phosphorylation events on NFP- and DMI3-induced signaling, the phosphoproteomes of nfp and dmi3 mutants were analyzed and global phosphorylation dynamics compared with the wild-type response. 13,993 unique localized phosphoisoforms were detected in at least one genotype, with individual genotypes containing 13,185, 10,198, and 10,062 isoforms for wild-type, nfp and dmi3 plants, respectively. The number of biological replicates across genotypes was not equal, making a direct comparison of significant changes difficult. Therefore, we compared the number of phosphorylation events changing greater than 1.5-fold post-NF treatment. Wild-type plants readily respond NF application, as the phosphorylation level of 6.3% of isoforms changed more than 1.5-fold. nfp plants demonstrate a reduced phosphorylation response with only 4.0% of isoforms altered in the same manner (Fig. 3A). In addition, nfp mutants contain a greater percentage of isoforms altered less than 1.2-fold (81% for nfp, 72% for wild-type), implicating NFP as a major contributor to the initiation of NF signaling. However, the presence of differential phosphorylation exhibited in nfp provides clear evidence for the involvement of additional NF receptors, prompting our transcriptomic approach discussed below. Interestingly, dmi3 appears hypersensitive to NF, as the phosphorylation status of 7.8% of isoforms was altered more than 1.5-fold after treatment. The increased phosphorylation response in dmi3 confirms its role in a negative feedback mechanism (18). In contrast, NF-induced transcriptional changes in dmi3 showed only one regulated gene, confirming that DMI3 is a key regulator of NF-induced transcription. Altogether, our data suggest that this negative feedback loop probably involves post-translational, but not transcriptional, mechanisms.
Fig. 3.
Global view of changes in the proteome and phosphoproteome from all the experiments combined. A, Pie charts displaying the distribution of phosphorylation changes for wild-type (WT), nfp and dmi3. Wild-type plants readily response to NF as 6.3% of phosphoisoform are altered more than 1.5-fold. Nfp displayed a lower response, 4.0%, but the presence of these changes provides evidence of a separate NF receptor sensing NF. Dmi3 more NF responses than wild-type plants, as the phosphorylation state of 7.8% of phosphoisoforms was altered more than 1.5-fold. B, Heatmap of phosphoisoforms altered in wild-type and not in nfp. Phophoisofroms exhibiting a fold change more than 1.35 in wild-type plants and less then 1.25 in nfp were grouped via hierarchical clustering. Functional analysis revealed phosphoisoforms, which demonstrate an up-regulation in phosphorylation upon NF treatment are enriched for the terms nucleotide binding and ATPase activity. C, Representative proteins significantly altered in wild-type and not in nfp. Six pohsphoisoforms that were significantly (p < 0.05, Student's t test, assuming equal variances) altered more than 1.35-fold in wild-type plants and less than 1.25-fold in nfp demonstrate the use of comparing wild-type and mutant measurements. These examples contain phosphoisoforms, which display both nfp dependent regulation and dmi3 dependent regulation.
To analyze phosphorylation changes directly related to NF signal transduction, we focused on phosphoisoforms that displayed a fold change greater than 1.35-fold in wild-type plants and less than 1.25-fold in nfp, indicating that these targets are downstream of the NFP receptor in the signaling cascade (Fig. 3B). Functional analysis of phosphorylation responses to NF revealed that phosphoisoforms in wild-type plants exhibiting increased phosphorylation were enriched for the GO terms nucleotide binding, ATPase activity and nucleotide triphosphate activity, whereas those phosphorylation changes that exhibited down-regulation in wild-type plants revealed no significant enrichment (Fig. 3B). To identify specific proteins involved in NF signaling, 64 phosphoisoforms significantly altered (p < 0.05) more than 1.35-fold in wild-type plants and less than 1.25-fold in nfp were analyzed. From these 64, 24 displayed similar fold changes in both wild-type and dmi3 plants, indicating that these are most likely affected before DMI3 activation. The other 40 isoforms were either not detected or exhibited expression in the dmi3 mutant different from wild-type plants, suggesting that these NF-induced phosphorylation changes are dependent on DMI3 activity. The phosphorylation response to NF for all three genotypes for six of the 64 candidates is displayed in Fig. 3C. Phosphorylation sites in two different amino acids within the same tryptic peptide, wall-associated receptor kinase-like 8 (WAKL8, Medtr5g085790.1), demonstrates two separate patterns of phosphorylation dynamics between mutants (Fig. 3C). Phosphorylation of s310 in WAKL8 is down-regulated in both wild-type and dmi3 plants upon NF treatment, whereas relatively unaltered in nfp, suggesting phosphorylation of this site is dependent on NFP, but not DMI3. s312 in WAKL8 displays increased phosphorylation in wild-type plants when compared with nfp and dmi3, suggesting this phosphorylation is DMI3-dependent. In addition, in both nfp and dmi3 mutants, the s213 phosphoisoform of eIF5, and two phosphoisoforms of DOCK-family proteins (MtSPIKE1 s1106 and DOCK7 s1306) display reduced phosphorylation levels upon NF treatment and in comparison with wild-type plants, suggesting regulation of these phosphosites downstream of DMI3. Although the s364 phosphoisoform of SNF1-related protein kinase is down-regulated upon NF treatment in both nfp and dmi3, it is reduced less in dmi3, suggesting NFP-dependent regulation.
NF Induce Rapid Changes in the Medicago Transcriptome
To monitor changes in gene expression one hour post-NF treatment, next generation RNA sequencing was performed in wild-type, nfp and dmi3. A total of 489.8 million 100 bp single-end reads (49 gigabases) were generated from the mRNA libraries. Individual sample sets ranged from 20.4 to 35.0 million reads (mean of 27.2 ± 3.8 M reads). Of these, an average of 76.3% ±3.5% mapped to a unique location on the genome—69.1% ±3.3% to annotated exons and 7.2% ±0.3% to nonexonic locations. The remaining reads mapped to multiple locations in the genome (9.1% ±3.1%) or did not map at all (14.6% ±0.7%). The variation in multireads was due primarily to per-sample variation in rRNA content, which ranged from 0.5 to 10.5% of total reads. A total of 25,237 genes from Mt3.5v4 averaged 10 or more mapped reads per kb transcript over all normalized samples as evidence of transcription, with an overall dynamic range of roughly 104 across this group of genes. A summary of RNA sequence data analyses mapped to the most recent Medicago genome assembly (Mt3.5 V4) is presented in supplemental Tables S4 and S5 and supplemental Fig. S8.
In wild-type plants, ∼136 genes were differentially regulated in response to NF (pooled dispersion, 5% FDR). Of these, 116 genes were significantly up-regulated and 20 were down-regulated. The major groups of genes differentially regulated in wild-type plants encode transcription factors, protein kinases, cell wall-modifying enzymes, defense-related proteins, hormone signaling proteins, transporters, and flavonoid biosynthesis enzymes (Fig. 4B; supplemental Table S6). As preliminary validation of our RNA sequencing data, we checked the expression of genes with known roles in NF signaling. NIN (Medtr5g099060) is an early marker for the activation of the NF signaling pathway in Medicago and Lotus, encoding a transcription factor required for both infection thread formation and cortical cell divisions (52, 53). Our data confirmed up-regulation of NIN expression one hour post-NF treatment (supplemental Table S6). Transcriptional regulators from the GRAS family (e.g. NSP1 and NSP2), as well as AP2/ERF-like transcription factors (e.g. ERN1, 2 and 3), also have known roles in early symbiotic signaling (19, 20, 54, 55). Our data confirms prior data that ERN1 (Medtr7g085810) is up-regulated one hour post-NF treatment (supplemental Table S6) (55). Genes encoding AP2/ERF-like transcription factors (Medtr7g085810, contig_82104, contig_52379, contig_16415, contig_71245, and contig_75657) were also up-regulated upon NF treatment (supplemental Table S6). We validated our next generation RNA sequencing approach using quantitative RT-PCR (qRT-PCR) for several additional candidates. Expression levels of genes encoding ethylene-responsive transcription factor 1B (contig_82104), ethylene-responsive transcription factor 1A (Medtr4g100380), tubulin beta chain (Medtr8g107250), nuclear transcription factor Y subunit B-3 (Medtr8g091720), pectinestarase (contig_73581), inorganic phosphate transporter 1, MtPT1 (Medtr1g043220), and TIR-NBS disease resistance protein (contig_72999) displayed the direction of changes (up- or down-regulated) similar to RNA sequencing measurements (supplemental Fig. S9A).
Fig. 4.
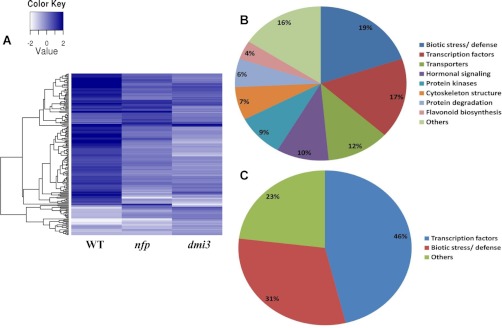
NF-induced transcriptional dynamics within 1 hour after NF-treatment in Medicago. A, Heat map of differentially regulated candidates in wild-type (WT) plants, nfp and dmi3 mutants identified in the transcriptomic data. B, Functional group of genes, which are differentially regulated in the roots of wild-type plants within 1 hour of NF-treatment. The major group of genes, which were differentially regulated, comprise biotic stress/defense, transcriptional factors, transporters, hormonal signaling, protein kinases etc. C, Functional group of genes that are differentially regulated in the roots of nfp mutants within one hour of NF-treatment. The two major groups of genes that were differentially regulated in nfp mutant include transcription factors and genes related to biotic stress/defense response.
NF-induced Transcriptional Changes in nfp Indicate the Existence of Cryptic NF Receptors
To our knowledge, no molecular or cellular response to NF has been observed in the nfp mutant and NFP was believed to be absolutely necessary for all the responses to NF in Medicago. However, our quantitative phosphoproteomics clearly revealed some responses in the nfp mutant, which prompted us to test this result by RNA sequencing. NF-induced transcriptional changes observed in wild-type plants were largely reduced but not abolished in the nfp mutant (Fig. 4A). Although 136 genes were differentially regulated in wild-type, 31 genes were differentially regulated in nfp, of which 20 were up-regulated. The two major groups of differentially-regulated genes in nfp include transcription factors and defense-related genes (Fig. 4C). Among the transcription factors, ethylene-responsive transcription factors (contig_61090, contig_82104, Medtr4g100380 and Medtr1g040430), bZIP transcription factor (AC146721_1015) and bHLH transcription factor (contig_14899) were significantly up-regulated. Defense-related genes coding for TIR-NBS disease resistance protein (contig_72999), disease resistance protein RPS4 (contig_71516), and glutaredoxin (contig_239949) were also up-regulated, whereas a gene encoding a Kunitz-type trypsin inhibitor alpha chain probably involved in defense (Medtr6g065460) was down-regulated. Because these NF-induced transcriptional changes in nfp were unexpected, we validated the expression of several candidate genes by qRT-PCR. Our results for Ctg_82104, Ctg_72999, Medtr4g100380, and Medtr4g112440 confirm regulation in wild-type and nfp observed by RNA sequencing upon NF treatment (supplemental Fig. S9B). These results confirm that NFP is responsible for transducing the majority of the NF signal, but also reveal clearly the existence of other NFP-independent NF receptor complexes.
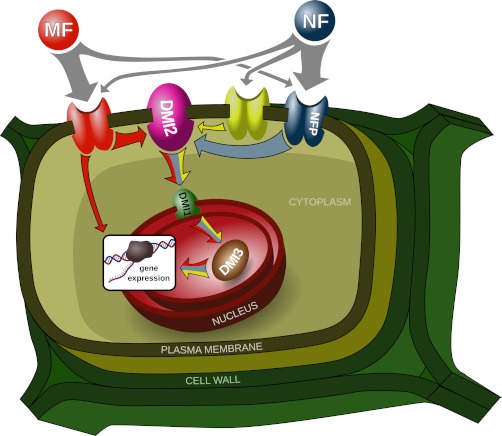
Given the structural similarities between Nod and Myc factors, we speculated that some of NF signal may be transduced by the mycorrhizal receptors. Therefore, we explored the transcriptional changes of genes that were regulated in nfp upon treatment with mycorrhizal signals (GSE). Contig_82104 and Medtr4g100380 were up-regulated in wild-type and nfp plants upon GSE treatment (supplemental Fig. S10A and S10B), indicating NFP-independent gene activation by the mycorrhizal pathway. However, GSE treatment did not result in up-regulation of contig_72999, which was up-regulated in nfp by NF-treatment in our RNA sequencing and qRT-PCR data (supplemental Fig. S9B and S10B). This indicates that some of the NFP-independent signal is not transduced by the Myc factor receptor, suggesting the existence of a third Myc factor- and NFP-independent NF receptor (Fig. 5).
Fig. 5.
Proposed model depicting putative LCO-receptor kinases on the plasma membrane and the cross-talks between different symbiotic signaling pathways in Medicago. NF are perceived with high stringency by NF receptor (NFP) and its putative heteromultimeric partner, an NFR1 homolog. This accounts for majority of signal transduction events. However, NF-induced, but NFP-independent signaling responses suggest the existence of other receptor complexes. Differential transcriptional regulation of these candidate genes by both NF and GSE in NFP-independent manner suggests the existence of Myc factor receptor, which is capable of perceiving NF. Interestingly, NFP-independent, but NF-specific transcriptional responses suggest the existence of NF-specific “cryptic” signaling receptor in Medicago. Although a majority of these transcriptional responses are dependent on DMI3, GSE-induced DMI3-independent transcriptional regulation of candidate genes, suggest the existence of parallel signaling pathway, which is independent of well established signaling pathway comprising DMI genes.
LYR1, an NFP homolog, up-regulated during AM symbiosis and has been suggested as potential candidate for Myc factor perception (8, 26, 56). We took a candidate reverse genetics approach to explore the possibility that LYR1 could mediate the transcriptional responses in nfp upon NF treatment. Using RNA interference, we silenced LYR1 in wild-type and nfp plants as described previously (42, 57). Transgenic roots containing the RNA interference construct were analyzed for transcriptional changes one hour post-NF treatment by qRT-PCR. Silencing LYR1 in wild-type and nfp plants did not abolish NF-induced up-regulation of contig_82104 transcript (supplemental Figs. S10A, 10B), indicating that differential regulation of this gene in response to NF is not mediated by LYR1. However, a negative result in these experiments may be misleading; expression analyses in a total lyr1 knock-out might provide more conclusive evidence of its potential role in NF- and/or Myc factor-induced transcriptional changes in Medicago.
DMI3 is a Key Regulator of NF-induced Transcriptional Changes in Medicago
Downstream of plasma membrane NF receptors, nuclear events play are essential for the transduction of NF signals. The nuclear envelope is the main calcium store for NF-induced calcium spiking and several key proteins (e.g. DMI1 and MCA8) are localized on nuclear membranes (13). DMI3 decodes calcium spiking signatures through its EF hands and transduces the signal downstream, probably by phosphorylation of specific targets (15, 58). We monitored NF-induced transcriptional changes in the dmi3 mutant. In contrast to the significant transcriptome changes observed in wild-type plants, the dmi3 mutant exhibited almost no change one hour post-NF treatment, confirming its central role in controlling NF-dependent transcriptional events (Fig. 4A; supplemental Table S6). Interestingly, the only genes showing a significant regulation in the dmi3 mutant encode well-characterized inorganic phosphate transporters. Medicago phosphate transporters MtPT1, MtPT2, and MtPT3 share ∼96% sequence identify at the nucleic acid level and their encoded proteins share 98% amino acid sequence identity (59, 60), making them impossible to distinguish by RNA sequencing. Our data indicate a significant down-regulation of these genes. A similar down-regulation of these genes and proteins was observed during AM symbiosis, suggesting that the NF-induced down-regulation may result from AM pathway activation (59, 61, 62). Using qRT-PCR, we confirmed that GSE treatment decreases Medtr1g043220 (MtPT1) expression similar to that observed with NF treatment (supplemental Figs. S9C, S10C). We validated this NF- and GSE-induced down-regulation of Medtr1g043220 in wild-type and nfp as well (supplemental Figs. S9A, 9B, S10A, 10B). Our data indicate that this event is DMI3- and NFP-independent, confirming this regulation as a probable consequence of AM pathway activation.
Data-sharing via the Medicago-Omics Repository (MORE)
The variety of experimental conditions used enables multiple levels of analysis, many of which escape the current scope of this article. To allow the research community to explore data not mentioned here, we created the Medicago-Omics Repository (MORE; http://more.biotech.wisc.edu/), a web-based resource containing transcriptomic, proteomic, and phosphoproteomic data collected during the course of this project. MORE allows users to view quantitative information for transcript, protein and post-translational modifications from multiple experiments simultaneously. Researchers are able to search for genes of interest and determine if gene expression, protein expression or post-translational modification levels are altered because of subcellular location, time, or growth conditions. MORE is also dynamic in that users will be assisted in uploading their data to the existing database. To allow offline analysis, all entries into the database are freely available for download.
DISCUSSION
Combining transcriptomics, proteomics, and phosphoproteomics approaches we have performed a comprehensive systems analysis of NF-induced symbiotic signaling response of legume roots. The collection of phosphoproteomic analyses presented here is a database representing high-throughput and large scale mass spectrometric experiments requiring many hours of instrument time on a high resolution tandem mass spectrometer (ESI-LTQ-orbitrap mass spectrometer). In the past, many proteomic studies in plants have used off-gel mass spectrometry analysis (63–68), and large-scale shotgun (untargeted) proteomic and phosphoproteomic technologies (69–72). Benschop et al. (73) used shotgun mass spectrometry analysis to quantify phosphoproteome changes in response to pathogen elicitors in Arabidopsis thaliana (Arabidopsis), yielding 1172 measurements. In 2010, we described the identification of 3457 phosphopeptides in Medicago roots (37), as well as the quantification of 4675 phosphopeptides in Arabidopsis (74). Prior to the results reported here, the largest phosphoproteomic study in plants identified 6919 phosphopeptides from rice (75). Our current report presents 15,335 unique phosphopeptides and demonstrates the potential value of large scale quantitative phosphoproteomic analysis in plants.
Whereas transcriptomic approaches have been used in the past to identify potential candidates regulating legume-rhizobia symbiosis, most of these studies explored transcriptional regulations occurring during later stages in the legume-rhizobia interaction, i.e. infection, nodule development and symbiotic nitrogen fixation spanning 1–32 days after inoculation with rhizobia (31, 32, 76–78). In contrast, our study sheds light on the early transcriptional dynamics that occur with-in the first hour of NF perception. The genes identified in our study play a role very early in the nodulation symbiotic signaling and may regulate the initial NF-induced cellular responses in host plants. Our current study used the deep coverage of next generation RNA sequencing to identify the Medicago transcripts regulated rapidly in response to NF. In addition, we took advantage of Medicago genetics by using mutants affected in NF perception (nfp) and transduction (dmi3), RNA interference in transgenic roots (LYR1), and additional symbiotic signals (GSE) in order to identify the different pathways regulating transcription in response to NF and their relative importance.
This large data set allowed for the identification of many candidates that may control the early molecular and cellular responses observed in response to NF and, ultimately, the establishment of legume nodulation. Our transcriptomics and phosphoproteomic data suggested the presence of at least two NF receptors independent of NFP as well as molecular markers that allow us to track down the corresponding pathways (Fig. 5). One of them is a receptor that also transduces mycorrhizal signals (GSE) but is probably not LYR1, and the other one is so far completely unknown. Our quantitative data also allowed for estimating the relative contribution of these pathways. NFP-dependent responses to NF represent about 80% of transcriptomic changes observed in wild-type plants, confirming that NFP is still the major signaling receptor in Medicago and that the other receptors play a less important role in NF perception (Fig. 5). Our integrated approach also helped us identify proteins potentially controlling downstream stages of NF signal transduction. Many differentially regulated transcripts, and proteins that are phosphorylated in response to NF belong to families involved in cell signaling, cell cycle, cell morphology, defense reactions, hormone signaling and cell wall remodeling. Many differentially regulated transcripts, and proteins that are phosphorylated in response to NF in the current study belong to families involved in cell signaling, cell cycle, cell morphology, defense reactions, hormone signaling, and cell wall remodeling.
Symbiotic Signaling and Cell Cycle Regulation
Protein Kinases
Receptor-like kinases play a major role in the perception and perpetuation of external environmental signals. Several receptor-like kinases are required for NF perception (NFP and LYK3) and signal transduction (DMI2/NORK). We identified sites differently phosphorylated on several receptor-like kinases (Medtr2g098910, Medtr5g085790, Medtr4g115630, Medtr3g076990, Medtr4g113100, and Medtr5g075630). Medtr2g098910 is, for instance, a close homolog of Arabidopsis ACR4, which is thought to control the initiation of cell divisions in the pericycle, for lateral root development and in the root tip meristem (79). This is a very interesting candidate for the control of NF-induced cell divisions in the cortex and pericycle, which give rise to the nodule primordium. Cell wall-associated receptor kinases (WAKs) belong to another group of plant receptor-like kinases, which play important roles in cell expansion, plant defense against pathogens, and tolerance to abiotic stresses (80, 81). WAKs physically link the plasma membrane to the cell wall matrix through the extracellular domain and mediate cellular events through their cytoplasmic kinase domain, hence acting as one of the most likely candidates participating in cell wall-cytoplasm signaling in plant response to external stimuli. In response to NF, WAK-like 8 (WAKL8, Medtr5g085790) seem to be differentially regulated at a significant level suggesting its potential role in NF signaling in legumes.
As mentioned previously, DMI3 is a calcium-/calmodulin-dependent protein kinase (CCaMK) that probably senses the nuclear calcium spikes through its EF hands and transduces this signal into phosphorylation changes in other nuclear proteins, such as transcription factors (15, 58). DMI3 was found to interact with and phosphorylate a nuclear protein with predicted coiled-coil domains called IPD3/CYCLOPS (16, 82, 83). We found a serine residue (s50) in IPD3 to have its phosphorylation level increased in vivo in the presence of NF, suggesting that this residue might be a direct target of DMI3 phosphorylation. This residue falls within the region of IPD3 identified to be phosphorylated in vitro by DMI3 (82). We had already highlighted this residue on IPD3 in our previous nonquantitative study surveying the Medicago phosphoproteome (37). We also identified a calcium-dependent protein kinase (CDPK, Medtr1g101630) and a calcium binding protein (Medtr8g107110) with sites differentially phosphorylated in response to NF. CDPKs have been identified as playing important roles in Medicago nodulation and root development (84, 85).
Mitogen-activated protein kinases (MAPKs) are involved in many aspects of plant development, in the response of plants to changes in their environment and in particular to pathogens (86, 87). MAPKs have been implicated in ethylene signaling (88) and ethylene is a potent inhibitor of NF signaling and, in particular, of calcium spiking (89, 90). More recently, MAPKs have been shown to be activated in response to exudates (Myc factors) of AM fungi (91). In our data set, MAPK-like proteins (Medtr8g093730 and Medtr5g094390) are significantly regulated in response to NF.
SNF1-related proteins, also called AMP-activated protein kinases (AMPK), are master regulators of energy metabolism and one of them shows a significantly increased level of phosphorylation in response to NF (Medtr6g0129900). These proteins have been shown, in particular, to inhibit the activity of 3-hydroxy-3-methylglutaryl-CoA reductases (HMGRs) by phosphorylation (92–94). An HMGR was found to interact with DMI2/NORK and is required for early NF signaling (57) although the function of this interaction is still unclear. In a separate study with Arabidopsis, we have also recently identified SNF1-related proteins whose phosphorylation levels are altered in response to abscisic acid (ABA) (74). Because ABA is a major regulator of NF signaling, nodule development and nitrogen fixation (95, 96), our observation suggests that this protein may be involved in the regulation of NF signal transduction by ABA. Several other protein kinases have also been found to be differentially phosphorylated in response to NF (Medtr4g128650, Medtr5g088400, and Medtr4g078290) but more targeted work is required to interpret these changes in the context of the NF signaling pathway.
Protein Phosphatases
Dephosphorylation of proteins plays essential roles in plant signaling pathways including ABA and auxin signaling but also in symbiotic signaling (74, 97). Several protein phosphatases 2C (PP2C) isoforms are themselves significantly phosphorylated (Medtr5g080680) or dephosphorylated (Medtr6g086970) after addition of NF. The expression of the dominant negative allele of a PP2C (abi1–1) from Arabidopsis in Medicago roots dominantly suppressed ABA signaling and enhanced nodulation (95). In L. japonicus, the expression of PP2C genes are induced in nodules and may play important roles at early and late stages of nodule development (97). Given the relationships between PP2C and ABA signaling, these PP2C proteins may also be involved in the crosstalk between ABA and NF signaling.
Transcription and Translation Factors
Several transcription factors have been shown to be regulated by NF. As mentioned previously, NSP1 and NSP2 are transcription factors of the GRAS family that are required for NF signaling. They interact together and bind to the promoters of early nodulin genes such as ENOD11, NIN, and ERN1 (19–21). ERN1 itself is an AP2-ERF transcription factor that also binds to a different region of the ENOD11 promoter along with two other AP2-ERF transcription factors, ERN2 and ERN3 (54, 55). Our phosphoproteomic study identified several transcription factors with significant increase (Medtr8g077920, Medtr3g089910, AC233675_22.1, Medtr5g038620, ABN08601.1, Medtr2g086140, Medtr1g023690) or decreased (Medtr2g060650, Medtr7g100790, AC234952_22.1) phosphorylation levels. Several of these transcription factors (Medtr5g038620, Medtr7g100790) contain IQ domains, which are reported to bind calmodulin and are involved in calcium signaling (98). These proteins may thus be activated in response to the calcium influx at the plasma membrane level or by the nuclear calcium spiking. The regulation of Medtr7g100790 by phosphorylation seems relatively complex because the same protein possesses sites more (s301) and less (s411, s413) phosphorylated in response to NF. Another of these transcription factors is a close homolog of Arabidopsis CIP7. This nuclear protein is an interactor of COP1 (Constitutive Photomorphogeneic 1) that possesses transcriptional activation activity without any obvious DNA binding motif and acts as positive regulator of light-regulated genes. CIP7 could provide a molecular basis for the inhibition of nodulation by light (99).
Similarly, after the perception of NF, several transcriptional regulators were differentially regulated at the transcript level in the root tissues. In addition to NIN, ERN1, and other ethylene responsive transcription factors mentioned earlier, genes encoding Myb family transcription factors (Medtr1g087540, contig_103831), bHLH transcription factors (Medtr4g087920, Medtr4g097920, Medtr5g014600), a NAC domain containing transcription factor (Medtr4g035590), and a bZIP transcription factor (Medtr3g117120) show significant differential regulation at the gene expression level. Two other bZIP transcription factors (MtATB2 in Medicago and ASTRAY in Lotus) have been found to play an important role in nodulation but probably at later stages of nodule development and senescence (100, 101).
Proteins involved in translation are also differentially phosphorylated (Medtr7g082940, Medtr6g021800, Medtr3g109550) in response to NF. For instance, Medtr7g082940 is a eukaryotic translation initiation factor 5 (eIF5) and possesses five phosphorylation sites (s207,s213, s431, s434, and s436), distributed in two different tryptic peptides, that are consistently less phosphorylated in presence of NF. In yeast, mammalian, and plant cells, the casein kinase 2 (CK2) controls cell cycle progression. This protein was shown to phosphorylate in vitro wheat and Arabidopsis eIF2a, eIF2b, eIF5, and wheat eIF3c (102, 103). Phosphosites s207, s431, s434, and s436 on Medicago eIF5 (Medtr7g082940) are conserved in Arabidopsis and wheat and these four sites correspond to those phosphorylated by CK2 in vitro (103). These phosphorylation sites on eIF5 are highly conserved from monocots to eudicots but are absent in yeast and mammalian proteins suggesting that they could be a plant-specific innovation. Interestingly, the fifth Medicago site (s213) is also conserved in Arabidopsis and wheat but this site was not found in the in vitro studies. All of this suggests that the reduction of phosphorylation on eIF5 that we observed might be related to the regulation of cell cycle. NF have been shown to regulate the cell cycle for the formation of both pre-infection threads and nodule primordium (104, 105). Our phosphoproteomic data suggest that eIF5 could be involved in these processes. Other phosphorylation sites found on eIF2ab and eIF3c in vitro by (103) were also quantified in our study but did not change significantly in response to NF treatment.
Other Protein Post-translational Modifications
Several proteins involved in protein ubiquitination have been identified recently in investigations of early and late stages of legume nodulation (106). Our study identified a RING finger RHF2A-like E3 ubiquitin-protein ligase (Medtr2g087820) with significantly decreased levels of phosphorylation. We also identified a RING finger SIZ1-like E3 SUMO-protein ligase with decreased levels of phosphorylation on a specific site. Nothing is known yet about the role of sumoylation in symbiotic signaling. These two types of protein modifications may be regulating the degradation, stability or sub-cellular localization of proteins involved in early responses to NF (107). However, our observation of little or no changes in protein levels of ca. 8000 proteins within the first hour suggests that either the ubiquitin/sumoyl-mediated protease systems do not become significantly stimulated until later in the response, or that the affected proteins are not within the 8000 proteins we measured (e.g. lower abundance transcription factors). At the transcript level, several members involved in protein degradation, such as RING finger family protein (contig_164276), F-box family protein (contig_48455 and contig_13936), U-box domain containing protein (Medtr5g083030) and several proteinases were also up-regulated by NF perception. Hence, these differentially regulated members involved in proteolytic activities may be involved in early NF signaling.
Cell Growth and Root Hair Deformations
Plant root hairs exhibit a characteristic polarized cell growth similar to the one observed in pollen tubes and fungal hyphae (108). Upon perception of NF, legume root hairs undergo a rapid calcium influx, an alkalinization of the cytoplasm, and a transient disorganization of the cytoskeleton followed by a growth re-orientation leading to characteristic root hair deformations (4, 109–111). When NF are applied locally on the root hair surface or by rhizobia, these deformations lead to a root hair curl that entraps the rhizobia (7). All of these mechanisms require a very dynamic reorganization of the cytoskeleton and vesicle trafficking. Many proteins involved in these processes displayed differential phosphorylation upon NF treatment. As mentioned previously, Dynamin 2B (DRP2B, Medtr4g030140) was found to present interesting changes in phosphorylation patterns (Fig. 2E) and probably plays a role in these vesicle trafficking processes.
In addition, a P-type plasma membrane proton pump ATPase (Medtr2g036650) shows a significant increase in phosphorylation level suggesting that this protein could be responsible for the observed alkalinization of the cytoplasm (109), especially because this was the only one of 12 members of the proton pump gene family whose protein was affected in the first hour. Another protein whose phosphorylation status was changed in the first hour (Medtr1g044570.1) is a close homolog or Arabidopsis CAX1-interacting protein 4. This nuclear protein was shown to regulate the activity of the CAX1 H+/Ca2+ antiporter (112). These proteins could be involved in the proton and calcium fluxes observed in the early response to NF (4, 110). Phosphorylation sites on a phosphoinositide phospholipase C (PLC) (Medtr3g070720) are significantly increased in presence of NF. PLCs catalyze the hydrolysis of phosphatidylinositol 4,5-bisphosphate in a calcium-dependent manner. The role of PLC activity was clearly shown in early responses to NF (113). For instance, PLC antagonists, neomycin and the aminosteroid PLC inhibitor U73122 inhibited NF-induced ENOD12 expression in Medicago. Medtr3g070720 seems therefore an excellent candidate to be involved in this NF-dependent pathway.
Small G proteins (guanine nucleotide-binding proteins) are known to be involved in the regulation of the phosphoinositide pathway, vesicle trafficking, root hair growth, and the early steps of legume nodulation (42, 108, 114, 115). The role of heterotrimeric G proteins in early symbiotic signaling was also proposed. In mammalian systems, Mastoparan is a well-characterized agonist of heterotrimeric G proteins. In Medicago, Mastoparan and its synthetic analog Mas7 was shown to induce calcium spiking and early nodulin expression (116). Even if the targets of this drug in plants remain unclear, a role for heterotrimeric G proteins in regulating calcium spiking remain a possibility. We identified an increased phosphorylation on a site of the alpha-2 subunit of heterotrimeric G protein (Medtr5g018510). The activity of these small and heteromeric G proteins is regulated by guanine nucleotide exchange factors (GEFs), GTPase-Activating Proteins (GAPs), guanine nucleotide dissociation inhibitors (GDIs), and other interacting proteins. We identified differential phosphorylation on many GEF and GAP proteins such as the RopGEF SPIKE1, a homolog of BIG1/2 (Brefeldin A-inhibited GEF) (Medtr4g124430), a GEF homologous to DOCK7 (Dedicator of cytokinesis 7) (Medtr8g056900) as mentioned previously (Fig. 2F), a putative RabGEF (Medtr3g073360), a putative RasGAP (Medtr2g037930), and a RasGAP-binding protein (Medtr4g083150). A protein potentially interacting with these small GTPases (Medtr5g007350) displays a decrease of phosphorylation in presence of NF.
Proteins probably involved in F-actin polymerization and belonging to the SCAR/WAVE (suppressor of cyclic AMP receptor/Wiskott-Aldrich syndrome Verprolin-homologous protein) complex were also regulated by phosphorylation (Medtr8g086300 and Medtr7g071440). This complex is known to regulate polar growth but also infection thread formation during later stages of this symbiosis (117, 118). Phosphorylation of proteins from this complex as well as interactions with small GTPases and their regulators regulates actin polymerization (119). A protein from the gelsolin superfamily and similar to villins (Medtr7g091460) displays a significant increase in phosphorylation level. Villins have been reported to regulate the organization of the actin cytoskeleton, cytoplasmic streaming, and bundling of actin filaments in root hairs (120). Activation of heterotrimeric and small G proteins dissociate these proteins from the barbed end of actin filaments (121). Medtr7g091460 may therefore be a key player connecting G protein activity to cytoskeleton reorganization in response to NF.
Proteins involved in vesicle trafficking were found to have their phosphorylation levels change, such as a vesicle-fusing ATPase (AC233659_1.1), a SNARE-interacting protein KEULE (Medtr4g102120), an AP-3 complex subunit delta (Medtr8g104380), a neurobeachin-like protein (Medtr7g075660), synaptogamins (Medtr8g035590 and Medtr1g094810), and a component of the exocyst complex (Medtr4g103540). Synaptogamins have been reported to be phosphorylated by the cell cycle regulator casein kinase 2 (CK2) like eIF5 (122). All of these observations support the presence of an extensive remodeling of cell cytoskeleton and vesicle trafficking in response to NF and mediated by phosphorylation.
Defense Reactions and Plant Hormones
The establishment of symbiotic associations requires a tight control of plant defense reactions. Similarities exist between responses to NF and responses to pathogen elicitors including the production of reactive oxygen species or proteins closely related to defense proteins (123, 124). However, the kinetic and the intensity of these responses to NF are different from those observed in response to pathogens (125). Two lipoxygenases (LOX, Medtr8g018520, and Medtr2g099570) and a putative Kunitz-type trypsin inhibitor (Medtr6g059730) display decreased phosphorylation levels in response to NF. Using antibodies, LOX have been detected in the nodule cortex and in the cytosol of uninfected nodule cells of the central tissue, but were absent in infected nodule cells and in vascular tissues (126). These proteins could be an indication of an early control of defense reactions mediated by NF. These lipoxygenases could also be related to jasmonic acid (JA) synthesis. JA is a potent inhibitor of responses to NF including calcium spiking and nodulin gene expression (127) and its production might be decreased to ensure the transduction of NF signals. In general, however, little is known about the role of these molecules during early NF signaling.
Similarly, stress and defense-related genes were also differentially regulated, at the transcript level in the first hour of NF-perception. In line with our observations, (28) reported the differential regulation of stress/defense-related genes 1 hour post inoculation with rhizobia. Such defense/disease-responses may be mediated not only by NF released by the rhizobia, but also by other bacterial signals. It is interesting to observe such defense-related responses when purified NF were applied at 10−8 m, a concentration that is known to activate symbiotic signaling but not an obvious defense reaction in legumes. Pathogen elicitors-induced defense responses and NF-induced symbiotic responses share certain common features, such as production of reactive oxygen species, nitric oxide production, and associated activation of redox balance machinery (125). In our transcriptomic study, a gene encoding a peroxidase (Medtr4g133800) is induced by NF but is distinct from the well-characterized Rhizobium meliloti-induced peroxidase (RIP; (124)). Among the defense-related genes, the up-regulation in the expression levels of salicylic acid carboxyl methyltransferase (SAMT, contig_54253 and contig_20507) are striking (16.61 and 14.41 fold increase respectively). SAMT catalyzes the formation of methylsalicylate, a major defense signal in plants. Similarly, genes encoding a TIR-NBS disease resistance-like protein (Medtr5g037700), pathogenesis-related protein 1 (PR1) (Medtr4g128750) and a disease resistance response protein (Medtr7g021300) were induced rapidly after NF perception. Induction of MtN1 (a homolog of cystein-rich pathogen inducible protein in pea) and PR10 (MtN13) have already been reported during early infection and root nodule development in Medicago (28, 123) but these are later stages of legume nodulation. Our data indicate that application of NF can trigger very rapid defense-like responses in the roots. These results highlight, once again, the similarities between responses to pathogens and symbionts (34).
Another major group of genes that are differentially regulated by NF-treatment include genes that are involved in hormone biosynthesis or hormonal responses. Auxin transport and auxin/cytokinin ratio play a major role in NF signaling and nodule formation (128). A gene encoding an auxin responsive SAUR protein (Medtr4g124700) is significantly up-regulated by NF. Similarly, genes encoding a cytokinin receptor histidine kinase (Medtr1g013360) and cytokinin-O-glucosyltransferases (Medtr7g070740 and Medtr5g072860) were also up-regulated by NF, confirming the involvement of cytokinin perception and transport during nodule organogenesis. In Medicago, RNA interference (RNAi) based-silencing of another cytokinin receptor MtCRE1 resulted in a strong reduction in rhizobia infection and nodule primordial formation (129). As mentioned previously, ethylene is a well-known negative regulator of symbiotic signaling and in particular, of calcium spiking (90). Medicago ethylene-insensitive skl mutant displays a 10-fold increase in nodule number (89). Several genes encoding 1-aminocyclopropane-1-carboxylate oxidases (contig_54169, contig_90027, contig_62339, and contig_74946), which catalyze the final step of ethylene biosynthesis, were differentially regulated. Similarly, the role of several ethylene responsive transcription factors (ERN1, 2, 3 and EFD) in early NF signaling is also well-established and in our data ERN1 and several other ethylene responsive transcription factors were significantly up-regulated. Altogether our data confirm that the regulation of ethylene signaling is critical during the early hours of NF signaling.
Cell Wall Remodeling
Cell wall degrading enzymes secreted by pathogens often act as pathogenesis factors in disease establishment. A similar strategy is employed, in the legume-rhizobia symbiosis, to facilitate bacterial entry. Cell wall degrading enzymes produced by the rhizobia play crucial roles in facilitating the entry of rhizobia through root hairs but also possibly infection thread progression and the release of bacteria into symbiosomes (130). In contrary to plant defense reactions, the plant itself loosens its cell wall in preparation for bacterial entry (131). Application of NF to legume root hairs induce a temporary swelling and it was shown that, during that phase, the cell wall exhibits a mottled aspect similar to the one observed in epidermal cells during root hair bulging (6). This indicates a transient relaxation of the cell wall induced by NF, even in the absence of rhizobia. In Medicago, expression of early nodulins, ENOD11 and ENOD12 is induced within 3–6 h following rhizobia inoculation (35). These genes are among the best characterized markers for the activation of the NF signaling pathway and were used as controls in our current study. ENOD11 and ENOD12 encode hydroxyproline-rich glycoproteins (HyPRPs) with relatively few tyrosine residues. The expression of ENOD11 and ENOD12 probably results in enhanced cell wall plasticity or in components of the infection thread matrix (35). Recently, a pectate lyase was shown to play a major role in root infection by the rhizobia. L. japonicus Nodulation Pectate Lyase (LjNPL) is induced in roots and root hairs by NF via the NF signaling pathway and the NIN transcription factor. In our study, a close homolog of MtNPL (Medtr3g086310) was up-regulated within 1 hour of NF perception. Other cell wall degrading enzymes such as polygalacturonases (contig_67431 and contig_83719) and pectinestarases (Medtr3g033690, contig_73581, Medtr3g010770, contig_51305) were also up-regulated. Hence, very rapidly and even before contact, legume root cells are paving the way for the accommodation of their symbiotic partner by remodeling the cell wall barrier. In conclusion, the combination of large-scale analyses of transcriptional, translational, and post-translational events has enabled a multifaceted examination of cellular responses to NF within one hour of treatment.
In conclusion, the combination of large-scale analyses of transcriptional, translational, and post-translational events has enabled a multifaceted examination of cellular responses to NF within 1 hour of treatment. Our transcriptomics data demonstrate that Medicago shows a limited but significant rapid response in changes of gene expression within one hour after NF-perception, and that symbiotic deficient mutants are lacking much of the NF-induced transcriptional dynamics, albeit not completely. The NF-induced transcriptional responses in nfp, combined with supporting evidence using Myc factor signaling perturbation suggest the presence of additional signaling receptors in Medicago other than NFP. Large-scale quantitative proteomic analyses indicate that protein levels remain relatively unaltered after NF treatment, but protein phosphorylation is actively regulated in all three genotypes within the first hour of NF-perception, with many changes in phosphorylation events occurring as early as 10 min after NF treatment. This integration of multiple-“omic” approaches has thus yielded an extensive list of candidate genes implicated in the NF signaling cascade. Few of these targets have documented roles in NF signaling pathway, whereas many novel candidates identified in our current study warrant targeted proteomic and biochemical validation. To enable the scientific community to explore further the data reported here, we have created the Medicago-Omics Repository (MORE; http://more.biotech.wisc.edu/), an online resource to obtain quantitative transcriptomic, proteomic, and phosphoproteomic data. To build a comprehensive resource, we invite researchers to upload quantitative and nonquantitative data relating to Medicago transcriptomics, proteomics, and phosphoproteomics. Hopefully, MORE will act as a central resource enabling researchers to query large scale data sets, facilitating future systems level studies in Medicago and other legumes.
Supplementary Material
Acknowledgments
We thank Thomas W. Bryan for technical support and Kari L. Forshey for critical reading of the manuscript.
Footnotes
* This work was supported by a grant from the National Science Foundation (NSF#0701846) to M.R.S., J.J.C., and J.M.A. C.M.R. was funded by an NSF Graduate Research Fellowship and NIH Traineeship (T32GM008505).
 This article contains supplemental Discussion, Figs. S1 to S11, and Tables S1 to S7.
This article contains supplemental Discussion, Figs. S1 to S11, and Tables S1 to S7.
1 The abbreviations used are:
- NF
- nod factors
- LCO
- lipochitooligosaccharides
- NFP
- nod factor perception
- DMI
- does not make infection
- AM
- abuscular mycorrhization
- FDR
- false discovery rate
- SCX
- strong cation exchange
- ETD
- electron transfer dissociation
- HCD
- high energy collisional activated dissociation
- GO
- gene ontology
- GSE
- germinating spore exudates.
REFERENCES
- 1. Hartwig U. A., Maxwell C. A., Joseph C. M., Phillips D. A. (1990) Chrysoeriol and luteolin released from alfalfa seeds induce nod genes in. Rhizobium meliloti. Plant Physiol. 92, 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dénarié J., Debellé F., Promé J. C. (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65, 503–535 [DOI] [PubMed] [Google Scholar]
- 3. Ehrhardt D. W., Wais R., Long S. R. (1996) Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85, 673–681 [DOI] [PubMed] [Google Scholar]
- 4. Felle H. H., Kondorosi E., Kondorosi A., Schultze M. (1998) The role of ion fluxes in nod factor signalling in Medicago sativa. Plant J. 13, 455–463 [Google Scholar]
- 5. Cárdenas L., Holdaway-Clarke T. L., Sánchez F., Quinto C., Feijó J. A., Kunkel J. G., Hepler P. K. (2000) Ion changes in legume root hairs responding to Nod factors. Plant Physiol. 123, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller D. D., Leferink-ten Klooster H. B., Emons A. M. (2000) Lipochito-oligosaccharide nodulation factors stimulate cytoplasmic polarity with longitudinal endoplasmic reticulum and vesicles at the tip in vetch root hairs. Mol. Plant Microbe Interact. 13, 1385–1390 [DOI] [PubMed] [Google Scholar]
- 7. Esseling J. J., Lhuissier F. G., Emons A. M. (2003) Nod factors-induced root hair curling: Continuous polar growth towards the point of nod factor application. Plant Physiol. 132, 1982–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arrighi J. F., Barre A., Ben Amor B., Bersoult A., Soriano L. C., Mirabella R., de Carvalho-Niebel F., Journet E. P., Ghérardi M., Huguet T., Geurts R., Dénarié J., Rouge P., Gough C. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amor B. B., Shaw S. L., Oldroyd G. E., Maillet F., Penmetsa R. V., Cook D., Long S. R., Dénarié J., Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34, 495–506 [DOI] [PubMed] [Google Scholar]
- 10. Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G. B. (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966 [DOI] [PubMed] [Google Scholar]
- 11. Wais R. J., Galera C., Oldroyd G., Catoira R., Penmetsa R. V., Cook D., Gough C., Dénarié J., Long S. R. (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 97, 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ané J. M., Kiss G. B., Riely B. K., Penmetsa R. V., Oldroyd G. E., Ayax C., Lévy J., Debellé F., Baek J. M., Kaló P., Rosenberg C., Roe B. A., Long S. R., Dénarié J., Cook D. R. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367 [DOI] [PubMed] [Google Scholar]
- 13. Capoen W., Sun J., Wysham D., Otegui M. S., Venkateshwaran M., Hirsch S., Miwa H., Downie J. A., Morris R. J., Ané J. M., Oldroyd G. E. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peiter E., Sun J., Heckmann A. B., Venkateshwaran M., Riely B. K., Otegui M. S., Edwards A., Freshour G., Hahn M. G., Cook D. R., Sanders D., Oldroyd G. E., Downie J. A., Ané J. M. (2007) The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 145, 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lévy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., Journet E. P., Ané J. M., Lauber E., Bisseling T., Dénarié J., Rosenberg C., Debellé F. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364 [DOI] [PubMed] [Google Scholar]
- 16. Messinese E., Mun J. H., Yeun L. H., Jayaraman D., Rouge P., Barre A., Lougnon G., Schornack S., Bono J. J., Cook D. R., Ané J. M. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of. Medicago truncatula. Mol. Plant Microbe Interact. 20, 912–921 [DOI] [PubMed] [Google Scholar]
- 17. Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B. W., Oldroyd G. E. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152 [DOI] [PubMed] [Google Scholar]
- 18. Oldroyd G. E., Mitra R. M., Wais R. J., Long S. R. (2001) Evidence for structurally specific negative feedback in the nod factor signal transduction pathway. Plant J. 28, 191–199 [DOI] [PubMed] [Google Scholar]
- 19. Kaló P., Gleason C., Edwards A., Marsh J., Mitra R. M., Hirsch S., Jakab J., Sims S., Long S. R., Rogers J., Kiss G. B., Downie J. A., Oldroyd G. E. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 20. Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science 308, 1789–1791 [DOI] [PubMed] [Google Scholar]
- 21. Hirsch S., Kim J., Muñoz A., Heckmann A. B., Downie J. A., Oldroyd G. E. D. (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gough C., Cullimore J. (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 24, 867–878 [DOI] [PubMed] [Google Scholar]
- 23. Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., Martinez E. A., Driguez H., Bécard G., Dénarié J. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63 [DOI] [PubMed] [Google Scholar]
- 24. Mukherjee A., Ané J. M. (2011) Germinating spore exudates from arbuscular mycorrhizal fungi: Molecular and developmental responses in plants and their regulation by ethylene. Mol. Plant Microbe Interact. 24, 260–270 [DOI] [PubMed] [Google Scholar]
- 25. Benedito V. A., Torres-Jerez I., Murray J. D., Andriankaja A., Allen S., Kakar K., Wandrey M., Verdier J., Zuber H., Ott T., Moreau S., Niebel A., Frickey T., Weiller G., He J., Dai X., Zhao P. X., Tang Y., Udvardi M. K. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513 [DOI] [PubMed] [Google Scholar]
- 26. Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., Benedito V. A., Mayer K. F., Gouzy J., Schoof H., Van de Peer Y., Proost S., Cook D. R., Meyers B. C., Spannagl M., Cheung F., De Mita S., Krishnakumar V., Gundlach H., Zhou S., Mudge J., Bharti A. K., Murray J. D., Naoumkina M. A., Rosen B., Silverstein K. A., Tang H., Rombauts S., Zhao P. X., Zhou P., Barbe V., Bardou P., Bechner M., Bellec A., Berger A., Berges H., Bidwell S., Bisseling T., Choisne N., Couloux A., Denny R., Deshpande S., Dai X., Doyle J. J., Dudez A. M., Farmer A. D., Fouteau S., Franken C., Gibelin C., Gish J., Goldstein S., González A. J., Green P. J., Hallab A., Hartog M., Hua A., Humphray S. J., Jeong D. H., Jing Y., Jocker A., Kenton S. M., Kim D. J., Klee K., Lai H., Lang C., Lin S., Macmil S. L., Magdelenat G., Matthews L., McCorrison J., Monaghan E. L., Mun J. H., Najar F. Z., Nicholson C., Noirot C., O'Bleness M., Paule C. R., Poulain J., Prion F., Qin B., Qu C., Retzel E. F., Riddle C., Sallet E., Samain S., Samson N., Sanders I., Saurat O., Scarpelli C., Schiex T., Segurens B., Severin A. J., Sherrier D. J., Shi R., Sims S., Singer S. R., Sinharoy S., Sterck L., Viollet A., Wang B. B., Wang K., Wang M., Wang X., Warfsmann J., Weissenbach J., White D. D., White J. D., Wiley G. B., Wincker P., Xing Y., Yang L., Yao Z., Ying F., Zhai J., Zhou L., Zuber A., Dénarié J., Dixon R. A., May G. D., Schwartz D. C., Rogers J., Quetier F., Town C. D., Roe B. A. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brechenmacher L., Lei Z., Libault M., Findley S., Sugawara M., Sadowsky M. J., Sumner L. W., Stacey G. (2010) Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 153, 1808–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lohar D. P., Sharopova N., Endre G., Peñuela S., Samac D., Town C., Silverstein K. A., VandenBosch K. A. (2006) Transcript analysis of early nodulation events in. Medicago truncatula. Plant Physiol. 140, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wan J., Torres M., Ganapathy A., Thelen J., DaGue B. B., Mooney B., Xu D., Stacey G. (2005) Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 18, 458–467 [DOI] [PubMed] [Google Scholar]
- 30. Asamizu E., Nakamura Y., Sato S., Tabata S. (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol. Plant Microbe Interact. 18, 487–498 [DOI] [PubMed] [Google Scholar]
- 31. Høgslund N., Radutoiu S., Krusell L., Voroshilova V., Hannah M. A., Goffard N., Sanchez D. H., Lippold F., Ott T., Sato S., Tabata S., Liboriussen P., Lohmann G. V., Schauser L., Weiller G. F., Udvardi M. K., Stougaard J. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One 4, e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maunoury N., Redondo-Nieto M., Bourcy M., Van de Velde W., Alunni B., Laporte P., Durand P., Agier N., Marisa L., Vaubert D., Delacroix H., Duc G., Ratet P., Aggerbeck L., Kondorosi E., Mergaert P. (2010) Differentiation of symbiotic cells and endosymbionts in medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 5, e9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitra R. M., Long S. R. (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 134, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Serna-Sanz A., Parniske M., Peck S. C. (2011) Phosphoproteome analysis of Lotus japonicus roots reveals shared and distinct components of symbiosis and defense. Mol. Plant Microbe Interact. 24, 932–937 [DOI] [PubMed] [Google Scholar]
- 35. Journet E. P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D. G., Gianinazzi-Pearson V. (2001) Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant-Microbe Interact. 14, 737–748 [DOI] [PubMed] [Google Scholar]
- 36. Catoira R., Galera C., de Billy F., Penmetsa R. V., Journet E. P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12, 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grimsrud P. A., den Os D., Wenger C. D., Swaney D. L., Schwartz D., Sussman M. R., Ané J. M., Coon J. J. (2010) Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 152, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Series B 57, 289–300 [Google Scholar]
- 42. Riely B. K., He H., Venkateshwaran M., Sarma B., Schraiber J., Ané J. M., Cook D. R. (2011) Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 65, 230–243 [DOI] [PubMed] [Google Scholar]
- 43. Thompson A., Scháfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 44. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 45. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 47. Wenger C. D., Lee M. V., Hebert A. S., McAlister G. C., Phanstiel D. H., Westphall M. S., Coon J. J. (2011) Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 8, 933–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wenger C. D., Phanstiel D. H., Lee M. V., Bailey D. J., Coon J. J. (2011) COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 50. Hsieh E. J., Hoopmann M. R., MacLean B., MacCoss M. J. (2010) Comparison of database search strategies for high precursor mass accuracy MS/MS data. J. Proteome Res. 9, 1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phanstiel D. H., Brumbaugh J., Wenger C. D., Tian S., Probasco M. D., Bailey D. J., Swaney D. L., Tervo M. A., Bolin J. M., Ruotti V., Stewart R., Thomson J. A., Coon J. J. (2011) Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods 8, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marsh J. F., Rakocevic A., Mitra R. M., Brocard L., Sun J., Eschstruth A., Long S. R., Schultze M., Ratet P., Oldroyd G. E. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J. A., Kawaguchi M., Kawasaki S. (2006) Positional cloning identifies lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Research 13, 255–265 [DOI] [PubMed] [Google Scholar]
- 54. Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D. G., de Carvalho-Niebel F. (2007) AP2-ERF transcription factors mediate nod factor dependent mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Middleton P. H., Jakab J., Penmetsa R. V., Starker C. G., Doll J., Kaló P., Prabhu R., Marsh J. F., Mitra R. M., Kereszt A., Dudas B., VandenBosch K., Long S. R., Cook D. R., Kiss G. B., Oldroyd G. E. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19, 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gomez S. K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E. B., Udvardi M. K., Harrison M. J. (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biology 9, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kevei Z., Lougnon G., Mergaert P., Horvath G. V., Kereszt A., Jayaraman D., Zaman N., Marcel F., Regulski K., Kiss G. B., Kondorosi A., Endre G., Kondorosi E., Ané J. M. (2007) 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19, 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimoda Y., Han L., Yamazaki T., Suzuki R., Hayashi M., Imaizumi-Anraku H. (2012) Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu H., Trieu A. T., Blaylock L. A., Harrison M. J. (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol. Plant Microbe Interact. 11, 14–22 [DOI] [PubMed] [Google Scholar]
- 60. Liu J., Versaw W. K., Pumplin N., Gomez S. K., Blaylock L. A., Harrison M. J. (2008) Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 283, 24673–24681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chiou T. J., Liu H., Harrison M. J. (2001) The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 25, 281–293 [DOI] [PubMed] [Google Scholar]
- 62. Grunwald U., Guo W., Fischer K., Isayenkov S., Ludwig-Müller J., Hause B., Yan X., Küster H., Franken P. (2009) Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 229, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lim C. W., Park J. Y., Lee S. H., Hwang C. H. (2010) Comparative proteomic analysis of soybean nodulation using a supernodulation mutant, SS2–2. Biosci. Biotechnol. Biochem. 74, 2396–2404 [DOI] [PubMed] [Google Scholar]
- 64. Lee M. V., Topper S. E., Hubler S. L., Hose J., Wenger C. D., Coon J. J., Gasch A. P. (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santelia D., Kötting O., Seung D., Schubert M., Thalmann M., Bischof S., Meekins D. A., Lutz A., Patron N., Gentry M. S., Allain F. H., Zeeman S. C. (2011) The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in arabidopsis. Plant Cell 23, 4096–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P. A., Roschitzki B., Hirsch-Hoffmann M., Hennig L., Kessler F., Gruissem W., Baginsky S. (2011) Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23, 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Elmore J. M., Liu J., Smith B., Phinney B., Coaker G. (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane proteome during arabidopsis immune signaling. Mol. Cell. Proteomics, In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2004) Phosphoproteomics of the arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen Y., Hoehenwarter W., Weckwerth W. (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 63, 1–17 [DOI] [PubMed] [Google Scholar]
- 70. Schütz W., Hausmann N., Krug K., Hampp R., Macek B. (2011) Extending SILAC to proteomics of plant cell lines. Plant Cell 23, 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reiland S., Finazzi G., Endler A., Willig A., Baerenfaller K., Grossmann J., Gerrits B., Rutishauser D., Gruissem W., Rochaix J. D., Baginsky S. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. U.S.A. 108, 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nakagami H., Sugiyama N., Ishihama Y., Shirasu K. (2012) Shotguns in the front line: Phosphoproteomics in plants. Plant Cell Physiol. 53, 118–124 [DOI] [PubMed] [Google Scholar]
- 73. Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 74. Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. El Yahyaoui F., Küster H., Ben Amor B., Hohnjec N., Pühler A., Becker A., Gouzy J., Vernié T., Gough C., Niebel A., Godiard L., Gamas P. (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 136, 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hernández G., Valdés-Lopez O., Ramírez M., Goffard N., Weiller G., Aparicio-Fabre R., Fuentes S. I., Erban A., Kopka J., Udvardi M. K., Vance C. P. (2009) Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151, 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Libault M., Farmer A., Joshi T., Takahashi K., Langley R. J., Franklin L. D., He J., Xu D., May G., Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99 [DOI] [PubMed] [Google Scholar]
- 79. De Smet I., Vassileva V., De Rybel B., Levesque M. P., Grunewald W., Van Damme D., Van Noorden G., Naudts M., Van Isterdael G., De Clercq R., Wang J. Y., Meuli N., Vanneste S., Friml J., Hilson P., Jürgens G., Ingram G. C., Inzé D., Benfey P. N., Beeckman T. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597 [DOI] [PubMed] [Google Scholar]
- 80. He Z. H., Fujiki M., Kohorn B. D. (1996) A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 271, 19789–19793 [DOI] [PubMed] [Google Scholar]
- 81. Li H., Zhou S. Y., Zhao W. S., Su S. C., Peng Y. L. (2009) A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69, 337–346 [DOI] [PubMed] [Google Scholar]
- 82. Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., Markmann K., White C., Schuller B., Sato S., Asamizu E., Tabata S., Murooka Y., Perry J., Wang T., Kawagachi M., Imaizumi-Anraku H., Hayashi M., Parniske M. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105, 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Horváth B., Yeun L. H., Domonkos A., Halász G., Gobbato E., Ayaydin F., Miró K., Hirsch S., Sun J., Tadege M., Ratet P., Mysore K. S., Ané J. M., Oldroyd G. E., Kalo P. (2011) Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24, 1345–1358 [DOI] [PubMed] [Google Scholar]
- 84. Ivashuta S., Liu J., Liu J., Lohar D. P., Haridas S., Bucciarelli B., VandenBosch K. A., Vance C. P., Harrison M. J., Gantt J. S. (2005) RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17, 2911–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gargantini P. R., Gonzalez-Rizzo S., Chinchilla D., Raices M., Giammaria V., Ulloa R. M., Frugier F., Crespi M. D. (2006) A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J. 48, 843–856 [DOI] [PubMed] [Google Scholar]
- 86. Tena G., Asai T., Chiu W. L., Sheen J. (2001) Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400 [DOI] [PubMed] [Google Scholar]
- 87. Wan J., Zhang S., Stacey G. (2004) Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant. Pathol. 5, 125–135 [DOI] [PubMed] [Google Scholar]
- 88. Ouaked F., Rozhon W., Lecourieux D., Hirt H. (2003) A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22, 1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Penmetsa R. V., Cook D. R. (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530 [DOI] [PubMed] [Google Scholar]
- 90. Oldroyd G. E., Engstrom E. M., Long S. R. (2001) Ethylene inhibits the Nod factor signal trasnduction pathway of Medicago truncatula. Plant Cell 13, 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Francia D., Chiltz A., Lo Schiavo F., Pugin A., Bonfante P., Cardinale F. (2011) AM fungal exudates activate MAP kinases in plant cells in dependence from cytosolic Ca2+ increase. Plant Physiol. Biochem. 49, 963–969 [DOI] [PubMed] [Google Scholar]
- 92. Dale S., Arró M., Becerra B., Morrice N. G., Boronat A., Hardie D. G., Ferrer A. (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-coa reductase (isoform Hmgr1) from arabidopsis-thaliana, and its inactivation by phosphorylation at Ser577 by brassica-oleracea 3-hydroxy-3-methylglutaryl-coa reductase kinase. Eur. J. Biochem. 233, 506–513 [DOI] [PubMed] [Google Scholar]
- 93. Halford N. G., Hey S., Jhurreea D., Laurie S., McKibbin R. S., Paul M., Zhang Y. (2003) Metabolic signalling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 54, 467–475 [DOI] [PubMed] [Google Scholar]
- 94. Ikeda Y., Koizumi N., Kusano T., Sano H. (2000) Specific binding of a 14–3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J. Biol. Chem. 275, 41528. [PubMed] [Google Scholar]
- 95. Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J. F., Harris J. M., Oldroyd G. E. (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in. Medicago truncatula. Plant Cell 20, 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tominaga A., Nagata M., Futsuki K., Abe H., Uchiumi T., Abe M., Kucho K., Hashiguchi M., Akashi R., Hirsch A. M., Arima S., Suzuki A. (2009) Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of. Lotus japonicus. Plant Physiol. 151, 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kapranov P., Jensen T. J., Poulsen C., de Bruijn F. J., Szczyglowski K. (1999) A protein phosphatase 2C gene, LjNPP2C1, from Lotus japonicus induced during root nodule development. Proc. Natl. Acad. Sci. U.S.A. 96, 1738–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Black D. J., Persechini A. (2011) In calmodulin-IQ domain complexes, the Ca2+-free and Ca2+-bound forms of the calmodulin C-lobe direct the N-lobe to different binding sites. Biochemistry 50, 10061–10068 [DOI] [PubMed] [Google Scholar]
- 99. Lee K. H., Larue T. A. (1992) Ethylene as a possible mediator of light- and nitrate-induced inhibition of nodulation of Pisum sativum L. cv sparkle. Plant Physiol. 100, 1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nishimura R., Ohmori M., Fujita H., Kawaguchi M. (2002) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc. Natl. Acad. Sci. U.S.A. 99, 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. D'haeseleer K., De Keyser A., Goormachtig S., Holsters M. (2010) Transcription factor MtATB2: About nodulation, sucrose and senescence. Plant Cell Physiol. 51, 1416–1424 [DOI] [PubMed] [Google Scholar]
- 102. Homma M. K., Wada I., Suzuki T., Yamaki J., Krebs E. G., Homma Y. (2005) CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 102, 15688–15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dennis M. D., Person M. D., Browning K. S. (2009) Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J. Biol. Chem. 284, 20615–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yang W. C., de Blank C., Meskiene I., Hirt H., Bakker J., van Kammen A., Franssen H., Bisseling T. (1994) Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bauer P., Ratet P., Crespi M. D., Schultze M., Kondorosi A. (1996) Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsENOD12A expression patterns in alfalfa roots. Plant J. 10, 91–105 [Google Scholar]
- 106. Hervé C., Lefebvre B., Cullimore J. (2011) How many E3 ubiquitin ligase are involved in the regulation of nodulation? Plant. Signal. Behav. 6, 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Miura K., Jin J. B., Hasegawa P. M. (2007) Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10, 495–502 [DOI] [PubMed] [Google Scholar]
- 108. Lee Y. J., Yang Z. (2008) Tip growth: Signaling in the apical dome. Curr. Opin. Plant Biol. 11, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Felle H. H., Kondorosi A., Kondorosi E., Schultze M. (1996) Rapid alkaliniztion in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 10, 295–301 [Google Scholar]
- 110. Shaw S. L., Long S. R. (2003) Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131, 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Timmers A. C., Vallotton P., Heym C., Menzel D. (2007) Microtubule dynamics in root hairs of Medicago truncatula. Eur. J. Cell Biol. 86, 69–83 [DOI] [PubMed] [Google Scholar]
- 112. Cheng N. H., Liu J. Z., Nelson R. S., Hirschi K. D. (2004) Characterization of CXIP4, a novel arabidopsis protein that activates the H+/Ca2+ antiporter, CAX1. FEBS Lett. 559, 99–106 [DOI] [PubMed] [Google Scholar]
- 113. den Hartog M., Musgrave A., Munnik T. (2001) Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: A role for phospholipase C and D in root hair deformation. Plant J. 25, 55–65 [DOI] [PubMed] [Google Scholar]
- 114. Yuksel B., Memon A. R. (2009) Legume small GTPases and their role in the establishment of symbiotic associations with rhizobium spp. Plant. Signal. Behav. 4, 257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu W., Chen A. M., Luo L., Sun J., Cao L. P., Yu G. Q., Zhu J. B., Wang Y. Z. (2010) Characterization and expression analysis of Medicago truncatula ROP GTPase family during the early stage of symbiosis. J. Integr. Plant. Biol. 52, 639–652 [DOI] [PubMed] [Google Scholar]
- 116. Sun J., Miwa H., Downie J. A., Oldroyd G. E. (2007) Mastoparan activates calcium spiking analogous to nod factor-induced responses in Medicago truncatula root hair cells. Plant Physiol. 144, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yokota K., Fukai E., Madsen L. H., Jurkiewicz A., Rueda P., Radutoiu S., Held M., Hossain M. S., Szczyglowski K., Morieri G., Oldroyd G. E., Downie J. A., Nielsen M. W., Rusek A. M., Sato S., Tabata S., James E. K., Oyaizu H., Sandal N., Stougaard J. (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by. Mesorhizobium loti. Plant Cell 21, 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Miyahara A., Richens J., Starker C., Morieri G., Smith L., Long S., Downie J. A., Oldroyd G. E. (2010) Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol. Plant Microbe Interact. 23, 1553–1562 [DOI] [PubMed] [Google Scholar]
- 119. Kim Y., Sung J. Y., Ceglia I., Lee K. W., Ahn J. H., Halford J. M., Kim A. M., Kwak S. P., Park J. B., Ho Ryu S., Schenck A., Bardoni B., Scott J. D., Nairn A. C., Greengard P. (2006) Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 442, 814–817 [DOI] [PubMed] [Google Scholar]
- 120. Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P. K., Shimmen T. (2000) The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of hydrocharis. Planta 210, 836–843 [DOI] [PubMed] [Google Scholar]
- 121. Cooper J. A., Schafer D. A. (2000) Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12, 97–103 [DOI] [PubMed] [Google Scholar]
- 122. Davletov B., Sontag J. M., Hata Y., Petrenko A. G., Fykse E. M., Jahn R., Sudhof T. C. (1993) Phosphorylation of synaptotagmin I by casein kinase II. J. Biol. Chem. 268, 6816–6822 [PubMed] [Google Scholar]
- 123. Gamas P., de Billy F., Truchet G. (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol. Plant Microbe Interact. 11, 393–403 [DOI] [PubMed] [Google Scholar]
- 124. Peng H. M., Dreyer D. A., VandenBosch K. A., Cook D. (1996) Gene structure and differential regulation of the rhizobium-induced peroxidase gene, rip1. Plant Physiol. 112, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cárdenas L., Martínez A., Sánchez F., Quinto C. (2008) Fast, transient and specific intracellular ROS changes in living root hair cells responding to nod factors (NFs). Plant J. 56, 802–813 [DOI] [PubMed] [Google Scholar]
- 126. Demchenko K., Zdyb A., Feussner I., Pawlowski K. (2012) Analysis of the subcellular localisation of lipoxygenase in legume and actinorhizal nodules. Plant. Biol. 14, 56–63 [DOI] [PubMed] [Google Scholar]
- 127. Sun J., Cardoza V., Mitchell D. M., Bright L., Oldroyd G., Harris J. M. (2006) Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 46, 961–970 [DOI] [PubMed] [Google Scholar]
- 128. Mathesius U. (2008) Auxin: At the root of nodule development? Functional Plant Biology 35, 651–668 [DOI] [PubMed] [Google Scholar]
- 129. Gonzalez-Rizzo S., Crespi M., Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with sinorhizobium meliloti. Plant Cell 18, 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mateos P. F., Jimenez-Zurdo J. I., Chen J., Squartini A. S., Haack S. K., Martinez-Molina E., Hubbell D. H., Dazzo F. B. (1992) Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl. Environ. Microbiol. 58, 1816–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. van Spronsen P. C., Bakhuizen R., van Brussel A. A., Kijne J. W. (1994) Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. Eur. J. Cell Biol. 64, 88–94 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.