Abstract
Primary biliary cirrhosis (PBC) is a chronic cholestatic liver disease of unknown etiology and is considered to be an autoimmune disease. Autoantibodies are important tools for accurate diagnosis of PBC. Here, we employed serum profiling analysis using a human proteome microarray composed of about 17,000 full-length unique proteins and identified 23 proteins that correlated with PBC. To validate these results, we fabricated a PBC-focused microarray with 21 of these newly identified candidates and nine additional known PBC antigens. By screening the PBC microarrays with additional cohorts of 191 PBC patients and 321 controls (43 autoimmune hepatitis, 55 hepatitis B virus, 31 hepatitis C virus, 48 rheumatoid arthritis, 45 systematic lupus erythematosus, 49 systemic sclerosis, and 50 healthy), six proteins were confirmed as novel PBC autoantigens with high sensitivities and specificities, including hexokinase-1 (isoforms I and II), Kelch-like protein 7, Kelch-like protein 12, zinc finger and BTB domain-containing protein 2, and eukaryotic translation initiation factor 2C, subunit 1. To facilitate clinical diagnosis, we developed ELISA for Kelch-like protein 12 and zinc finger and BTB domain-containing protein 2 and tested large cohorts (297 PBC and 637 control sera) to confirm the sensitivities and specificities observed in the microarray-based assays. In conclusion, our research showed that a strategy using high content protein microarray combined with a smaller but more focused protein microarray can effectively identify and validate novel PBC-specific autoantigens and has the capacity to be translated to clinical diagnosis by means of an ELISA-based method.
Primary biliary cirrhosis (PBC)1 is a chronic, progressive cholestatic autoimmune liver disease (1) characterized by immune-mediated injury of the small and medium-sized bile ducts, which leads to fibrosis, cirrhosis, and eventual liver failure (2–5). PBC predominantly affects middle-aged women, and the female/male ratio has been reported to be 8–10:1 (6, 7). Both genetic and environmental risk factors have been identified for PBC, and the disease is believed to be triggered by a combination of the two (8). Although the annual incidence of PBC remains relatively low, at 0.7–49 per million (3), it is generally believed to be under-diagnosed by the currently available epidemiological instruments used in the clinic (3, 9, 10).
There are three criteria commonly used for diagnosis of PBC: (a) abnormally high biochemical profiles of serum alkaline phosphatase and γ-glutamyl transpeptidase; (b) the presence of anti-mitochondrial antibodies (AMA) in serum, especially of anti-M2 autoantigen complex antibodies; and (c) histological features of nonsuppurative destructive cholangitis. The major hallmark of PBC is considered to be a high titer of AMA, because it has a high sensitivity for PBC (>90%) (2, 8). Targets of M2 autoantibodies have been identified as members of the 2-oxo acid dehydrogenase complex located at the inner membrane of the mitochondria, including the E2 subunits of the pyruvate dehydrogenase complex (PDC-E2), the 2-oxoglutarate dehydrogenase complex (OGDC-E2), the branched chain 2-oxoacid dehydrogenase complex (BCOADC-E2), the E3 binding protein of pyruvate dehydrogenase complex (E3BP), and the E1α and E1β subunits of pyruvate dehydrogenase complex (PDC-E1α and PDC-E1β) (11–15). Although previous studies had characterized AMA as highly specific to PBC and observable long before clinical diagnosis (2, 16), a more recent study challenged this idea by demonstrating that 28 of 69 (40.9%) acute liver failure patients also reacted positively with the major mitochondrial autoantigens (PDC-E2, BCOADC-E2, and OGDC-E2) by immunoblot (17). In addition, further reports emerged indicating that AMA are frequently detected in patients with primary Sjögren's syndrome, scleroderma, and autoimmune hepatitis (18, 19), as well as in some patients afflicted with infectious diseases, such as tuberculosis and viral hepatitis (20–23). Furthermore, no evidence has yet proven that AMA-M2 is correlated with either PBC severity or its pathophysiological process (24–26).
The anti-nuclear antibodies (ANA) represent another type of important serum biomarkers for PBC diagnosis and have been reported to be achieving positive rates of 50–70% (27–30). Three major immunofluorescence patterns of ANA have been described for PBC, and some of their contributing autoantigens have also been identified. Nuclear envelope pattern (anti-nuclear envelope antibodies) refers to the anti-nuclear pore glycoprotein 210 (Gp210), anti-nuclear pore glycoprotein p62 (p62), and anti-lamin B receptor (LBR) antibodies (31); the multiple nuclear dot pattern associates with anti-nuclear body protein Sp140 and nuclear autoantigen Sp100 antibodies (32). Anti-centromere antibodies (ACA) mainly target CENP-B (33). Recent studies have shown that ANA, especially the anti-Gp210 antibody and the ACA, are correlated with severe disease course and poor prognosis of PBC (34–36). The sensitivity of anti-Gp210 for PBC has been reported to be 20–30% (31, 37). However, similar to the AMA, ANA have been clinically detected in sera of patients with other autoimmune diseases and even in a remarkable proportion (1 of 20) of sera from healthy individuals (38). ACA are widely accepted as relatively specific biomarkers of limited cutaneous systemic sclerosis (SSc) (39) but again have been frequently detected in a number of other conditions, including primary Sjögren's syndrome, systematic lupus erythematosus (SLE) (40, 41), and rheumatoid arthritis (RA) (41), and occasionally detected in certain cancers, such as breast cancer (42) and non-Hodgkin's lymphoma (43). The diagnosis of PBC can be further complicated by the clinical and biochemical features of liver abnormalities shared by other autoimmune diseases, such as autoimmune hepatitis (AIH), SSc, RA, and SLE (44–47). Although the detection of PBC-specific autoantibodies may help to distinguish PBC from other autoimmune diseases (37), the diagnostic role of PBC-specific ANA has not been fully developed (48). Therefore, the identification of new autoantibodies as noninvasive biomarkers remains a priority of PBC research (49).
The conventional methods to screen for novel autoantibodies in PBC patients are cumbersome and time-consuming (50). The recently developed functional protein microarrays were designed to survey thousands of potential antigens in a single experiment and have facilitated rapid and cost effective identification of novel biomarkers (51–53). Although human protein microarrays have become a robust tool for screening autoantibodies in sera from patients with various autoimmune diseases (52–60) and cancers (56, 61), the major limitation is the comprehensiveness of these previously used human protein microarrays. Here, we employed a recently developed human proteome microarray with ∼80% coverage of the human proteome to screen for novel PBC-specific biomarkers. The resulting PBC-associated candidate proteins were then used to construct a focused PBC microarray, for additional validation using a much larger cohort. Two of the validated autoantigens were further converted to the traditional ELISA-based assay to demonstrate their utility for clinical diagnosis.
EXPERIMENTAL PROCEDURES
Serum Samples
All of the human serum samples were collected at Peking Union Medical College Hospital between 2006 and 2010. The samples were collected from 297 PBC patients (54.2 ± 11.5 years, 92.5% female), 53 AIH patients (46.7 ± 17.6 years, 84.9% female), 112 hepatitis B virus (HBV) patients (42.6 ± 14.8 years, 42.9% female), 54 hepatitis C virus (HCV) patients (51.0 ± 13.2 years, 40.7% female), 122 RA patients (47.2 ± 13.3 years, 85.2% female), 86 SLE patients (33.0 ± 11.7 years, 93.0% female), 123 SSc patients (45.0 ± 12.1 years, 92.7% female), and 87 healthy controls (40.6 ± 14.1 years, 47.1% female). PBC patients were diagnosed according to the criteria established by the American Association for the Study of Liver Diseases (62). All PBC serum samples were tested for AMA detection by indirect immunofluorescence assay with Hep2 cells (Euroimmune, Lübeck, Germany) and for the AMA-M2 antibodies detection by the anti-M2–3E ELISA kit (Euroimmune). Furthermore, any PBC patient whose sera produced negative results for AMA and/or M2 were confirmed as having PBC by liver biopsy and meeting other criteria. Of the 191 PBC serum samples used for validation with PBC microarray, 167 were clinically characterized as AMA-positive and 163 as M2-positive. In addition, 66 and 44 PBC sera were characterized as anti-nuclear envelope antibody- and ACA-positive, respectively. The remaining 550 serum samples representing the various disease controls were diagnosed according to the respective general criteria used for each disease. The study was approved by the Ethics Committee of Peking Union Medical College Hospital.
Human Proteome Microarrays
The human proteome microarray used in the first phase of this study was composed of about 17,000 unique human full-length proteins and was constructed in Dr. Zhu's laboratory at Johns Hopkins University Medical School (63). This novel human proteome microarray contained 48 blocks arranged in a 25 × 32 array layout. Each of the recombinant human proteins was printed in duplicate, as were the control probes (printing buffer, human IgG, etc.). All of the recombinant human proteins were generated by the Saccharomyces cerevisiae expression system and carried an N-terminal GST tag.
The quality of the microarray was measured by using mouse anti-GST monoclonal antibody and confirmed with Cy5-labeled anti-mouse IgG antibody. In brief, the microarray was first incubated with blocking buffer (3% BSA in 0.1% (v/v) PBS plus 0.1% (v/v) Tween 20) at 37 °C for 1 h, after which 180 μl of mouse anti-GST monoclonal antibody (1:1000 dilution; Beijing Protein Innovation Co. Ltd., Beijing, China) was added and further incubated under a glass coverslip (LifterSlip; Erie Science Company, Portsmouth, NH) at 37 °C for 1 h. After washing three times with 1× PBS plus Tween 20 by gentle shaking for 10 min each, the microarray was incubated with 180 μl of Cy5-labeled goat anti-mouse IgG antibody (1:1000; Jackson Laboratory, Bar Harbor, ME) in the dark at 37 °C for 1 h. Subsequently, the microarray was washed three times with 1× PBS plus Tween 20 and then three times with double-distilled H2O. To fully remove the double-distilled H2O, the microarray was centrifuged for 5 min at 100 × g in a 50-ml centrifuge tube. Finally, the microarray was scanned with the LuxScanTM 10K Microarray Scanner (BioCapital, Beijing, China), and the probe signals were acquired using GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA). We considered the probes detectable when their signal to noise ratios for both duplicates were over 3. SPSS 17.0 software (Chicago, IL) was used to calculate the percentage of detectable probes and the coefficient factor of duplicate spots.
Sera Profiling Using the Human Proteome Microarrays
The procedure of sera assay was similar to that described above for the mouse anti-GST antibody assay, but with the following modifications. First, after blocking, the 1:1,000 diluted patient sera were incubated with the microarrays. Second, after washing away the sera, the 1:1,000 diluted Cy5-labeled goat anti-human IgG antibody was applied.
After the microarray was scanned and probes' signal intensities were acquired, positive calling in each microarray was conducted according to the procedure previously described by Hu et al. (64). PBC-specific autoantigen candidates were identified according to the following criteria: (a) Fisher's exact test on positive incidence showing statistical significance between PBC and control samples (p < 0.05) or (b) positive rate above 15% in PBC sera and 0% in controls.
Construction of the PBC Microarray
Twenty-one PBC-specific autoantigen candidates and nine other PBC-related antigens, which had been identified either in clinical use or by experimental studies reported in the literature, were prepared and printed on the PBC-focused microarray. All of the antigens were expressed and purified according to the method previously described (65). The recombinant proteins were eluted into protein microarray printing buffer (30% glycerol, 50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm Triton X-100, 30 mm reduced glutathione, and 1 mm DTT). The purified antigens, together with negative and positive controls (printing buffer, GST, human IgG, mouse IgG, and nucleoprotein of influenza) were printed in duplicate within 12 identical probe areas on each OPEpoxySlideTM (CapitalBio Corp., Beijing, China) to prepare the PBC-focused microarray. The printed low density protein microarrays were stored at 4 °C under vacuum until use.
Sera Assay with PBC Microarray
The PBC microarrays were subjected to 191 PBC and 321 control sera. First, the arrays were warmed to room temperature (∼30 min) and then a 12-hole rubber gasket (CapitalBio Corp.) was applied to each array to form 12 individual chambers. The subsequent assay process was identical to that described for the human proteome microarray assay, with the exception of the volume of fluid for each chamber being 30 μl. After incubation with the Cy5-labeled anti-human IgG, the rubber gaskets were removed carefully, and the wells were washed. The arrays were scanned by the LuxScanTM 10K microarray scanner using the same parameters for the larger array, and all of the signal intensities were acquired with the GenePix Pro 6.0 software. The serum samples with extremely high GST signal intensity (beyond the 99% spectrum of normal distribution) were considered to be GST-positive and were excluded from further analysis. The average signal intensity from healthy sera plus five times their standard deviations were set as the cutoff. The reproducibility of PBC-focused microarray was demonstrated by repeated hybridization experiments with randomly selected PBC (n = 12) and control (n = 12) sera, which resulted in a high correlation coefficient value (0.991) with p value equaled 0.000.
ELISA
The purified recombinant proteins were further verified prior to their use in validation analysis. Specifically, the ORF constructs of the newly identified antigens were verified by sequencing, and the identities of their protein products were confirmed by mass spectrometry. Then the verified recombinant proteins were coated onto 96-well plates at 4 °C overnight. Nonspecific binding was blocked by incubating with 200 μl of PBS plus Tween 20 containing 1% BSA/well at 4 °C overnight. The following day, the wells were incubated with human sera (1:100) at 37 °C for 1 h and then washed three times with 400 μl/well of PBS plus Tween 20. Subsequently, 100 μl of horseradish peroxidase-labeled mouse anti-human IgG monoclonal antibody (1:1,000; Beijing Protein Innovation Co. Ltd.) was added to each well. After three washes with 300 μl/well of PBS plus Tween 20, 100 μl of tetramethybenzidine substrate solution (Sigma-Aldrich) was added and incubated for 90 s at room temperature. The reaction was terminated by addition of 50 μl of 2 n H2SO4/well, and immunoreactivity was measured by reading the A450. The sera with extremely high GST reactions (beyond the 99% spectrum of normal distribution) were considered GST-positive and were excluded from further analysis. We set A450 >0.4 as the cutoff for positive hits.
Immunoblot Analysis
Two-hundred nanograms of hexokinase-1 (HK1) (isoform I, 128.5 kDa) was expressed by yeast with the GST-His6 tag, 200 ng of KLHL7 (65 kDa) expressed by Escherichia coli with the His6 tag, and 50 ng of GST-His6 tag (26 kDa) were resolved by 12% SDS-PAGE at 20 mA until the smallest band of the prestained protein marker (7 kDa) (New England Biolabs, Ipswich, MA) reached the bottom of the gel. The separated proteins were electrotransferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA) at 350 mA for 1.5 h. After blocking nonspecific binding sites with 5% nonfat milk powder diluted in 1× PBS plus Tween 20, the membranes were incubated with sera (1:400) or mouse anti-GST antibody (1:2,000) at 37 °C for 1 h, followed by three washes with 1× PBS plus Tween 20 for 10 min each. Then the membranes were incubated with horseradish peroxidase-conjugated anti-human IgG or anti-mouse IgG at 37 °C for 1 h. After three washes with 1× PBS plus Tween 20 for 10 min each, the immunoreactive bands were detected by the addition of enhanced chemiluminescence reagents (Beijing Applygen Ltd., Beijing, China) and were visualized by ImageQuant (GE Healthcare).
Statistics
All statistical analysis was performed with SPSS 17.0 software. p values were calculated by the chi-squared test or Fisher's exact test when suitable.
RESULTS
Identification of PBC-associated Autoantigens Using the Human Proteome Microarrays
To globally identify the PBC-associated autoantigens, we employed the human proteome microarrays containing about 17,000 human proteins to perform serum profiling of samples collected from PBC patients and other liver diseases and healthy controls. Because the human proteins on the microarrays were expressed and purified from yeast as N-terminally tagged GST fusion proteins, the quality of the microarray was able to be evaluated by applying the anti-GST antibody. The assay indicated that 97.6% of the proteins were detectable by anti-GST signals, and the microarray had a high correlation coefficient (0.978) between duplicate spots, which suggested that it was of high quality (supplemental Fig. 1).
To identify novel biomarkers more efficiently, we employed a two-phase strategy, as previously described (52). Phase I included a prescreening with a small sample size on the high content proteome microarray, whereas phase II was a large scale validation on a smaller but more focused microarray. In Phase I, we selected 26 PBC patients and 20 control samples, with the latter including 5 AIH, 5 HBV, and 10 healthy samples. These serum samples were individually applied as probes to the human proteome microarrays, followed by detection of bound human autoantibodies using a Cy5-conjugated anti-human IgG antibody (Fig. 1).
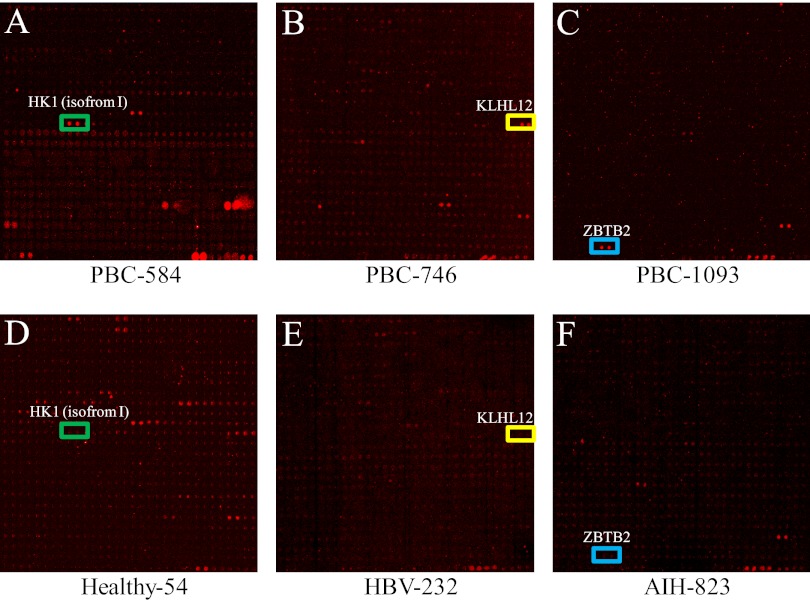
Fig. 1.
Probing of the human proteome microarray with PBC and control sera. 26 PBC and 20 control serum samples were diluted 1:1000 and individually incubated with the human proteome microarray, followed by the addition of Cy5-conjugated anti-human IgG antibody. Chips were dried and scanned to acquire images of positive immunoreactions. Representative areas of the images are shown. A, B, and C show that some PBC sera (PBC-584, PBC-746, and PBC-1093) were recognized by HK1 (isoform 1), KLHL12, and ZBTB2, respectively, on the human proteome microarray. The colored box indicates positive candidate autoantigens. D, E, and F show that the control serum samples were negative for the HK1 (isoform 1), KLHL12, and ZBTB2 proteins, respectively.
To identify potential PBC-associated autoantigens, we used Genepix Pro 6.0 to acquire the resultant signal intensities of all protein spots in each assay and identified the positives within each microarray (see details under “Experimental Procedures”). The total number of positive hits on each microarray ranged from 70 to 709 for the PBC sera (347 ± 191) and from 74 to 1,440 for the control sera (429 ± 306). Using Fisher's exact test, we determined that seven proteins, including two members of the M2 autoantigen complex, BCOADC-E2 and PDC-E1α, showed significant association with the PBC patients (p < 0.05). In addition, 16 other proteins, including another member of the M2 autoantigen complex, PDC-E1β, were found to be positive in more than 15% of the PBC patient sera, whereas none of them were scored as positive in the control samples (supplemental Table 1).
Validation of PBC-associated Autoantigens with PBC Microarray
To validate the potential autoantigens identified on the comprehensive human proteome microarray, and to determine their respective sensitivities and specificities for PBC diagnosis, we purified 21 of the 23 candidates, along with the other three members of the M2 complex (PDC-E2, E3BP, and OGDC-E2) and six additional autoantigens previously reported in PBC-related studies (Gp210, p62, CENP-B, Sp100, Sp140, and LBR) (32, 35, 66–69), to fabricate a PBC-focused microarray composed of 30 antigens. We then probed this new PBC-focused microarray with different cohorts of a much larger sample size, which included 191 PBC patients, 43 AIH patients, 55 HBV patients, 31 HCV patients, 48 RA patients, 45 SLE patients, 49 SSc patients, and 50 healthy controls (Fig. 2).
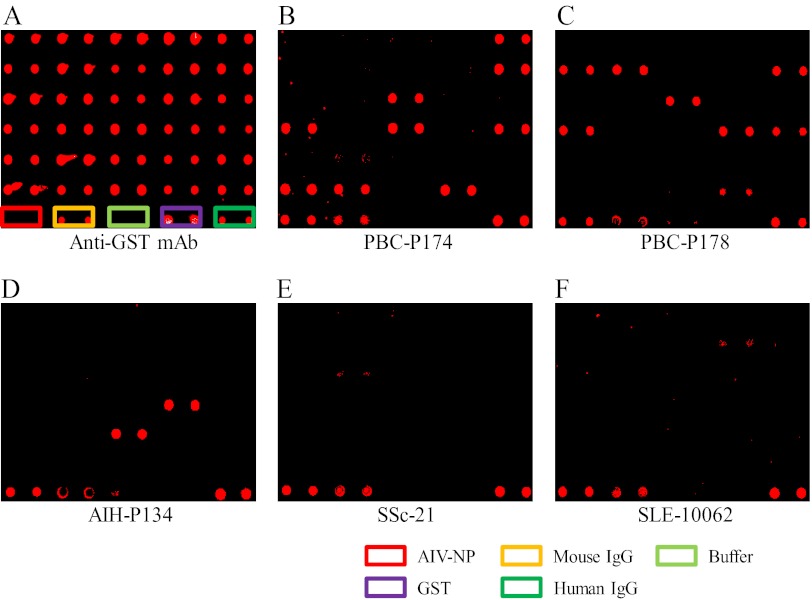
Fig. 2.
Preparation of PBC microarray and its probing with large cohorts of serum samples. The 21 PBC autoantigen candidates and nine other reported PBC autoantigens were prepared and printed, together with five controls (boxes of different colors in the last row), onto epoxy slides in duplicate to generate the PBC-focused microarray. Serum samples (B–F) or mouse anti-GST (A) antibody were applied to the array, followed by Cy5-labeled goat anti-human IgG antibody or Cy5-labeled goat anti-mouse IgG antibody to detect immunoreactivities.
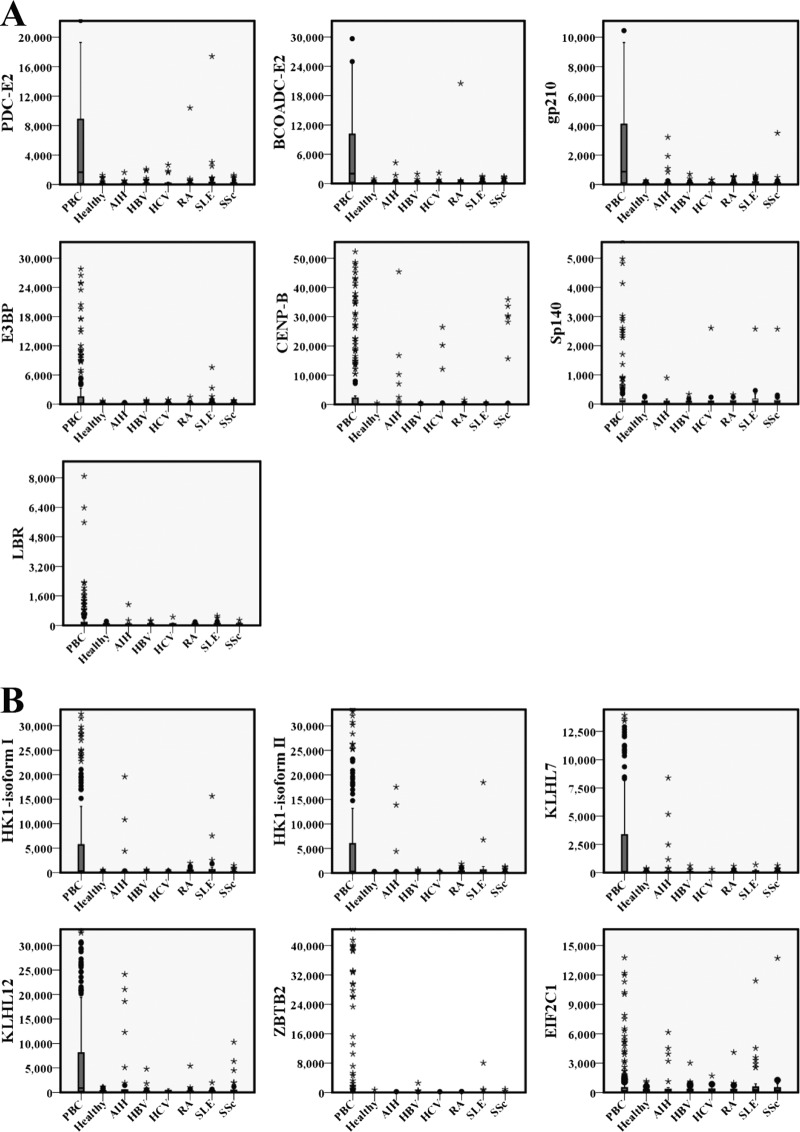
Using a z score of >5 (see “Experimental Procedures”) as the cutoff value, we analyzed the positive rates of each antigen in different groups. We first examined the sensitivity of these 30 antigens by comparing the results from the 191 PBC and 50 healthy samples (Table I and supplemental Table 2). The positive rates for all 30 antigens in the healthy controls were ≤2%. In contrast, 13 of the PBC candidate proteins, six of which were newly identified, were scored as positive in at least 29 (>15%) of the PBC samples. At this cutoff, the difference of positive rates between the PBC patients and healthy controls for all 13 autoantigens were statistically significant (p < 0.05). Fig. 3 shows the box plot analysis of signal intensities from the 13 proteins to provide a more intuitive view.
Table I. Positive numbers and rates for 30 antigens in various cohorts of serum samples.
| Autoantigens | PBC (n = 191) | Non-PBC (n = 321)a | Healthy (n = 50) | AIH (n = 43) | HBV (n = 55) | HCV (n = 31) | RA (n = 48) | SLE (n = 45) | SSc (n = 49) |
|---|---|---|---|---|---|---|---|---|---|
| M2 autoantigens | |||||||||
| BCOADC-E2 | 119 (62.3%) | 11 (3.4%) | 1 (2%) | 2 (4.7%) | 1 (1.8%) | 1 (3.2%) | 1 (2.1%) | 2 (4.4%) | 3 (6.1%) |
| PDC-E2 | 99 (51.8%) | 10 (3.1%) | 0 (0%) | 1 (2.3%) | 2 (3.6%) | 3 (9.7%) | 1 (2.1%) | 3 (6.7%) | 0 (0.0%) |
| OGDC-E2 | 16 (8.4%) | 6 (1.9%) | 0 (0%) | 0 (0.0%) | 1 (1.8%) | 0 (0.0%) | 1 (2.1%) | 2 (4.4%) | 2 (4.1%) |
| E3BP | 64 (33.5%) | 12 (3.7%) | 1 (2%) | 0 (0.0%) | 1 (1.8%) | 2 (6.5%) | 1 (2.1%) | 5 (11.1%) | 2 (4.1%) |
| PDC-E1β | 24 (12.6%) | 1 (0.3%) | 0 (0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.0%) |
| PDC-E1α | 18 (9.4%) | 4 (1.2%) | 0 (0%) | 1 (2.3%) | 1 (1.8%) | 0 (0.0%) | 1 (2.1%) | 0 (0.0%) | 1 (2.0%) |
| Nuclear envelope autoantigens | |||||||||
| Gp210 | 112 (58.6%) | 20 (6.2%) | 0 (0%) | 4 (9.3%) | 2 (3.6%) | 2 (6.5%) | 4 (8.3%) | 6 (13.3%) | 2 (4.1%) |
| p62 | 14 (7.3%) | 5 (1.6%) | 0 (0%) | 0 (0.0%) | 0 (0.0%) | 1 (3.2%) | 0 (0.0%) | 3 (6.7%) | 1 (2.0%) |
| LBR | 30 (15.7%) | 4 (1.2%) | 0 (0%) | 1 (2.3%) | 0 (0.0%) | 1 (3.2%) | 0 (0.0%) | 2 (4.4%) | 0 (0.0%) |
| ACA autoantigen | |||||||||
| CENP-B | 59 (30.9%) | 19 (5.9%) | 0 (0%) | 6 (14.0%) | 0 (0.0%) | 4 (12.9%) | 2 (4.2%) | 1 (2.2%) | 6 (12.2%) |
| Multiple nuclear dots autoantigens | |||||||||
| Sp100 | 27 (14.1%) | 9 (2.8%) | 1 (2%) | 0 (0.0%) | 3 (5.5%) | 2 (6.5%) | 0 (0.0%) | 3 (6.7%) | 0 (0.0%) |
| Sp140 | 29 (15.2%) | 6 (1.9%) | 0 (0%) | 2 (4.7%) | 0 (0.0%) | 1 (3.2%) | 0 (0.0%) | 2 (4.4%) | 1 (2.0%) |
| Newly identified PBC-related autoantigens | |||||||||
| HK1 isoform I | 89 (46.6%) | 33 (10.3%) | 0 (0%) | 3 (7.0%) | 2 (3.6%) | 0 (0.0%) | 11 (22.9%) | 13 (28.9%) | 4 (8.2%) |
| HK1 isoform II | 84 (44.0%) | 30 (9.3%) | 0 (0%) | 3 (7.0%) | 1 (1.8%) | 0 (0.0%) | 10 (20.8%) | 12 (26.7%) | 4 (8.2%) |
| KLHL12 | 77 (40.3%) | 16 (5.0%) | 0 (0%) | 7 (16.3%) | 2 (3.6%) | 0 (0.0%) | 1 (2.1%) | 1 (2.2%) | 5 (10.2%) |
| KLHL7 | 67 (35.1%) | 8 (2.5%) | 0 (0%) | 4 (9.3%) | 1 (1.8%) | 0 (0%) | 1 (2.1%) | 1 (2.2%) | 1 (2.0%) |
| ZBTB2 | 32 (16.8%) | 5 (1.6%) | 1 (2%) | 0 (0.0%) | 1 (1.8%) | 0 (0.0%) | 0 (0.0%) | 2 (4.4%) | 1 (2.0%) |
| EIF2C1 | 29 (15.2%) | 15 (4.7%) | 0 (0%) | 4 (9.3%) | 1 (1.8%) | 1 (3.2%) | 1 (2.1%) | 7 (15.6%) | 1 (2.0%) |
| NXN | 28 (14.7%) | 12 (3.7%) | 0 (0%) | 2 (4.7%) | 1 (1.8%) | 0 (0.0%) | 4 (8.3%) | 2 (4.4%) | 3 (6.1%) |
| RPS19 | 26 (13.6%) | 13 (4.0%) | 0 (0%) | 3 (7.0%) | 1 (1.8%) | 0 (0.0%) | 1 (2.1%) | 6 (13.3%) | 2 (4.1%) |
| ANXA10 | 25 (13.1%) | 3 (0.9%) | 0 (0%) | 1 (2.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (4.4%) | 0 (0.0%) |
| SNX9 | 22 (11.5%) | 3 (0.9%) | 0 (0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (6.7%) | 0 (0.0%) |
| EIF4H | 21 (11.0%) | 11 (3.4%) | 0 (0%) | 2 (4.7%) | 1 (1.8%) | 1 (3.2%) | 0 (0.0%) | 5 (11.1%) | 2 (4.1%) |
| SPATA5 | 19 (9.9%) | 8 (2.5%) | 0 (0%) | 1 (2.3%) | 2 (3.6%) | 0 (0.0%) | 1 (2.1%) | 1 (2.2%) | 3 (6.1%) |
| ATCAY | 11 (5.8%) | 3 (0.9%) | 1 (2%) | 1 (2.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.0%) |
| DDX55 | 10 (5.2%) | 3 (0.9%) | 0 (0%) | 0 (0.0%) | 1 (1.8%) | 0 (0.0%) | 0 (0.0%) | 1 (2.2%) | 1 (2.0%) |
| KEAP1 | 6 (3.1%) | 5 (1.6%) | 0 (0%) | 1 (2.3%) | 1 (1.8%) | 0 (0.0%) | 0 (0.0%) | 3 (6.7%) | 0 (0.0%) |
| TRA16 | 3 (1.6%) | 0 (0.0%) | 0 (0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| IL1A | 1 (0.5%) | 3 (0.9%) | 0 (0%) | 0 (0.0%) | 0 (0.0%) | 1 (3.2%) | 1 (2.1%) | 0 (0.0%) | 1 (2.0%) |
| DR1 | 0 (0.0%) | 1 (0.3%) | 1 (2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
a Non-PBC including healthy, AIH, HBV, HCV, RA, SLE, and SSc.
Fig. 3.
Box plots of the PBC microarray signal distribution for 13 PBC autoantigens in various serum groups. All of the 512 cases were classified according to clinical diagnosis of PBC, AIH, HBV, HCV, RA, SLE, SSc, or healthy. The signal distributions detected for each of the seven clinical or reported PBC-specific autoantigens (A, PDC-E2, BCOADC-E2, Gp210, E3BP, LBR, CENP-B, and Sp140) and six newly identified autoantigens (B, HK1 isoform I, HK1 isoform II, KLHL7, KLHL12, ZBTB2, and EIF2C1) are displayed. The rectangles indicate the interquartile range, and the bar within the rectangle indicates the median value. The bars above and below the rectangles define the 1.5 interquartile range outlier ranges. All of the extreme outliers beyond the 1.5× interquartile range + median are shown as black dots.
Of the six known members of the M2 complex, BCOADC-E2, PDC-E2, and E3BP showed strong positive signals in PBC patient sera, producing positive rates of 62.3, 51.83, and 33.51%, respectively. The positive rates for the other three members, OGDC-E2, PDC-E1α, and PDC-E1β, were only 8.4, 9.4, and 12.6%, respectively. The positive rates of some of the M2 components (PDC-E2, OGDC-E2, and E3BP) were somewhat lower than previously reported (14), which could be because we only detected the presence of the IgG isotype, whereas the previous studies also measured other isotypes. When only one of the six M2 components was required to attain a positive score for PBC diagnosis, the positive rate increased to 82.2% (157 of 191) in PBC sera, similar to that of 84.8% (162 of 191), determined with a commercial anti-M2–3E kit (Euroimmun Corp.) that is commonly used in clinics. Detailed information for M2 detection in the 191 PBC sera by both methods is listed in supplemental Table 3. More importantly, the results of our microarray-based method were comparable with those obtained with the commercial kit at a 91.6% matched ratio. These results confirmed that our microarray system is reliable.
For the PBC antigens reported in the literature, CENP-B and Sp140 showed positive rates of 30.9 and 15.2% in our assays, respectively, similar to the previously reported rates of 26–29% (35, 36) and 15% (32), respectively. In addition, Gp210 and LBR showed strong positive rates in PBC patient sera, with positive ratios of 58.6 and 15.7%, respectively, whereas much lower positive rates of 22.3–44% (35, 67, 70–73) and 1–9% (31, 73–75), respectively, were previously reported. It is plausible that the immunoblotting technique used in these previous reports was not as sensitive as the microarray-based assays used in this study.
More importantly, we were able to confirm six new antigens that showed rather high positive rates in PBC samples: HK1 isoform I (46.6%), HK1 isoform II (44.0%), Kelch-like protein 12 (KLHL12) (40.3%), Kelch-like protein 7 (KLHL7) (35.1%), zinc finger and BTB domain-containing protein 2 (ZBTB2) (16.8%), and eukaryotic translation initiation factor 2C, subunit 1 (EIF2C1) (15.2%). Because PBC is a liver-specific autoimmune disease and because other liver diseases tend to exhibit similar symptoms at the first clinical presentation (76), we next examined the specificity of these new PBC autoantigens among other types of liver diseases, including AIH and viral hepatitis. By comparing the results of PBC sera with those from sera of 43 AIH and 86 viral hepatitis (55 HBV and 31 HCV) (Table I), we found that all but EIF2C1 showed significantly reduced positive rates (p < 0.01) in the sera from AIH patients, as compared with the sera from PBC patients; similarly, all proteins showed significantly reduced positive rates (p < 0.001) in the sera from viral hepatitis patients. Therefore, the majority, if not all, of these newly identified autoantigens were highly specific for PBC.
The ability of the newly identified autoantigens to differentiate PBC from AIH prompted us to examine their abilities to differentiate PBC from other autoimmune diseases. We therefore surveyed their autoimmunity in 142 serum samples collected from systemic autoimmune disease patients, including 48 RA patients, 45 SLE patients, and 49 SSc patients (Table I). As compared with the PBC results, all five antigens showed significantly reduced positive rates in both RA (p < 0.01) and SSc (p < 0.05) sera. Although to a lesser extent for SLE samples, the HK1 isoform I, KLHL12, and KLHL7 still showed significant differentiation power (p < 0.05). Therefore, these newly identified autoantigens have the potential to be used for distinguishing PBC from other autoimmune diseases.
Development of New Biomarkers for Clinical Diagnosis
The ultimate application of novel biomarkers is for clinical diagnosis, preferably in the form of an ELISA, because it is still one of the most popular methods used in clinics. Because the sequences of isoforms I and II of HK1 are identical, except for a stretch of 21 amino acids at the N terminus, the two antigens performed similarly in sera detection assays; therefore, we selected only isoform I for further development as a biomarker. In addition, because EIF2C1 showed a similar positive ratio in SLE and PBC patients, we did not pursue further development of this potential biomarker. Thus, we only applied HK1-isoform I, KLHL7, KLHL12, and ZBTB2 to an ELISA-based platform and tested them against 495 of the serum samples used in the PBC-focused microarray and 439 additional samples, for a total of 934 samples, 297 of which were PBC-diagnosed. OD distribution for KLHL12 and ZBTB2 were shown in Fig. 4. However, all tested samples showed similar OD values for HK1-isoform I and KLHL7 (data not shown).
Fig. 4.
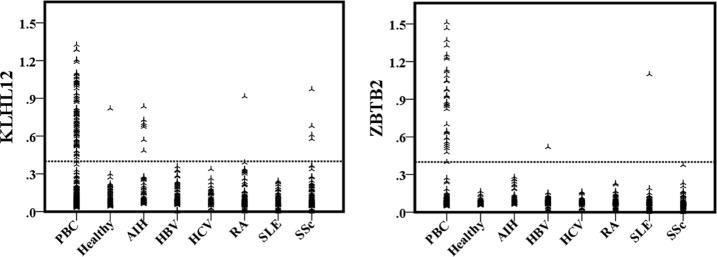
Plots of ELISA signal distribution for the autoantigens KLHL12 and ZBTB2 in various serum groups. All 934 cases were classified according to clinical diagnosis of PBC, AIH, HBV, HCV, RA, SLE, SSc, and healthy. The signal distributions of KLHL12 and ZBTB2 reacting with the serum samples in each case are displayed.
Using an A450 of >0.4 as the cutoff value, we analyzed the positive rates for each group and compared these results with those obtained using the microarray platform. The observed positive detection rates for KLHL12 and ZBTB2 ELISA were 29.6 and 11.2%, respectively, slightly lower than the rates obtained with the microarray platform, indicating that the latter is probably more sensitive (Table II).
Table II. ELISA results for KLHL12 and ZBTB2 in 934 serum samples.
| Disease | Cases | KLHL12 | ZBTB2 |
|---|---|---|---|
| PBC | 297 | 88 (29.6%) | 35 (11.2%) |
| Non-PBC | 637 | 13 (2.0%) | 2 (0.3%) |
| Healthy | 87 | 1 (1.2%) | 0 (0.0%) |
| AIH | 53 | 7 (13.2%) | 0 (0.0%) |
| HBV | 112 | 0 (0.0%) | 1 (0.9%) |
| HCV | 54 | 0 (0.0%) | 0 (0.0%) |
| RA | 122 | 1 (0.8%) | 0 (0.0%) |
| SLE | 86 | 0 (0.0%) | 1 (1.2%) |
| SSc | 123 | 4 (3.3%) | 0 (0.0%) |
Autoantibodies to HK1 and KLHL7 Were Detected in PBC Sera by Immunoblot
Although HK1 isoform I and KLHL7 showed 46.6 and 35.1% positive rates, respectively, for PBC samples in the protein microarray-based assays (Table I), we were unable to use them to differentiate PBC samples from other disease samples by ELISA. To determine whether the positive signals observed in the microarray assays were artifacts of unknown nature, we conducted immunoblot analysis using recombinant HK1 isoform I and KLHL7 proteins purified from yeast and bacteria, respectively. We also included GST-His6 protein as a negative control because HK1 and KLHL7 proteins were tagged with either the GST-His6 or His6 epitope. When four PBC serum samples that showed positive reactivity with HK1 or KLHL12 on the protein microarrays were tested by immunoblot, both proteins were readily recognized by the autoantibodies in each case. As expected, no significant signals were observed for the negative controls (Fig. 5). These results suggested that the protein microarray platform has a much higher detection limit than the traditional ELISA platform, consistent with our previously published observation of detecting SARS-CoV antigens in serum samples (77).
Fig. 5.
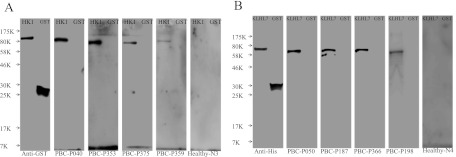
Validation of autoantibodies against HK1 isoform I and KLHL7 by immunoblotting. A, blotted recombinant GST-HK1 (128 kDa) and GST-His6 protein (26 kDa) were probed with anti-GST mAb and PBC microarray-identified HK1-positive PBC sera samples. B, blotted recombinant His6-KLHL7 (65 kDa) and GST-His6 protein were probed with anti-His monoclonal antibody and PBC microarray-identified KLHL7-positive PBC sera samples.
DISCUSSION
PBC was first identified as an autoimmune disease in the 1960s, based on clinical observations of a high titer of AMA in PBC patient sera (78–80). Since then, AMA, especially the anti-M2 complex autoantibodies, have become established as one of three standard diagnostic criteria for PBC. Despite the fact that research studies using a wide range of investigative strategies have identified and validated many other PBC autoantigens (12, 50, 68, 69, 81–84), M2 are still considered the hallmark feature of the disease. However, accumulating evidence has indicated that AMA, including anti-M2 autoantibodies, are frequently detected in patients with other non-PBC autoimmune diseases (85), such as SLE (86, 87), AIH (19, 22), SSc (18), primary Sjögren's syndrome (20), and even in some infectious diseases, such as tuberculosis (23) and viral hepatitis (21). In addition, there are still about 10–15% PBC patients with M2 negativity requiring biopsy and other clinic features for correct diagnosis. As a result, recent research efforts have aimed to discover novel PBC-specific biomarkers, but these attempts have been impeded by the conventional methods of autoantigen identification, which are not amenable to high throughput or comprehensive screening (50, 73, 88, 89). In this study, we used protein microarray technology to perform unbiased, proteome-wide identification of PBC-associated autoantigens, from which six novel proteins with at least 15% sensitivity for PBC were identified, including KLHL7, KLHL12, ZBTB2, HK1 isoform I, HK1 isoform II, and EIF2C1.
KLHL7, KLHL12, and ZBTB2 are all nuclear proteins and share a BTB (for BR-C, ttk, and bab) or POZ (for pox virus and zinc finger) domain. The BTB/POZ motif is widely described as an evolutionarily conserved protein-protein interaction motif that has been found in species ranging from flies to mammals (90, 91) and is often present at the N termini of zinc finger transcription factors. Previous studies have suggested that the BTB/POZ domain is required for interaction with cullin-3 and for the formation of the ubiquitin-protein E3 ligase complex, which could interact with the dishevelled homolog 3 protein DVL3 upon activation of the Wnt signaling pathway by WNT3A (92).
Besides the BTB domain, KLHL12 and KLHL7 also share six Kelch motif repeats (90, 91). The Kelch motif generally contains 50 amino acids that form a four-stranded β-sheet “blade,” whereas repeated Kelch motifs form a larger structure called a β-propeller that contains multiple potential protein-protein contact sites (93). KLHL12 and KLHL7 have been identified as autoantigens in Sjögren's syndrome by means of SEREX technology and determined to have 23 and 17% sensitivities, respectively, but they have not been found in RA, SLE, or healthy subjects (94). In our test of large cohorts of patients with PBC or other types of autoimmune diseases, KLHL12 and KLHL7 were both identified as new biomarkers for PBC diagnosis and showed rather high sensitivities of 40.3 and 35.1%, respectively. Consistent with previous reports, only a few (2 of 48 RA and 2 of 45 SLE) or no (0 of 50 healthy) serum samples in this study were found to be serologically reactive to KLHL12 or KLHL7.
ZBTB2, a transcription factor of the POZ and Kruppel (better known as POK) subfamily, is characterized by four C2H2-type zinc fingers. It has been demonstrated to function as a potential proto-oncogenic master control gene in the p53 pathway by directly interacting with the zinc finger domains of p53. In addition, ZBTB2 acts as a potent p21 transcription repressor, a function that requires its BTB domain for interactions with other co-repressors, such as BCoR, N-CoR, and SMRT (95). Although it has not been previously reported as an autoantigen for any autoimmune diseases, this study provides convincing evidence that ZBTB2 can serve as a useful PBC-specific autoantigen with 11.8% sensitivity and 99.7% specificity. The fact that the BTB domain of ZBTB2 is in common with those of KLHL7 and KLHL12 suggests that there might be a common epitope residing in the BTB domain. Further experiments are required to pinpoint this epitope.
HK1 is a member of the hexokinase family, which phosphorylates glucose to produce glucose-6-phosphate. The HK1 isoform I is localized to the outer mitochondrial membrane in tissues that are strictly dependent on glucose utilization for their physiologic functions, including the brain, erythrocytes, platelets, lymphocytes, and fibroblasts (96). In our microarray-based serum profiling assay, we found that two of the four AMA+/M2− sera were reactive to HK1. Because of its mitochondrial location, HK1 positivity may partially explain why some sera demonstrate AMA positivity and M2 negativity. Compared with isoform I, the HK1 isoform II lacks the N-terminal porin-binding domain, which is responsible for targeting the protein to the mitochondrial outer membrane; thus, isoform II is localized mainly in the cytosol (98). Because of the high sequence similarity shared between these two isoforms, it is expected that they are targeted by at least some of the same autoantibodies. Indeed, the signal intensities of these two isoforms were highly correlated. Additionally, the HK1 has been identified as an autoantigen associated with autistic children, and anti-HK1 autoantibodies were shown to impair growth and induce apoptosis in cultured human neuroblastoma cells (97). It would be interesting to determine whether HK1-positive patients also suffer from similar nervous system-associated impairments.
In addition, the eukaryotic translation initiation factor EIF2C1 was also identified as a new autoantigen that was recognized by 15.2% PBC sera. However, it was not specific for PBC, because 15.6% of SLE sera and 9.3% of AIH sera also showed positive immunoreactivity.
To begin to evaluate the potential clinical utility of our newly identified PBC autoantigens, we performed correlation analysis and found that most of the newly identified antigens did not significantly correlate with known antigens in PBC patients (supplemental Table 4). Then we calculated the diagnosis sensitivity and specificity achieved with different combinations of the autoantigens. The diagnosis sensitivity of the six M2 components was improved from 82.2 to 92.2% when combined with the six newly identified autoantigens. When the above 12 antigens were combined with another six reported PBC-associated antigens, the sensitivity was further improved to 94.8%. However, the combinations did not improve specificity, which was decreased from 89.7 to 73.5 and 66.7%, respectively.
Currently, M2 autoantigen-based tests are the most frequently used method for PBC diagnosis. However, an M2-negative result does not absolutely rule out PBC diagnosis, and these patients must undergo biopsy and careful analysis of many other clinical features to obtain an accurate diagnosis. Therefore, identification of novel biomarkers for M2-negative PBC patients will reduce the physical impact on these patients and associated costs of invasive testing and subjective evaluation. When we combined the newly identified antigen KLHL12 with Gp210, we could readily detect positive signals in 47.8% of M2-negative PBC patients with a specificity of 90.4%, similar to that of the M2-based diagnosis for PBC patients (89.7%).
In conclusion, we have identified six new autoantigens with at least 15% positivity in PBC serum samples by using a protein microarray-based approach. This two-phase strategy combines a proteome-wide screen for novel autoantigens followed by a stringent validation step using additional cohorts to ensure the success of identification of useful autoantigens for a particular disease. Successful application of these autoantigens in a clinic friendly ELISA suggests that these new autoantigens may serve as useful serological biomarkers for diagnosis of PBC, especially for M2-negative PBC patients.
Supplementary Material
Footnotes
* This work was supported in part by Grants 2008BAI59B02 and 2008BAI59B03 from the National Science Technology Pillar Program in the 11th Five-Year Plan (to Y.-Z. L.); National Natural Science Foundation of China Grants 30471617, 30640084, 30872331, and 81072486 (to Y.-Z. L.); Key Clinical Program of the Ministry of Health Grant 2010–2012 (to X. Z.); Ministry of Science and Technology of the People's Republic of China Grants 2006AA02A311 and 2009CB522204 (to L. Wu); and National Institutes of Health Grants RR020839 and CA125807 (to H. Z.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PBC
- primary biliary cirrhosis
- AMA
- anti-mitochondrial antibodies
- ANA
- anti-nuclear antibodies
- PDC-E2
- E2 subunits of pyruvate dehydrogenase complex
- OGDC-E2
- E2 subunits of the 2-oxoglutarate dehydrogenase complex
- BCOADC-E2
- E2 subunits of the branched chain 2-oxoacid dehydrogenase complex
- E3BP
- E3 binding protein of pyruvate dehydrogenase complex
- PDC-E1α
- E1α subunit of pyruvate dehydrogenase complex
- PDC-E1β
- E1β subunit of pyruvate dehydrogenase complex
- Gp210
- nuclear pore glycoprotein 210
- p62
- nuclear pore glycoprotein p62
- LBR
- lamin B receptor
- Sp140
- nuclear body protein Sp140
- Sp100
- nuclear autoantigen Sp100
- ACA
- anti-centromere antibodies
- CENP-B
- centromere protein B
- SSc
- systemic sclerosis
- SLE
- systematic lupus erythematosus
- RA
- rheumatoid arthritis
- AIH
- autoimmune hepatitis
- HBV
- hepatitis B virus
- HCV
- hepatitis C virus
- HK1
- hexokinase-1
- KLHL12
- Kelch-like protein 12
- KLHL7
- Kelch-like protein 7
- ZBTB2
- zinc finger and BTB domain-containing protein 2
- EIF2C1
- eukaryotic translation initiation factor 2C, subunit 1
- BTB
- BR-C, ttk, and bab domain
- POZ
- pox virus and zinc finger domain.
REFERENCES
- 1. Selmi C., Bowlus C. L., Gershwin M. E., Coppel R. L. (2011) Primary biliary cirrhosis. Lancet 377, 1600–1609 [DOI] [PubMed] [Google Scholar]
- 2. Kaplan M. M., Gershwin M. E. (2005) Primary biliary cirrhosis. N. Engl. J. Med. 353, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 3. Prince M. I., James O. F. (2003) The epidemiology of primary biliary cirrhosis. Clin. Liver Dis. 7, 795–819 [DOI] [PubMed] [Google Scholar]
- 4. Hohenester S., Oude-Elferink R. P., Beuers U. (2009) Primary biliary cirrhosis. Semin. Immunopathol. 31, 283–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Milkiewicz P. (2008) Liver transplantation in primary biliary cirrhosis. Clin. Liver Dis. 12, 461–472 [DOI] [PubMed] [Google Scholar]
- 6. Gershwin M. E., Selmi C., Worman H. J., Gold E. B., Watnik M., Utts J., Lindor K. D., Kaplan M. M., Vierling J. M. (2005) Risk factors and comorbidities in primary biliary cirrhosis: A controlled interview-based study of 1032 patients. Hepatology 42, 1194–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Invernizzi P., Pasini S., Selmi C., Gershwin M. E., Podda M. (2009) Female predominance and X chromosome defects in autoimmune diseases. J. Autoimmun. 33, 12–16 [DOI] [PubMed] [Google Scholar]
- 8. Poupon R. (2010) Primary biliary cirrhosis: A 2010 update. J. Hepatol. 52, 745–758 [DOI] [PubMed] [Google Scholar]
- 9. Wong G. L., Hui A. Y., Wong V. W., Chan F. K., Sung J. J., Chan H. L. (2005) A retrospective study on clinical features and prognostic factors of biopsy-proven primary biliary cirrhosis in Chinese patients. Am. J. Gastroenterol. 100, 2205–2211 [DOI] [PubMed] [Google Scholar]
- 10. Kumagi T., Alswat K., Hirschfield G. M., Heathcote J. (2008) New insights into autoimmune liver diseases. Hepatol. Res. 38, 745–761 [DOI] [PubMed] [Google Scholar]
- 11. Fussey S. P., Bassendine M. F., Fittes D., Turner I. B., James O. F., Yeaman S. J. (1989) The E1 α and β subunits of the pyruvate dehydrogenase complex are M2′d′ and M2′e′ autoantigens in primary biliary cirrhosis. Clin. Sci. 77, 365–368 [DOI] [PubMed] [Google Scholar]
- 12. Fussey S. P., Bassendine M. F., James O. F., Yeaman S. J. (1989) Characterisation of the reactivity of autoantibodies in primary biliary cirrhosis. FEBS Lett. 246, 49–53 [DOI] [PubMed] [Google Scholar]
- 13. Van de Water J., Cooper A., Surh C. D., Coppel R., Danner D., Ansari A., Dickson R., Gershwin M. E. (1989) Detection of autoantibodies to recombinant mitochondrial proteins in patients with primary biliary cirrhosis. N. Engl. J. Med. 320, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 14. Leung P. S., Coppel R. L., Ansari A., Munoz S., Gershwin M. E. (1997) Antimitochondrial antibodies in primary biliary cirrhosis. Semin. Liver Dis. 17, 61–69 [DOI] [PubMed] [Google Scholar]
- 15. Lindenborn-Fotinos J., Baum H., Berg P. A. (1985) Mitochondrial antibodies in primary biliary cirrhosis: Species and nonspecies specific determinants of M2 antigen. Hepatology 5, 763–769 [DOI] [PubMed] [Google Scholar]
- 16. Miyakawa H., Tanaka A., Kikuchi K., Matsushita M., Kitazawa E., Kawaguchi N., Fujikawa H., Gershwin M. E. (2001) Detection of antimitochondrial autoantibodies in immunofluorescent AMA-negative patients with primary biliary cirrhosis using recombinant autoantigens. Hepatology 34, 243–248 [DOI] [PubMed] [Google Scholar]
- 17. Leung P. S., Rossaro L., Davis P. A., Park O., Tanaka A., Kikuchi K., Miyakawa H., Norman G. L., Lee W., Gershwin M. E. (2007) Antimitochondrial antibodies in acute liver failure: Implications for primary biliary cirrhosis. Hepatology 46, 1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chung L., Utz P. J. (2004) Antibodies in scleroderma: Direct pathogenicity and phenotypic associations. Curr. Rheumatol. Rep. 6, 156–163 [DOI] [PubMed] [Google Scholar]
- 19. O'Brien C., Joshi S., Feld J. J., Guindi M., Dienes H. P., Heathcote E. J. (2008) Long-term follow-up of antimitochondrial antibody-positive autoimmune hepatitis. Hepatology 48, 550–556 [DOI] [PubMed] [Google Scholar]
- 20. Nardi N., Brito-Zerón P., Ramos-Casals M., Aguiló S., Cervera R., Ingelmo M., Font J. (2006) Circulating auto-antibodies against nuclear and non-nuclear antigens in primary Sjogren's syndrome: Prevalence and clinical significance in 335 patients. Clin. Rheumatol. 25, 341–346 [DOI] [PubMed] [Google Scholar]
- 21. Ramos-Casals M., Pares A., Jara L. J., Solans R., Viñas O., Vázquez P., Sánchez-Tapias J. M., Rodés J., Font J. (2005) Antimitochondrial antibodies in patients with chronic hepatitis C virus infection: Description of 18 cases and review of the literature. J. Viral. Hepat. 12, 648–654 [DOI] [PubMed] [Google Scholar]
- 22. Montano-Loza A. J., Carpenter H. A., Czaja A. J. (2008) Frequency, behavior, and prognostic implications of antimitochondrial antibodies in type 1 autoimmune hepatitis. J. Clin. Gastroenterol. 42, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 23. Klein R., Wiebel M., Engelhart S., Berg P. A. (1993) Sera from patients with tuberculosis recognize the M2a-epitope (E2-subunit of pyruvate dehydrogenase) specific for primary biliary cirrhosis. Clin. Exp. Immunol. 92, 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Invernizzi P., Crosignani A., Battezzati P. M., Covini G., De Valle G., Larghi A., Zuin M., Podda M. (1997) Comparison of the clinical features and clinical course of antimitochondrial antibody-positive and -negative primary biliary cirrhosis. Hepatology 25, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 25. Jones D. E. (2008) Pathogenesis of primary biliary cirrhosis. Clin. Liver Dis. 12, 305–321 [DOI] [PubMed] [Google Scholar]
- 26. Selmi C., Zuin M., Gershwin M. E. (2008) The unfinished business of primary biliary cirrhosis. J. Hepatol. 49, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Worman H. J., Courvalin J. C. (2003) Antinuclear antibodies specific for primary biliary cirrhosis. Autoimmun. Rev. 2, 211–217 [DOI] [PubMed] [Google Scholar]
- 28. Yang W. H., Yu J. H., Nakajima A., Neuberg D., Lindor K., Bloch D. B. (2004) Do antinuclear antibodies in primary biliary cirrhosis patients identify increased risk for liver failure? Clin. Gastroenterol. Hepatol. 2, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 29. Rigopoulou E. I., Davies E. T., Pares A., Zachou K., Liaskos C., Bogdanos D. P., Rodes J., Dalekos G. N., Vergani D. (2005) Prevalence and clinical significance of isotype specific antinuclear antibodies in primary biliary cirrhosis. Gut 54, 528–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muratori L., Granito A., Muratori P., Pappas G., Bianchi F. B. (2008) Antimitochondrial antibodies and other antibodies in primary biliary cirrhosis: Diagnostic and prognostic value. Clin. Liver Dis. 12, 261–276 [DOI] [PubMed] [Google Scholar]
- 31. Worman H. J. (2007) Nuclear envelope protein autoantigens in primary biliary cirrhosis. Hepatol. Res. 37, (Suppl. 3) S406–S411 [DOI] [PubMed] [Google Scholar]
- 32. Granito A., Yang W. H., Muratori L., Lim M. J., Nakajima A., Ferri S., Pappas G., Quarneti C., Bianchi F. B., Bloch D. B., Muratori P. (2010) PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am. J. Gastroenterol. 105, 125–131 [DOI] [PubMed] [Google Scholar]
- 33. Parveen S., Morshed S. A., Nishioka M. (1995) High prevalence of antibodies to recombinant CENP-B in primary biliary cirrhosis: Nuclear immunofluorescence patterns and ELISA reactivities. J. Gastroenterol. Hepatol. 10, 438–445 [DOI] [PubMed] [Google Scholar]
- 34. Wesierska-Gadek J., Penner E., Battezzati P. M., Selmi C., Zuin M., Hitchman E., Worman H. J., Gershwin M. E., Podda M., Invernizzi P. (2006) Correlation of initial autoantibody profile and clinical outcome in primary biliary cirrhosis. Hepatology 43, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 35. Nakamura M., Kondo H., Mori T., Komori A., Matsuyama M., Ito M., Takii Y., Koyabu M., Yokoyama T., Migita K., Daikoku M., Abiru S., Yatsuhashi H., Takezaki E., Masaki N., Sugi K., Honda K., Adachi H., Nishi H., Watanabe Y., Nakamura Y., Shimada M., Komatsu T., Saito A., Saoshiro T., Harada H., Sodeyama T., Hayashi S., Masumoto A., Sando T., Yamamoto T., Sakai H., Kobayashi M., Muro T., Koga M., Shums Z., Norman G. L., Ishibashi H. (2007) Anti-gp210 and anti-centromere antibodies are different risk factors for the progression of primary biliary cirrhosis. Hepatology 45, 118–127 [DOI] [PubMed] [Google Scholar]
- 36. Gao L., Tian X., Liu B., Zhang F. (2008) The value of antinuclear antibodies in primary biliary cirrhosis. Clin. Exp. Med. 8, 9–15 [DOI] [PubMed] [Google Scholar]
- 37. Bogdanos D. P., Komorowski L. (2011) Disease-specific autoantibodies in primary biliary cirrhosis. Clin. Chim. Acta 412, 502–512 [DOI] [PubMed] [Google Scholar]
- 38. Invernizzi P. (2007) Antinuclear antibodies: General introduction. In: Shoenfeld M. D. Y., Gershwin M. M. E., Meroni M. D. P. L., eds. Autoantibodies, 2nd Ed., pp. 129–133, Elsevier, Burlington, MA [Google Scholar]
- 39. Fritzler M. J., Rattner J. B., Luft L. M., Edworthy S. M., Casiano C. A., Peebles C., Mahler M. (2011) Historical perspectives on the discovery and elucidation of autoantibodies to centromere proteins (CENP) and the emerging importance of antibodies to CENP-F. Autoimmun. Rev. 10, 194–200 [DOI] [PubMed] [Google Scholar]
- 40. Nakano M., Ohuchi Y., Hasegawa H., Kuroda T., Ito S., Gejyo F. (2000) Clinical significance of anticentromere antibodies in patients with systemic lupus erythematosus. J. Rheumatol. 27, 1403–1407 [PubMed] [Google Scholar]
- 41. Russo K., Hoch S., Dima C., Varga J., Teodorescu M. (2000) Circulating anticentromere CENP-A and CENP-B antibodies in patients with diffuse and limited systemic sclerosis, systemic lupus erythematosus, and rheumatoid arthritis. J. Rheumatol. 27, 142–148 [PubMed] [Google Scholar]
- 42. Atalay C., Dogan L., Atalay G. (2010) Anti-CENP-B antibodies are associated with prolonged survival in breast cancer. Future Oncol. 6, 471–477 [DOI] [PubMed] [Google Scholar]
- 43. Bencimon C., Salles G., Moreira A., Guyomard S., Coiffier B., Bienvenu J., Fabien N. (2005) Prevalence of anticentromere F protein autoantibodies in 347 patients with non-Hodgkin's lymphoma. Ann. N.Y. Acad. Sci. 1050, 319–326 [DOI] [PubMed] [Google Scholar]
- 44. Youssef W. I., Tavill A. S. (2002) Connective tissue diseases and the liver. J. Clin. Gastroenterol. 35, 345–349 [DOI] [PubMed] [Google Scholar]
- 45. Siegel J. L., Luthra H., Donlinger J., Angulo P., Lindor K. (2003) Association of primary biliary cirrhosis and rheumatoid arthritis. J. Clin. Rheumatol. 9, 340–343 [DOI] [PubMed] [Google Scholar]
- 46. Rigamonti C., Shand L. M., Feudjo M., Bunn C. C., Black C. M., Denton C. P., Burroughs A. K. (2006) Clinical features and prognosis of primary biliary cirrhosis associated with systemic sclerosis. Gut 55, 388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Assassi S., Fritzler M. J., Arnett F. C., Norman G. L., Shah K. R., Gourh P., Manek N., Perry M., Ganesh D., Rahbar M. H., Mayes M. D. (2009) Primary biliary cirrhosis (PBC), PBC autoantibodies, and hepatic parameter abnormalities in a large population of systemic sclerosis patients. J. Rheumatol. 36, 2250–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Invernizzi P., Selmi C., Ranftler C., Podda M., Wesierska-Gadek J. (2005) Antinuclear antibodies in primary biliary cirrhosis. Semin. Liver Dis. 25, 298–310 [DOI] [PubMed] [Google Scholar]
- 49. Mayo M. J. (2008) Natural history of primary biliary cirrhosis. Clin. Liver Dis. 12, 277–288 [DOI] [PubMed] [Google Scholar]
- 50. Gershwin M. E., Mackay I. R., Sturgess A., Coppel R. L. (1987) Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. J. Immunol. 138, 3525–3531 [PubMed] [Google Scholar]
- 51. Stoevesandt O., Taussig M. J., He M. (2009) Protein microarrays: High-throughput tools for proteomics. Expert Rev. Proteomics 6, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song Q., Liu G., Hu S., Zhang Y., Tao Y., Han Y., Zeng H., Huang W., Li F., Chen P., Zhu J., Hu C., Zhang S., Li Y., Zhu H., Wu L. (2010) Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J. Proteome Res. 9, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen C. S., Sullivan S., Anderson T., Tan A. C., Alex P. J., Brant S. R., Cuffari C., Bayless T. M., Talor M. V., Burek C. L., Wang H., Li R., Datta L. W., Wu Y., Winslow R. L., Zhu H., Li X. (2009) Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol. Cell. Proteomics 8, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hueber W., Kidd B. A., Tomooka B. H., Lee B. J., Bruce B., Fries J. F., Sønderstrup G., Monach P., Drijfhout J. W., van Venrooij W. J., Utz P. J., Genovese M. C., Robinson W. H. (2005) Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 52, 2645–2655 [DOI] [PubMed] [Google Scholar]
- 55. Quintana F. J., Farez M. F., Viglietta V., Iglesias A. H., Merbl Y., Izquierdo G., Lucas M., Basso A. S., Khoury S. J., Lucchinetti C. F., Cohen I. R., Weiner H. L. (2008) Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 105, 18889–18894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Babel I., Barderas R., Díaz-Uriarte R., Martínez-Torrecuadrada J. L., Sánchez-Carbayo M., Casal J. I. (2009) Identification of tumour-associated autoantigens for the diagnosis of colorectal cancer in serum using high-density protein microarrays. Mol. Cell. Proteomics 8, 2382–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Auger I., Balandraud N., Rak J., Lambert N., Martin M., Roudier J. (2009) New autoantigens in rheumatoid arthritis (RA): Screening 8268 protein arrays with sera from patients with RA. Ann. Rheum. Dis. 68, 591–594 [DOI] [PubMed] [Google Scholar]
- 58. Vigil A., Ortega R., Nakajima-Sasaki R., Pablo J., Molina D. M., Chao C. C., Chen H. W., Ching W. M., Felgner P. L. (2010) Genome-wide profiling of humoral immune response to Coxiella burnetii infection by protein microarray. Proteomics 10, 2259–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vigh-Conrad K. A., Conrad D. F., Preuss D. (2010) A protein allergen microarray detects specific IgE to pollen surface, cytoplasmic, and commercial allergen extracts. PLoS ONE 5, e10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vermeulen N., de Béeck K. O., Vermeire S., Van Steen K., Michiels G., Ballet V., Rutgeerts P., Bossuyt X. (2011) Identification of a novel autoantigen in inflammatory bowel disease by protein microarray. Inflamm. Bowel Dis. 17, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 61. Madoz-Gúrpide J., Kuick R., Wang H., Misek D. E., Hanash S. M. (2008) Integral protein microarrays for the identification of lung cancer antigens in sera that induce a humoral immune response. Mol. Cell. Proteomics 7, 268–281 [DOI] [PubMed] [Google Scholar]
- 62. Heathcote E. J. (2000) Management of primary biliary cirrhosis. The American Association for the Study of Liver Diseases practice guidelines. Hepatology 31, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 63. Jeong J. S., Jiang L., Albino E., Marrero J., Rho H. S., Hu J., Hu S., Vera C., Bayron-Poueymiroy D., Rivera-Pacheco Z. A., Ramos L., Torres-Castro C., Qian J., Bonaventura J., Boeke J. D., Yap W. Y., Pino I., Eichinger D. J., Zhu H., Blackshaw S. (2012) Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol. Cell. Proteomics 11, 10.1074/mcp.O111.016253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 66. Guldner H. H., Szostecki C., Grötzinger T., Will H. (1992) IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 149, 4067–4073 [PubMed] [Google Scholar]
- 67. Bandin O., Courvalin J. C., Poupon R., Dubel L., Homberg J. C., Johanet C. (1996) Specificity and sensitivity of gp210 autoantibodies detected using an enzyme-linked immunosorbent assay and a synthetic polypeptide in the diagnosis of primary biliary cirrhosis. Hepatology 23, 1020–1024 [DOI] [PubMed] [Google Scholar]
- 68. Wesierska-Gadek J., Hohenuer H., Hitchman E., Penner E. (1996) Autoantibodies against nucleoporin p62 constitute a novel marker of primary biliary cirrhosis. Gastroenterology 110, 840–847 [DOI] [PubMed] [Google Scholar]
- 69. Lin F., Noyer C. M., Ye Q., Courvalin J. C., Worman H. J. (1996) Autoantibodies from patients with primary biliary cirrhosis recognize a region within the nucleoplasmic domain of inner nuclear membrane protein LBR. Hepatology 23, 57–61 [DOI] [PubMed] [Google Scholar]
- 70. Luettig B., Boeker K. H., Schoessler W., Will H., Loges S., Schmidt E., Worman H. J., Gershwin M. E., Manns M. P. (1998) The antinuclear autoantibodies Sp100 and gp210 persist after orthotopic liver transplantation in patients with primary biliary cirrhosis. J. Hepatol. 28, 824–828 [DOI] [PubMed] [Google Scholar]
- 71. Bauer A., Habior A. (2007) Measurement of gp210 autoantibodies in sera of patients with primary biliary cirrhosis. J. Clin. Lab. Anal. 21, 227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stinton L. M., Swain M., Myers R. P., Shaheen A. A., Fritzler M. J. (2011) Autoantibodies to GW bodies and other autoantigens in primary biliary cirrhosis. Clin. Exp. Immunol. 163, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsangaridou E., Polioudaki H., Sfakianaki R., Samiotaki M., Tzardi M., Koulentaki M., Panayotou G., Kouroumalis E., Castanas E., Theodoropoulos P. A. (2010) Differential detection of nuclear envelope autoantibodies in primary biliary cirrhosis using routine and alternative methods. BMC Gastroenterol. 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nickowitz R. E., Wozniak R. W., Schaffner F., Worman H. J. (1994) Autoantibodies against integral membrane proteins of the nuclear envelope in patients with primary biliary cirrhosis. Gastroenterology 106, 193–199 [DOI] [PubMed] [Google Scholar]
- 75. Miyachi K., Hankins R. W., Matsushima H., Kikuchi F., Inomata T., Horigome T., Shibata M., Onozuka Y., Ueno Y., Hashimoto E., Hayashi N., Shibuya A., Amaki S., Miyakawa H. (2003) Profile and clinical significance of anti-nuclear envelope antibodies found in patients with primary biliary cirrhosis: A multicenter study. J. Autoimmun. 20, 247–254 [DOI] [PubMed] [Google Scholar]
- 76. Kim W. R., Ludwig J., Lindor K. D. (2000) Variant forms of cholestatic diseases involving small bile ducts in adults. Am. J. Gastroenterol. 95, 1130–1138 [DOI] [PubMed] [Google Scholar]
- 77. Zhu H., Hu S., Jona G., Zhu X., Kreiswirth N., Willey B. M., Mazzulli T., Liu G., Song Q., Chen P., Cameron M., Tyler A., Wang J., Wen J., Chen W., Compton S., Snyder M. (2006) Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U.S.A. 103, 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Walker J. G., Doniach D., Roitt I. M., Sherlock S. (1965) Serological tests in diagnosis of primary biliary cirrhosis. Lancet 1, 827–831 [DOI] [PubMed] [Google Scholar]
- 79. Doniach D., Roitt I. M., Walker J. G., Sherlock S. (1966) Tissue antibodies in primary biliary cirrhosis, active chronic (lupoid) hepatitis, cryptogenic cirrhosis and other liver diseases and their clinical implications. Clin Exp Immunol 1, 237–262 [PMC free article] [PubMed] [Google Scholar]
- 80. Goudie R. B., Macsween R. N., Goldberg D. M. (1966) Serological and histological diagnosis of primary biliary cirrhosis. J. Clin. Pathol. 19, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yeaman S. J., Fussey S. P., Danner D. J., James O. F., Mutimer D. J., Bassendine M. F. (1988) Primary biliary cirrhosis: Identification of two major M2 mitochondrial autoantigens. Lancet 1, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 82. Courvalin J. C., Lassoued K., Worman H. J., Blobel G. (1990) Identification and characterization of autoantibodies against the nuclear envelope lamin B receptor from patients with primary biliary cirrhosis. J. Exp. Med. 172, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Szostecki C., Guldner H. H., Netter H. J., Will H. (1990) Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 145, 4338–4347 [PubMed] [Google Scholar]
- 84. Bloch D. B., de la Monte S. M., Guigaouri P., Filippov A., Bloch K. D. (1996) Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem. 271, 29198–29204 [DOI] [PubMed] [Google Scholar]
- 85. Hu C. J., Zhang F. C., Li Y. Z., Zhang X. (2010) Primary biliary cirrhosis: What do autoantibodies tell us? World J. Gastroenterol. 16, 3616–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li C. H., Xu P. S., Wang C. Y., Zhang Y., Zou G. L. (2006) The presence of anti-mitochondrial antibodies in Chinese patients with liver involvement in systemic lupus erythematosus. Rheumatol. Int. 26, 697–703 [DOI] [PubMed] [Google Scholar]
- 87. Matsushita M., Matsudaira R., Ikeda K., Nawata M., Tamura N., Takasaki Y. (2009) Anti-proteasome activator 28α is a novel anti-cytoplasmic antibody in patients with systemic lupus erythematosus and Sjogren's syndrome. Mod. Rheumatol. 19, 622–628 [DOI] [PubMed] [Google Scholar]
- 88. Berg P. A., Klein R. (1992) Antimitochondrial antibodies in primary biliary cirrhosis and other disorders: Definition and clinical relevance. Dig. Dis. 10, 85–101 [DOI] [PubMed] [Google Scholar]
- 89. Feuchtinger M., Christ S., Preuss B., Dengjel J., Duman S., Stevanovic S., Klein R. (2009) Detection of novel non-M2-related antimitochondrial antibodies in patients with anti-M2 negative primary biliary cirrhosis. Gut 58, 983–989 [DOI] [PubMed] [Google Scholar]
- 90. Zollman S., Godt D., Privé G. G., Couderc J. L., Laski F. A. (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 91, 10717–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bardwell V. J., Treisman R. (1994) The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 8, 1664–1677 [DOI] [PubMed] [Google Scholar]
- 92. Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 93. Adams J., Kelso R., Cooley L. (2000) The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 10, 17–24 [DOI] [PubMed] [Google Scholar]
- 94. Uchida K., Akita Y., Matsuo K., Fujiwara S., Nakagawa A., Kazaoka Y., Hachiya H., Naganawa Y., Oh-Iwa I., Ohura K., Saga S., Kawai T., Matsumoto Y., Shimozato K., Kozaki K. (2005) Identification of specific autoantigens in Sjogren's syndrome by SEREX. Immunology 116, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jeon B. N., Choi W. I., Yu M. Y., Yoon A. R., Kim M. H., Yun C. O., Hur M. W. (2009) ZBTB2, a novel master regulator of the p53 pathway. J. Biol. Chem. 284, 17935–17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bianchi M., Crinelli R., Serafini G., Giammarini C., Magnani M. (1997) Molecular bases of hexokinase deficiency. Biochim. Biophys. Acta 1360, 211–221 [DOI] [PubMed] [Google Scholar]
- 97. Gonzalez-Gronow M., Cuchacovich M., Francos R., Cuchacovich S., Del Pilar Fernandez M., Blanco A., Bowers E. V., Kaczowka S., Pizzo S. V. (2010) Antibodies against the voltage-dependent anion channel (VDAC) and its protective ligand hexokinase-I in children with autism. J. Neuroimmunol. 227, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Murakami K., Piomelli S. (1997) Identification of the cDNA for human red blood cell-specific hexokinase isozyme. Blood 89, 762–766 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.