Abstract
Mass spectrometry-based proteomics is increasingly being used in biomedical research. These experiments typically generate a large volume of highly complex data, and the volume and complexity are only increasing with time. There exist many software pipelines for analyzing these data (each typically with its own file formats), and as technology improves, these file formats change and new formats are developed. Files produced from these myriad software programs may accumulate on hard disks or tape drives over time, with older files being rendered progressively more obsolete and unusable with each successive technical advancement and data format change. Although initiatives exist to standardize the file formats used in proteomics, they do not address the core failings of a file-based data management system: (1) files are typically poorly annotated experimentally, (2) files are “organically” distributed across laboratory file systems in an ad hoc manner, (3) files formats become obsolete, and (4) searching the data and comparing and contrasting results across separate experiments is very inefficient (if possible at all). Here we present a relational database architecture and accompanying web application dubbed Mass Spectrometry Data Platform that is designed to address the failings of the file-based mass spectrometry data management approach. The database is designed such that the output of disparate software pipelines may be imported into a core set of unified tables, with these core tables being extended to support data generated by specific pipelines. Because the data are unified, they may be queried, viewed, and compared across multiple experiments using a common web interface. Mass Spectrometry Data Platform is open source and freely available at http://code.google.com/p/msdapl/.
Mass spectrometry-based proteomics is a rapidly developing technology that is increasingly being applied to biological and biomedical research. Many software applications have been developed to analyze mass spectrometry data and each of these programs typically use (as input) and produce (as output) files of various formats. Not only are these formats often specific to certain analysis software, but it is often specific to certain versions of that software as well. And as technology advances, these file formats often change or entirely new formats are introduced. Data viewers, converters, and analysis software are developed or updated to support these new formats, rendering older files progressively more obsolete and unlikely to be supported as input to current programs.
In an effort to improve data portability and address this issue of many disparate proprietary file formats, important work has gone into the development of standardized and open data formats. Chief among this work are the XML-based data formats mzML (1), mzIdentML (2), mzData (3) (deprecated), mzXML (4), and pepXML (5); although non-XML formats have also been proposed (MGF (http://www.matrixscience.com/help/data_file_help.html), MS2 and SQT (6), and SQLite.(7)). Effort has also been put toward developing a universal application programming interface, dubbed mzAPI (8), to allow third-party applications direct access to proprietary proteomics formats. As a proof of concept, mzAPI has been used to develop both a desktop environment (9) and web-based environment (10) for accessing and viewing proteomics data. Although these efforts are well-developed and portable, they are less than ideal as a data archival and search format; and they suffer from drawbacks intrinsic to any file-based data solution:
Files are often spread across file systems, hard drives, and tape backups in an ad hoc manner, making them hard to locate.
Files may be poorly annotated with regards to experimental conditions, software parameters, or even the software used to generate the data.
File formats become obsolete and eventually unsupported by current tools.
Visualization tools and analysis software must be installed and maintained to view or interrogate the data files.
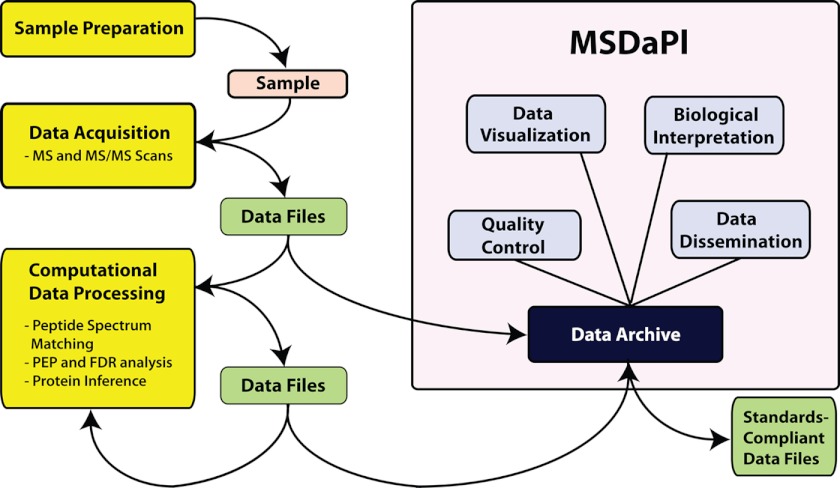
Government and journal rules regarding public data dissemination have led to the development of online proteomics data repositories such as PRIDE (11), Peptide Atlas (12), Global Proteome Machine (GPM) (13), Human Proteinpedia (14), and the Yeast Research Center (YRC) PDR (15) all of which provide unified web-based interfaces to underlying proteomics data stored in relational databases. Although these resources are very important in terms of data distribution, they are not typically designed to be part of a laboratory's archival and workflow process. Tranche (16), a distributed storage and archival network for proteomics data, provides an effective means of archiving and distributing data, but it distributes only files and requires that the end-user have access to visualization and analysis tools capable of working with files of that specific format before they may be of use. LabKey Server (17) and its accompanying proteomics module (formerly known as CPAS) is a comprehensive and robust proteomics pipeline and workflow management system. It is highly customizable and includes options for configuring and running multistep proteomics pipelines, and includes options for viewing the output of the respective analysis programs. Here we present the Mass Spectrometery Data Platform (MSDaPl)1, a proteomics data management system that, instead of driving proteomics workflows, focuses on long-term archiving, searching, evaluating, and performing simple analysis of the data that result from the workflows and may be used to compliment systems such as the LabKey Server (Fig. 1).
Fig. 1.
Where MSDaPl fits into the broad proteomics workflow. MSDaPl is not intended to be a workflow manager, but a platform for archiving, interpreting, and disseminating the data generated by each step of the proteomics workflow.
At its core MSDaPl uses a unified relational database architecture that supports storing data generated by multiple pipelines, and may be extended to support more. MSDaPl includes import algorithms for major proteomics pipelines, and once in the database, there is no longer any dependence on specific file formats. In addition, the data are searchable and may be compared across separate experiments (even if the experimental data were generated by different pipelines). Built on the database is a web application designed for searching the data archive, evaluating the quality of tandem MS (MS/MS) runs, and viewing results. To ensure data portability for subsequent re-analysis by additional proteomics software and inclusion in public repositories, data may be exported as mzML and mzIdentML XML, regardless of the original input format. The web application is developed in Java, and as such, is cross-platform. MSDaPl is open source and may be freely downloaded at http://code.google.com/p/msdapl/.
Data Architecture
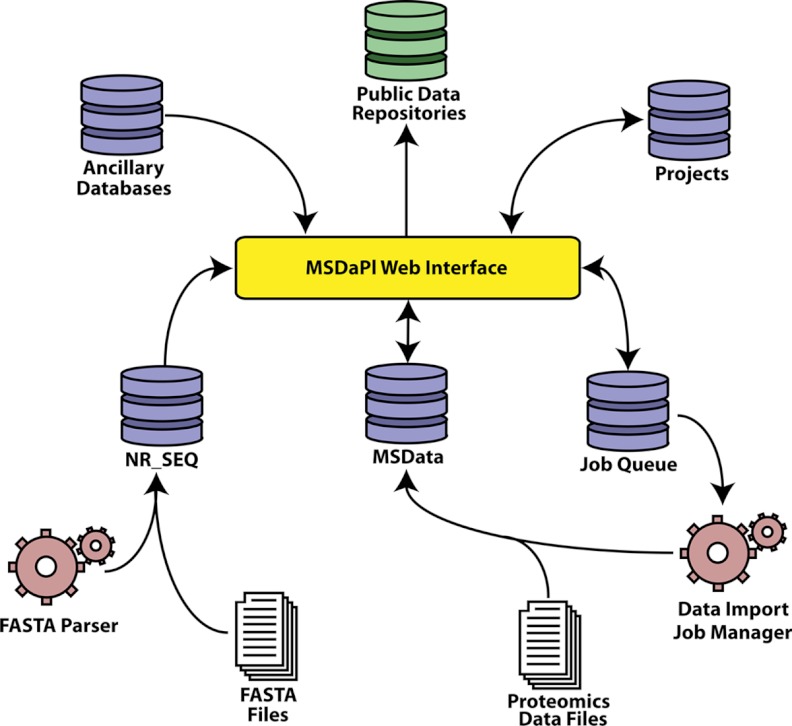
MSDaPl is supported by four primary databases (referred to here as “msData,” “NR_SEQ,” “Job Queue,” and “Projects Database”) and several ancillary databases created to provide biological context to MS/MS results (Fig. 2). The structured query language (SQL) required to generate these databases for the MySQL Relational Database Management System is available with the MSDaPl distribution at http://code.google.com/p/msdapl/.
Fig. 2.
An overview of the different databases and programs that support MSDaPl. Arrowheads in the diagram indicate the flow of information, database icons represent the separate databases, and the gears indicate the support programs responsible for processing and inserting data into the databases. Data from all databases are viewable in the web interface, and two support programs are responsible for processing FASTA sequence database files and the proteomics data files. All results may be exported in a format suitable for insertion into public databases.
MsData Database
At the heart of MSDaPl is a database schema, dubbed “mzData,” designed to be independent of any specific mass spectrometry pipeline. This is accomplished by conceptually separating typical mass spectrometry analysis into three fundamental areas: (1) the raw mass spectra, (2) peptide searches (including post-translational modifications), and (3) protein inference. Data describing each of these three areas (regardless of the particular analysis pipeline) will likely have common attributes, as they are describing the same kind of analysis. These common attributes are abstracted into a core set of tables in each of these three areas, and these core tables are populated for every MS run, peptide search, or protein inference result set loaded into the database. Because all data are represented homogeneously in these core tables, these core attributes may be searched and displayed using a common interface and results compared and contrasted across experiments and disparate pipelines.
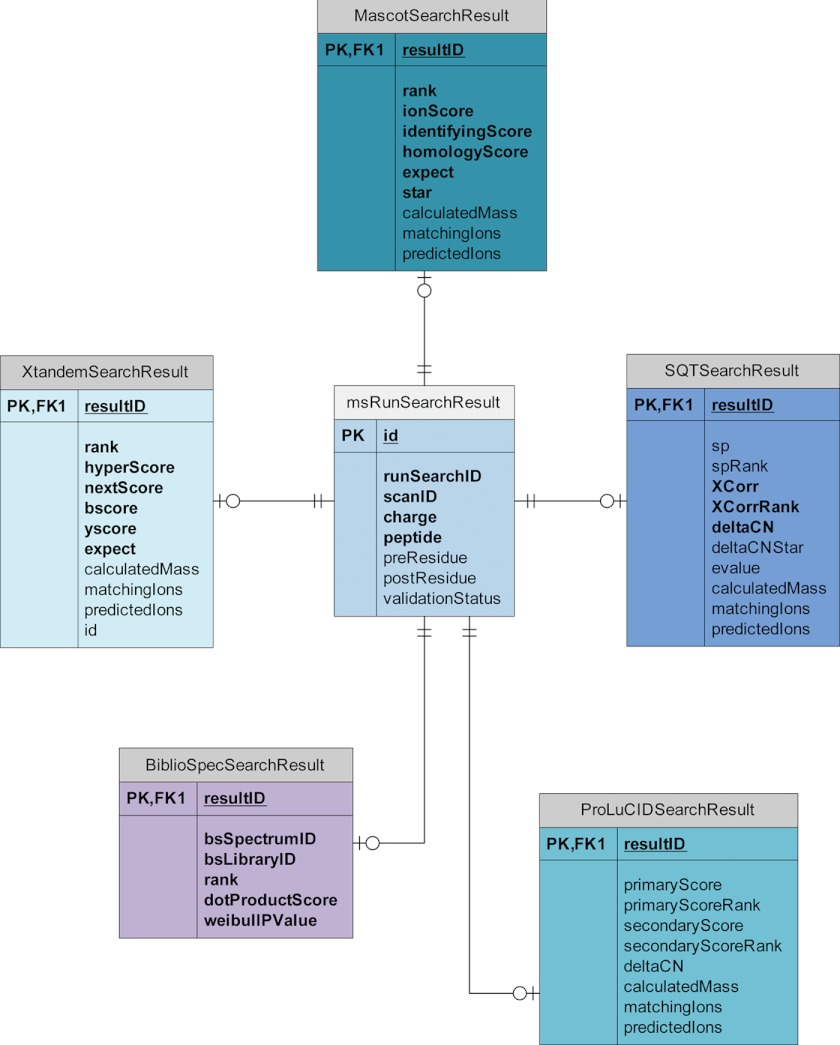
The core tables may be extended in order to encapsulate data specific to particular mass spectrometry analysis programs (Fig. 3). This is done by creating a new table in the schema specific to an analysis program where each row may be considered a horizontal extension of the rows from the core table. For example, a row representing a result in the peptide search results core table may have a corresponding row in the Mascot (18) results table that acts as a logical extension of this table. This row in the Mascot table would contain data specific to Mascot that describe that search result. Adding support for new pipelines to the database then becomes a matter of extending the core database schema with tables that encapsulate program-specific information. (Note that, although adding support to the database for new programs is relatively simple, importing the data will require development of new code that understands the format—a task simplified somewhat by implementing the import interface defined in MSDaPl.) Support for data generated by the following programs currently exists in the database:
SEQUEST (19)
Mascot
X!Tandem (20)
Trans Proteomic Pipeline (PeptideProphet and ProteinProphet) (21)
Tide (22)
ProLuCID (23)
Percolator (24)
Fig. 3.
A diagram illustrating how core tables in the msData database schema may be extended to support data of that type generated by separate programs. In this example, the table responsible for storing a peptide search result is extended by tables containing columns specific to Mascot, X!Tandem, Sequest, ProLuCID, and BiblioSpec. The connections between the tables indicate exactly one row in the core search result table may have zero or one rows in one of the extension tables.
NR_SEQ Database
MSDaPl uses a database dubbed “NR_SEQ” that allows for unambiguously referencing proteins by internally assigned identification numbers. NR_SEQ is designed around a nonredundant protein sequence table, and a protein is defined as a unique protein sequence with a particular National Center for Biotechnology Information taxonomy identification number (25). Each protein may have multiple references (including names, descriptions, external URLs, and other information) from many data sources, and each reference is linked to both the protein and the respective data source. Of particular relevance to MSDaPl, these data sources may be FASTA sequence files (26), and these references may serve as a mapping between the accession strings used to identify sequences in FASTA files and internally assigned protein identification numbers used to identify those proteins independently of any particular FASTA file.
Proteomics data files typically refer to proteins by the accession strings present in the FASTA file used to perform the analysis. The sequence databases used to generate these files may change the sequence associated with accession strings or the accession strings associated with sequences over time. Thus, attempting to identify the same protein across experiments that used different FASTA files by mapping accession strings from one database to another (or even accession strings between multiple versions of the same database) is an inherently unreliable process (assuming the user even used a FASTA file supported by the mapping). To address this problem, when data are uploaded to MSDaPl these accession strings are mapped to protein identification numbers in the NR_SEQ database by looking up the accession string in the protein reference table for the respective FASTA file. These protein identifier numbers are then stored as experimental results in the “msData” database (in addition to the original accession strings), which ensures that MSDaPl unambiguously and reliably refers to the same protein the same way across all experiments—regardless of which database (or version of that database) was used to create a FASTA file. Consequently, users may perform protein-level analysis across experiments without attempting to map accession strings from one database to another. Additionally, current names, descriptions, and annotations for proteins may be displayed using preferred protein databases instead of what may be have been used to generate the FASTA file. In order to achieve this mapping, the FASTA file must be parsed into the database previous to uploading data. This parsing is described in more detail under “Software Architecture.”
Projects Database
MSDaPl uses a projects database that is designed to facilitate collaboration and data organization by associating uploaded data with experiments, and experiments with projects. This database includes support for authentication and limiting access to projects (and data associated with those projects) to specific users. The projects database ensures that data may always be viewed in the context of a project, providing meaningful experimental context (including contact information) to the data that may be lost when files are stored on disk.
Job Queue
The “Job Queue” database exists to store data upload requests made by users of the web application. Because proteomics data files are typically too large to be uploaded directly via a web interface, requests to upload data are placed in this database and a separate job queue manager application periodically scans this database for upload requests and processes pending uploads. This process is described in more detail in the “Job Queue and Data Importers” section.
Ancillary Databases
In order to provide views and analysis in a biological context, such as Gene Ontology (27) enrichment analysis, MSDaPl makes use of several ancillary databases created by mirroring external protein annotation databases. These databases may be downloaded from the MSDaPl distribution site.
Software Architecture
The software developed for MSDaPl comprises a web application running on top of the databases described above, a back end job queue and data importers designed for uploading MS/MS results to the database, and a FASTA parsing program designed to map FASTA headers to protein identifiers in the database. All software, including source code, is available at the MSDaPl download site at http://code.google.com/p/msdapl/.
MSDaPl Web Application
Technology
The MSDaPl web application was developed using Java and Java Server Pages using the Struts web application framework. It is intended to run using Apache Tomcat, an open source servlet container also written in Java. Because it is Java-based, the MSDaPl web application may be deployed to any server capable of running Java, including Windows, MacOS, Linux, and others. The web interface has been developed using standard web technologies and is compatible with all current web browsers.
Organization
Central to the organization of MSDaPl is the concept of a project. All users and data are assigned to projects, and only users assigned to any given project may view data associated with that project. All access to MSDaPl requires authentication and the front page lists the projects for which the user is listed as a researcher. Project information (including a title, abstract, progress report, publications, comments, grants, and users associated with the project) may be edited by any user listed as a researcher of a project by using the “Edit Project” button present on any project page. Users may also edit their own information, such as their name, contact information, username, and password, by using the “Account” tab present at the top of all pages.
To upload data, users may use the “Upload Data” button present on any project page. This link leads to a form where the user provides basic experimental annotation and indicates the location of the data. Because data files associated with proteomics experiments are typically too large to be directly uploaded via HTTP, the data from this form are captured to the database and processed by a “job queue manager” program running as a daemon on the server (or on a different server). This job queue manager is responsible for downloading the data and importing it into the MSDaPl database schema. This software is described below in the “Job Queue and Data Importers” section.
Once data are imported into the database they may be accessed from the project page. Each data upload request is organized into distinct experiments, and multiple experiments may be uploaded to each project. For each experiment, the user may directly view the peptide search results (as a list of peptides, associated statistics, and links to view the underlying spectrum) or protein inference results (as a list of identified proteins, associated statistics, underlying peptides, and their underlying spectra). All spectra may be viewed using the integrated Lorikeet spectrum viewer (http://code.google.com/p/lorikeet/), which requires the use of no third party software or plugins. Users may also download all results from a given experiment as a standards-compliant mzData and mzIdentML XML files suitable for data dissemination and submittal to public proteomics data repositories.
Select Features
MSDaPl offers tools for viewing, validating, and interpreting MS/MS data that take advantage its relational database model. Select features are described below.
Protein List Filtering and Comparison Across Experiments
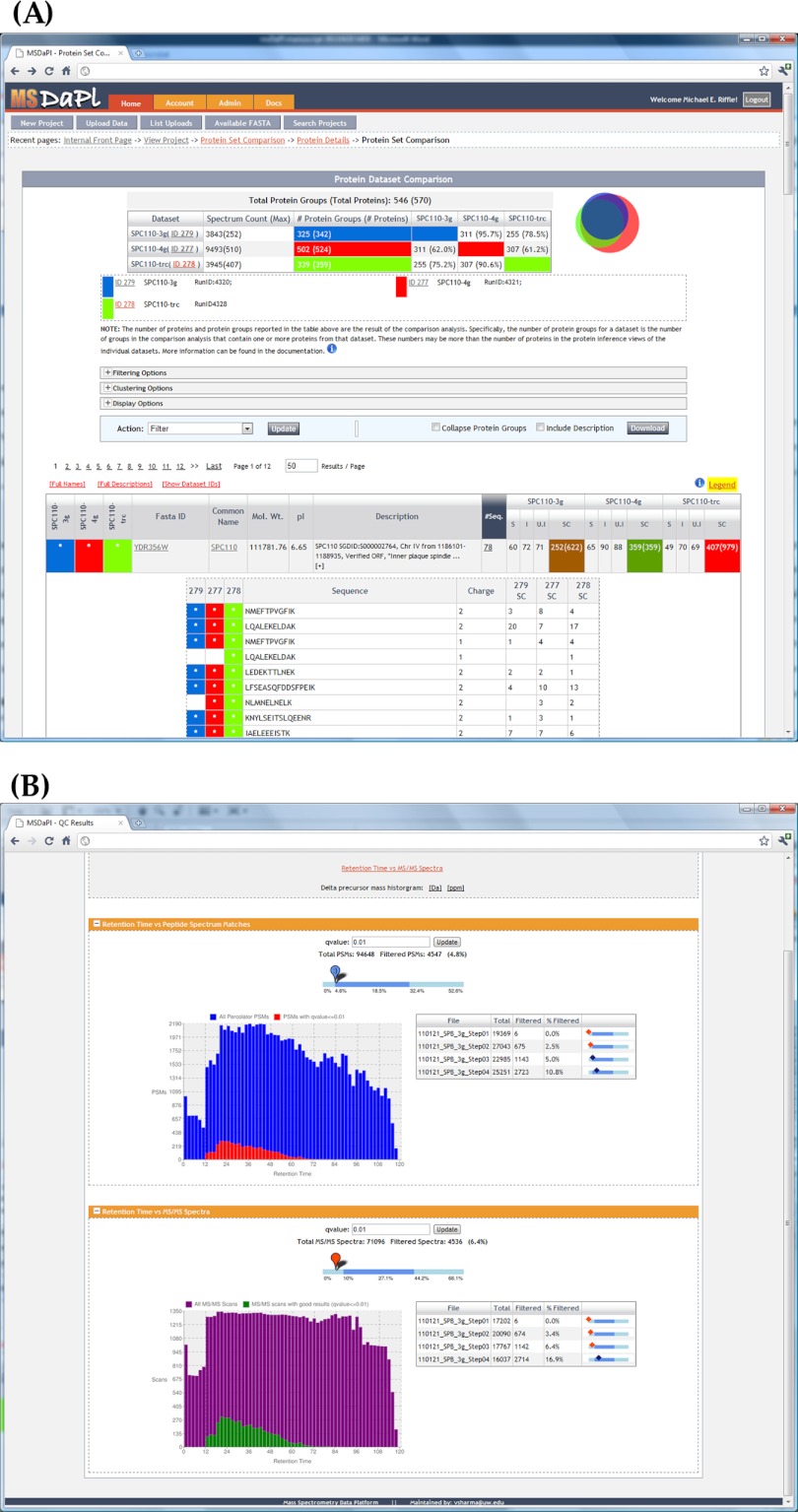
MSDaPl provides tools for filtering and viewing protein lists from experiments, including tools for comparing and contrasting protein lists across multiple experiments (Fig. 4A). For a given experiment, users may filter protein results by the confidence of the identifications, protein physical properties, GO annotations, names, descriptions, and peptide sequence.
Fig. 4.
Screenshots from the MSDaPl web application interface. A, A view of the experiment comparison interface in which three experiments are being compared. Each experiment is assigned a color, which is used consistently throughout the page. The top table and Venn diagram indicate the degree of overlap between the three experiments. The bottom table shows the results for a single protein with the optional view of the peptides used to identify the protein and their presence (including abundance) or absence in each of the experiments. B, A view of the quality control interface in MSDaPl. The top graph indicates the total number of PSMs and the proportion of quality PSMs to total PSMs found as a function of retention time, and the bottom graph indicates the total number of scans and the proportion of scans with quality matches to total scans as a function of retention time.
When comparing results across experiments, users are presented with summary statistics describing numbers of proteins found in each experiment (including a Venn diagram depicting the intersection of proteins among all experiments) and a list of proteins found in all experiments that includes visual cues indicating in which experiment each protein was found. Users may filter this protein list using not only the filtering options described above, but may also filter the list based on presence (or absence) of the proteins in any of the experiments being compared. Specifically, the user may choose to only include proteins from specific experiments if it's (1) present in all other experiments being compared (AND), (2) present or absent in other experiments (OR), (3) not present in other experiments being compared (NOT), or (4) only include proteins present in one of the experiments being compared, but not the other (XOR).
The protein list may also be sorted according to hierarchical clustering of the normalized spectrum counts, which provides a method for comparing relative protein abundance across the experiments. The clustering may be viewed as a PDF-formatted heatmap or as an interactive Hypertext Markup Language heatmap where the user may click on rows of the heatmap to “zoom” to that protein's row in the main protein list or view a bar graph depicting that protein's spectrum counts across the experiments being compared.
Note that the results may be compared across experiments, and indeed, across totally separate projects where disparate software pipelines and FASTA files were used in the analysis. There are no restrictions on specific pipelines or naming databases that must be used in order to perform this analysis.
Gene Ontology Analysis
To help apply biological context to protein inference results, MSDaPl provides two types of Gene Ontology (GO) analysis to users. The first type provides a pie graph and list of the most represented GO terms based on the GO annotations for the proteins found in a given run. The second type provides a pie graph and list of the most statistically enriched GO terms based on an enrichment analysis that calculates a p value for each GO term based on the hypergeometric distribution given the number of proteins found in the run, number of proteins in the respective organism annotated with the given GO term, the number of proteins in the run annotated with the given go term, and the total number of annotated proteins for the respective organism. As of this writing, GO analysis is limited to S. cerevisiae, C. elegans, D. melanogaster, and H. sapiens.
Protein Inference
An implementation of the parsimonious protein identification method in the IDPicker (28) algorithm for protein inference has been integrated into MSDaPl. For a given peptide search, all peptides that meet user-supplied cutoff criteria are assembled into a parsimonious protein list that the user may browse, compare, or contrast with other protein inference results (including results that have been imported from ProteinProphet), or perform analysis such as testing for GO term enrichment. As of this writing, protein inference can only be performed on peptides and statistics generated by the Percolator algorithm; however, supporting other algorithms is a relatively simple matter of developing an interface for filtering peptides based on statistics produced by those other algorithms, and is very likely to be included in future development.
Quality Control
For experiments containing peptide search analysis that contain peptide spectrum match (PSM)-level q-value scores (currently only Percolator is supported), users may click the “Statistics” link in the peptide search result section to view graphs that provide a metric for the quality of the mass spectrometry experiment (Fig. 4B). The graphs indicate the proportion of PSMs with q-values better than a given cutoff to all PSMs and the proportion of MS/MS scans that resulted in a quality PSM to all MS/MS scans.
Job Queue and Data Importers
Software for importing data into MSDaPl has been developed and is included in the MSDaPl distribution. The import system has been designed as a job queue system, where users of the MSDaPl web interface submit requests to upload data and these requests are saved to the database as jobs to be completed by a separate Java program. This program is responsible for querying for new jobs, transferring the data, importing the data to the MSDaPl database, and notifying the user when complete. The pending and completed jobs may be viewed and managed via the web interface. The import program currently supports files with the following formats:
XML (mzML, mzXML, mzIdentML, pepXML (5), protXML (5), and Percolator)
MS1, MS2, and compressed MS2 (CMS2)
SQT
The import libraries for each of these formats implement a common interface that may be implemented by other developers to add support for other file formats to the import system. Additionally, jobs may be submitted to the job queue system directly via web services in order to better support command-line programs or automated proteomics pipelines.
FASTA Parser
As described in the “Data Architecture” section, MSDaPl makes use of a database (“NR_SEQ”) to map the accession strings present in specific FASTA files to internal protein identification numbers, in order to unambiguously refer to the same protein across experiments and to provide current and preferred naming and annotations for those proteins in reported results. To accomplish this mapping, the FASTA file used in the analysis must be parsed into the database previous to the data being uploaded. A Java program for parsing FASTA files and storing the results in the “NR_SEQ” database is included with MSDaPl. It is important to note that the program uses the name of the file (e.g. “yeast_orfs_20120301.fa”) to identify specific FASTA files. Data uploads to the MSDaPl database also use the name of the FASTA file used in the experiment to determine how to map accession strings to protein identification numbers. It, therefore, becomes critical that any new or alternative version of a FASTA file used to search data have a unique filename, otherwise there is no longer a guarantee that an accession string for a particular filename will map to a unique sequence.
Current Usage
MSDaPl was developed in collaboration with two research consortia, the YRC (http://www.yeastrc.org/) and the University of Washington Proteomics Resource (http://www.proteomicsresource.washington.edu/). Each of these groups comprise multiple proteomics laboratories, each with distinct software analysis pipelines and data-format support requirements. In these environments, MSDaPl has been used to store, disseminate, or analyze data from many mass spectrometry experiments. To date, the amount of data in the YRC instance of MSDaPl includes 1927 experiments, which includes 11,109 MS/MS runs, 88 million scans, and 495 million PSMs. The University of Washington Proteomics Resource instance of MSDaPl includes 2055 experiments, which includes 9207 runs, 152 million scans, and 725 million PSMs. Both of these installations grow regularly, and thus far, the data design has scaled sufficiently to support the respective amounts of data. Readers concerned with scalability may use these numbers as a guide for MSDaPl's suitability for their data.
Future Directions
New features in MSDaPl are predominately driven by requests by users and collaborating proteomics laboratories. Current emphasis is on developing improved support for protein and peptide quantification, which will be designed similarly to other types of data in MSDaPl: first developing core support for quantification data that is independent of specific pipelines, then extending this core support to include data generated by specific pipelines. In addition, it is our intent to expose the data in MSDaPl to third-party bioinformatics applications via web services that will allow them to search, filter, read, or write to the database. Finally, a system to allow users to define custom queries of the database from the web interface will be explored.
Conclusion
MSDaPl is a collaborative data management, data analysis, and data dissemination tool for mass spectrometry-based proteomics. It has been designed to address the shortcomings of a file-based approach and is user-friendly, robust, open-source, and cross-platform. MSDaPl includes support for many of the most popular proteomics pipelines, and thus far, development for any given pipeline has been purely driven by user demand. The software has been designed to be modular and extendible. And it is our hope that by releasing MSDaPl to the community as an open-source project, users will help expand MSDaPl by contributing support for more proteomics tools and file formats.
Footnotes
* This work is supported by grants P41 RR11823 from the National Center for Research Resources and P41 GM103533 from the National Institute of General Medical Studies from the National Institutes of Health; and the University of Washington Proteomics Resource (UWPR95794).
1 The abbreviations used are:
- MSDaPl
- mass spectrometery data platform
- MS/MS
- tandem mass spectrometry
- XML
- extensible markup language
- PSM
- peptide spectrum match.
REFERENCES
- 1. Deutsch E. W. (2010) Mass spectrometer output file format mzML. Methods Mol. Biol. 604, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eisenacher M. (2011) mzIdentML: an open community-built standard format for the results of proteomics spectrum identification algorithms. Methods Mol. Biol. 696, 161–177 [DOI] [PubMed] [Google Scholar]
- 3. Orchard S., Montechi-Palazzi L., Deutsch E. W., Binz P. A., Jones A. R., Paton N., Pizarro A., Creasy D. M., Wojcik J., Hermjakob H. (2007) Five years of progress in the Standardization of Proteomics Data 4th Annual Spring Workshop of the HUPO-Proteomics Standards Initiative April 23–25, 2007 Ecole Nationale Superieure (ENS), Lyon, France. Proteomics 7, 3436–3440 [DOI] [PubMed] [Google Scholar]
- 4. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 5. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd. (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 7. Webber J. T., Askenazi M., Marto J. A. (2011) mzResults: an interactive viewer for interrogation and distribution of proteomics results. Mol. Cell. Proteomics 10, M110.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) Multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Askenazi M., Webber J. T., Marto J. A. (2011) mzServer: web-based programmatic access for mass spectrometry data analysis. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.003988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vizcaino J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenyö D., Eriksson J., Beavis R. (2010) Mass spectrometric protein identification using the global proteome machine. Methods Mol. Biol. 673, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goel R., Muthusamy B., Pandey A., Prasad T. S. (2011) Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol 48, 87–95 [DOI] [PubMed] [Google Scholar]
- 15. Riffle M., Malmström L., Davis T. N. (2005) The Yeast Resource Center Public Data Repository. Nucleic Acids Res. 33, D378–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith B. E., Hill J. A., Gjukich M. A., Andrews P. C. (2011) Tranche distributed repository and ProteomeCommons.org. Methods Mol. Biol. 696, 123–145 [DOI] [PubMed] [Google Scholar]
- 17. Nelson E. K., Piehler B., Eckels J., Rauch A., Bellew M., Hussey P., Ramsay S., Nathe C., Lum K., Krouse K., Stearns D., Connolly B., Skillman T., Igra M. (2011) LabKey Server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics 12, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 19. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 20. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 21. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diament B. J., Noble W. S. (2011) Faster SEQUEST searching for peptide identification from tandem mass spectra. J. Proteome Res. 10, 3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu T., Venable J. D., Park S. K., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., Yates J. R. (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics 5, S174 [Google Scholar]
- 24. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 25. Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Federhen S., Feolo M., Fingerman I. M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Lu Z., Madden T. L., Madej T., Maglott D. R., Marchler-Bauer A., Miller V., Mizrachi I., Ostell J., Panchenko A., Phan L., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Slotta D., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Wang Y., Wilbur W. J., Yaschenko E., Ye J. (2011) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 39, D38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lipman D. J., Pearson W. R. (1985) Rapid and sensitive protein similarity searches. Science 227, 1435–1441 [DOI] [PubMed] [Google Scholar]
- 27. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang B., Chambers M. C., Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]