Abstract
Morphine has long been known to have immunosuppressive properties in vivo, but the molecular and immunologic changes induced by it are incompletely understood. To explore how these changes interact with lentiviral infections in vivo, animals from two nonhuman primate species (African green monkeys and pigtailed macaques) were provided morphine and studied using a systems biology approach. Biological specimens were obtained from multiple sources (e.g. lymph node, colon, cerebrospinal fluid, and peripheral blood) before and after the administration of morphine (titrated up to a maximum dose of 5 mg/kg over a period of 20 days). Cellular immune, plasma cytokine, and proteome changes were measured and morphine-induced changes in these parameters were assessed on an interorgan, interindividual, and interspecies basis. In both species, morphine was associated with decreased levels of Ki-67+ T-cell activation but with only minimal changes in overall T-cell counts, neutrophil counts, and NK cell counts. Although changes in T-cell maturation were observed, these varied across the various tissue/fluid compartments studied. Proteomic analysis revealed a morphine-induced suppressive effect in lymph nodes, with decreased abundance of protein mediators involved in the functional categories of energy metabolism, signaling, and maintenance of cell structure. These findings have direct relevance for understanding the impact of heroin addiction and the opioids used to treat addiction as well as on the potential interplay between opioid abuse and the immunological response to an infective agent.
With 20 million heroin addicts estimated worldwide and countless others addicted to prescription opioids, drug addiction remains a large public health problem. Addicted individuals are susceptible to a broad array of infectious complications and, in the United States alone, drugs of abuse contribute directly to a third of all HIV infections (1, 2). A better understanding of the underlying mechanisms of drug-induced pathology may allow more informed application of opioid replacement therapy as well as the development of additional novel therapeutic interventions.
Opioids induce effects across a multitude of biological systems in vivo and these are not easily categorized into meaningful patterns using standard experimental approaches. Although opioids are known to have effects on immune responses in humans, these effects have only been studied in a limited manner and with conflicting results. For instance, opioids directly suppress phagocytic function in vitro (3), with little to no direct in vitro effects on T cells, natural killer (NK)1 cells, and B cells (4–6). By contrast, in vivo administration of morphine has been shown to depress delayed-type hypersensitivity reaction, cytotoxic T-cell activity, T-cell antigen expression, antibody production, NK cell activity, and neutrophil activity (7–11).
Methadone and buprenorphine, opioids commonly prescribed to relieve symptoms during heroin withdrawal, have also been reported to impact the immune system in potent but diverse ways. Methadone, for instance, has been variously reported to improve (12), impair (11, 13), or have no effect (14) on immune function. Similarly, buprenorphine has been variously reported to improve (15–17) or to suppress (7, 8, 13) immune function, or to have no effect at all (10, 18). Such conflicting information makes it difficult to establish best practices for medical intervention of opioid abuse. By example, it is possible that common opioid replacements such as methadone and buprenorphine have dissimilar effects on biological systems, such that it would be optimal to provide one instead of the other to certain opioid-addicted patients. Similarly, in settings wherein opioids are used for pain relief, it would be preferable to understand which may be immunosuppressive and likely to enhance susceptibility to infection in vulnerable patient populations. Given the existing conflicting and/or insufficient information, biological signatures associated with these drugs could reveal new avenues for therapeutics development. Additionally, such signatures may lead to the discovery of common pathways of addiction amenable to therapeutic intervention. These possibilities prompt a need for a comprehensive analysis of the effects of opioid exposure that can best be carried out using the range of new analytics that comprise a systems biology approach.
To date, only a limited number of studies have investigated the proteomic response to morphine. These studies have predominantly focused on in vivo brain tissues and structures from morphine-treated rodents (19–28) whereas a few have measured the proteomic response in morphine-treated cell cultures from humans (29, 30), rats (31, 32), and Chinese hamster ovaries (33). The majority of the studies (12 out of 15) used two-dimensional gel electrophoresis for protein separation, providing information about changes in protein size and isoelectric point, but with limited throughput, proteome coverage, and dynamic range. On the basis of these results, a recent meta-analysis (34) generated a consolidated database (termed the “morphinome”) composed of 236 differentially expressed morphine-responsive proteins in the central nervous system. However, it is clear from the literature that morphine affects multiple organs that communicate with one another to produce a coordinated response within an individual. Therefore, we chose to obtain a broader integrative view useful for understanding the overall impact of opioids on an organism.
In the current study, morphine was administered over a 20-day study period to six nonhuman primates (NHPs) of two different species (African green monkeys (AGMs) and pigtailed macaques (PTs)), chosen because of their differential susceptibility to pathogenesis induced by the simian immunodeficiency virus (SIV). The data reported here represent a baseline for a follow-on analysis of the morphine effects on the course of SIV infection in these species. Overall, we comprehensively sampled inguinal lymph node, colon, CSF, and peripheral blood, and measured the immunomic and proteomic changes within these various biological compartments before and after the administration of morphine. We report a series of changes that are induced, directly or indirectly, by morphine in each species of NHP.
EXPERIMENTAL PROCEDURES
Study Design
All animal and in vitro procedures were performed using standard protocols and according to guidelines approved by the University of Washington Environmental Health and Safety Committee, the Occupational Health Administration, the Primate Center Research Committee, and the Institutional Animal Care and Use Committee.
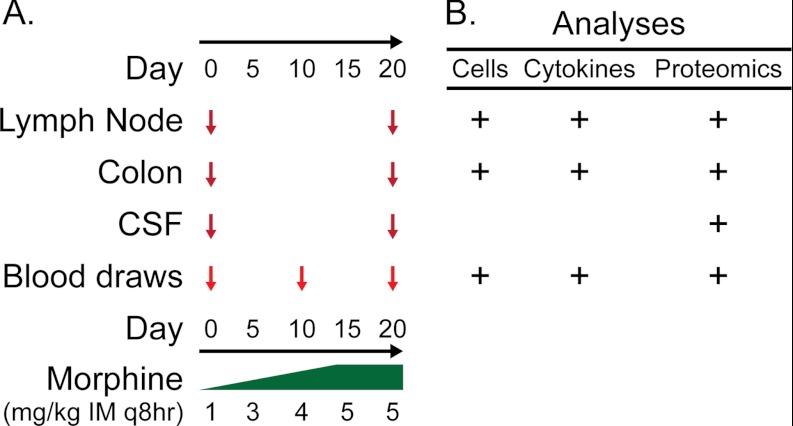
Three African green monkeys (Chlorocebus sabaeus) and three pigtailed macaques (Macaca nemestrina) were housed and followed longitudinally for 85 days at the Washington National Primate Research Center under the supervision of dedicated veterinary staff. The data collected in the initial 20 day morphine arm of this study are reported here, during which time morphine was titrated up to a maximal dose of 5 mg/kg intramuscularly every 8 h as outlined in Fig. 1. Peripheral blood was sampled at day 0, 10, and 20, whereas iliac lymph node biopsies, colonic mucosal biopsies, and cerebrospinal fluid samples were obtained at days 0 and 20 according to guidelines approved by the University of Washington Environmental Health and Safety Committee, the Occupational Health Administration, the Primate Center Research Review Committee, and the Institutional Animal Care and Use Committee. NHPs were sedated for all procedures that required handling using ketamine HCl at a dose of 10–15 mg/kg or tiletamine zolazepam at a dose of 2–8 mg/kg with atropine 0.04 mg/kg. Samples were either shipped overnight or flash frozen and stored at −80 °C on-site until shipping.
Fig. 1.
Study design. Morphine was administered starting at 1 mg/kg intramuscularly every 8 h (mg/kg IM q8hr) and titrated upwards to a maximum dose of 5 mg/kg IM q8hr by study day 20. Animals were sampled across a variety of biologic compartments (A). The tissues and fluids obtained were subjected to cell phenotypic, cytokine, and proteomic analyses (B).
Flow Cytometry
Panels of antibodies used for multiparameter flow cytometry are listed in supplementary Table S1 (including clone, manufacturer, and conjugate). FACS analysis was performed on a four-laser BD LSR-II flow cytometer (BD Biosciences). Complete blood counts (CBC) were determined by absolute counting of 50 μl whole blood after overnight shipping using Trucount absolute counting tubes (BD Biosciences). Peripheral blood mononuclear cells were isolated by density centrifugation using Histopaque®-1077 (Sigma Aldrich, Saint Louis, MO) and then stained using a variety of phenotyping panels (supplementary Table S1). Colonic mucosa was mechanically disrupted, digested with collagenase type II (Sigma Aldrich), strained, and washed for the isolation of mucosal mononuclear cells. Lymph nodes were mechanically disrupted and washed to isolate mononuclear cells (35). Phenotyping was performed by cell surface staining and, when necessary, by intracellular staining following manufacturer's instructions (BD Biosciences and eBiosciences, San Diego, CA) (supplementary Table S1). FACS data were analyzed using FlowJo software v9.1 (TreeStar, Ashland, OR) with standard gating strategies and then transferred into analysis and graphic software including Excel (Microsoft, Mountain View, CA), SPICE (kindly provided by Dr. M. Roederer of the Vaccine Research Center, NIAID/NIH, Bethesda, MD), and GraphPad Prism5 (San Diego, CA).
Plasma Cytokine Analyses
Plasma was isolated after overnight shipment and subsequently stored at −80 °C prior to analysis. Cytokines and lipopolysaccharide-binding protein (LBP) levels were measured using Cytokine Bead Array for non-human primates (BD Biosciences, San Jose, CA), IL-12/23p40 Monkey ELISA (U-Cytech CM, Utrecht, The Netherlands), IFN-α human ELISA kit (PBL Biomedical Laboratories, Piscataway, NJ), and lipopolysaccharide-binding protein (HK315) ELISA kit (Hycult Biotechnology, Uden, The Netherlands) all according to manufacturer's instructions. Cytokine Bead Array data were acquired on a LSR-II flow cytometer (BD) and analyzed using FlowJo Software (Treestar). ELISA data were acquired using a Spectramax spectrophotometer (Molecular Devices, Sunnyvale, CA) by measuring absorbance at 450 nanometers. Data were further analyzed using Excel (Microsoft, Mountain View, CA) and GraphPad Prism5 (San Diego, CA).
Sample Preparation for Proteome Analysis
Samples were denatured in 8 m urea. A bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL, USA) was performed to determine protein concentration. Samples were reduced with 10 mm dithiothreitol for 1 h at 37 °C, and then diluted 10-fold with 50 mm NH4HCO3, pH 7.8. Trypsin digestion was performed with sequencing grade-modified porcine trypsin prepared according to the manufacturer (Promega, Madison, WI). Trypsin was added to all protein samples at a 1:50 (wt/wt) trypsin-to-protein ratio for 3 h at 37 °C, and samples desalted by solid-phase extraction (Discovery DSC-18, SUPELCO, Bellefonte, PA). Peptides were eluted from the solid-phase extraction column using 80% acetonitrile: 20% water with 0.1% trifluoroacetic acid, and dried by SpeedVac SC 250 Express (Thermo Savant, Holbrook, NY).
Whole blood was collected in EDTA collection tubes, followed by centrifugation at 1500 rcf for 15 min at 4 °C. 5 ml top plasma layer was collected, treated with approximately one-half tablet of EDTA-free Complete Mini Protease Inhibitor Mixture (Roche Diagnostics, Indianapolis, IL), and stored at −20 °C. Subsequently, albumin and IgG depletion was performed using the ProteoExtract Albumin/IgG removal kit (EMD Biosciences, San Diego, CA), following the manufacturer's protocol. Briefly, EDTA-plasma was diluted fivefold in cold 1X PBS, and then passed through the removal column resin bed by gravity flow into a collection tube. The removal column was subsequently rinsed twice with 1× cold PBS, with the flow-through added to the same collection tube. Depleted plasma was stored as 1.5 ml aliquots at −80 °C until sample preparation with urea and trypsin as described above. Sample processing conditions were the same for individual tissue/biofluids compartments across AGM and PT groups.
NanoRPLC Separations and MS(/MS) Data Acquisition
The method used in this coupled a constant-pressure (5000-lb/in2) reversed-phase capillary liquid chromatography system (75-μm inside diameter, 360-μm outside diameter, 65-cm capillary, C-18 solid phase; Polymicro Technologies Inc., Phoenix, AZ) to a VelosLTQ-Orbitrap mass spectrometer (MS; Thermo Scientific, Waltham, MA) using an electrospray ionization source manufactured in-house, as previously reported (36–38). The instrument was operated in data-dependent mode with an m/z range of 400–2000. The 10 most abundant ions from the MS analysis were selected for MS/MS analysis using a normalized collision energy setting of 35%. A dynamic exclusion of 1 min was used to avoid repetitive analysis of the same abundant precursor ion. The heated capillary was maintained at 200 °C, and the ESI voltage was held at 2.2 kV (36–38).
Proteome Analysis
Neither AGMs nor PTs have a fully sequenced genome. The rhesus macaque (Macaca mulatta) is the closest relative with a fully sequenced genome (39). The rhesus macaque's Ensemble protein list (40) (http://www.ensembl.org) was obtained from InParanoid (M.mulatta.fa 21-Apr-2009) (41–43), containing 21,905 nonredunant protein sequences (http://omics.pnl.gov), and used by SEQUEST software (Version v.27, Rev 12, Thermo Fisher Scientific, Waltham MA) to search the MS/MS spectral data. Although the genomes of AGM and PT are not fully sequenced, the Macaca mulatta protein list has previously been used in other NHP studies (44, 45), and there is high sequence similarity between members of the Primate taxon. Porcine trypsin was added to the database as an expected contaminant. No cleavage specificity was defined in the database searching, and no fixed or variable modifications were considered in the MS/MS analysis. Tandem mass spectral analysis peaks were extracted with ExtractMSn (version 5.0, Thermo Scientific) for subsequent analysis using 3.0 and 1.0 Da for parent and fragment ion tolerances, respectively. For label-free quantification of peptide abundance, we employed the accurate mass and time (AMT) tag strategy (46) and for AMT tag database creation, raw SEQUEST results were filtered on: (1) Xcorr ≥ 1.9 for charge state +1, (2) Xcorr ≥ 2.2 for charge state +2, and (3) 3.75 for charge state +3.
Homologous proteins, including protein isoforms, are typically identified by the same peptide or group of peptides. It is generally impossible to unambiguously assign such peptides to a specific protein. Signature peptides, defined as peptides unique to a single isoform, can provide differentiating information that can be used to filter ambiguous peptide assignments. However, the use of signature peptides falsely assumes that other isoforms, whose sequence may not code for identifiable signature peptides, do not exist in the sample. To address this, identified peptides were mapped to proteins and then manually grouped into protein families. Raw data and collated peptide identification information are available to the community as supplemental data.
High resolution LC-MS features were deconvoluted by Decon2Ls (version 1.0.2, using default parameters) and aligned to the AMT tag database using VIPER (version 3.45 using default parameters), and the theoretical mass and observed normalized elution times (NET) for each peptide. Peptide alignment results were further refined with a mass error < 2 ppm and FDR < 5% (47). Relative peptide abundance was assigned using peak intensity. Data analysis was facilitated by a number of both published (48–52) and unpublished in-group development tools that are freely available for download at http://omics.pnl.gov.
Statistical Analysis
Outlier data sets were identified using a peptide-centric approach that considers five statistical metrics to generate a robust Mahalanobis distance score (53). Three data sets resulted in extreme peptide abundance distributions (p value < 0.001), and were removed from the analysis. Relative peptide abundances measurements were scaled in DanteR (version 0.1.1) (R Forge: https://r-forge.r-project.org/projects/rgtk2extras/). A model-based statistical approach was used to impute missing peptide abundance values (54) and a protein-level ANOVA was applied to identify statistically significant changes (p < 0.01) in protein expression levels. To adjust for multiple comparisons, a Benjamini-Hochberg correction was applied to p values (55). Relative protein abundances were obtained by averaging scaled peptide abundance values, and used for heatmap visualizations and Pearson product-moment correlation analyses. Statistical analysis on cellular and cytokine data was performed using paired t-tests in R (http://www.r-project.org/).
Pathway Analysis
The rhesus macaque genome provided limited functional annotation for both PT and AGM data sets, and software applications typically used for mammalian functional enrichment, including DAVID (56, 57), InGenuity IPA, and Panther (58), currently have limited or no support for NHP identifiers. Therefore, rhesus macaque Ensembl protein identifiers were converted to human identifiers with the biomaRt package implemented in R (59), using Ensembl Genes 61 database and the Macaca mulatta genes (MMUL_1.0) data sets. Human identifiers were queried for pathway analysis in DAVID (56, 57) and InGenuity IPA for functional protein annotation. Due to complexities inherent to pathway analysis and differences in the functional organization between different databases, such as gene ontology (GO) and IPA, functional assignments were manually consolidated. The biomaRt package was also used to convert rhesus macaque Ensembl protein identifiers to rat UniProt identifiers to query the Morphinome database (34) for proteins previously associated with morphine administration.
RESULTS
Overall Summary of Proteomic Response to Morphine Treatment
To determine the systemic response to morphine, AGMs and PTs were treated with increasing doses of morphine (ranging from 1–5 mg/kg over a period of 20 days) and monitored, as outlined in Fig. 1. Samples were obtained from lymph node, colon, and CSF on days 0 and 20 and from peripheral blood on days 0, 10, and 20, and these samples were subjected to immunomics and proteomics analyses. A high throughput, label-free quantification (AMT tag) approach resulted in the confident identification and quantification of 1771 unique proteins from lymph node (1196 proteins; 4896 peptides), colon (1225 proteins; 5034 peptides), CSF (253 proteins; 2341 peptides), and plasma (141 proteins; 2049 peptides). Human homologs were identified for 1548 NHP proteins (87%). Morphine treatment resulted in significant changes (p ≤ 0.01) in the abundance of 689 proteins in the tissues and fluids analyzed: 422 (35%) in the lymph node, 215 (18%) in the colon, 113 (45%) in CSF, and 56 (40%) in plasma.
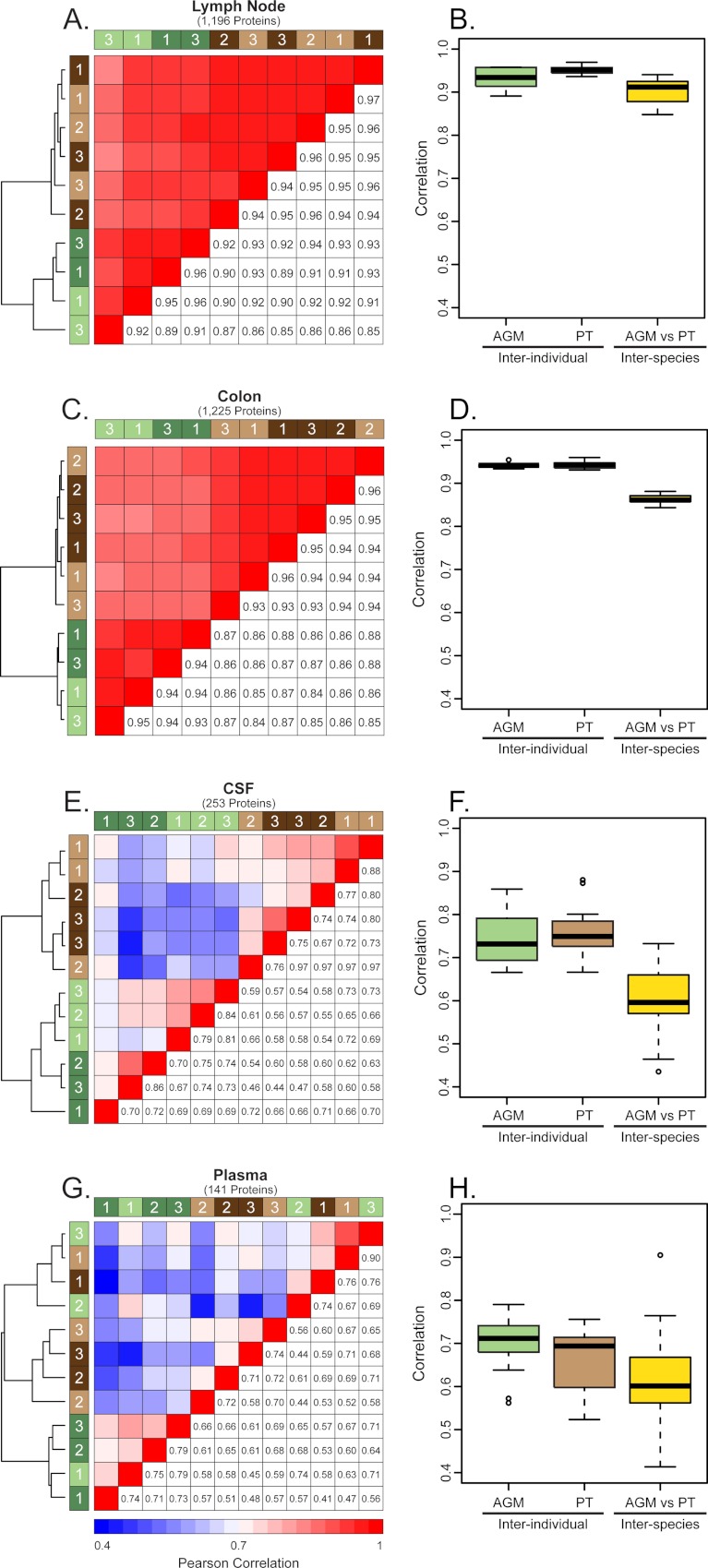
Correlation analyses were performed to measure inter-individual and inter-species variation in tissue and fluid compartments before and after treatment with morphine. The highest correlation between and within species was observed in the lymph node (0.92 ± 0.03 [mean correlation R value ±standard deviation]) and colon (0.90 ± 0.04) (Figs. 2B and 2D, respectively). Comparing the tissues, the colon had the greatest inter-individual correlation (AGMs: 0.94 ± 0.007 and PTs: 0.94 ± 0.008) and the lowest inter-species correlation (0.86 ± 0.01) (Fig. 2D). By contrast, the lowest inter-individual and inter-species correlation was observed in the CSF (0.67 ± 0.10) and plasma (0.63 ± 0.10) (Figs. 2F and 2H, respectively). Overall, greater variation in morphine-induced protein expression was observed in the CSF and plasma compared with the lymph node and colon. In the results presented below, the global response to morphine will be discussed in the context of individual compartments, i.e. lymph node, colon, CSF, and peripheral blood.
Fig. 2.
Inter-individual and inter-species proteomic variation. Correlation measurements of inter-individual and inter-species variation based on relative protein abundances between lymph node (A, B), colon (C, D), CSF (E, F), and plasma (G, H). For the heatmaps (A, C, E, and G), the number of proteins considered in each analysis is represented in parentheses below the compartment title. Green bars represent AGMs and brown bars represent PTs, with light and dark bars indicating pre- and postmorphine, respectively. The numbers within the colored bars indicate represents the animal number. Pearson correlation values between NHP individuals are reported in the lower right cells and upper left cells are color-coded to graphically represent the correlation, with red representing strong correlation and blue representing weak correlation. Hierarchical clustering was performed and the results are indicated by the dendrogram at the left of each heatmap. Boxplots (B, D, F, and H) indicate the interquartile ranges of correlation values. The median is shown by the horizontal black line, with vertical whiskers representing the range. “AGM” and “PT” indicate the inter-individual correlation, both pre- and postmorphine, whereas “AGM versus PT” illustrates the inter-species variation.
Morphine Suppressed T-cell Activation in the Lymph Node
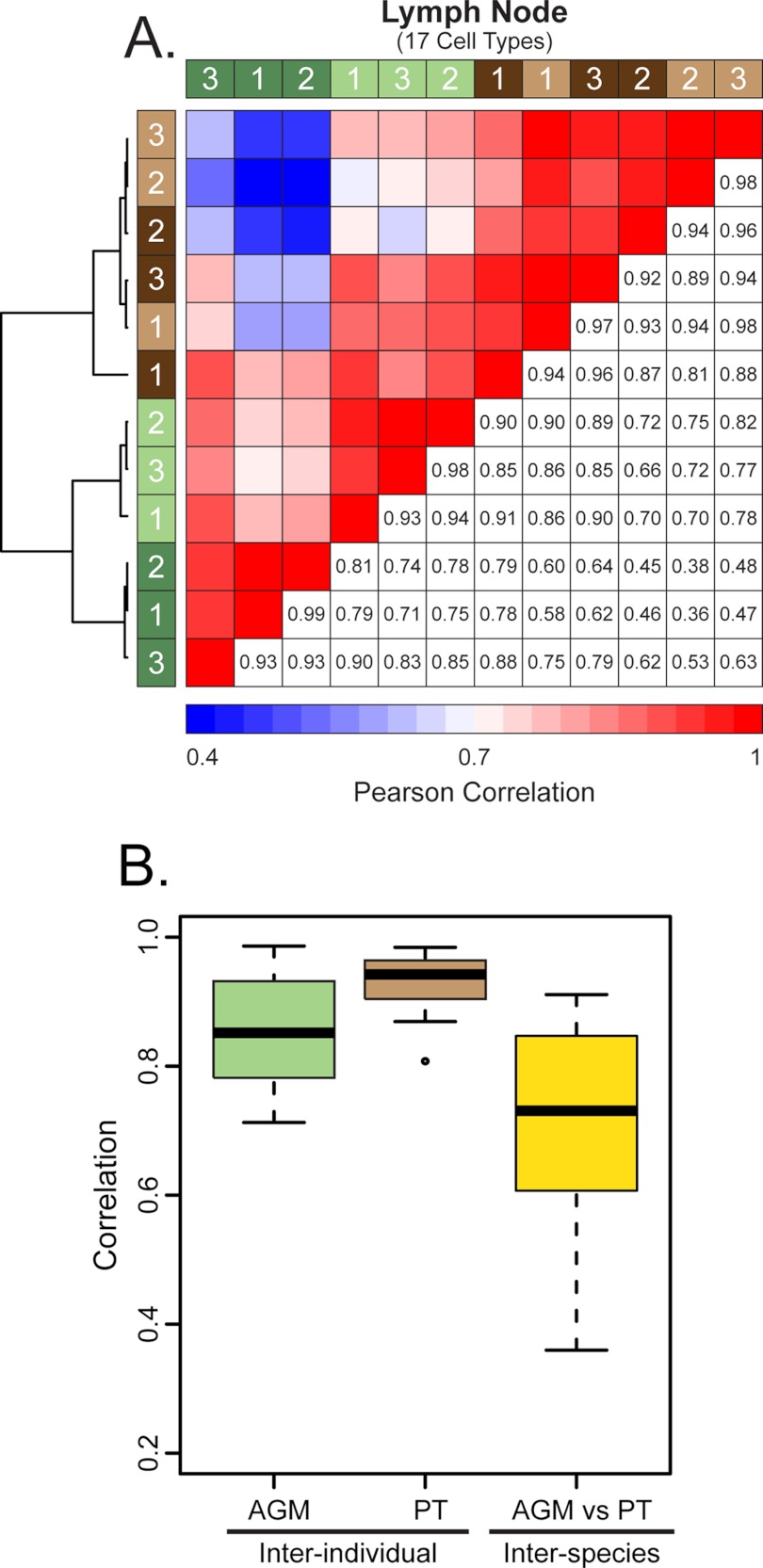
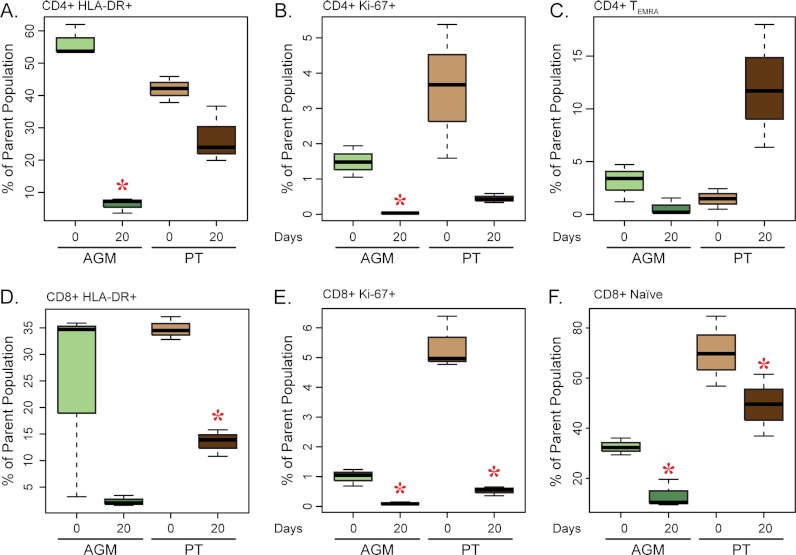
Similar to the proteomics results, the immunomics of the lymph node showed a bifurcation in the clustering dendrogram between AGMs and PTs, indicating that the greatest level of variance in the immunomics data was between the two species rather than between individuals of each (Fig. 3). In contrast to the PTs, all three premorphine biological replicates from the AGMs clustered together on a separate branch relative to the postmorphine samples. Although there were no significant changes in the percentage of discrete T-cell subpopulations between the two species after morphine treatment (supplemental Fig. S1), there were significant decreases in T-cell activation, as reflected by the fraction of CD4+ HLA-DR+, CD4+ Ki-67+, CD8+ HLA-DR+, and CD8+ Ki-67+ as well as of CD8+ naïve T cells (Fig. 4). PTs showed a trend of increasing CD8+ TEMRA cells during morphine exposure that was not apparent for the AGMs (Fig. 4F). Although not statistically significant, a decrease in the frequency of classical CD14+CD16− monocyte was also observed after morphine treatment in both species (supplemental Fig. S1).
Fig. 3.
Correlation measurements of inter-individual and inter-species variation based on cell counts within the lymph node. A, is a correlation heatmap of Pearson correlation values, with the number of cell types determined in the analysis above the heatmap in parentheses. Green bars represent AGMs and brown bars represent PTs, with light and dark bars indicating pre- and postmorphine, respectively. The numbers within the colored bars indicate the animal number. Pearson correlation values between NHP individuals are reported in the lower right cells and upper left cells are color-coded to graphically represent the correlation, with red representing strong correlation and blue representing weak correlation. Hierarchical clustering was performed and the results are indicated by the dendrogram at the left of each heatmap. The boxplots Panel B indicate the interquartile ranges of correlation values. The median is shown by the horizontal black line, with vertical whiskers representing the range. “AGM” and “PT” indicate the inter-individual correlation, both pre- and postmorphine, whereas “AGM versus PT” illustrates the inter-species variation.
Fig. 4.
Analysis of activated T cells in the lymph node. Boxplots representing the percent distribution of CD4+ HLA-DR+ (A), CD4+ Ki-67+ (B), CD4+ TEMRA (C), CD8+ HLA-DR+ (D), CD8+ Ki-67+ (E), and CD8+ Naïve (F) cells. The dark bar within the box represents the median, the box indicates the interquartile range, and the whiskers show the overall range. AGM values are indicated by light and dark green boxes for pre- and postmorphine, respectively. PT values are represented by light and dark brown boxes for pre- and postmorphine, respectively. Red asterisks indicate a significant change (p < 0.05) in cell count during morphine administration as determined by a paired t test.
The magnitude of the lymph node proteomic response, in terms of number of proteins significantly altered in abundance, was greater in the AGMs compared with PTs, with 326 and 175 proteins altered, respectively (supplemental Fig. S2A). There were 79 proteins that were differentially expressed in both species (supplemental Fig. S2). Although a small subset of proteins increased in abundance during morphine treatment, 267 (82%) and 166 (95%) of the proteins showed significant decreases in abundance in the AGMs and PTs, respectively, mirroring the suppressed immunomics parameters of T-cell activation observed in the blood and lymph node.
Opioids have been reported to perturb metabolic processes in the central nervous system (23, 60) and proteomic analysis of the lymph node revealed 52 metabolic proteins (43 in the AGMs and 23 in the PTs) that were decreased in abundance (supplemental Fig. S3). Of note, 14 of these proteins were associated with energy metabolism (including AHCY, ATP5B, (28) DBI, ENO1, FASN, MDH2, PKM2, PRDX1, TKT, TPI1, and VCP) and were significantly decreased in both AGMs and PTs. Energy pathways with suppressed protein levels include glycolysis (12 proteins), pyruvate metabolism (6 proteins), oxidative phosphorylation (5 proteins), pentose phosphate pathway (4 proteins), the tricarboxylic acid cycle (3 proteins), and glycerolipid metabolism (3 proteins) (supplemental Fig. S4). Concomitant with suppression of proteins involved in energy metabolism was a decrease in the abundance of proteins involved in the inflammatory process [e.g. COX2 (cyclooxygenase 2), COTL1 (coactosin-like 1), and IL-16 (interleukin-16)] and of those involved in leukocyte recruitment response to inflammation [e.g. RAC1 (ras-related C3 botulinum toxin substrate 1), TLN1 (talin 1), and RHOB (ras homolog gene family, member B)].
The 422 significantly altered proteins in the lymph nodes of AGMs and PTs (supplemental Fig. S2) were queried against the previously-published morphinome database (34). Rat UniProt identifiers were assigned to 372 macaque proteins using BioMart and 37 of these proteins were present in the morphinome database (supplemental Table S2). These proteins included heat-shock proteins (HSPA5, HSPA9, and HSPBL2), multiple energy metabolism proteins (ENO2, ENO3, MDH2, TKT, DLD, TPI1, and AKR1B1), and proteins associated with maintaining structural integrity (LMNA, VCP, CAPZB, YWHAZ, and DPYSL2).
Discordant Response in the Colon
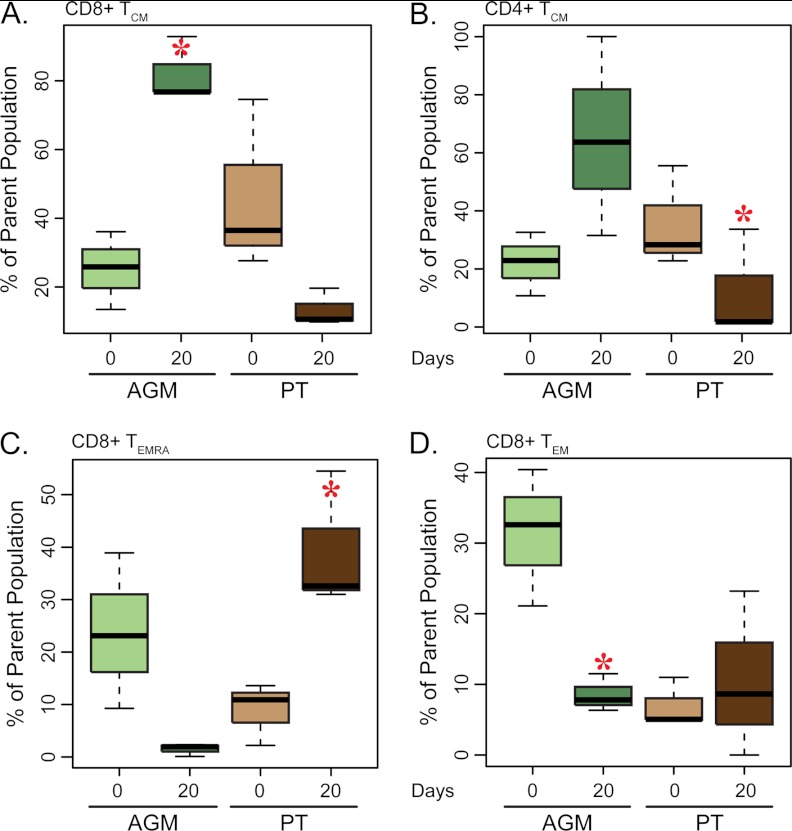
The percentage of CD8+ T cells was increased within the colonic mucosa of AGMs after morphine administration, whereas PTs had a slight decrease in CD8+ T-cell percentages over the same study period (supplemental Fig. S5). In contrast to AGMs, which showed a significant increase (p < 0.05) in CD8+ T-cell maturation toward central memory (TCM), PTs had a considerable, yet non-significant, decrease in CD8+ TCM levels (Fig. 5A). PTs did show a significant decrease in CD4+ TCM cells (Fig. 5B). Discordance was also observed in CD8+ TEMRA and TEM cells. AGMs showed considerable, yet nonsignificant decrease, in CD8+ TEMRA cells, whereas PTs had a significant increase (Fig. 5C). CD8+ TEM cells showed a significant decrease in percentage in the AGMs, but the PT response was highly variable (Fig. 5D).
Fig. 5.
Analysis of T-cell subpopulations in the colon. Boxplot representing the percent distribution of CD8+ Temra (A), CD8+ Tcm (B), CD8+ Tem (C), and CD4+ Tcm (D) cells. The dark bar within the box represents the median, the box indicates the interquartile range, and the whiskers show the overall range. AGM values are indicated by light and dark green boxes for pre- and postmorphine, respectively. PT values are represented by light and dark brown boxes for pre- and postmorphine, respectively. Red asterisks indicate a significant change (p < 0.05) in cell count during morphine administration as determined by a paired t test.
Protein abundance changes in the colon resulting from morphine treatment were measured in colon biopsies from AGMs and PTs. The proteomic response of the colon to morphine showed 215 proteins that changed significantly in relative abundance, 103 and 143 in AGMs or PTs (supplemental Fig. S6A), respectively while 31 (30%) proteins were significantly altered in both species (supplemental Fig. S6B). From the 31 proteins that showed a significant change in abundance, only three shared directionality in the NHPs: profilin 1 (PFN1) and annexin A1 (ANXA1) were decreased and peptide YY (PYY) was increased. Similar to observations in the lymph node, the predominant response in AGMs was a decrease in protein abundance. From the 72 (70%) proteins that showed a significant change exclusively in the AGMs, 49 of these proteins were decreased (supplemental Fig. S6C). In contrast, of the 109 (76%) proteins that showed a significant change exclusively in the PTs, the majority, 87 proteins, were increased in abundance (supplemental Fig. S6D). Paralleling the immunomics measurements, the proteomics results showed discordant and heterogeneous responses between the AGMs and the PTs at the colonic mucosa.
To map the 215 proteins to the morphinome database, rat UniProt accession identifiers were assigned to 192 macaque proteins. These rat identifiers were queried against the morphinome database, identifying 14 proteins significantly changed in the colon and previously reported in the literature (34) (supplemental Table S1).
CSF Proteomic Response
Morphine exposure resulted in altered expression of 113 CSF proteins, 107 in the AGMs and 17 in the PTs (supplemental Fig. S7A). From the 113 significant proteins in the CSF, 15 showed a significant change in both AGMs and PTs (supplemental Fig. S7B). These 15 proteins included several proteins previously shown to be altered by morphine, including GOT1 (aspartate aminotransferase) (34). Although some proteins (including VIM, ECM1, and APOA2) showed reduced relative abundance in the CSF of AGMs during morphine administration, the predominant effect was an increase in abundance in AGMs (supplemental Fig. S7C). Some proteins were exclusively altered in AGMs, including NCAM1 (neural cell adhesion molecule 1) and NTM (neurotrimin). Significant increases of AGA (l-asparginase), OPCML (opioid binding protein/cell adhesion molecule-like), NRCAM (neuronal cell adhesion molecule), and NPY (neuropeptide Y) were exclusively found in AGM CSF and not in PT CSF. AGA is an enzyme that cleaves N-linked oligosaccharides from glycoproteins and has been linked to morphine-dependent immunosuppression in lymphoid cells (61, 62). OPCML is a GPI-anchored protein that exhibits selectivity for mu opioid receptor (MOR) ligands and acts as a potent tumor suppressor protein (63). NRCAM is an adhesion protein typically expressed in neurons linked to reward and memory, and is involved in polysubstance abuse (64). A significant decrease of TTBK2 (tau tubulin kinase 2) protein was measured in both AGMs and PTs. TTBK2 is a serine-threonine kinase that putatively phosphorylates tau and tubulin proteins. Dysfunction of TTBK2 has been linked to Alzheimer's disease (65) and spinocerebellar ataxia type 11 (SCA11) (66), a neurodegenerative disease characterized by progressive ataxia and atrophy of the cerebellum and brainstem. Chronic morphine treatment of rats has been associated with an increase in NPY expression in the hypothalamus (67), indicative of increased food intake and decreased physical activity. NPY has also been linked to morphine-mediated immunosuppression of NK cell activity (68).
AGMs Experienced Larger Decreases in Peripheral Blood T-cell Activation State After Morphine Treatment
In the case of the peripheral blood, CD4+ T-cell percentages remained constant while CD8+ T-cell percentages slightly increased by day 20 in both AGMs and PTs (supplemental Fig. S8A). Mirroring these changes, the percent of CD4−CD8− (DN%) T cells decreased in both species by day 20 (Fig. 6A). The percentages of regulatory T cells (Treg) were not significantly altered by morphine administration (supplemental Fig. S8A). Most significant was the decrease in activated T cells that was associated with the morphine treatment period. By day 20, for instance, both species (and especially the AGMs) had a statistically significant decrease in the fraction of circulating Ki-67+CD4+ T cells (Fig. 6B) and both also showed decrease in the fraction of circulating HLA-DR+CD4+ T cells (supplemental Fig. S8A). Of note, AGMs started with a higher baseline activation state in all T-cell compartments. Additionally, the fraction of circulating naïve T-cell levels was decreased in both species after morphine treatment (supplemental Fig. S8A). In terms of circulating myeloid cells, PTs had a decrease in the frequency of nonclassical CD14dimCD16− monocytes (Fig. 6C) and a relative increase in the frequency of in classical CD14hiCD16− monocytes, whereas AGMs had a decrease in the frequency of the latter population (Fig. 6D). Although the expression of HLA-DR remained the same in PTs, HLA-DR expression decreased on the AGM non-classical monocyte population (data not shown). There were no statistically significant changes in plasma cytokine measures after morphine treatment (supplemental Fig. S9). There was, however, a trend toward a decrease in pro-inflammatory cytokine IL-12 levels in the plasma of AGMs and a decrease in IFN-gamma levels in the plasma of PTs.
Fig. 6.
Analysis of T cell and myeloid subpopulations in the peripheral blood. Boxplot representing the percent distribution of CD4−CD8− (A) and CD4+ Ki-67+ (B) T cells, CD14dim/CD16− nonclassical monocytes (C), CD14+/CD16− classical monocytes (CD14 high) (D), CD8+ Ki-67+ (E), and CD8+ TEM (F) cells. The dark bar within the box represents the median, the box indicates the interquartile range, and the whiskers show the overall range. AGM values are indicated by light and dark green boxes for pre- and postmorphine, respectively. PT values are represented by light and dark brown boxes for pre- and postmorphine, respectively. Red asterisks indicate a significant change (p < 0.05) in cell count from day 0 to day 20 as determined by a paired t test.
Similar to the immunomics, discordant responses were observed in the plasma proteome analysis between AGMs and PTs, with AGMs showing a greater proteomic response to morphine (two increased, 49 decreased) compared with PTs (19 increased) (supplemental Fig. S10A). Of note, several complement proteins (e.g. C3, C4A, and C7) were significantly decreased in the AGMs and significantly increased in PTs. Clusterin (CLU, formerly known as cytolysis inhibitor), which is involved in the complement pathway, was significantly decreased in AGMs and increased in PTs (supplemental Fig. S10B). Morphine has been reported to both enhance (69) and to suppress (70) complement receptor-mediated phagocytosis. Conflicting reports in the literature and the discordance in complement proteins observed between AGMs and PTs suggest that the differential complement response to morphine may be species-dependent (supplemental Figs. S10C, S10D).
DISCUSSION
The biological impact of opioid use is only partly understood, yet the global public health impact of these drugs is quite large. Clearly, the effects of in vitro experiments measure only the direct effects of these drugs without taking into account indirect and secondary pathways that may propagate opioid-associated impacts within a biological system. Here, we have provided a comprehensive characterization of the in vivo effects of opioids in the context of two different NHP species. We have found that morphine produces an immunosuppressive effect in the lymph node of both NHPs, paralleled by decreases in activation levels of T cells in the peripheral blood of AGMs and PTs. Substantial decreases in energy metabolism proteins were concurrently measured within the lymph node proteome. These findings suggest that the observed decreases in energy metabolism proteins may be contributing to the decreased T-cell activation within lymph node and peripheral blood. Findings across gut mucosa, CSF, and plasma were inconsistent between the species, suggesting that protein changes that are highly tissue- and species-specific may accompany the response to morphine in vivo. These findings provide a framework for understanding the effects of morphine across a highly relevant but complicated in vivo system and have identified protein pathways of interest that may be playing key roles in immune-inhibitory actions of opioids.
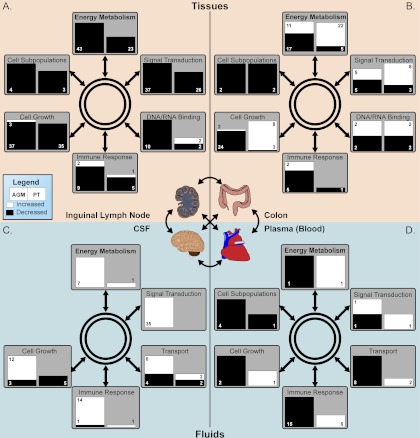
Overall, distinct responses to morphine were observed in the proteomic data across the collected lymph node, colon, CSF, and peripheral blood samples. For instance, although protein expression was largely suppressed in expression in the lymph nodes of both AGMs and PTs (Figs. 7 and supplemental Fig. S2), protein abundances were predominantly increased in the CSF of AGMs but minimally impacted in PTs (Figs. 7 and supplemental Fig. S2). It is this variation in response between NHP species that is the most intriguing. Numerous studies have focused on the immunomodulatory effect of morphine in NHPs (typically the rhesus macaque) and the effect that chronic morphine exposure exerts during simian immunodeficiency virus (SIV)/simian-human immunodeficiency virus (SHIV) infection by increasing viral pathogenesis (71–73). Given the large discrepancy in the response to morphine between AGMs and PTs, this study raises the question of how diverse NHP species may respond to SIV/SHIV infections during different opioid administration regiments.
Fig. 7.
Model schema of morphine immunosuppressive effects across various biological compartments in NHP. Samples were divided between tissues (A and B) and fluids (C and D). The tissue samples were inguinal lymph node (A) and colon (B) biopsies whereas the fluid samples were composed of CSF (C) and plasma (D). The left and right sides of the colored box indicate significant changes in AGMs or PTs, respectively. The size of the boxes are scaled to represent the number of proteins significantly altered within the functional category and numbers indicate the number of proteins whose abundance is significantly changed or the number of cellular subpopulations significantly altered in the case of “Cell Subpopulations.” White represents a significant increase and black a significant decrease.
This study directly compares the responses to morphine in multiple organs of two NHP species by integrating proteomic and immunomic response profiles. In terms of inter-species correlations, the collected tissues showed higher correlation between both individuals and species, while the fluid compartments (i.e. CSF and blood) were more variable. Fewer proteins were observed in the fluids compared with the tissues and the overall distribution of protein abundance values showed greater variability in the fluids compared with the tissues (supplemental Figs. S11C and S11D versus supplemental Figs. S11A and S11B), suggesting that this increased variability was not because of a few outlier protein abundances but rather due to an inherent characteristic of fluid compartments (74). Plasma also provided the least amount of protein identifications and subsequently was the least informative of all the biological compartments. Additional abundant protein depletion steps were used to achieve a higher dynamic range of detection for plasma, which could be a contributing factor to this variability, whereas the NHP CSF was taken cleanly through preparation with minimal processing. However, plasma results still reflected a clear differentiation of protein expression between species with predominant down-regulation in AGMs compared with PTs (Figs. 7 and supplemental Fig. S10). These observations highlight the perspective gained by broadening the proteome analysis across diverse compartments where indirect effects of the core morphine response may be evident.
In the lymph nodes, the vast majority of significantly altered proteins (82% in AGMs and 95% in PTs) decreased in abundance. This decrease in protein abundance likely contributes to the immunosuppressive effects reported during morphine treatment (75) and parallels the decreased T-cell activation observed in the NHPs (Fig. 7). Several functional pathways were altered during morphine treatment, but cellular energy metabolism was by far the most affected. Many key proteins involved in energy metabolism (e.g. MDH2, TKT, DLD, TPI1, ENO2, and ENO3) have previously been reported to decrease in response to morphine (34). For instance, MDH2 had a twofold decrease in rat brains (22). Similar profile trends between brain and lymph node potentially indicate mechanistic commonalities in the response to morphine. In contrast, MDH2 was significantly increased in PT colon, as colon appears to produce a distinct response to morphine compared with lymph node and the neural system. TPI, which was decreased in the lymph node, has also been shown to be decreased in rat forebrain upon morphine exposure (28). TPI and MDH2 play important roles in glycolysis and the citric acid cycle, respectively. A number of studies have identified links between immunosuppression and changes in energy metabolism, which fuels defense functions (76, 77). In fact other drugs such as cannabinoids are known to reduce energy metabolism and possess immunosuppressive properties (78). The impact by morphine on energy metabolism may represent a centralized functional hub in the lymph node that contributes immunosuppression.
While the current experiment was not designed to distinguish between direct and indirect effects of morphine, a significant decrease in the members of the energy metabolism pathway could result in decreased functional activity and thereby immunosuppression. Several other pathways also showed restricted protein expression during morphine, including signal transduction, immune response, cell growth, and DNA/RNA binding proteins. Notably, the decreased abundance of proteins involved in these pathways paralleled the suppressed levels of T-cell activation in both the lymph node and peripheral blood.
In comparison with the lymph node, the colon showed a more heterogeneous response in the AGMs, displaying predominantly decreases in abundances (70% decreased), in contrast to PTs, which showed predominantly increases in abundances (76% increased). The colonic proteomic response to morphine appears to be predominantly species-specific, and further efforts are warranted to determine the extent of variability within and between these two NHP species. In regard to the known effects of morphine on the gut, opioid receptors (including the MOR) are present throughout the gastrointestinal tract (79, 80). Morphine can produce side effects at the gastrointestinal tract, including opioid bowel dysfunction, which occurs upon activation of the MOR (81). Such dysfunction can become so severe that many terminally ill patients choose to discontinue opioid treatment to attenuate their gastrointestinal discomfort. However, different proteomic profiles between AGMs and PTs could serve as important models for understanding and intervening morphine gastrointestinal side-effects as well as for better understanding outcomes after SIV infection.
As mentioned above, the decreases in T-cell activation levels in the various biologic compartments between the two NHP species reflect the suppressive impact of morphine on energy metabolism pathways. For example, PTs and AGMs both experienced declines in Ki-67+ T cells in peripheral blood and lymph node. The impact on T-cell maturation subsets was more variable between the two NHP species. For example, PTs experienced increases in the more terminally differentiated TEMRA subset in lymph node and colon, while AGMs experienced increases in central memory T-cell subsets at the colon. These differences may be linked to the proteomic variability observed between the two species or linked to fundamental biologic differences among the two species. Indeed, when studied in the context of SIV infection, these two NHP species meet diametrically apposed outcomes, where PTs suffer a pathogenic infection and AGMs coexist with minimal inflammation and pathology. The decreases observed in the current study in T-cell activation levels is hypothesized to be favorable for decreasing the target cell pool necessary for SIV/HIV replication while increases in central memory subsets is hypothesized to be a favorable change, by increasing the number of effector cells poised to mount effective immune responses against offending pathogens. Whether these changes persist after lentiviral infection, and how this baseline perturbation of the biological system impacts SIV outcome will be subject of a subsequent report.
Indeed, the major thrust of our findings is consistent with the majority of other published studies revealing both direct and indirect immunosuppression resulting from opioid therapy. Various studies have sought to further explore the complicated mechanisms underlying morphine-associated immunosuppression, but few have described the molecular changes associated with morphine activity in vivo. Here, we show that morphine is associated with a decrease in T-cell activation in two different species of NHPs, particularly within the lymph node and the peripheral blood. Paralleling this effect was a large effect on the energy metabolism protein pathways. These findings suggest that energy metabolism changes measured within lymph node may be a marker or a causal change driving decreased T-cell activation levels. Understanding these effects in more detail, and their potential impact on various disease outcomes remains an area of interest. Finally, these findings highlight the need for alternative analgesics in the clinical setting that do not produce the same peripheral morphine-associated side effects, including immunosuppression and gastrointestinal dysfunction. Future studies directed at identifying the mechanism(s) for these peripheral responses and the use of morphine analogs and antagonists will be instrumental in the effort to identify such alternative analgesics.
Acknowledgments
We thank Drs. Stephen Dominy and Elinore McCance-Katz at the University of California San Francisco for helpful discussions and insights. We would also like to acknowledge Drs. Brian Agricola and Leon Flanary, and the staff at the Washington National Primate Research Center for excellent supervision and care of the animals used in this study, following the guidelines established by the Animal Care and Use Committee (ACUC); Emilie Jalbert, Rick Dunham and Karim Sacre (Division of Experimental Medicine, University of California, San Francisco) for help in processing NHP sample shipments; and Dr. M. Babette Fontenot and Dr. Dana Hasselschwert (Univeristy of Louisiana at Layfayette, New Iberia Research Center) for allowing the purchase of blood samples from their AGM and PT colony for purposes of assay development.
Footnotes
* This work was supported in part by funds from NIH National Center for Research Resources (Grant P41 GM103493 to RDS and Grant P51 RR000166 to WaNPRC) and NIH National Institute on Drug Abuse (Grant P01 DA026134 to RDS). This research was performed in the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory). Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RLO-1830. GMO was supported in part by NIH training grants (T32 AI060530 and T32 AI007641) and is a participant of the NIH Loan Repayment Program. EYC was supported by T32 AI07140. JMM is a recipient of the NIH Director's Pioneer Award Program, part of the NIH Roadmap for Medical Research.
 This article contains supplemental Figs S1 to S11 and Tables S1 and S2.
This article contains supplemental Figs S1 to S11 and Tables S1 and S2.
1 The abbreviations used are:
- NK
- natural killer
- NHP
- nonhuman primate
- AGM
- African green monkey
- PT
- pigtailed macaque
- SIV
- simian immunodeficiency virus
- AMT
- accurate mass and Time
- MOR
- mu opioid receptor.
REFERENCES
- 1. CDC (2003) HIV diagnoses among injection-drug users in states with HIV surveillance–25 states, 1994–2000. MMWR Morb. Mortal. Wkly. Rep. 52, 634–636 [PubMed] [Google Scholar]
- 2. CDC (2006) Methamphetamine use and HIV risk behaviors among heterosexual men–preliminary results from five northern California counties, December 2001-November 2003. MMWR Morb. Mortal. Wkly. Rep. 55, 273–277 [PubMed] [Google Scholar]
- 3. Szabo I., Rojavin M., Bussiere J. L., Eisenstein T. K., Adler M. W., Rogers T. J. (1993) Suppression of peritoneal macrophage phagocytosis of Candida albicans by opioids. J. Pharmacol. Exp. Ther. 267, 703–706 [PubMed] [Google Scholar]
- 4. Messmer D., Hatsukari I., Hitosugi N., Schmidt-Wolf I. G., Singhal P. C. (2006) Morphine reciprocally regulates IL-10 and IL-12 production by monocyte-derived human dendritic cells and enhances T cell activation. Mol. Med. 12, 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho W. Z., Guo C. J., Yuan C. S., Douglas S. D., Moss J. (2003) Methylnaltrexone antagonizes opioid-mediated enhancement of HIV infection of human blood mononuclear phagocytes. J. Pharmacol. Exp. Ther. 307, 1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bussiere J. L., Adler M. W., Rogers T. J., Eisenstein T. K. (1993) Effects of in vivo morphine treatment on antibody responses in C57BL/6 bgJ/bgJ (beige) mice. Life Sci. 52, PL43–48 [DOI] [PubMed] [Google Scholar]
- 7. Gomez-Flores R., Weber R. J. (2000) Differential effects of buprenorphine and morphine on immune and neuroendocrine functions following acute administration in the rat mesencephalon periaqueductal gray. Immunopharmacology 48, 145–156 [DOI] [PubMed] [Google Scholar]
- 8. Carrigan K. A., Saurer T. B., Ijames S. G., Lysle D. T. (2004) Buprenorphine produces naltrexone reversible alterations of immune status. Int. Immunopharmacol. 4, 419–428 [DOI] [PubMed] [Google Scholar]
- 9. Carpenter G. W., Carr D. J. (1995) Pretreatment with beta-funaltrexamine blocks morphine-mediated suppression of CTL activity in alloimmunized mice. Immunopharmacology 29, 129–140 [DOI] [PubMed] [Google Scholar]
- 10. Sacerdote P., Franchi S., Gerra G., Leccese V., Panerai A. E., Somaini L. (2008) Buprenorphine and methadone maintenance treatment of heroin addicts preserves immune function. Brain Behav. Immun. 22, 606–613 [DOI] [PubMed] [Google Scholar]
- 11. Tubaro E., Avico U., Santiangeli C., Zuccaro P., Cavallo G., Pacifici R., Croce C., Borelli G. (1985) Morphine and methadone impact on human phagocytic physiology. Int. J. Immunopharmacol. 7, 865–874 [DOI] [PubMed] [Google Scholar]
- 12. Zajicova A., Wilczek H., Holan V. (2004) The alterations of immunological reactivity in heroin addicts and their normalization in patients maintained on methadone. Folia Biol. 50, 24–28 [DOI] [PubMed] [Google Scholar]
- 13. Hutchinson M. R., Somogyi A. A. (2004) (S)-(+)-methadone is more immunosuppressive than the potent analgesic (R)-(–)-methadone. Int. Immunopharmacol. 4, 1525–1530 [DOI] [PubMed] [Google Scholar]
- 14. Sacerdote P. (2006) Opioids and the immune system. Palliat. Med. 20, s9–15 [PubMed] [Google Scholar]
- 15. D'Elia M., Patenaude J., Hamelin C., Garrel D. R., Bernier J. (2003) No detrimental effect from chronic exposure to buprenorphine on corticosteroid-binding globulin and corticosensitive immune parameters. Clin. Immunol. 109, 179–187 [DOI] [PubMed] [Google Scholar]
- 16. Van Loveren H., Gianotten N., Hendriksen C. F., Schuurman H. J., Van der Laan J. W. (1994) Assessment of immunotoxicity of buprenorphine. Lab. Anim. 28, 355–363 [DOI] [PubMed] [Google Scholar]
- 17. Marone G., Stellato C., Mastronardi P., Mazzarella B. (1993) Mechanisms of activation of human mast cells and basophils by general anesthetic drugs. Ann. Fr. Anesth. Reanim. 12, 116–125 [DOI] [PubMed] [Google Scholar]
- 18. McLachlan C., Crofts N., Wodak A., Crowe S. (1993) The effects of methadone on immune function among injecting drug users: a review. Addiction 88, 257–263 [DOI] [PubMed] [Google Scholar]
- 19. Bierczynska-Krzysik A., Bonar E., Drabik A., Noga M., Suder P., Dylag T., Dubin A., Kotlinska J., Silberring J. (2006) Rat brain proteome in morphine dependence. Neurochem. Int. 49, 401–406 [DOI] [PubMed] [Google Scholar]
- 20. Bierczynska-Krzysik A., Pradeep John J. P., Silberring J., Kotlinska J., Dylag T., Cabatic M., Lubec G. (2006) Proteomic analysis of rat cerebral cortex, hippocampus and striatum after exposure to morphine. Int. J. Mol. Med. 18, 775–784 [PubMed] [Google Scholar]
- 21. Li K. W., Jimenez C. R., van der Schors R. C., Hornshaw M. P., Schoffelmeer A. N., Smit A. B. (2006) Intermittent administration of morphine alters protein expression in rat nucleus accumbens. Proteomics 6, 2003–2008 [DOI] [PubMed] [Google Scholar]
- 22. Kim S. Y., Chudapongse N., Lee S. M., Levin M. C., Oh J. T., Park H. J., Ho I. K. (2005) Proteomic analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res. Mol. Brain Res. 133, 58–70 [DOI] [PubMed] [Google Scholar]
- 23. Chen X. L., Lu G., Gong Y. X., Zhao L. C., Chen J., Chi Z. Q., Yang Y. M., Chen Z., Li Q. L., Liu J. G. (2007) Expression changes of hippocampal energy metabolism enzymes contribute to behavioural abnormalities during chronic morphine treatment. Cell Res. 17, 689–700 [DOI] [PubMed] [Google Scholar]
- 24. Shui H. A., Ho S. T., Wang J. J., Wu C. C., Lin C. H., Tao Y. X., Liaw W. J. (2007) Proteomic analysis of spinal protein expression in rats exposed to repeated intrathecal morphine injection. Proteomics 7, 796–803 [DOI] [PubMed] [Google Scholar]
- 25. Li Q., Zhao X., Zhong L. J., Yang H. Y., Wang Q., Pu X. P. (2009) Effects of chronic morphine treatment on protein expression in rat dorsal root ganglia. Eur. J. Pharmacol. 612, 21–28 [DOI] [PubMed] [Google Scholar]
- 26. Yang L., Sun Z. S., Zhu Y. P. (2007) Proteomic analysis of rat prefrontal cortex in three phases of morphine-induced conditioned place preference. J. Proteome Res. 6, 2239–2247 [DOI] [PubMed] [Google Scholar]
- 27. Morón J. A., Abul-Husn N. S., Rozenfeld R., Dolios G., Wang R., Devi L. A. (2007) Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol. Cell. Proteomics 6, 29–42 [DOI] [PubMed] [Google Scholar]
- 28. Prokai L., Zharikova A. D., Stevens S. M., Jr. (2005) Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: a quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J. Mass Spectrom. 40, 169–175 [DOI] [PubMed] [Google Scholar]
- 29. Moulédous L., Neasta J., Uttenweiler-Joseph S., Stella A., Matondo M., Corbani M., Monsarrat B., Meunier J. C. (2005) Long-term morphine treatment enhances proteasome-dependent degradation of G beta in human neuroblastoma SH-SY5Y cells: correlation with onset of adenylate cyclase sensitization. Mol. Pharmacol. 68, 467–476 [DOI] [PubMed] [Google Scholar]
- 30. Neasta J., Uttenweiler-Joseph S., Chaoui K., Monsarrat B., Meunier J. C., Moulédous L. (2006) Effect of long-term exposure of SH-SY5Y cells to morphine: a whole cell proteomic analysis. Proteome Sci. 4, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bodzon-Kulakowska A., Bierczynska-Krzysik A., Drabik A., Noga M., Kraj A., Suder P., Silberring J. (2005) Morphinome–proteome of the nervous system after morphine treatment. Amino Acids 28, 13–19 [DOI] [PubMed] [Google Scholar]
- 32. Suder P., Bodzon-Kulakowska A., Mak P., Bierczynska-Krzysik A., Daszykowski M., Walczak B., Lubec G., Kotlinska J. H., Silberring J. (2009) The proteomic analysis of primary cortical astrocyte cell culture after morphine administration. J. Proteome Res. 8, 4633–4640 [DOI] [PubMed] [Google Scholar]
- 33. Xu H., Wang X., Zimmerman D., Boja E. S., Wang J., Bilsky E. J., Rothman R. B. (2005) Chronic morphine up-regulates G alpha12 and cytoskeletal proteins in Chinese hamster ovary cells expressing the cloned mu opioid receptor. J. Pharmacol Exp. Ther. 315, 248–255 [DOI] [PubMed] [Google Scholar]
- 34. Bodzon-Kulakowska A., Kulakowski K., Drabik A., Moszczynski A., Silberring J., Suder P. (2011) Morphinome–a meta-analysis applied to proteomics studies in morphine dependence. Proteomics 11, 5–21 [DOI] [PubMed] [Google Scholar]
- 35. Kruisbeek A. M. (2001) Isolation of mouse mononuclear cells. Curr. Protoc. Immunol. Chapter 3, Unit 3 1 [DOI] [PubMed] [Google Scholar]
- 36. Marginean I., Kelly R. T., Moore R. J., Prior D. C., LaMarche B. L., Tang K., Smith R. D. (2009) Selection of the optimum electrospray voltage for gradient elution LC-MS measurements. J. Am. Soc. Mass. Spectrom. 20, 682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelly R. T., Page J. S., Zhao R., Qian W. J., Mottaz H. M., Tang K., Smith R. D. (2008) Capillary-based multi nanoelectrospray emitters: improvements in ion transmission efficiency and implementation with capillary reversed-phase LC-ESI-MS. Anal. Chem. 80, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Page J. S., Kelly R. T., Tang K., Smith R. D. (2007) Ionization and transmission efficiency in an electrospray ionization-mass spectrometry interface. J. Am. Soc. Mass Spectrom. 18, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 39. Gibbs R. A., Rogers J., Katze M. G., Bumgarner R., Weinstock G. M., Mardis E. R., Remington K. A., Strausberg R. L., Venter J. C., Wilson R. K., Batzer M. A., Bustamante C. D., Eichler E. E., Hahn M. W., Hardison R. C., Makova K. D., Miller W., Milosavljevic A., Palermo R. E., Siepel A., Sikela J. M., Attaway T., Bell S., Bernard K. E., Buhay C. J., Chandrabose M. N., Dao M., Davis C., Delehaunty K. D., Ding Y., Dinh H. H., Dugan-Rocha S., Fulton L. A., Gabisi R. A., Garner T. T., Godfrey J., Hawes A. C., Hernandez J., Hines S., Holder M., Hume J., Jhangiani S. N., Joshi V., Khan Z. M., Kirkness E. F., Cree A., Fowler R. G., Lee S., Lewis L. R., Li Z., Liu Y. S., Moore S. M., Muzny D., Nazareth L. V., Ngo D. N., Okwuonu G. O., Pai G., Parker D., Paul H. A., Pfannkoch C., Pohl C. S., Rogers Y. H., Ruiz S. J., Sabo A., Santibanez J., Schneider B. W., Smith S. M., Sodergren E., Svatek A. F., Utterback T. R., Vattathil S., Warren W., White C. S., Chinwalla A. T., Feng Y., Halpern A. L., Hillier L. W., Huang X., Minx P., Nelson J. O., Pepin K. H., Qin X., Sutton G. G., Venter E., Walenz B. P., Wallis J. W., Worley K. C., Yang S. P., Jones S. M., Marra M. A., Rocchi M., Schein J. E., Baertsch R., Clarke L., Csuros M., Glasscock J., Harris R. A., Havlak P., Jackson A. R., Jiang H., Liu Y., Messina D. N., Shen Y., Song H. X., Wylie T., Zhang L., Birney E., Han K., Konkel M. K., Lee J., Smit A. F., Ullmer B., Wang H., Xing J., Burhans R., Cheng Z., Karro J. E., Ma J., Raney B., She X., Cox M. J., Demuth J. P., Dumas L. J., Han S. G., Hopkins J., Karimpour-Fard A., Kim Y. H., Pollack J. R., Vinar T., Addo-Quaye C., Degenhardt J., Denby A., Hubisz M. J., Indap A., Kosiol C., Lahn B. T., Lawson H. A., Marklein A., Nielsen R., Vallender E. J., Clark A. G., Ferguson B., Hernandez R. D., Hirani K., Kehrer-Sawatzki H., Kolb J., Patil S., Pu L. L., Ren Y., Smith D. G., Wheeler D. A., Schenck I., Ball E. V., Chen R., Cooper D. N., Giardine B., Hsu F., Kent W. J., Lesk A., Nelson D. L., O'Brien W E., Prufer K., Stenson P. D., Wallace J. C., Ke H., Liu X. M., Wang P., Xiang A. P., Yang F., Barber G. P., Haussler D., Karolchik D., Kern A. D., Kuhn R. M., Smith K. E., Zwieg A. S. (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 [DOI] [PubMed] [Google Scholar]
- 40. Chen Y., Cunningham F., Rios D., McLaren W. M., Smith J., Pritchard B., Spudich G. M., Brent S., Kulesha E., Marin-Garcia P., Smedley D., Birney E., Flicek P. (2010) Ensembl variation resources. BMC Genomics 11, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berglund A. C., Sjölund E., Ostlund G., Sonnhammer E. L. (2008) InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 36, D263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Brien K. P., Remm M., Sonnhammer E. L. (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33, D476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Remm M., Storm C. E., Sonnhammer E. L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 44. Brown J. N., Palermo R. E., Baskin C. R., Gritsenko M., Sabourin P. J., Long J. P., Sabourin C. L., Bielefeldt-Ohmann H., Garcia-Sastre A., Albrecht R., Tumpey T. M., Jacobs J. M., Smith R. D., Katze M. G. (2010) Macaque Proteome Response to Highly Pathogenic Avian Influenza and 1918 Reassortant Influenza Virus Infections. J. Virol. 84, 12058–12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown J. N., Estep R. D., Lopez-Ferrer D., Brewer H. M., Clauss T. R., Manes N. P., O'Connor M., Li H., Adkins J. N., Wong S. W., Smith R. D. (2010) Characterization of Macaque Pulmonary Fluid Proteome during Monkeypox Infection: Dynamics of Host Response. Mol. Cell. Proteomics 9, 2760–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimmer J. S., Monroe M. E., Qian W. J., Smith R. D. (2006) Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 25, 450–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanley J. R., Adkins J. N., Slysz G. W., Monroe M. E., Purvine S. O., Karpievitch Y. V., Anderson G. A., Smith R. D., Dabney A. R. (2011) A statistical method for assessing Peptide identification confidence in accurate mass and time tag proteomics. Anal. Chem. 83, 6135–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jaitly N., Monroe M. E., Petyuk V. A., Clauss T. R., Adkins J. N., Smith R. D. (2006) Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 78, 7397–7409 [DOI] [PubMed] [Google Scholar]
- 49. Kiebel G. R., Auberry K. J., Jaitly N., Clark D. A., Monroe M. E., Peterson E. S., Tolić N., Anderson G. A., Smith R. D. (2006) PRISM: a data management system for high-throughput proteomics. Proteomics 6, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 50. Monroe M. E., Tolić N., Jaitly N., Shaw J. L., Adkins J. N., Smith R. D. (2007) VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 51. Monroe M. E., Shaw J. L., Daly D. S., Adkins J. N., Smith R. D. (2008) MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 32, 215–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petritis K., Kangas L. J., Yan B., Monroe M. E., Strittmatter E. F., Qian W. J., Adkins J. N., Moore R. J., Xu Y., Lipton M. S., Camp D. G., 2nd, Smith R. D. (2006) Improved peptide elution time prediction for reversed-phase liquid chromatography-MS by incorporating peptide sequence information. Anal. Chem. 78, 5026–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matzke M. M., Waters K. M., Metz T. O., Jacobs J. M., Sims A. C., Baric R. S., Pounds J. G., Webb-Robertson B. J. (2011) Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics 27, 2866–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karpievitch Y., Stanley J., Taverner T., Huang J., Adkins J. N., Ansong C., Heffron F., Metz T. O., Qian W. J., Yoon H., Smith R. D., Dabney A. R. (2009) A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinformatics 25, 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Benjamini Y., Hochberg Y. (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. 57, 289–300 [Google Scholar]
- 56. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 57. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 58. Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J. A., Doremieux O. (2003) PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. (2005) BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 [DOI] [PubMed] [Google Scholar]
- 60. Dodge P. W., Takemori A. E. (1969) Effect of morphine on cerebral glycolytic intermediates and enzymes of rats in vitro. Biochem. Pharmacol. 18, 1873–1882 [DOI] [PubMed] [Google Scholar]
- 61. Gragera R. R., de Miguel E., Muñiz E., Arenas-Díaz G., Alonso M. J., Gomez de Segura I. A., Martínez-Rodriguez R. (1995) L-aspartate aminotransferase and L-asparaginase in rat lymph node: a histoenzymological and immunohistochemical study. Eur. J. Histochem. 39, 195–200 [PubMed] [Google Scholar]
- 62. Prager M. D., Derr I. (1971) Metabolism of asparagine, aspartate, glutamine, and glutamate in lymphoid tissue: basis for immunosuppression by L-asparaginase. J. Immunol. 106, 975–979 [PubMed] [Google Scholar]
- 63. Cui Y., Ying Y., van Hasselt A., Ng K. M., Yu J., Zhang Q., Jin J., Liu D., Rhim J. S., Rha S. Y., Loyo M., Chan A. T., Srivastava G., Tsao G. S., Sellar G. C., Sung J. J., Sidransky D., Tao Q. (2008) OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE 3, e2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ishiguro H., Liu Q. R., Gong J. P., Hall F. S., Ujike H., Morales M., Sakurai T., Grumet M., Uhl G. R. (2006) NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology 31, 572–584 [DOI] [PubMed] [Google Scholar]
- 65. Sato S., Cerny R. L., Buescher J. L., Ikezu T. (2006) Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 98, 1573-1584 [DOI] [PubMed] [Google Scholar]
- 66. Edener U., Kurth I., Meiner A., Hoffmann F., Hubner C. A., Bernard V., Gillessen-Kaesbach G., Zühlke C. (2009) Missense exchanges in the TTBK2 gene mutated in SCA11. J. Neurol. 256, 1856–1859 [DOI] [PubMed] [Google Scholar]
- 67. Ferenczi S., Núñez C., Pintér-Kübler B., Földes A., Martín F., Márkus V. L., Milanés M. V., Kovács K. J. (2010) Changes in metabolic-related variables during chronic morphine treatment. Neurochem. Int. 57, 323–330 [DOI] [PubMed] [Google Scholar]
- 68. Saurer T. B., Ijames S. G., Lysle D. T. (2006) Neuropeptide Y Y1 receptors mediate morphine-induced reductions of natural killer cell activity. J. Neuroimmunol. 177, 18–26 [DOI] [PubMed] [Google Scholar]
- 69. Lipovsky M. M., Gekker G., Hu S., Hoepelman A. I., Peterson P. K. (1998) Morphine enhances complement receptor-mediated phagocytosis of Cryptococcus neoformans by human microglia. Clin. Immunol. Immunopathol. 87, 163–167 [DOI] [PubMed] [Google Scholar]
- 70. Welters I. D., Menzebach A., Goumon Y., Langefeld T. W., Teschemacher H., Hempelmann G., Stefano G. B. (2000) Morphine suppresses complement receptor expression, phagocytosis, and respiratory burst in neutrophils by a nitric oxide and mu(3) opiate receptor-dependent mechanism. J. Neuroimmunol. 111, 139–145 [DOI] [PubMed] [Google Scholar]
- 71. Rivera-Amill V., Silverstein P. S., Noel R. J., Jr., Kumar S., Kumar A. (2010) Morphine and rapid disease progression in nonhuman primate model of AIDS: inverse correlation between disease progression and virus evolution. J. Neuroimmune Pharmacol. 5, 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carr D. J., France C. P. (1993) Immune alterations in morphine-treated rhesus monkeys. J. Pharmacol. Exp. Ther. 267, 9–15 [PubMed] [Google Scholar]
- 73. Bokhari S. M., Hegde R., Callen S., Yao H., Adany I., Li Q., Li Z., Pinson D., Yeh H. W., Cheney P. D., Buch S. (2011) Morphine Potentiates Neuropathogenesis of SIV Infection in Rhesus Macaques. J. Neuroimmune Pharmacol. 6, 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Good D. M., Thongboonkerd V., Novak J., Bascands J. L., Schanstra J. P., Coon J. J., Dominiczak A., Mischak H. (2007) Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. J. Proteome Res. 6, 4549–4555 [DOI] [PubMed] [Google Scholar]
- 75. Odunayo A., Dodam J. R., Kerl M. E., DeClue A. E. (2010) Immunomodulatory effects of opioids. J. Vet. Emerg. Crit. Care 20, 376–385 [DOI] [PubMed] [Google Scholar]
- 76. Frühbeck G., Gómez-Ambrosi J., Muruzábal F. J., Burrell M. A. (2001) The adipocyte: a model for integration of endocrine and metabolic signaling in energy metabolism regulation. Am. J. Physiol. Endocrinol Metab. 280, E827–847 [DOI] [PubMed] [Google Scholar]
- 77. Fox C. J., Hammerman P. S., Thompson C. B. (2005) Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5, 844–852 [DOI] [PubMed] [Google Scholar]
- 78. Guzmán M., Sánchez C. (1999) Effects of cannabinoids on energy metabolism. Life Sci. 65, 657–664 [DOI] [PubMed] [Google Scholar]
- 79. Polak J. M., Bloom S. R., Sullivan S. N., Facer P., Pearse A. G. (1977) Enkephalin-like immunoreactivity in the human gastrointestinal tract. Lancet 1, 972–974 [DOI] [PubMed] [Google Scholar]
- 80. Frantzides C. T., Condon R. E., Schulte W. J., Cowles V. (1990) Effects of morphine on colonic myoelectric and motor activity in subhuman primates. Am. J. Physiol. 258, G247–252 [DOI] [PubMed] [Google Scholar]
- 81. DeHaven-Hudkins D. L., DeHaven R. N., Little P. J., Techner L. M. (2008) The involvement of the mu-opioid receptor in gastrointestinal pathophysiology: therapeutic opportunities for antagonism at this receptor. Pharmacol. Ther. 117, 162–187 [DOI] [PubMed] [Google Scholar]