Abstract
This study reports the comprehensive comparison of 15N metabolic labeling and label free proteomic strategies for quantitation, with particular focus on plant proteomics. Our investigation of proteome coverage, dynamic range and quantitative precision for a wide range of mixing ratios and protein loadings aim to aid the investigators in the decision making process during experimental design. One of the main characteristics of the label free strategy is the applicability to all starting material, which is a limitation to the metabolic labeling. However, particularly at mixing ratios up to 10-fold the 15N metabolic labeling proved to be more precise. Contrary to usual practice based on the results from this study, we suggest that nonequal mixing ratios in metabolic labeling could further increase the proteome coverage for quantitation. On the other hand, the label free strategy, in combination with low protein loading allows the extension of the dynamic range for quantitation and it is more precise at very high ratios, which could be important for certain types of experiments.
Quantitative comparative analyses of proteomes and their dynamic changes under various growth conditions and stimuli has become a widely used approach in experimental and systems biology. Quantitative proteome analysis is particularly important when the functional roles of proteins in biological contexts are being addressed. This has been greatly aided by the development of soft ionization methods for macromolecules (1, 2) and with ongoing developments in high accuracy mass spectrometer instrument technology (3, 4). Thus, analysis of thousands of proteins in a high-throughput manner is now almost a routine task. In parallel, completion of annotated genome sequences for a range of model organisms has opened these for large-scale proteome studies (5). In plant science, the expansion of sequencing efforts to key crop plant species provides a solid basis for efficient interpretation of acquired peptide mass spectra for proteome wide studies also in commercially important crop plants. Various methods and workflows for large-scale quantitative proteome profiling have been developed in the past years (reviewed in (6–8)). Among them, metabolic labeling using stable isotope labeled amino acids in mammalian cell cultures (9) or full 15N-labeling for autotrophic organisms (5, 10) are now widely used. In addition, more recently, label-free methods involving computational alignment of ion chromatograms based on accurate mass and retention time of detected ions have been developed and are increasingly used either in studies on tissue not accessible to metabolic labeling or because of their simple and cheap experimental design (6, 11–16). In addition, chemical labeling using differential mass tags today allows quantitative comparisons in multiplexed samples (17).
Previous studies comparing methods for relative quantitation of proteins in a mixture with known protein abundance ratios, concluded that it was the researcher's experience that was decisive for correct quantitation and that no method, as such, out-performed others (18). In a more systematic analysis, the two-dimensional-DIGE method was compared with the chemical labeling strategies cleavable isotope-coded affinity tags (cICAT)1 and isobaric tags for relative and absolute quantification (iTRAQ) (19) concluding that two-dimensional-gel based quantitation and chemical labeling were complementary strategies. However, in many cases, comparative analysis of methods has been carried out using a less complex mixture of known standard proteins rather than using particular mixing ratios of truly complex proteomes. A comparison of quantitation by stable isotope labeling with amino acids (SILAC) with spectrum counting using mammalian cell lines as an example revealed higher sensitivity of quantitation in the metabolic labeling but higher sequence coverage of identified proteins with the label-free method (20).
Particularly for studying the proteome of plant species, several issues during sample preparation have to be addressed that may also influence the choice of quantitation strategy: Because plant cells are surrounded by a cell wall, the plant proteins usually are only accessible after rigorous mechanical destruction of the cells by mortar and pestle or a ball mill leading to potential losses of protein yield. Particularly when working on green tissue, a highly skewed protein abundance distribution has to be considered, with ribulose-bisphophate carboxylase being the most highly abundant protein in leaf tissue with an at least 2000-fold higher abundance than some of the kinases or transcription factors (21). Furthermore, the plant cell is not only shaped by the cell wall but also by the large central vacuole that helps to build the turgor pressure. This organelle takes up a large proportion of the cellular volume. Therefore the protein content per fresh weight in mature plant tissue can be rather low. Finally, some plant species and tissues, such as senescent leaves, flowers or stressed plants, often have a high content of secondary metabolites—polymeric carbohydrates, phenolic compounds, isoprenoids and alkaloids. These classes of small molecules are usually very hard to separate from protein or peptides during sample preparation and can provide a serious challenge to subsequent mass spectrometric analysis.
Because of their autotrophic nature, for quantitative comparisons in plants, full labeling with 15N-inorganic salts has in the past years been established by various groups as a widely used strategy for quantitative comparisons (reviewed in: (22)). However, the method fails to cover the entire spectrum of tissues used in modern plant science. For example, full metabolic labeling is not readily applicable to the most natural and widely used starting material, namely the soil-grown plant. In order to compensate for this deficiency the experimenter will have to use different quantitation strategies (e.g. label-free or chemical labeling), or spike in common isotope-labeled reference tissue (23, 24).
Despite this vast choice of analytical methods for quantitative proteome profiling, the performance quality regarding proteome coverage, quantitative precision and dynamic range has not been systematically evaluated for plant-derived protein extracts on modern high mass accuracy instruments. In order to provide a solid basis for decision on quantitative strategies in plant proteomics, we performed an in-depth comparison between the metabolic 15N labeling and the label free methods using conditions of wide range of protein amounts per sample and comparative ratios of complex protein mixtures.
EXPERIMENTAL PROCEDURES
Metabolic Labeling of Cell Cultures, Sample Mixing, and Protein Extraction
Metabolically 15N-labeled and unlabeled Arabidopsis cell cultures were grown and harvested as described (10). On average, the 15N-labeling efficiency of the proteins was at 96.7 atom%, ranging from 91.2 atom% to 99.4 atom%, as estimated based on the ratio of the monoisotopic (M) peak and the M-1 peak (25).
The cells were grown in medium with 10 mm potassium nitrate as the sole nitrogen source, which was supplied either in normal form (natural abundance of nitrogen, yielding unlabeled cells) or in 98 atom% enriched 15N form (yielding metabolically labeled cells). 15N-enriched salts were obtained from Sigma-Aldrich. Frozen cells were manually ground in liquid nitrogen and extracted in 50 mm Tris- HCl pH 7.5, 1% Nonidet P-40, and a protease inhibitor mixture (Complete Tabs, Roche). The cell debris was pelleted by centrifugation 10 min at 4000rpm and 4 °C. The supernatant was used further. The protein content was determined using the Bradford solution (Bio-Rad, Hercules, CA).
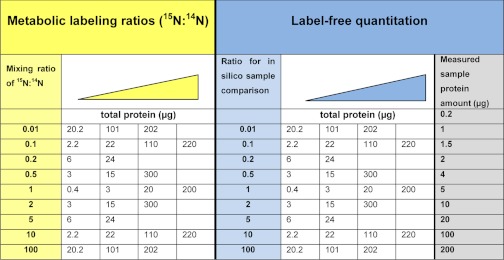
In the case of the metabolic labeling approach, 15N-labeled and unlabeled samples were mixed in specific ratios at different total protein content (Table I). Details on mixed protein amounts from labeled and unlabeled protein are listed in supplemental Table S1. In the case of the label free approach, the samples were aliquoted based on protein content also covering a 100-fold range of total protein (Table I). For each combination, three technical replicates were analyzed.
Table I. Overview of the samples included in the methods comparison. For each quantitative strategy (metabolic labeling and label-free) a range of mixing ratios has been used at different total protein amounts. The triangles indicate the increasing protein loading for each ratio. In both cases, the ratios varying with a factor 10 have been represented with four protein loadings whereas intermediate ratios were represented with two protein loadings. In label-free samples, the samples with grey background were actually measured, and the ratios listed resulted from combinations of these measurements in the data analysis.
Sample Preparation for Mass Spectrometry
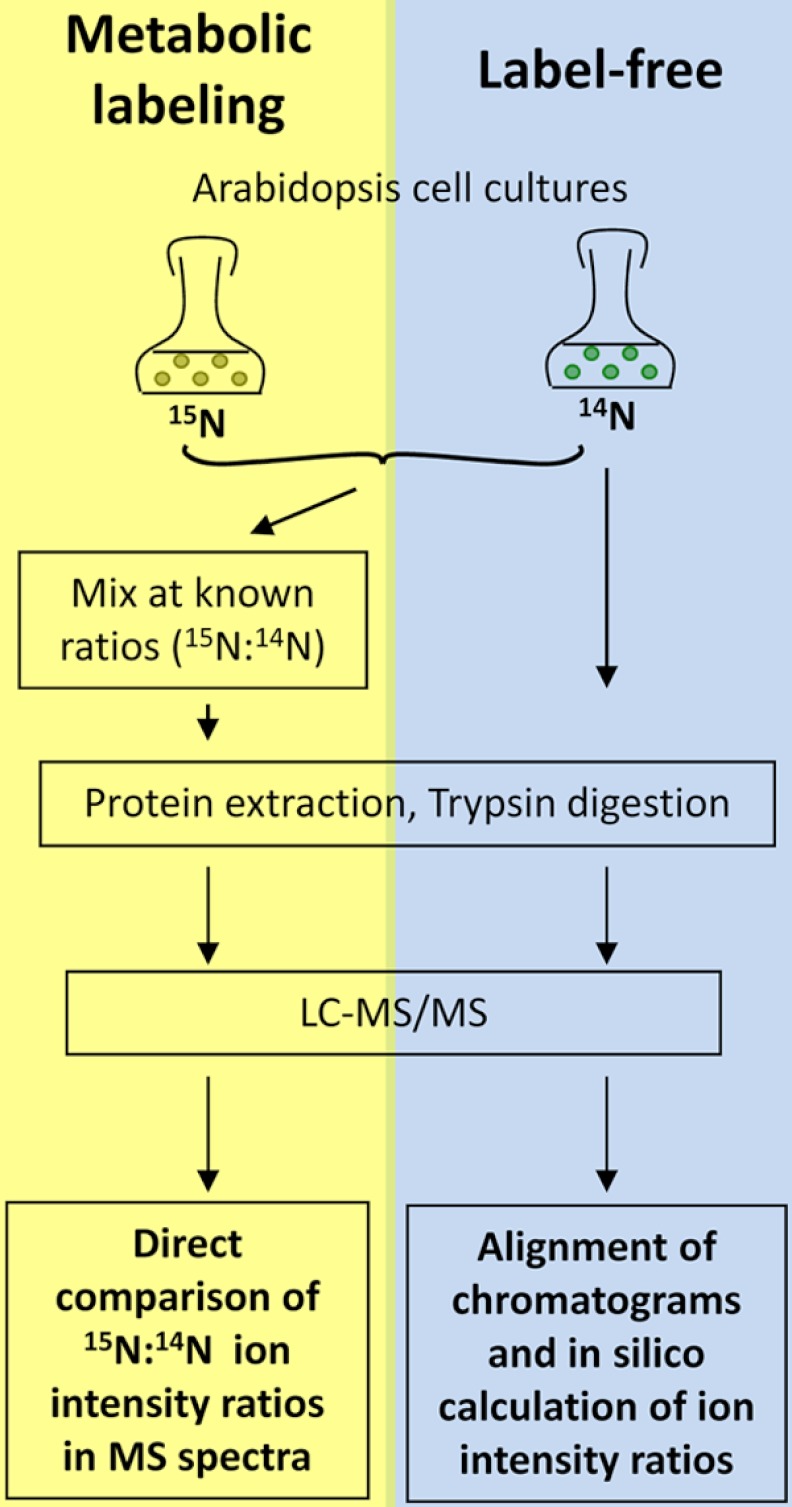
Protein mixtures were acetone precipitated and then in-solution digested as described (10). In brief, proteins were denatured using 6 m urea, 2 m thiourea, pH 8. After reduction and alkylation of cysteine residues by iodoacetamide, protein was digested at room temperature for 3 h by LysC (Wako, Japan). Samples were then diluted fourfold in 10 mm TrisHCl pH 8 before additional overnight digestion at room temperature with sequencing grade trypsin (Promega, Germany). In general, the workflow of sample preparation and mass spectrometric analysis is outlined in Fig. 1.
Fig. 1.
Workflow of the method comparison. Metabolic labeling: Labeled and unlabeled cell cultures were mixed at different protein ratios prior to protein digestion and mass spectrometric analysis. Intensity ratios were directly determined from the acquired MS spectra. Label-free comparison: Different total amounts of protein was digested and analyzed. Total ion chromatograms were then aligned and intensity ratios were calculated from different raw files.
Mass Spectrometric Analysis and Protein Identification
Tryptic peptide mixtures were analyzed by LC/MS/MS using nanoflow Easy-nLC (Thermo Scientific) as an HPLC-system and an LTQ-Orbitrap hybrid mass spectrometer (Thermo Scientific) as a mass analyzer. Peptides were eluted from a 100 mm long analytical column (Reprosil C18, Dr. Maisch GmbH, Tübingen, Germany) with 75 μm inner diameter on a gradient using 0.5% acetic acid as aqueous phase (solution A) and 0.5% acetic acid in 80% acetonitrile as organic phase (solution B) and a flow-rate of 250 nL/min. Peptide elution followed a two-step gradient of 71 min of linear increase of solution B from 5% to 30%, then an increase of solution B from 30% to 60% within 14 min, followed by a 10 min wash with 90% solution B. Eluting peptides were sprayed directly into the LTQ-Orbitrap mass spectrometer. Peptides were identified by information-dependent acquisition of fragmentation spectra of multiple-charged ions. Up to five data dependent MS/MS spectra were recorded in the linear ion trap for each full scan spectrum acquired within the m/z range between 300 and 1500 at a resolution of 60000 FWHM in the Orbitrap. Overall cycle time was approximately 1 s, target ions for the Orbitrap were set to 106 and 30,000 for the linear ion trap. For fragmentation collision-induced dissociation was chosen, collision energy was 35 eV.
Fragment MS/MS spectra from raw files were extracted as DTA-files and then merged to peak lists using default settings of DTASuperCharge version 1.18 (msquant.alwaysdata.net) with a tolerance for precursor ion detection of 50 ppm. The DTA-files of comprising a label free technical replicate set were combined into a single file, which was submitted to the database search. In case of the metabolic labeling, samples forming the same ratio in one replicate set were grouped together and submitted to the database search. Spectra were searched against a nonredundant version of the Arabidopsis protein database (Version: TAIR 9, 33595 entries, released 2009-06-19) using the mascot algorithm version 2.2.2. (Matrix Science, UK, www.matrixscience.com). The database was supplemented with typical contaminants (trypsin, keratins, lysC, and BSA). Identified contaminants were excluded from further quantitative analysis. In all cases, search parameters included carbamidomethylation as a fixed modification for cysteines, and methionine oxidation as a variable modification. Identified peptides were required to be fully tryptic peptides, whereas two missed cleavages were allowed. Precursor mass tolerance was set to 10 ppm, MS/MS tolerance was set to 0.8 Da. For the metabolic labeling samples, “15N metabolic labeling” was chosen as a quantitative method during Mascot database searching, allowing identification of 15N-labeled and unlabeled peptides within the same database search. For samples from label-free comparison, no quantification method was selected.
In general, only peptide identifications with a length of more than five amino acids were considered. Peptides were accepted without manual interpretation if they displayed a Mascot score greater than 29 as defined by Mascot p < 0.01 significance threshold. Peptide assignment to protein groups was done according to the Mascot default settings. Thus, each redundant peptide was primarily assigned to the highest scoring protein. Isoforms of protein only appear in the tables as separate protein entry if they were assigned at least one unique peptide.
Samples for label-free quantitation were also analyzed by MaxQuant version 1.1.1.14 (26, 27) to study the effect of retention time correlation on the number of quantified protein pairs. The same settings for database, modifications and enzyme cleavage and peptide length were used as described above. Peptide false-discovery rate and protein false-discovery rate were set to 0.01. Two analyses with MaxQuant were carried out, either with or without selection of “match between runs.” MaxQuant data were only used for counting the number of identified proteins.
Calculation of 15N Enrichment
As already described, the enrichment of heavy nitrogen within the metabolically labeled proteome can be linked to the relative intensities of the Monoisotopic (M) and M-1 satellite peaks in the MS spectrum of a peptide (25). Thus to estimate the percentage of heavy nitrogen enrichment we used the following equation ((M – 1)/(M))*100 where M is the intensity of the monoisotopic peak of the analyzed peptide in the full scan. This calculates the percentage of M – 1 from the Monoisotopic peak. The enrichment was calculated by deducing the (M – 1)% from 100, which would be the case when complete labeling is achieved. Average 15N enrichment was calculated from 20 randomly selected peptides.
Quantitative Analysis: 15N Metabolic Labeling
Ratios between labeled and unlabeled forms of each tryptic peptides were calculated in MSQuant version 1.5.a22 (released 2008-05-30; msquant.sourceforge.net). Quantitative information was taken from extracted ion chromatograms of labeled and unlabeled form of each identified peptide. Thereby, co-elution of both peptide forms was made a requirement.
Intensity ratios of labeled 15N-form to unlabeled 14N-form of each identified peptide were averaged across all peptides belonging to the same protein within one experimental set. Nonproteotypic peptides were excluded from quantitative analysis if they displayed ratios significantly different from proteotypic peptides of the same protein (28, 29). Peptides meeting the criteria for sequence identification, but for which only 14N forms or only 15N forms were quantified and therefore no ratio could be calculated, were counted as identified, but not as quantified. Because quantitative information was extracted from full scan spectra obtained in the Orbitrap mass analyzer with very low level of noise, no minimum threshold was set for quantitation (30). The obtained ratios were log2 transformed and median normalization was carried out as described (31). Average for each peptide ion species were calculated across technical replicates.
Raw data files (supplemental Table S1A) have been submitted to the Tranche database (www.proteomecommons.org/tranche) and can be accessed using the following hash: bEA4aOFnbQ5MxP0vljxz7+gmFBH4HC3pEACNS63nh9610rkhqjQHHT2/d1JGXhHPrNG5kb+X9h2HyUbr3YjWp/QlHUYAAAAAAABD8A== (15N-labeling dataset).
Quantitative Analysis: Label-free Comparison
Label-free relative quantitation was performed as described (28, 32). Ion intensity extraction, based on ion chromatograms of the raw data files was done using MSQuant (version 1.4.3; released 2008-05-30; msquant.alwaysdata.net). For each peptide ion species (i.e. each m/z value), the ion intensity within each sample was normalized to the total ion intensity of all peptide ions in that sample. A normalization factor was calculated by dividing the protein amount in the sample (in μg) by the average protein amount in the label free replica set. Ion intensity ratios between different samples were then calculated based on the normalized peptide ion intensities. Protein abundance ratios were calculated by averaging respective peptide ion intensity ratios. Nonproteotypic peptides were excluded from quantitative analysis if they displayed ratios significantly different from proteotypic peptides of the same protein (28, 29). Average for each peptide ion species were calculated across technical replicates.
In order to investigate the influence of retention time correlation the label free samples were processed separately using the MaxQuant software version 1.1.1.14 (26) with and without the “mach between runs” function. This data was used only for comparing the number of identified peptides with and without retention time correlation, but not for the assessment of precision and dynamic range.
Raw data files (supplemental Table S1B) have been submitted to the Tranche database (www.proteomecommons.org/tranche) and can be accessed using the following hash: lJ/g1iB4s+TrLKf49IQ78sZBOkIg46kU87iWXD9/Ah1ZButZPsiGHoawtxgrpcFf+fPGe10Zf5TI7PlDLdjhKOvZ0jMAAAAAAAAXRQ== (label-free dataset).
Calculations and Statistics
Data normalization and outlier tests were carried out using in-house R-scripts and Excel spreadsheets. SigmaPlot version 11.0 (Systat Software Inc.) was used for the generation of the presented plots.
RESULTS
The aim of this study was to perform a robust comparative analysis of metabolic 15N-labeling and label-free quantitation across a wide range of mixing ratios derived from plant protein extracts. Parallel to variation of the mixing ratio, we also analyzed samples with a wide range of total protein amount. We were particularly interested in addressing method-specific differences in proteome coverage, quantitative precision and the covered quantitative dynamic range.
Proteome Coverage
In general, and independent of the labeling strategy, the highest numbers of proteins were identified in samples containing about 20 to 100 μg total protein amount. Among the three replicates, 1189 and 1186 proteins were identified in 100 μg and 20 μg label free samples respectively, and around 1281 and 1272 protein groups were also identified in the 202 and 101 μg metabolic labeling replicas (supplemental Table S2). In samples of much lower protein content, predominantly highly abundant proteins were identified (supplemental Fig. S1A) and thus the sensitivity of the mass spectrometric detection became limiting. In contrast, particularly in mixtures for metabolic labeling, the number of identified peptides and proteins did not always increase further in samples of very high total protein amounts, and in some cases even decreased (supplemental Table S2). This is possibly because of the limiting sampling speed of the classic Orbitrap XL mass spectrometer. The increased protein loading lead to a slight broadening of chromatographic peaks (supplemental Fig. S1B), which in turn may have resulted in sampling of fewer ion species. When comparing the reciprocal 15N/14N mixtures, there was a tendency for higher numbers of protein and peptide identifications if the light isotope was more abundant in the mixture (supplemental Table S2). This is consistent with findings that fully 15N-labeled peptides sometimes are assigned an incorrect satellite isotope peak as precursor ion, which then can result in no or incorrect identification efficiency in fully 15N labeled proteomes (33). In our experiments, plant material was 15N labeled to an average of 96.7 atom%, which still produces respective 14N-enriched satellite iosotope peaks (33).
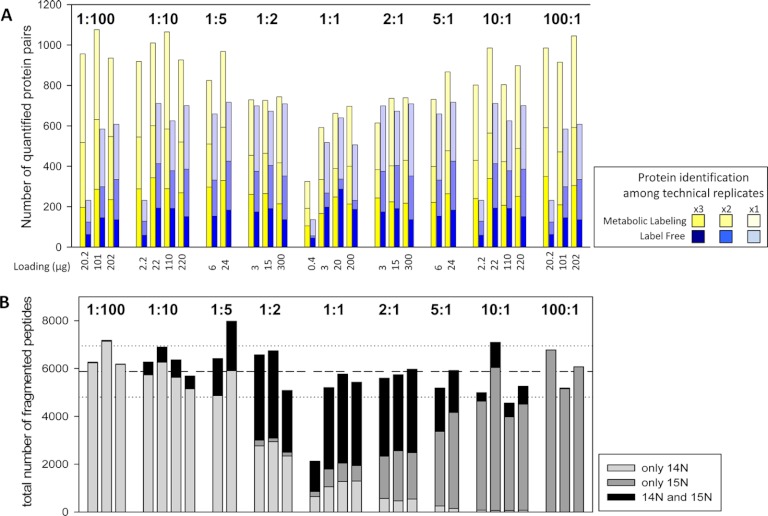
It is not the total numbers of peptide and protein identifications that are critical for quantitative comparisons, but rather the number of peptide pairs that can be used for quantitation. Interestingly, for the 15N metabolic labeling samples, the highest numbers of proteins were identified and quantified in samples with at least a fivefold difference in mixing ratio (Fig. 2A). This finding was consistent across the ranges of total protein present in each mixture. In the 1:1 mixtures consisting of equal amounts of labeled and unlabeled proteomes, the number of quantified proteins was lowest compared with samples with higher mixing ratios. This could be explained by a high rate of MS/MS fragmentations on both isotope forms of the same peptides. Thus, although on average the total number of acquired fragment spectra was similar in all sample mixtures (except for those with very low protein amount), the high rate of fragment spectra sampling both isotope partners decreased the total number of identified proteins. In contrast, in high mixing ratios, the tendency for only one isotope partner to be selected for fragmentation was higher, therefore more unique peptides mapping to different protein groups were sampled (Fig. 2B).
Fig. 2.
Numbers of quantified proteins. A, The number of protein pairs quantified in different mixing ratios and in samples of different total protein content. B, Total number of fragment spectra acquired in all three replicates of each sample as sorted for the unlabeled peptide only, the labeled peptide only or for both peptide forms. Dashed line represents the average of total fragment spectra acquired across all sample, dotted lines represent standard deviation.
In general, the number of quantified proteins correlated with the number of identified proteins. For metabolic labeling about 82% of all peptides identified by a fragment spectrum could be assigned a labeled or unlabeled counterpart for calculation of an abundance ratio (supplemental Fig. S3). In contrast, in the label-free samples, the overlap of peptide pairs based on MS/MS events was lower, particularly in those samples with extreme comparative ratios. On average, only 46% of all identified proteins could be directly quantified by label-free approach.
For label-free quantitation, the number of quantified proteins was independent of the ratio comparison, but showed a tendency for higher number of quantitation pairs in samples with higher protein amount. Furthermore, the number of peptide pairs for label-free quantitation could be increased on average by 25% once peaks without MS/MS fragmentation were included based on accurate mass and retention time alignment. This was particularly important for increasing the quantitative coverage in samples with low protein amounts, in which ion chromatogram alignment increased the quantitative coverage by 125% (supplemental Fig. S2, supplemental Table S3).
Dynamic Range for Quantitation
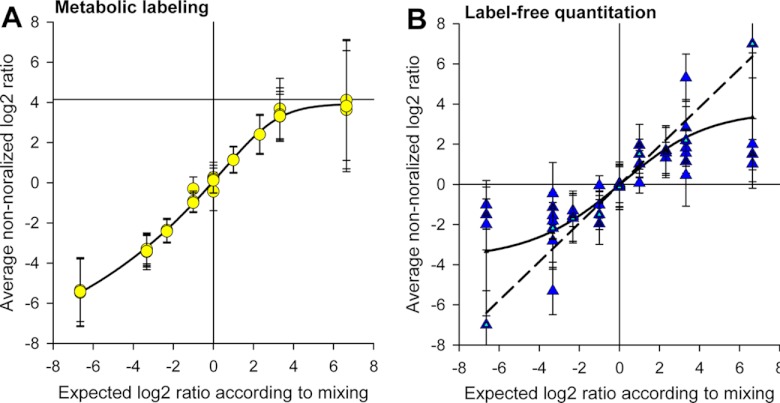
The average nonnormalized ratios for 15N metabolic labeling showed linearity between mixing ratios of 1:10 and 10:1, whereas nonlinear behavior was observed for the higher or lower mixing ratios (Fig. 3A). There was no influence of total sample protein amount on the linear range of 15N metabolic quantitation. This is in agreement with earlier studies also observing a linear quantitative range of within 10-fold mixing ratios on an Orbitrap mass spectrometer (30).
Fig. 3.
Measured and expected ratios. A, In metabolic labeling, saturation of quantitation occurs at mixing ratios greater than 10 or smaller than 0.1. B, Label-free quantitation at different total protein amounts. Large deviations occur at high total protein amounts.
For label-free quantitation, the protein amount was critical for the linear dynamic range (Fig. 3B). Low protein amounts displayed a larger linear range than high protein amounts. In samples with up to 15 μg of total protein, close to linear quantitation was possible even at very high ratios (dashed line in Fig. 3B). In general, the measured ion intensities increased proportionally to the supplied protein amount in the range between 0.2 μg and 10 μg. At higher protein amounts from 100 μg or higher, ion intensities became saturated and therefore the linear range of quantitation decreased (sigmoid curve in Fig. 3B).
Precision of Quantitation
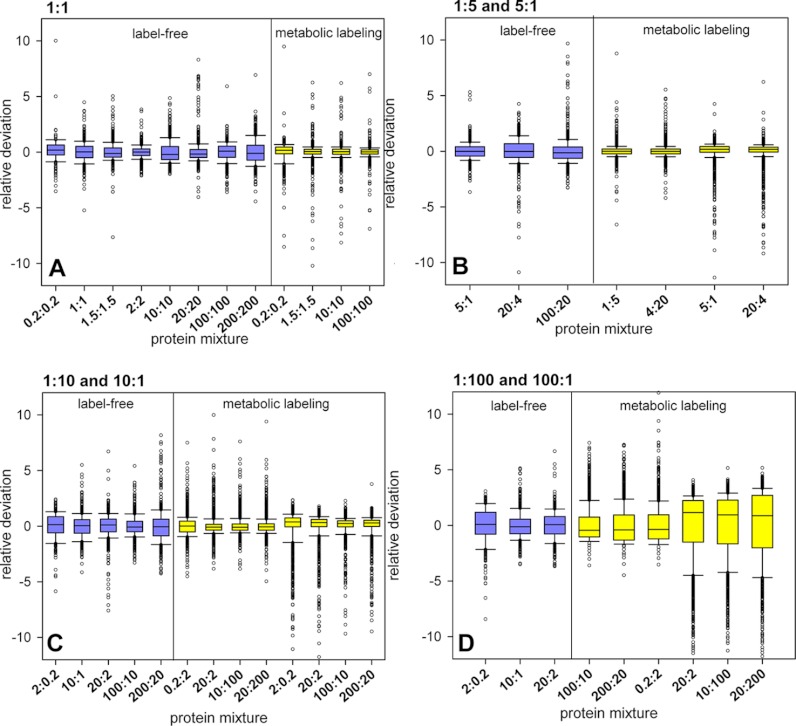
To study the precision of quantitation in each method at the different mixing ratios and total protein amounts analyzed, we plotted the deviations of measured peptide abundance ratios from expected values as boxplots (Fig. 4). As the incorporation of the heavy label was estimated to 96.7 atom% (±3) we did not account for the effect of incomplete labeling (33). For metabolic 15N labeling, in 1:1 ratios the distributions were symmetric and deviation from expected mixing ratio was rather low (Fig. 4A). Up to a mixing ratio of about 1:5 and 5:1 the ratio distribution was broader in label-free comparisons than in metabolic labeling (Fig. 4B). At high (1:10 and 10:1) and very high (1:100 and 100:1) mixing ratios, we observed an increasingly skewed ratio distribution for metabolic labeling, particularly in those samples containing high proportions of 15N-labeled protein. Although average deviation at mixing ratios of 1:10 and 10:1 in metabolic labeling was not different from label-free samples (Fig. 4C), in the 1:100 and 100:1 the label-free quantitation yielded higher precision compared with the 15N metabolic labeling (Fig. 4D).
Fig. 4.
Boxplot of the relative deviation from expected mixing ratio for metabolic labeling and label-free comparison at different total protein amounts. A, 1:1 mixtures. B, 1:5 and 5:1 mixtures. C, 1:10 and 10:1 mixtures. D, 1:100 and 100:1 mixtures.
A direct comparison of measured abundance ratios in label-free and in 15N metabolic labeling samples revealed that quantitative precision decreases with higher mixing ratios and with higher protein amounts. 15N metabolic-labeling was more precise than the label-free approach, with lower scatter at low mixing ratios up to 10-fold. Scatter around the expected ratio in these cases was higher for label-free quantitation (supplemental Fig. S4). However, for 15N metabolic labeling, a strong deviation from expected ratios was observed at high sample protein amount and at comparative ratios greater than 10-fold, and particularly with high proportion of 15N-labeled protein.
Influence of Peptide Characteristics in Quantitative Proteomics
Particularly in very high comparative ratios (Fig. 4D) we observed unequal distribution of peptides around the expected ratio in the 15N metabolically labeled mixtures. Peptide intensity had the strongest influence on the deviation from the expected ratio (Fig. 5A), with low intensity peptides showing the largest deviation. Furthermore, peptides with lower number of nitrogen atoms had a tendency for larger deviations from expected ratios (Fig. 5B), whereas deviation from expected ratio was not dependent on mass to charge ratio or total peptide molecular weight. Peptide ion intensity depends on peptide abundance in the sample as well as the peptide ionization properties. In that respect, we found that peptides with higher numbers of nitrogen atoms or in general longer peptides usually resulted in higher ion intensities (Fig. 5C).
Fig. 5.
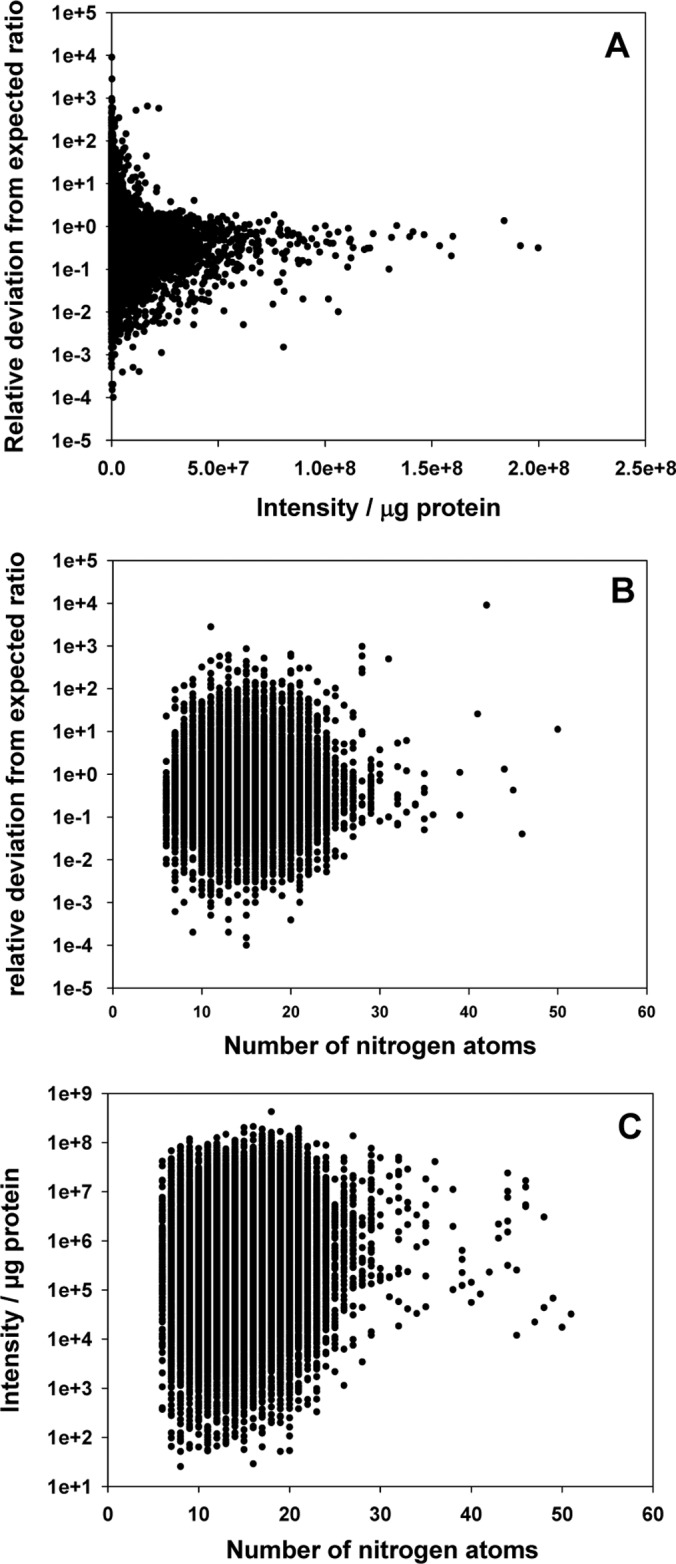
The influence of peptide properties on deviation from expected ratios. A, Peptide intensity. B, Number of nitrogen atoms. C, Relationship between number of nitrogen atoms and peptide intensity.
DISCUSSION
Proteome Coverage Depends on Experimental Setup, Total Protein Loading, and Mixing Ratio
The experimental setup will have limitations to the number of proteins that can efficiently be resolved on a column at a specific protein loading (34). For our standard setup using a single 100 mm C18-column and a 120 min separation on a linear gradient, we found the best peptide identification rate using between 20 μg and 100 μg of protein (supplemental Table S2, Fig. 2).
A main finding of our comparison of protein coverage, dynamic range and quantitative precision of 15N metabolic labeling and label-free quantitation was that in 15N metabolic labeling lowest numbers of identified and quantified proteins were yielded at a 1:1 mixture (Fig. 2). This was because of increased fragmentation of both labeled and unlabeled forms of the same peptide. This finding may have direct implications for experimental designs involving mixtures of labeled and unlabeled material. Although in our study the number of peptide pairs for quantification increased up to the 1:5 and 5:1 mixtures, it is important to mention that in our present experimental design the 1:2 and 2:1 samples had a large gap in the protein amounts investigated with total protein amount leaping from 15 μg (10 μg and 5 μg) to 300 μg (200 μg and 100 μg). The former protein amount is slightly below, whereas the latter is way above the optimal protein range for maximum peptide identifications on our LC-MS/MS setup as discussed above (34). Possibly, using mixing ratios deviating from the usually used 1:1 mixtures (e.g. 3:1 or 1:3) in biological experiments may already be advantageous with regards to total numbers of proteins quantified. Data normalization and identification of significant treatment-dependent alterations in protein abundance could still be carried out as described (31). In particular, if a reciprocal experimental design is used (35, 36), but also for use as a universal standard (24), significant treatment effects can efficiently be identified also in mixtures deviating from 1:1. The comparatively low efficiency of protein identification and quantitation in 1:1 mixtures observed here, may be less apparent if longer chromatographic gradients are being used leading to overall higher protein identifications (34). However, longer gradient times might result in a bottleneck in sample measurements in many laboratories. On the other hand, fragmentation of both isotope pairs can increase the confidence in peptide identifications, and thus may be beneficial particularly when protein modifications are analyzed. Particularly in mixtures with high proportions of 15N, the protein identifications can be reduced by formation of isotope cluster satellite peaks (33).
The 15N metabolic labeling method clearly outperformed the label free approach with regards to the number of quantified peptide pairs. This is clearly because of the inherent method characteristic of finding peptide pairs as co-eluting peaks with defined mass difference for quantitation rather than based on mass and elution time in cases in which only one of the peptides ions of the pair has been identified by MS/MS (reviewed in (22)). However, our estimation shows that when retention time correlation is used in label free quantitation the number of quantified proteins increased and became similar to that in 15N metabolic labeling in which no retention time correlation was applied (supplemental Table S3). Because of practical reasons we studied the effect of retention time correlation using the MaxQuant software (26). As MaxQuant yielded more identifications than MSQuant (37) even without the use of matching between runs, these numbers could not be compared directly to the MSQuant results. Therefore, we focused on the percentage of change between the two settings. It is beyond the scope of this study to compare the performance of the two quantitation softwares, particularly because MaxQuant cannot be used for quantitation of 15N metabolically labeled samples. Of course, our estimation assumes that upon the performance of retention time correlation, both quantitation programs will identify a similar number of peptides based on the elution time and mass. Should retention time correlation be applied to metabolically labeled data, we expect that the number of quantified proteins would further increase if the total protein amount remains within the optimal settings for the LC-MS/MS system. For label-free quantitation, matching between retention times and mass is essential for high coverage of quantitation, particularly at high mixing ratios.
Both Methods Show Linear Behavior for Two Orders of Magnitude, But the Label-free Quantitation Could be Extended to Four Orders of Magnitude with Low Protein Loading
Looking at the dynamic range of these two methods it was interesting to find that the total protein amount in the sample has a strong influence on the linear range in the label-free approach, but no influence at all on the metabolic labeling. On one side this meant that the metabolic labeling was more robust regarding total protein loading, on the other hand it also showed that if lower total protein loadings were used with the label-free approach, the linear dynamic range could be extend up to four orders of magnitude. Nonlinear behavior in label-free quantitation at high protein amounts loaded onto the column could partly also be attributed to column overloading.
In case of 15N metabolic labeling, the saturation of the linear range was also dependent on whether the heavy or the light isotope represents the greater proportion in the sample. In practice, where a reciprocal labeling experimental design is followed (36), this needs to be considered more carefully. Particularly at high mixing ratios, proteins quantified with the 14N form as a more intense partner, display a larger linear range than proteins with 15N form as a more intense partner.
Precision at Lower Mixing Ratios is Greater in Metabolic Labeling Approach, but at High Mixing Ratios it is Greater in Label-free Samples
It has already been shown that the linear quantitation powers of the 15N metabolic labeling is good until the 1:10 and 10:1 mixing ratios (38). However, we were not aware of the stepwise decrease of precision from the 1:1 mixture to the 10:1 and 1:10 ratios in 15N metabolic labeling. Beyond a mixing ratio of 5:1 and 1:5 the scatter becomes similar to what we observed for label-free quantitation (Fig. 4). Gradually building up from the 5:1 reciprocal samples, this nonnormal distribution reaches the extreme in the very high mixing ratio. Especially in these very high mixing ratios, we observed a strong underestimation of the measured abundance ratios for many of the proteins. This nonequal distribution was particularly striking for cases in which the heavy sample was the predominant component of the mixture, which leads to the assumption that the heavy nitrogen labeled peptide ion and its characteristic isotopomer envelope could be, at least partially, responsible for this behavior (33, 39), particularly as average 15N incorporation only yielded 96.7 atom%. Incorrect precursor mass assignment (33) for 15N labeled precursors was shown to lead to incorrect quantitation and underestimation particularly at high mass ions.
Peptide Intensity Has the Strongest Influence on the Deviation from the Expected Ratio for Both Methods, Closely Followed by the Number of Nitrogen Atoms Per Amino Acid
The identification of the peptide properties that influence quantitation is of utmost importance, as it can result in increased confidence upon manual inspection of peptides or for the development of correction factors and error models. In both, samples from label-free quantitation and 15N-metabolic labeling, we found that on average peptides with low intensity had a strong deviation from the expected ratio (31). As the intensity among other factors will depend on total protein loading, this explained the variations in standard deviations we found among the various total protein mixtures, resulting in the same ratio (e.g. Fig. 4C). We could furthermore link the number of nitrogen atoms per amino acid to the deviation from expected ratio, with proteins of high nitrogen content showing a tendency for smaller deviation from expected ratio. This in our opinion is the main explanation of the characteristic underestimation of the 15N metabolic labeling ratios, especially in situations in which the labeled partner is the predominant component in the sample. We showed that the peptides with low deviation of 15N to 14N ratios had a tendency for higher ion intensity. There was a trend for these peptides to be longer and contain more nitrogen atoms. Although in agreement to previous studies we did observe about 10% less protein peptide identifications for fully 15N labeled peptides, we did not observe a great difference in precursor mass accuracy. Possibly, because of strict mass accuracy filters in database search of 10 ppm, peptides with incorrect precursor assignment (33), did not end up in our result lists. In our observation, the main source for under-estimation of ratios with high proportion of 15N results from quantitation on the monoisotopic peak only. Particularly when metabolic labeling results in noncomplete 15N-icorporation a number of satellite peaks also occur for each peptide. These additional peaks result in a decreased height for the fully labeled monoisotopic peak, which then can lead to underestimation of the 15N-labeled peak intensity.
CONCLUSION
Our findings indicate that both methods under comparison, label-free quantitation and 15N metabolic labeling have a number of advantages under certain experimental conditions and constraints. A summary of the performance of the two quantitation methods at the mixing ratios and protein amounts tested in this study is presented in Fig. 6. With this study we provide information helping investigators in deciding on the optimal experimental design for their purposes and for making optimal use of the method of choice by considering three major factors influencing quantitation quality, namely proteome coverage, dynamic range and quantitative precision. Small ratio differences were more precisely detected in 15N metabolic labeling, whereas precision for large ratios was higher in label-free methods.
Fig. 6.
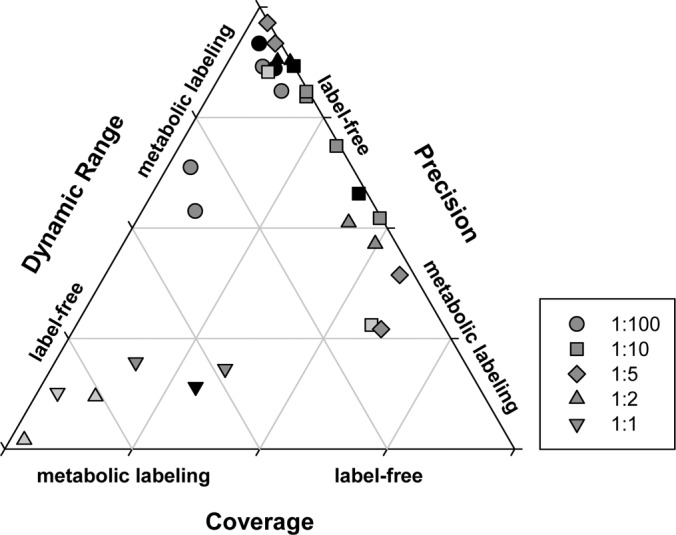
Ternary plot to assess performance of quantitation by label-free method or 15N-metabolic labeling based on proteome coverage, quantitative precision and dynamic range for different mixing ratios. Shadings of gray indicate increasing total protein loading: light gray less than 10 μg; dark gray, 10–100 μg; black, more than 100 μg protein.
The label free strategy has the unsurpassed advantage of being applicable to all starting material, which is a limitation to the 15N metabolic labeling particularly in plants. However under certain conditions the 15N metabolic labeling provides greater quantitative precision, often at the cost of quantitative coverage, depending on mixing ratio and protein amount loaded. We suggest that nonequal mixing ratios could further increase the number of quantified proteins while maintaining high precision. This finding will potentially also be applicable to other labeling strategies, such as SILAC or iTRAQ. Additionally, 15N metabolic labeling allows easier data processing, as no complex normalization is required before peptide ratios are processed. On the other hand, the label free strategy, in combination with low protein loading allows the extension of the dynamic range for quantitation, which could be important for investigation of certain types of biological questions.
Supplementary Material
Footnotes
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
1 The abbreviations used are:
- cICAT
- cleavable isotope-coded affinity tags
- iTRAQ
- isobaric tags for relative and absolute quantification
- SILAC
- stable isotope labeling with amino acids
- LTQ
- linear trap quadrupole.
REFERENCES
- 1. Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 2. Hillenkamp F., Karas M. (1990) Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 193, 280–295 [DOI] [PubMed] [Google Scholar]
- 3. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 5. Gouw J. W., Krijgsveld J., Heck A. J. (2010) Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 7. Schulze W. X., Usadel B. (2010) Quantitation in mass-spectrometry-based proteomics. Annu. Rev. Plant Biol. 61, 491–516 [DOI] [PubMed] [Google Scholar]
- 8. Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 9. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 10. Engelsberger W. R., Erban A., Kopka J., Schulze W. X. (2006) Metabolic labeling of plant cell cultures with K15NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooper B., Feng J., Garrett W. M. (2010) Relative, label-free protein quantitation: Spectral counting error statistics from nine replicate MudPIT samples. J. Am. Soc. Mass Spectrom. 21, 1534–1546 [DOI] [PubMed] [Google Scholar]
- 12. Griffin N. M., Yu J., Long F., Oh P., Shore S., Li Y., Koziol J. A., Schnitzer J. E. (2010) Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 14. Wong J. W., Cagney G. (2010) An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol. Biol. 604, 273–283 [DOI] [PubMed] [Google Scholar]
- 15. Zhang B., VerBerkmoes N. C., Langston M. A., Uberbacher E., Hettich R. L., Samatova N. F. (2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5, 2909–2918 [DOI] [PubMed] [Google Scholar]
- 16. Hoehenwarter W., van Dongen J. T., Wienkoop S., Hummel J., Erban A., Steinfath M., Sulpice R., Regierer B., Kopka J., Geigenberger P., Weckwerth W. (2008) A rapid approach for phenotype-screening and database independent detection of cSNP/protein polymorphism using mass accuracy precursor alignment. Proteomics 8, 4214–4225 [DOI] [PubMed] [Google Scholar]
- 17. Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. (2008) Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turck C. W., Falick A. M., Kowalak J. A., Lane W. S., Lilley K. S., Phinney B. S., Weintraub S. T., Witkowska H. E., Yates N. A. (2007) The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: relative protein quantitation. Mol. Cell. Proteomics 6, 1291–1298 [DOI] [PubMed] [Google Scholar]
- 19. Wu W. W., Wang G., Baek S. J., Shen R. F. (2006) Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D Gel- or LC-MALDI TOF/TOF. J. Proteome Res. 5, 651–658 [DOI] [PubMed] [Google Scholar]
- 20. Collier T. S., Randall S. M., Sarkar P., Rao B. M., Dean R. A., Muddiman D. C. (2011) Comparison of stable-isotope labeling with amino acids in cell culture and spectral counting for relative quantification of protein expression. Rapid Commun. Mass Spectrom. 25, 2524–2532 [DOI] [PubMed] [Google Scholar]
- 21. Piques M., Schulze W. X., Höhne M., Usadel B., Gibon Y., Rohwer J., Stitt M. (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5, E1–E17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arsova B., Kierszniowska S., Schulze W. X. (2012) The use of heavy nitrogen in quantitative proteomics experiments in plants. Trends Plant Sci. 17, 102–112 [DOI] [PubMed] [Google Scholar]
- 23. Sato T., Ishihama Y., Oda Y. (2007) Quantitative proteomics of mouse brain and specific protein-interaction studies using stable isotope labeling. Methods Mol. Biol. 359, 53–70 [DOI] [PubMed] [Google Scholar]
- 24. Mühlhaus T., Weiss J., Hemme D., Sommer F., Schroda M. (2011) Quantitative shotgun proteomics using a uniform 15N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.004739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schaff J. E., Mbeunkui F., Blackburn K., Bird D. M., Goshe M. B. (2008) SILIP: a novel stable isotope labeling method for in planta quantitative proteomic analysis. Plant J. 56, 840–854 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 28. Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Direct proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 29. Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 30. Venable J. D., Wohlschlegel J., McClatchy D. B., Park S. K., Yates J. R., 3rd (2007) Relative quantification of stable isotope labeled peptides using a linear ion trap-Orbitrap hybrid mass spectrometer. Anal. Chem. 79, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ting L., Cowley M. J., Hoon S. L., Guilhaus M., Raftery M. J., Cavicchioli R. (2009) Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol. Cell. Proteomics 8, 2227–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 33. Gouw J. W., Tops B. B., Mortensen P., Heck A. J., Krijgsveld J. (2008) Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal. Chem. 80, 7796–7803 [DOI] [PubMed] [Google Scholar]
- 34. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Fröhlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kierszniowska S., Seiwert B., Schulze W. X. (2009) Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-β-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8, 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kierszniowska S., Walther D., Schulze W. X. (2009) Ratio-dependent significance thresholds in reciprocal 15N-labeling experiments as a robust tool in detection candidate proteins responding to biological treatment. Proteomics 9, 1916–1924 [DOI] [PubMed] [Google Scholar]
- 37. Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 38. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. I. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 39. Nelson C. J., Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.