Abstract
The most common mutation in cystic fibrosis (CF) is a deletion of Phe at position 508 (ΔF508-CFTR). ΔF508-CFTR is a trafficking mutant that is retained in the ER, unable to reach the plasma membrane. To identify compounds and drugs that rescue this trafficking defect, we screened a kinase inhibitor library enriched for small molecules already in the clinic or in clinical trials for the treatment of cancer and inflammation, using our recently developed high-content screen technology (Trzcinska-Daneluti et al. Mol. Cell. Proteomics 8:780, 2009). The top hits of the screen were further validated by (1) biochemical analysis to demonstrate the presence of mature (Band C) ΔF508-CFTR, (2) flow cytometry to reveal the presence of ΔF508-CFTR at the cell surface, (3) short-circuit current (Isc) analysis in Ussing chambers to show restoration of function of the rescued ΔF508-CFTR in epithelial MDCK cells stably expressing this mutant (including EC50 determinations), and importantly (4) Isc analysis of Human Bronchial Epithelial (HBE) cells harvested from homozygote ΔF508-CFTR transplant patients. Interestingly, several inhibitors of receptor Tyr kinases (RTKs), such as SU5402 and SU6668 (which target FGFRs, VEGFR, and PDGFR) exhibited strong rescue of ΔF508-CFTR, as did several inhibitors of the Ras/Raf/MEK/ERK or p38 pathways (e.g. (5Z)-7-oxozeaenol). Prominent rescue was also observed by inhibitors of GSK-3β (e.g. GSK-3β Inhibitor II and Kenpaullone). These results identify several kinase inhibitors that can rescue ΔF508-CFTR to various degrees, and suggest that use of compounds or drugs already in the clinic or in clinical trials for other diseases can expedite delivery of treatment for CF patients.
Cystic fibrosis (CF)1 is a disease characterized by defective epithelial ion transport. In the lung airways, reduced Cl− transport caused by defective Cystic Fibrosis Transmembrane conductance Regulator (CFTR), coupled with increased Na+ absorption caused by elevated activity of the Epithelial Na+ Channel (ENaC), result in dehydration and thickening of the mucosal fluid (1–4). This predisposes patients to bacterial colonization, repeated pulmonary infections, and ultimately death. CF is associated with a wide-spread defect in the secretory processes of all secretory epithelia, including abnormalities in airways, gastrointestinal and genitourinary tracts, and elevated sweat electrolyte concentrations.
CF is caused by mutations in the cystic fibrosis gene (CFTR). CFTR encodes a 1480 amino acid polypeptide, called CFTR, which functions as a chloride channel in epithelial membranes (4–6). Besides its function as a chloride channel, CFTR regulates other apical membrane conductance pathways, such as the Epithelial Na+ Channel, ENaC (1), and bicarbonate secretion (7). The CFTR protein in healthy individuals is found in the apical membrane of epithelial cells, which lines the airways, gastrointestinal tract, and other exocrine ducts in the body.
Although many (∼1900) mutations in CFTR have been identified to date (www.genet.sickkids.on.ca/cftr), the most common mutation found in >70% of patients of European ancestry is a deletion of phenylalanine at position 508 (ΔF508-CFTR) (8, 9). The F508 deletion, located in the nucleotide binding domain 1 (NBD1) of CFTR, alters the folding and prevents the full maturation of the ΔF508-CFTR protein, which is subsequently degraded in the proteasome very early during biosynthesis. This abnormal folding of the ΔF508-CFTR mutant is thought to be responsible for its improper cellular localization. As ΔF508-CFTR is a trafficking-impaired mutant that is retained in the ER, its level at the apical membrane is reduced dramatically, precluding proper Cl− secretion, which leads to CF (10–13). Efforts to enhance exit of ΔF508-CFTR from the ER and its trafficking to the plasma membrane are therefore of utmost importance for the development of treatment for this disease. Indeed, over the past few years several groups have identified a few small molecules that can correct the trafficking and functional defects of the ΔF508 mutant, including corrector (corr)-3a and corr-4a, carboplatin, sildenafil, or its analogs glafenine, VX-325, VX-640, and in particular, the promising compound VX-809 (14–20). However, although VX-809 was recently tested in a phase II clinical trial, its effectiveness in alleviating the lung disease of CF patients was rather limited, underscoring the urgent need to identify new correctors (21). We had previously developed a high-content screen aimed at identifying proteins and small molecules that correct the trafficking defect of ΔF508-CFTR using human HEK293 MSR GripTite cells that stably express ΔF508-CFTR (22). Using this approach we recently performed a kinase inhibitor screen to identify kinases that, when inhibited, rescue ΔF508-CFTR. Here we describe a screen of a kinase inhibitor library biased toward compounds that are already in the clinic or in clinical trials for the treatment of other diseases, such as cancer and inflammation. Our screen identified several small molecule kinase inhibitors (and their signaling cascades) that rescue ΔF508-CFTR function, with some of these compounds already in clinical trials, thus potentially accelerating their use for the treatment of CF.
EXPERIMENTAL PROCEDURES
Media and Reagents
Dulbecco's Modified Eagle's Medium (DMEM), F12 nutrient mixture, Dulbecco's Phosphate Buffered Saline (D-PBS) with and without calcium or magnesium, fetal bovine serum (FBS), trypsin, G418, Blasticidin, and Zeocin were obtained from Invitrogen (Carlsbad, CA). SuperSignal West Femto Maximum Sensitivity kit was from Pierce (Rockford, IL), and Affinipure goat anti-mouse antibody (Cat.#115005062) was from Jackson ImmunoResearch (West Grove, PA). The small molecules kinome library was obtained from the Ontario Institute for Cancer Research (OICR-see below). The mouse M3A7 anti-CFTR monoclonal antibody was obtained from Millipore (Billerica, MA), and the anti-β-actin monoclonal antibody was from Sigma (A5441). Mouse anti-HA.11 monoclonal antibody was from Covance (MMS-101R), and Alexa Fluor 647-labeled goat anti-mouse antibody was from Invitrogen (A21236). The small molecules kinase inhibitors used for validation of the compound kinome screen were from Tocris (Bristol, UK), Selleck Chemicals and EMD Chemicals (San Diego, CA). shRNA TRC clones for FGFR1 knockdown were a kind gift from Dr. Jason Moffat (University of Toronto).
Small Molecules Kinase Inhibitor Library
The OICR Kinase Inhibitor Cassette that was screened contains 231 compounds that are reported to inhibit at least 68 kinases (supplemental Table S1). These inhibitors were purchased from a panel of more than 20 different vendors, or synthesized when not commercially available. The library was designed to cover as many targets and drug-like compounds as possible. In cases where there are multiple compounds targeting the same primary kinase, it was anticipated that having multiple chemotypes with different properties and selectivity profiles would enrich the screening set. Approximately 25% of the library consists of inhibitors that have made it into the clinic, an additional 25% being compounds in different phases of discovery (lead generation or optimization), and the remaining 50% are tool compounds that have not been advanced to the clinic but are known to be active inhibitors against various kinase targets.
Cells
HEK293 MSR GripTite (293MSR-GT) cells stably expressing ΔF508-CFTR or wild type CFTR (WT-CFTR) protein (22) were stably transfected with eYFP(H148Q/I152L) cDNA in pcDNA3.1/zeo vector using calcium phosphate. At 24 h post-transfection, the cells were seeded onto 5 × 10 cm dishes at various densities (in order to easily pick individual clones) and selected under 100 μg/ml Zeocin. Individual clones were picked and expanded. Expression of WT-CFTR or ΔF508-CFTR was validated by immunoblotting using M3A7 anti-CFTR monoclonal antibodies as previously described (22). Expression of eYFP(H148Q/I152L) was validated by fluorescent microscopy. 293MSR-GT cells stably co-expressing eYFP(H148Q/I152L) and ΔF508-CFTR or WT-CFTR protein were cultured in DMEM medium supplemented with 10% FBS, 1× nonessential amino acids, 0.6 mg/ml G418, 10 μg/ml Blasticidin, and 50 μg/ml Zeocin, at 37 °C, 5% CO2 in humidified atmosphere. Baby Hamster Kidney (BHK) cells stably expressing wild type (CFTR-3HA) or mutant (ΔF508-CFTR-3HA) protein with the triple hemagglutinin (3HA) tag at the ectodomain were a kind gift from D. Y. Thomas (McGill University, Montreal). The cells were propagated as monolayer cultures in DMEM-F12 medium (1:1) supplemented with 5% FBS and 0.5 mm Methotrexate at 37 °C, 5% CO2. Madin Darby Canine Kidney (MDCK) cells stably expressing ΔF508-CFTR protein were cultured in DMEM supplemented with 10% FBS, 1×PenStrep and 5 μg/ml Blasticidin at 37 °C, 5% CO2. Before the short-circuit current (Isc) studies, MDCK cells were grown on permeable millicell inserts (12 mm, Millipore) for 4 days and then treated with 10 μm kinase inhibitors for 48 h. Primary human bronchial epithelial (HBE) cells homozygous for ΔF508-CFTR or WT-CFTR were kindly provided by Dr. P. Karp at the University of Iowa Cell Culture Facility, and propagated on collagen-coated permeable millicell inserts (12 or 6.5 mm, Millipore) as previously described (23). Prior to the Ussing chamber assay the ΔF508-CFTR inserts were treated with 10 μm kinase inhibitors or 0.2% DMSO (vehicle control) for 48 h at 37 °C.
Cellomics YFP Halide Exchange Screen
Cellomics halide exchange assay was performed as described previously (22). Briefly, 50,000 293MSR-GT cells (stably expressing eYFP(H148Q/I152L) (14) and ΔF508-CFTR) per well were seeded in the 96-well plates. The next day ΔF508-CFTR cells were treated (in triplicate) with 10 μm small molecule kinome library, 0.2% DMSO (vehicle control), or corr-4a (positive control) at 37 °C, or incubated at 27 °C (positive control). A 10 μm dose was chosen based on a preliminary screen data (not shown) as a dose that covers a wide range of inhibiting concentrations but is not toxic to ΔF508-CFTR cells. After 48 h of incubation the medium was replaced with 152 μl of chloride solution (137 mm NaCl, 2.7 mm KCl, 0.7 mm CaCl2, 1.1 mm MgCl2, 1.5 mm KH2PO4, 8.1 mm Na2HPO4, pH 7.1), in the absence or presence of FIG (25 μm Forskolin, 45 μm IBMX, 50 μm Genistein) at 37 °C. After 20 min incubation, 92 μl of iodide buffer (137 mm NaI, 2.7 mm KCl, 0.7 mm CaCl2, 1.1 mm MgCl2, 1.5 mm KH2PO4, 8.1 mm Na2HPO4, pH 7.1) was added (final I− concentration 52 mm). Using the Cellomics VTI (Thermo Fisher), and a modified target activation algorithm, objects (individual cells or sometimes clusters of cells) were defined by eYFP(H148Q/I152L) fluorescence intensity, and the decrease in fluorescence intensity over 24-s time course, at 30 °C, 5% CO2 was recorded. The number of primary objects was used as an indicator of cell toxicity (cell death). Valid wells contained between 70 and 300 objects per field. After collecting and analyzing data, a second run of the screen was performed with compounds preselected based on the first run (∼100 compounds, each in triplicate).
Data Analysis
Compounds with a difference in fluorescence intensity between unstimulated (−FIG) and stimulated (+FIG) samples lower than 0.08 were rejected after the first run of the screen. The rest of the compounds were subjected to the secondary screen. Only the compounds that exhibited a difference in average fluorescence intensity between unstimulated and stimulated cells of at least 0.10 were further analyzed (supplemental Table S2). Compounds that displayed a difference in average fluorescence intensity of at least 0.17 were considered Tier I hits. Compounds that showed a difference in average fluorescence intensity lower than 0.17 were considered Tier II hits. Representative compounds of both groups were selected for further validation of the ΔF508-CFTR rescue.
shRNA Knockdown of FGFR1
Prior to the Cellomics halide exchange assay ΔF508-CFTR cells (stably expressing eYFP(H148Q/I152L) were transfected with FGFR1 or luciferase (nonsilencing control) shRNA constructs using Lipofectamine 2000, according to the manufacturer's instructions. Medium was changed 6 h after transfection, and ΔF508-CFTR cells were placed at 37 °C, 5% CO2. 48 h after transfection the cells were incubated with media containing Puromycin (5 μg/ml, 3 days). Cellomics halide exchange assay was performed as described above.
Knockdowns were validated by two-step RT-qPCR. Total RNA was isolated using the RNeasy 96 kit (Qiagen, Dorking, Surrey, UK), and cDNA was prepared using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Real time PCR reactions were performed using Platinum® SYBR® Green qPCRSuperMix-UDG (Invitrogen) and CFX96 Real-Time System (BioRad). Primers were obtained from Integrated DNA Technologies. For standard curves, real time PCR was performed on a fivefold dilution series DNA.
Validation of Rescue of the ΔF508-CFTR Mutant
Immunoblotting
The rescue of ΔF508-CFTR was validated by Western blotting as described previously (22). Briefly, prior to immunoblotting ΔF508-CFTR cells were treated with 15 μm kinase inhibitors or 0.3% DMSO (vehicle control) for 48 h at 37 °C, or incubated for 48 h at 27 °C (positive control). Cells were then rinsed in cold PBS and lysed in lysis buffer (50 mm Hepes pH7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 10% glycerol (v/v), 1% Triton X-100 (v/v), 2 mm phenylmethylsulfonyl fluoride, 2× PAL inhibitors). Proteins were resolved on SDS-PAGE, transferred to nitrocellulose membranes and immunoblotted with anti-CFTR monoclonal antibodies (M3A7, 1:1000) or anti-β-actin antibodies (1:10000). Membranes were washed with 5% Blotto, incubated with HRP-conjugated goat anti-mouse antibody (1:5000), and washed with PBST. Signal was detected with SuperSignal West Femto reagent.
Flow Cytometry
The rescue of ΔF508-CFTR was also validated by flow cytometry as described previously (22). Briefly, at 48 h after adding 10 μm kinase inhibitor or 0.2% DMSO (vehicle control), BHK cells were trypsinized, washed, and resuspended in ice-cold FACS buffer (PBS supplemented with 2% FBS). To stain CFTR at the cell surface, cells were incubated with anti-HA.11 monoclonal antibody (1:25) or AF647-labeled goat anti-mouse antibody (1:200) as a control, for 1 h at 4 °C. Subsequently, the cells were washed with the cold FACS buffer and incubated with AF647-conjugated goat anti-mouse antibody (1:200) at 4 °C for 1 h. They were then washed as above and resuspended in FACS buffer with 1 μg/ml propidium iodide (PI). The flow-cytometric analysis was performed using LSRII System (BD Biosciences). The data from 10,000 live (propidium iodide negative) cells were collected and analyzed with FlowJo v.7.6.4 software. Cell toxicity, as defined as >10% of cells staining positive for PI, was only observed for Ki8751 treatment. Alsterpaullone treatment resulted in altered cellular morphology (increased cell granularity and size) but not toxicity.
Short-circuit Current (Isc) Measurements in Ussing Chambers
Cell inserts or Snapwells, seeded with polarized MDCK or HBE cells (expressing ΔF508 or WT -CFTR), were mounted on an Ussing chamber apparatus (Physiological Instruments, San Diego, CA) and studied under voltage clamp conditions as previously described (23–25). Briefly, ENaC channels were inhibited with 10 μm amiloride (Sigma), and non-CFTR chloride channels were blocked with 250 μm DNDS (4,4′-dinitrostilbene-2,2′-disulfonate, Sigma). CFTR currents were then stimulated using 25 μm Forskolin, 25 μm IBMX, and 50 μm Genistein (FIG), and after the indicated time (min) inhibited using 15–50 μm GlyH-101 (Gly). Data were recorded and analyzed using Analyzer 2.1.3. Dose-response analyses (EC50) for the top kinase inhibitor hits were carried out with increasing inhibitor doses between 1 nm to 10 μm, applied to MDCK cells stably expressing ΔF508-CFTR. A few of the tested compounds (PKC412, GDC0941, PD184352, Go6976, Alsterpaullone, Kenpaullone) were toxic to MDCK cells, resulting in loss of cell monolayer integrity and loss of resistance, detected in the Ussing chambers. These were thus excluded from the data analysis.
RESULTS
Identification of Kinase Inhibitors that Correct ΔF508-CFTR Function Using the YFP High-content Assay
To systematically identify proteins and pathways that are responsible for correction of the trafficking defect of ΔF508-CFTR we previously developed a high-content functional assay or screen that allows for the identification of proteins and small molecules that correct ΔF508-CFTR function in multiple individual cells simultaneously, using Cellomics VTI Array Scan technology (22). For this, we generated cell lines in which halide-sensitive eYFP(H148Q/I152L) mutant (14) was stably transfected into HEK293 MSR GripTite (293MSR-GT) cells stably expressing wild type CFTR (WT-CFTR) or ΔF508-CFTR (22). Expression of WT-CFTR and ΔF508-CFTR in these cells was verified by immunoblotting with antibodies to CFTR (supplemental Fig. S1A), and expression of eYFP(H148Q/I152L) was verified by fluorescence microscopy (supplemental Fig. S1B).
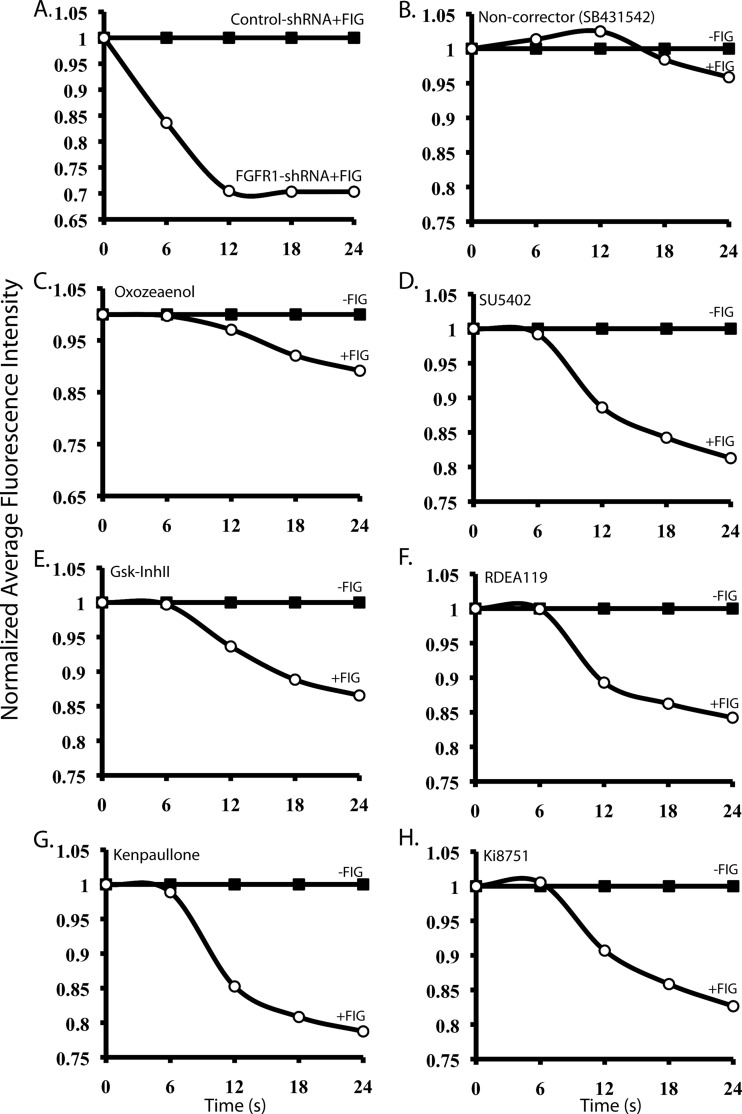
We screened 231 kinase inhibitors representing FDA-approved drugs, compounds presently in clinical trials mainly for cancer treatment and inflammation, or their derivatives, and other compounds (see Methods). In the Cellomics halide exchange assay 293MSR-GT cells stably co-expressing the Cl− sensitive eYFP(H148Q/I152L) mutant and ΔF508-CFTR (ΔF508-CFTR cells) were treated with 10 μm of each kinase inhibitor from the library at 37 °C for 48 h. Aside from temperature-rescued (27 °C) and corr-4a-treated ΔF508-CFTR cells, (5Z)-7-oxozeaenol, SU5402, or Kenpaullone-treated ΔF508-CFTR cells were used as additional positive controls in the assay. These positive controls were selected based on the hits of a previously performed Cellomics esiRNA kinome screen to identify kinases and associated proteins that when knocked-down rescue ΔF508-CFTR (which will be described in a separate manuscript). (5Z)-7-oxozeaenol, SU5402, and Kenpaullone are small molecule kinase inhibitors that mimic the effect of knockdown of several of the kinases identified in the esiRNA kinome screen. After 48-hour incubation, cells were stimulated for 20 min with Forskolin (25 μm)/IBMX (45 μm)/Genistein (50 μm) mixture (FIG). They were then exposed to low Cl−/high I− solution and fluorescence quenching of the cells caused by Cl−/I− exchange (presumably via CFTR) was recorded and quantified over time by the Cellomics VTI Array Scan. Fig. 1A shows ΔF508-CFTR rescue following shRNA-mediated knockdown of FGFR1, one of the kinases targeted by numerous “hit” compounds discovered in our screen (e.g. SU5402). Fig. 1 (C-H) depicts examples of several “hit” kinase inhibitors that when incubated with ΔF508-CFTR cells exhibited correction of the mutant CFTR function, whereas Fig. 1B provides an example of kinase inhibitor (SB431542) that did not correct ΔF508-CFTR function, shown here for comparison. The list of the hits, defined as those exhibiting difference in average fluorescence intensity between unstimulated (−FIG) and stimulated (+FIG) cells of at least 0.10, is provided in Table I and supplemental Table S2. This hit list reveals inhibitors of kinases that belong to the important cellular signaling pathways such as the (1) Ras/Raf/MEK/ERK (e.g. SU5402, SU6668, EKI-785/CL-387,785, PD0325901, PD173074, RDEA-119/AR-119/BAY869766, (5Z)-7-oxozeaenol, GW5074), (2) Wnt/GSK-3β (e.g. GSK-3β Inhibitor II, Kenpaullone, Alsterpaullone), (3) TAK1/p38 (e.g. (5Z)-7-oxozeaenol, SKF86002), or (4) PI3K/Akt/mTOR (e.g. PI-103, FPA124, 10-DEBC). Interestingly, we noticed that inhibition of activity of several receptor Tyr kinases (RTKs) (or of their downstream signaling), especially the FGFRs, with small molecules (e.g. SU5402, SU6668, PD173074, EKI-785/CL-387,785, Ki8751) led to a robust rescue of ΔF508-CFTR (Fig. 1). This suggests that FGFRs (and other RTKs) normally suppress maturation of this mutant.
Fig. 1.
Representative hits of the high-content screen. Average normalized fluorescence intensity values of ΔF508-CFTR cells (which co-express eYFP(H148Q/I152L) that were (A) transfected with shRNA for FGFR1, or (B) treated with 10 μm SB431542 (noncorrector), (C) (5Z)-7-Oxozeaenol, (D) SU5402, (E) GSK-3β Inhibitor II, (F), RDEA119, (G) Kenpaullone, (H), Ki8751, and grown at 37 °C. After 48 h (5 days in case of shRNA knockdown of FGFR1) cells were stimulated with FIG (25 μm Forskolin, 45 μm IBMX and 50 μm Genistein), and quenching of fluorescence during Cl−/I− exchange of 70–300 cells was quantified simultaneously and recorded. FGFR1 knockdown was 90% (as determined by RT-qPCR). One representative shRNA clone out of 8 is shown.
Table I. Hit compounds and their validation.
Hits were validated by immunoblotting in 293MSR-GT cells (WB), flow cytometry in BHK cells (Flow), and short circuit current (Isc) analysis in Ussing chambers on epithelial MDCK cells (MDCK) or on primary Human Bronchial Epithelial (HBE) cells harvested from lungs of ΔF508/ΔF508 homozygote patients undergoing lung transplant. 293MSR-GT, BHK, and MDCK cells were stably expressing ΔF508-CFTR. For MDCK, (+) indicates rescue, (−) no observed rescue and (*) indicates increased toxicity in MDCK cells. For HBE, (+) indicates rescue observed in a sample from a patient, with (/) separating between samples from different patients, (−) indicates no observed rescue. For the flow experiments, (+) indicates >10% rescue, (±) 5–10% rescue, (−) indicates no rescue, (*) indicates increased cell toxicity and (#) indicates morphological changes observed in the treated BHK cells. For the immunoblotting experiments, (+) indicates strong rescue of ΔF508-CFTR (manifested as increase in amount of band C in comparison to vehicle-alone control), (±) poor rescue, (−) no rescue, and (*) indicates increased toxicity in 293MSR-GT cells. For dose response experiments (EC50), MDCK cells were treated with increasing concentrations (1 nM to 10 μm range) of select compounds prior to Isc analysis in Ussing chambers, (§) indicates compounds that rescue ΔF508-CFTR function at 10 μm only.
| Compound | Pathway | Target | CAS number | PubChem ID | Clinical Trials | MDCK | EC50 (MDCK) [nM] | HBE | Flow (BHK) | WB (293GT) |
|---|---|---|---|---|---|---|---|---|---|---|
| (5Z)-7-oxozeaenol | Ras/Raf/MEK/ERK or Tak1/p38 | ERK1/2, TAK1 MAP3K | 66018-38-0 | CID 9799061 | Phase I, II (E6201) | + | 60 | + +/+ + +/− +/+/+ | + | + |
| SU6668 (Orantinib) | Ras/Raf/MEK/ERK | PDGFRβ, VEGFR2, FGFR1, EGFR | 252916-29-3 | CID 5329099 | Phase I | + | 1047 | + +/+ | + | * |
| SU5402 | Ras/Raf/MEK/ERK | VEGFR2, FGFR1, PDGFRβ | 215543-92-3 | CID 5289418 | + | 12.9 | +/+/+/− /+ + | + | + | |
| EKI-785, CL-387,785 | Ras/Raf/MEK/ERK | EGFR | 194423-06-8 | CID 2776 | + | 124.6 | −/+ | ± | + | |
| FPA 124 | PI3K/Akt/mTOR | Akt/PKB | 902779-59-3 | CID 16034833 | + | § | +/+ | − | − | |
| CID 9566171 | ||||||||||
| Gsk-3β Inhibitor II | Wnt/GSK-3β | GSK-3β | 478482-75-6 | CID 6539732 | + | 127.8 | + +/+ | + | − | |
| AZD0530, Saracatinib, NSC-735464 | Src-Bcr-Abl activates: Ras/Raf/MEK/ERK (proliferation); JAK/STAT (proliferation); PI3K/Akt (mitochondrion) pathways | Src/Abl | 379231-04-6 | CID 10302451 | Phase II | + | 165.7 | + + | − | ±* |
| 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine (Lopac-C-8863) | T-cell receptor signaling; phosphorylates PKC and PI3K; activates Ras/Raf/MEK/ERK pathway | Lck | 213743-31-8 | CID 6603792 | + | 147.4 | +/+ + | − | ±* | |
| PD173074 | Ras/Raf/MEK/ERK | FGFR1 | 219580-11-7 | CID 1401 | + | § | +/+ | − | − | |
| PD0325901 | Ras/Raf/MEK/ERK | MEK1/2 | 391210-10-9 | CID 9826528 | Phase I | + | 6.1 | + +/+ − | + | − |
| PI-103 | PI3K/Akt/mTOR | p110 PI3Ks, mTORC1/2, DNA-PK | 371935-74-9 | CID 9884685 | + | 16.4 | −/+ | + | − | |
| RDEA-119, AR-119, BAY869766 | Ras/Raf/MEK/ERK | MEK1/2 | 923032-37-5 | CID 44182295 | Phase I, II | + | 41 | +/− | + | − |
| SKF-86002 | Tak1/p38 | p38 MAP kinase | 72873-74-6 | CID 5228 | + | 140.5 | +/− | ± | − | |
| GW5074 | Ras/Raf/MEK/ERK | Raf1 | 220904-83-6 | CID 5924208 | − | − | +/+ | + | − | |
| Kenpaullone | Wnt/GSK-3β | GSK-3β, cdks, Lck | 142273-20-9 | CID 3820 | −* | −* | + + + +/+ | − | + | |
| Alsterpaullone | Wnt/GSK-3β | GSK-3β, cdks, Lck | 237430-03-4 | CID 5005498 | −* | −* | −/+ | +# | * | |
| Ki8751 | VEGFR2 activates: Ras/Raf/MEK/ERK pathway via PKC, Akt/PKB pathway via PI3K | VEGFR2, PDGFRα, FGFR2 | 228559-41-9 | CID 11317348 | − | − | − +/+/+ | +*# | + | |
| 10-DEBC (Akt specific inhibitor X) | PI3K/Akt/mTOR | Akt/PKB | 201788-90-1 | CID 16760284 | − | − | +/− | − | + |
Validation of the Hits
Maturation of ΔF508-CFTR
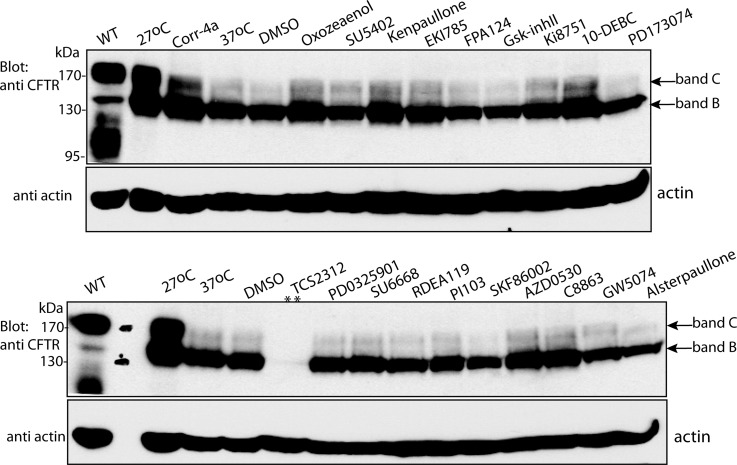
To further analyze and validate the hits, we employed alternative methods to demonstrate rescue of ΔF508-CFTR by the identified kinase inhibitors. For this we selected 41 representative compounds that inhibited kinases from the signaling pathways identified in our screen (e.g. Ras/Raf/MEK/ERK, Wnt/GSK-3, PI3K/Akt/mTOR, TAK1/p38 signaling pathways) (Table I and supplemental Table S2). First, we tested for the appearance of a mature ΔF508-CFTR protein represented by band C in an immunoblot. Fig. 2 shows that ΔF508-CFTR migrates primarily as a 140–150 kDa protein (band B) when analyzed by SDS-PAGE, whereas the mature wild type CFTR protein migrates primarily as a 170–180 kDa protein (band C). Treatment of ΔF508-CFTR cells with the indicated compounds (at 15 μm) led to the appearance of the mature band C in some of them, similar to that observed following low temperature (27 °C) treatment (albeit not as robustly).
Fig. 2.
Effect of select kinase inhibitors on ΔF508-CFTR maturation analyzed by immunoblotting. 293MSR-GT cells stably expressing ΔF508-CFTR were treated with 15 μm kinase inhibitors or 0.3% DMSO (vehicle control), as indicated, grown at 37 °C for 48 h, and the appearance of the mature protein, band C, monitored by immunoblotting with anti-CFTR antibodies. Band B represents the immature protein. DMSO represents vehicle-alone control, 27 °C represents temperature rescue of ΔF508-CFTR at 27 °C, 37 °C represents untreated ΔF508-CFTR control, and WT represents WT-CFTR. Top panels depict the anti-CFTR immunoblot and bottom panels depict actin (loading) control. ** represents cellular toxicity.
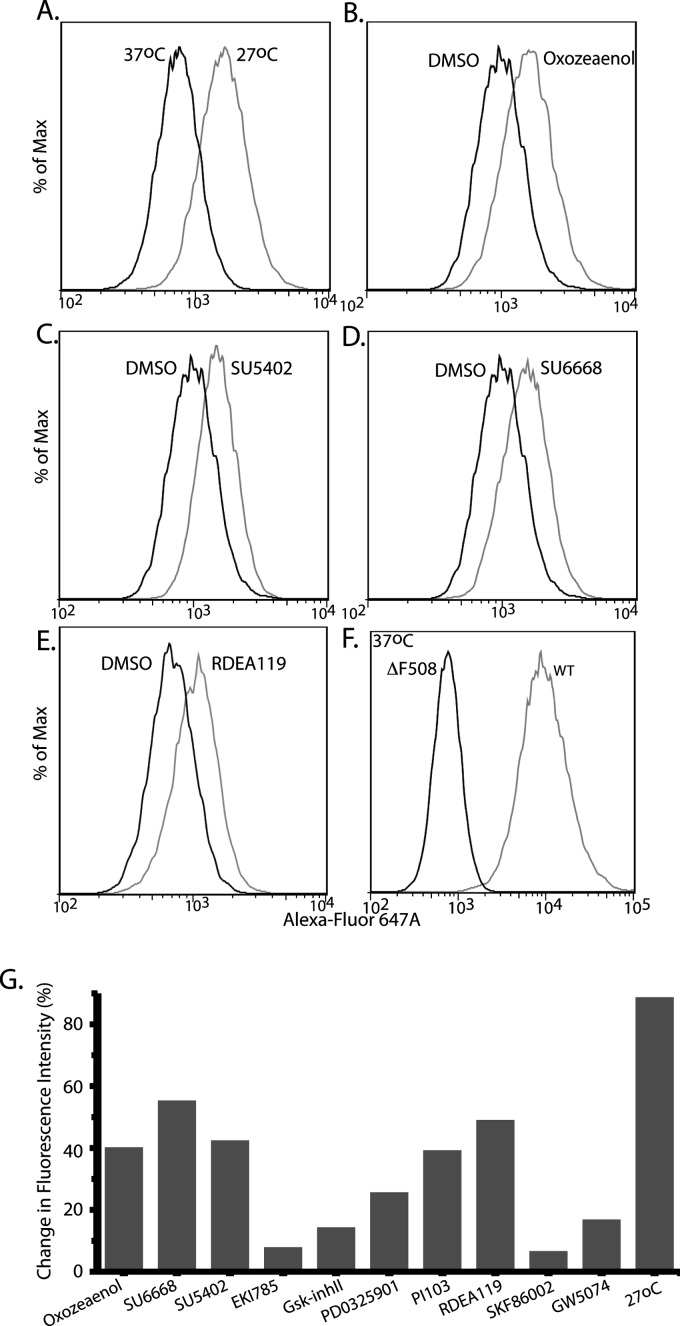
As 293MSR-GT cells showed increased sensitivity toward some of the analyzed compounds (see supplemental Table S2) because of drug toxicity, we were unable to successfully test these compounds by immunoblotting. Moreover, we wanted to directly demonstrate the presence of ΔF508-CFTR at the plasma membrane. Therefore, we tested the appearance of ΔF508-CFTR protein at the plasma membrane of nonpermeabilized BHK cells (which are not as sensitive to those compounds as the 293MSR-GT cells) using flow cytometry. BHK cells stably expressing ΔF508-CFTR-3HA were treated with 10 μm kinase inhibitors or 0.2% DMSO (vehicle control), or grown at 27 °C (positive control) for 48 h. Flow cytometry was then performed on nonpermeabilized cells following immunostaining for the HA epitope located at the ectodomain of ΔF508-CFTR, to quantify the amount of cell-surface ΔF508-CFTR. Fig. 3 depicts different degrees of ΔF508-CFTR cell surface expression obtained in cells treated with (5Z)-7-oxozeaenol, SU5402, SU6668, RDEA-119/AR-119/BAY869766 and other compounds.
Fig. 3.
Effect of kinase inhibitors on cell surface expression of ΔF508-CFTR analyzed by flow cytometry. BHK cells stably expressing ΔF508-CFTR-3HA were placed at (A) 27 °C (positive control) for 48 h, or (B) treated with 10 μm (5Z)-7-oxozeaenol, (C) SU5402, (D) SU6668, or (E) RDEA-119/AR-119/BAY869766, at 37 °C. (F) BHK cells stably expressing WT-CFTR. Flow cytometry was then performed on nonpermeabilized cells following immunostaining for the HA epitope located at the ectodomain of ΔF508-CFTR or WT-CFTR, to quantify the amount of cell-surface CFTR in the analyzed cells. (G) Summary of increase in cell surface expression of ΔF508-CFTR (% change in fluorescence intensity) of the hits analyzed by flow cytometry (two independent experiments, 10,000 live cells per treatment per experiment).
Functional Analysis of Correction of ΔF508-CFTR by the Kinase Inhibitors
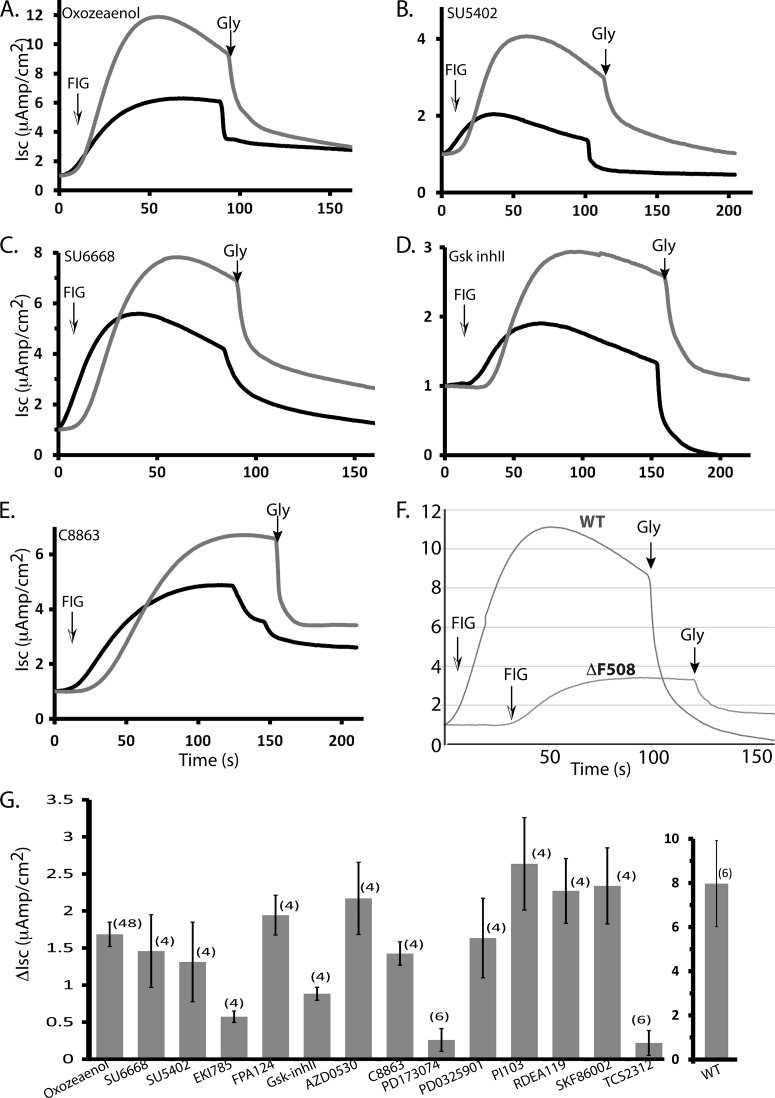
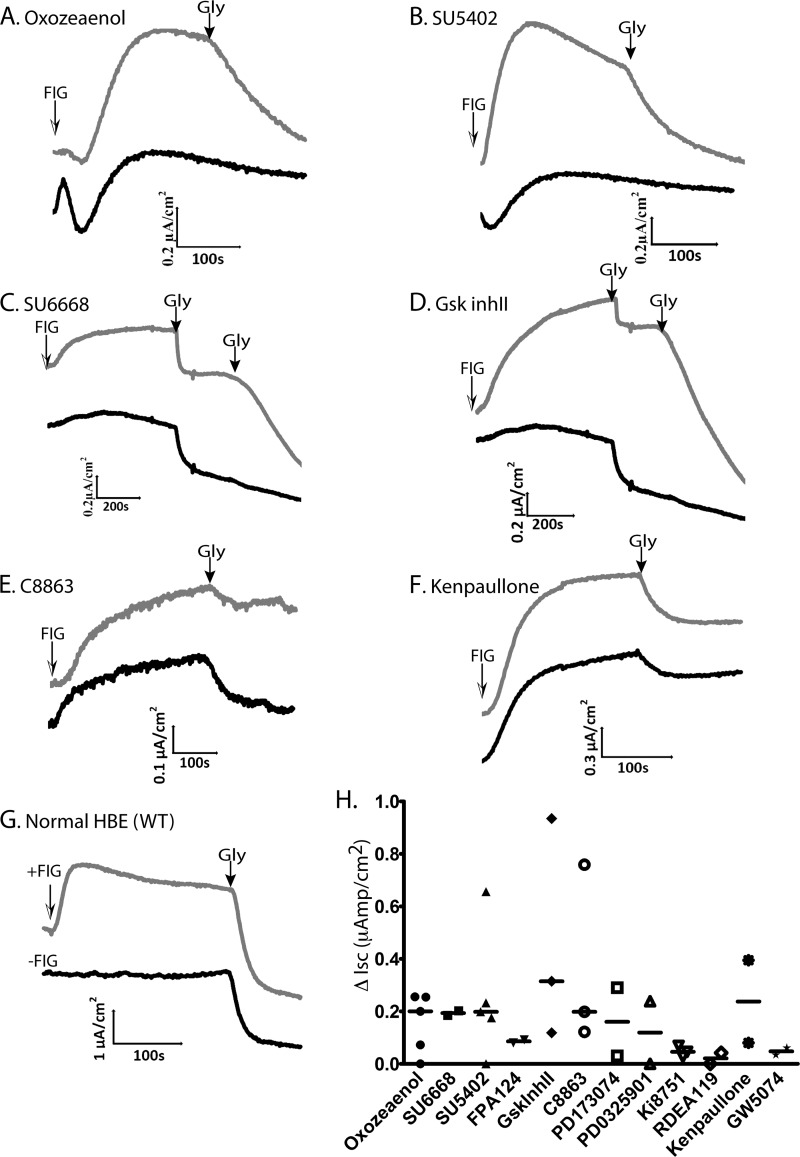
To determine whether treatment with the above kinase inhibitors can lead to correction of ΔF508-CFTR function, we performed short-circuit current (Isc) analysis in Ussing chambers on epithelial MDCK cells that stably express ΔF508-CFTR (supplemental Fig. S1C) to assess CFTR chloride channel activity. MDCK cells were treated with 10 μm kinase inhibitor or 0.2% DMSO (vehicle control) and grown at 37 °C for 48 h. The effect of treatment with representative compounds (e.g. (5Z)-7-oxozeaenol, SU5402, SU6668, GSK-3β Inhibitor II, 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine/C8863), and others on ΔF508-CFTR trafficking and function (i.e. chloride channel activity) is shown in Fig. 4 (A–E), and summarized in Fig. 4G, demonstrating various degrees of functional rescue of ΔF508-CFTR with several of these hit compounds. We noted small channel activity (“leakage”) of untreated ΔF508-CFTR at 37 °C (Fig. 4F), suggesting some escape from the ER of this mutant. DMSO alone did not add further to this “leakage,” indicating that at the concentration used in our experiments (0.2%), it does not contribute to rescue of ΔF508-CFTR (This is different from the observed rescue by 2% DMSO reported earlier (26)).
Fig. 4.
Effect of compounds treatment on the ΔF508-CFTR channel activity in the MDCK cells stably expressing ΔF508-CFTR. Representative short-circuit current (Isc) traces of MDCK ΔF508-CFTR monolayers treated with vehicle (DMSO) alone, or 10 μm (A) (5Z)-7-oxozeaenol, (B) SU5402, (C) SU6668, (D) GSK-3β Inhibitor II, (E) 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine (C8863), for 48 h prior to analysis in Ussing chambers. ENaC sodium channels were inhibited with 10 μm amiloride; non-CFTR chloride channels were blocked with 250 μm DNDS. CFTR currents were stimulated with FIG (25 μm Forskolin, 25 μm IBMX and 50 μm Genistein) at time 0 and after the indicated times (arrows) inhibited using 15 μm GlyH-101 (Gly). F, Representative short-circuit currents mediated by MDCK cells that stably express WT-CFTR. G, Summary of the increase in short-circuit currents (ΔIsc) in MDCK cells stably expressing ΔF508-CFTR that were treated by the analyzed compounds (relative to DMSO vehicle control alone). Data are mean ± S.E. (n): number of experiments.
Effect of Kinase Inhibitors on ΔF508-CFTR Chloride Channel Activity in Primary Human Bronchial Epithelial (HBE) Cells Harvested from a CF Patient
Although the above compounds appear to rescue ΔF508-CFTR in tissue culture cells, it is essential to determine if they can also rescue function in HBE cells harvested from CF patients. To this end, we investigated the effect of treatment with select validated kinase inhibitors in primary cultures of HBE cells obtained from transplant patients homozygous for the ΔF508-CFTR mutation. The effect of compound treatment was compared with control (vehicle alone) on monolayers obtained from the same patient, which allowed us to eliminate the influence of patient-to-patient variability. Fig. 5 (A–F) shows examples from several ΔF508/ΔF508 patients (as well as two control “normal” individuals expressing WT-CFTR, panel G), demonstrating enhanced activity of the mutant CFTR after treatment of cells with (5Z)-7-oxozeaenol, SU5402, SU6668, GSK-3β Inhibitor II, 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine (C8863), and Kenpaullone. Other compounds exhibited very little or variable rescue (Fig. 5H). These findings suggest that cell surface expression of ΔF508-CFTR is enhanced in HBE cells by delivering small molecule kinase inhibitors designed to correct the trafficking or maturation defect of this mutant protein, although from these latter results we cannot preclude the possibility that these compounds also potentiate ΔF508-CFTR activity once at the plasma membrane. All validation data are summarized in Table I and supplemental Table S2.
Fig. 5.
Effect of compounds treatment on ΔF508-CFTR activity in primary Human Bronchial Epithelial (HBE) cells harvested from lungs of ΔF508/ΔF508 homozygote patients undergoing lung transplant. Representative short-circuit currents (Isc) mediated by ΔF508-CFTR human bronchial epithelial (HBE) monolayers treated with vehicle (DMSO) alone, or 10 μm (A) (5Z)-7-oxozeaenol, (B) SU5402, (C) SU6668, (D) GSK-3β Inhibitor II, (E) 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine (C8863), (F) Kenpaullone, for 48 h prior to analysis in Ussing chambers. ENaC sodium channels were inhibited with 10 μm amiloride; non-CFTR chloride channels were blocked with 250 μm DNDS. CFTR currents were stimulated with FIG (25 μm Forskolin, 25 μm IBMX and 50 μm Genistein) as indicated, and after the indicated times (black arrows) inhibited using 15 μm (panels A, B, E, F) or 50 μm (panels C, D) GlyH-101 (Gly). In panels C and D, half of the Gly solution (25 μm) was added twice sequentially, as indicated. G, Representative short-circuit currents mediated by HBE cells from non-CF controls (WT-CFTR). H, Summary of increase in short-circuit currents (ΔIsc) in HBE cells stably expressing ΔF508-CFTR that were treated by analyzed compounds. Data from individual patients are shown. Where several replica were tested from the same patient (see Table I), the average value is shown. Bars represent median values. The baseline currents (before amiloride addition) ranged between 6–20 μAmp/cm2 for WT-HBE and 19–40 μAmp/cm2 for ΔF508-CFTR HBE. After adding amiloride, the currents for both WT and ΔF508-CFTR HBE were ∼0–3 μAmp/cm2.
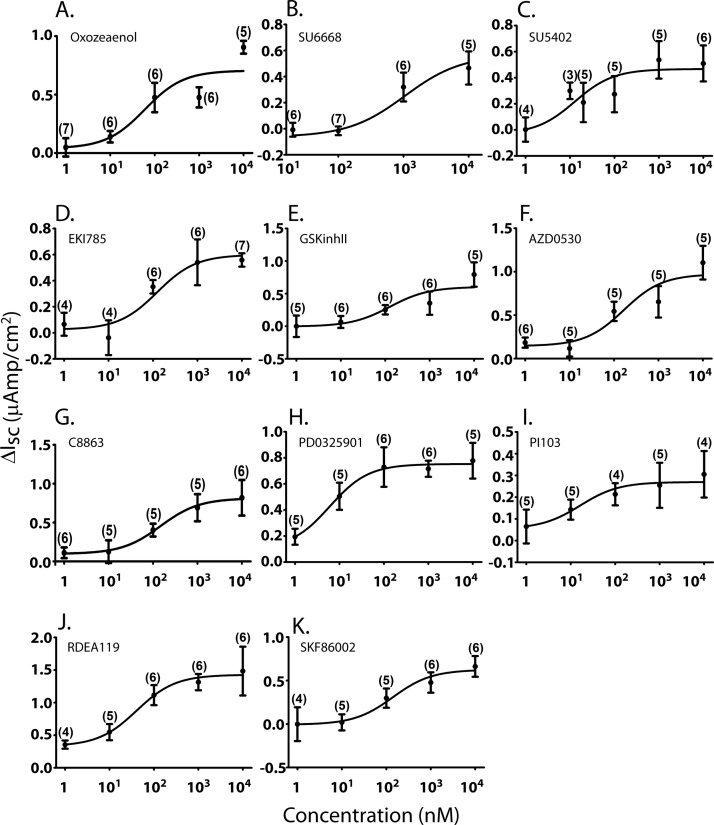
Dose Response Curves of Rescue of ΔF508-CFTR in MDCK Cells Treated with Select Kinase Inhibitors
To assess the effect of increasing doses of kinase inhibitors on correcting ΔF508-CFTR function, we treated MDCK cells (stably expressing ΔF508-CFTR) with increasing concentrations (1 nm to 10 μm range) of the top hit compounds prior to Isc analysis in Ussing chambers (Fig. 6 and Table I). The estimated half maximal effective concentrations (EC50) of the analyzed compounds were in the nanomolar range (with an exception of SU6668). PD17304 and FPA124 showed rescue of ΔF508-CFTR function only at a concentration of 10 μm. Rescue of ΔF508-CFTR by kinase inhibitors was also seen in other independently derived MDCK clones stably expressing ΔF508-CFTR (e.g. supplemental Fig. S2 for (5Z)-7-oxozeaenol). Because of limited supply of cells from patients, we could not perform dose-response analyses of these compounds on HBE cells.
Fig. 6.
Dose response curves of select kinase inhibitors for rescue of ΔF508-CFTR expressed in the MDCK cells. Average increase in short-circuit currents (ΔIsc) of MDCK cell monolayers stably expressing ΔF508-CFTR (relative to DMSO vehicle control alone) treated for 48 h with 1, 10, 20, 100, 200, 1000, and 10,000 nm of the top inhibitor compounds, indicated in panels (A–K). Data are mean ± S.E. of (n) samples.
DISCUSSION
In this paper we report novel correctors (kinase inhibitors) of the ΔF508-CFTR defect. In addition to performing the functional kinase inhibitor screen and functional validation using Isc in Ussing chambers, we carried out biochemical and flow cytometry analyses to demonstrate maturation and trafficking of ΔF508-CFTR to the plasma membrane. Nevertheless, we cannot currently preclude the possibility that at least some of the kinase inhibitors we identified may also potentiate activity of rescued ΔF508-CFTR.
The inhibitors present in the kinase inhibitor library chosen for our screen comprised a substantial number of compounds that are already used in the clinic or are in clinical trials for the treatment of other diseases, such as cancer and inflammation. Thus, potential use of any “hits” from the screen, once validated, can be moved to clinical trials for CF more quickly. Indeed, some of the identified kinase inhibitors that rescue ΔF508-CFTR are already in clinical trials for the treatment of other diseases. For example, E6201, a (5Z)-7-Oxozeaenol derivative, is now in clinical trials for the treatment of cancer (phase I (ClinicalTrials.gov identifier: NCT00794781)) and Psoriasis (phase II (ClinicalTrials.gov identifiers: NCT01268527, NCT00539929)) (27, 28). SU6668 (Orantinib) is currently in clinical trials for advanced solid tumors (phase I completed (ClinicalTrials.gov identifier: NCT00024206)) and AZD0530 (Saracatinib) is in phase II clinical trials for prostate, pancreatic, breast, colorectal, bone, and ovarian cancers (ClinicalTrials.gov identifiers: NCT01267266, NCT00735917, NCT00558272, NCT00397878, NCT00610714). RDEA119/BAY869766 (in combination with Sorafenib (ClinicalTrials.gov identifiers: NCT01204177, NCT00785226) or Gemcitabine (ClinicalTrials.gov identifier: NCT01251640)) and PD0325901 (in combination with PF-04691502 (ClinicalTrials.gov identifier: NCT01347866)) are in clinical trials in late-stage cancer patients (phase II and phase I, respectively). Therefore, our identification of the kinase inhibitors that rescue ΔF508-CFTR, which are already in clinical trials for other diseases, can accelerate use of these compounds for the treatment of CF patients, most of whom carry the ΔF508 mutation.
Our results reveal that in HBE cells from CF patients, CFTR activity of ΔF508-CFTR treated with several of the hit compounds (relative to the vehicle-treated controls) was ∼10–30% of untreated WT-CFTR (although in some cases ∼50% rescue was observed). It should be kept in mind that even a partial correction of the ΔF508-CFTR defect can be very beneficial, because several reports proposed that as low as 10–25% rescue of CFTR activity may be sufficient to restore airway epithelial function (29, 30), and CF patients and mice with residual CFTR activity exhibit milder disease than those lacking it altogether (31–33). Also, recent studies showed that a more complex approach is required to fully restore normal ΔF508-CFTR biogenesis (34). The proposed correction strategy employs a two-step folding model in which the correction of both ΔF508-CFTR NBD1 stability and the NBD1-MSD2 domain interaction are necessary. These findings may explain the limited efficacy of ΔF508-CFTR correctors currently undergoing clinical trials, and suggest that a combination drug therapy for a complete correction of the ΔF508-CFTR defect may be required.
The vast majority of the top hit compounds reported in our paper rescue ΔF508-CFTR at nanomolar concentrations. The estimated half maximal effective concentration (EC50) for the analyzed molecules were calculated from the data of functional assays (Isc) using Ussing chambers, and they should not be directly compared with IC50 values obtained from biochemical experiments. To better illustrate this point, SU6668 (Orantinib) corrects ΔF508-CFTR function with EC50 of ∼1 μm (Table I). In in vitro assays, SU6668 inhibits PDGFRβ, VEGFR2, and FGFR1 autophosphorylation at IC50 values of 8 nm, 21 nm, and 1.2 μm, respectively (35). For comparison, in cellular assays SU6668 inhibits VEGF- and PDGF-dependent signaling with IC50 values of 0.5 and 1 μm, respectively. The SU6668 pharmacokinetic analysis performed on animal plasma samples revealed an inhibition of VEGFR2 phosphorylation at concentrations of ≥ 1 μg/ml, whereas the pharmacokinetic profile of patients with advanced solid tumors showed the maximal plasma concentrations (Cmax) of 3.0 μg/ml (day 1) and 2.0 μg/ml (day 22) at a fed dose of 200 mg/m2/day (36, 37).
Our identification of several kinase inhibitors that promote rescue of ΔF508-CFTR suggests that these kinases normally inhibit maturation or trafficking of this CFTR mutant, probably by affecting the function of specific chaperones. Although a large body of literature describes destabilization of numerous activated kinases and oncogenes by Hsp90 inhibitors (38), the effect of kinases and their downstream signaling proteins on chaperones has not been extensively studied.
Several inhibitors (e.g. SU5402, SU6668, EKI-785/CL-387,785, PD173074, and Ki8751) that rescued ΔF508-CFTR inhibit receptor tyrosine kinases (RTKs) (Table I). The indolinone-containing compound SU5402 is a potent Vascular Endothelial Growth Factor Receptor (VEGFR) and Fibroblast Growth Factor Receptor (FGFR) inhibitor whereas its analog, SU6668 (Orantinib), is more selective toward Platelet Derived Growth Factor Receptor (PDGFR) (39, 40). PD173074 targets mainly FGFR3 and FGFR1 (41, 42), Ki8751 is a potent inhibitor of VEGFR2 (43), and EKI-785 (CL-387,785) is an irreversible inhibitor of EGFR (44). In accord with the rescuing effect of these RTK inhibitors, especially FGFR1, we found that shRNA-mediated knockdown of FGFR1 (Fig. 1A) also led to rescue of ΔF508-CFTR, providing further evidence for the suppressive role of at least some RTKs on maturation of ΔF508-CFTR. How FGFRs (or other RTKs) regulate maturation of this mutant protein is unknown.
In addition to the RTK inhibitors described above, we identified other small molecules that inhibit downstream effectors of these receptors, several of them targeting the MAPK pathways. For example, (5Z)-7-Oxozeaenol (ERK2 and TAK1 inhibitor) and GW5074 (Raf1 inhibitor) also rescued ΔF508-CFTR, as did the MEK1/2 inhibitors RDEA-119 (BAY869766) and PD0325901 (45, 46) (Table I). MEK1/2 are part of the Ras/Raf/MEK/ERK signaling pathway and are critical for transducing signals to ERK1/2 (47, 48). Interestingly, STAT1, which we showed promotes rescue of ΔF508-CFTR (22) was recently demonstrated to inhibit the Ras/Raf/MEK/ERK pathway (49), lending further support to the notion that this pathway inhibits ΔF508-CFTR maturation.
ERK1/2 were shown to be potent inhibitors of Heat Shock Factor 1 (HSF-1) activity and thus have an inhibitory effect on production of heat shock proteins (Hsps) such as Hsp70 (50–52). Inactive, monomeric HSF-1 exists in a complex with either Hsp70 (53) or Hsp90 (54). This repressed state of HSF-1 is maintained through inhibitory phosphorylation of the specific serine residues by ERK1/2, GSK-3, PKCα, and PKCζ kinases (51, 52). ERK1/2 role in the negative regulation of HSF-1 activity is mediated by its phosphorylation of HSF-1 on Ser307. This initial phosphorylation marks HSF-1 for a secondary phosphorylation on Ser303, which represses HSF-1 function. Ser303 is phosphorylated by glycogen synthase kinase 3 (GSK-3), which inactivates this transcription factor and inhibits subsequent expression of Hsps (51). Thus, HSF1 function is antagonized by ERK1/2 in concert with GSK-3 kinase activity.
GSK-3 isoforms α and β, the key components in Wnt and insulin signaling pathways, are constitutively active serine/threonine kinases involved in the regulation of a wide range of cellular factors and responses (55). Interestingly, our screen identified GSK-3 inhibitors that rescued ΔF508-CFTR, such as paullone derivatives (e.g. Kenpaullone) and GSK-3β Inhibitor II (56, 57). GSK-3 has an inhibitory effect on both activation and DNA binding of HSF-1 (58, 59). It was previously shown that activation of HSF-1 has an inducing effect on expression of Hsp70, Hsp60, Hsp40, Hsp27, and CRYAB (Hsp20 family) (60–65). Because Hsps facilitate folding and inhibit protein denaturation (66), it is possible that derepression of HSF-1 through inhibition of ERK1/2 or GSK-3, followed by elevated expression of Hsps, could potentially lead to a rescue of ΔF508-CFTR protein. In agreement, Hsp70 was shown to enhance ΔF508-CFTR maturation (67); also, our previous study identified numerous chaperones as correctors of the ΔF508-CFTR defect, including the Hsp70-related protein HSPA4 and CRYAB (22). Furthermore, it was shown that Hsp90 maintains the stability of GSK-3β, which is Hsp90 client protein (54, 68). The effect of the Hsp90 chaperone system on maturation of ΔF508-CFTR was recently demonstrated. Both Hsp90 inhibitors (e.g. geldanamycin; also known to activate HSF-1) and siRNA knockdown of the Hsp90 cochaperone Aha1 were demonstrated to rescue ΔF508-CFTR function by decreasing degradation of ΔF508-CFTR by the proteasome (22, 69–71).
As noted in Table I, three of our hits (FPA-124, PI-103, and 10-DEBC) target PI3K/Akt/mTOR signaling. So far, it is not known whether or how this pathway may regulate ΔF508-CFTR maturation. Although Akt was shown to promote cAMP-mediated trafficking of WT-CFTR to the plasma membrane (72), its possible effect on trafficking of ΔF508-CFTR that escaped the ER is unknown. Moreover, in some cases (e.g. AQP2, Norepinephrine transporter) Akt signaling actually decreases cell surface expression of plasma membrane proteins (73, 74). Thus, how PI3K/Akt/mTOR inhibitors may promote rescue of ΔF508-CFTR awaits future investigation.
In summary, our work here has demonstrated the rescue of ΔF508-CFTR by several kinase inhibitors in 293MSR-GT cells, BHK cells, epithelial MDCK cells and, importantly, in primary HBE cells from CF patients. Because some of our identified kinase inhibitors that rescued ΔF508-CFTR are already used in the clinic or are in clinical trials for the treatment of cancer or inflammatory disease, their potential testing or use for treatment of CF patient carrying the ΔF508 mutation (i.e. the majority of CF patients) can be greatly expedited. Moreover, these kinase inhibitors may be useful for the treatment of other “trafficking diseases,” in which proteins are stuck in the ER much like ΔF508-CFTR.
Supplementary Material
Acknowledgments
We thank Dr. P. Karp and the Iowa Tissue Culture Facility for HBE cells, the Ontario Institute for Cancer Research (OICR), Toronto, for the kinase inhibitors library, Dr. Jason Moffat for shRNA for FGFR1, and SIDNET facility (The Hospital for Sick Children) for technical support.
Footnotes
* This work was supported by the Canadian CF Foundation/Cystic Fibrosis Canada (CCFF/CFC), the Canadian Institute of Health Research (CIHR) and the Canadian Foundation for Innovation (CFI) (to DR). DR is a recipient of a CRC chair (Tier I) from the CFI.
 This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
1 The abbreviations used are:
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- RTK
- receptor tyrosine kinase
- Hsp
- heat shock protein
- GSK-3
- glycogen synthase kinase 3
- HBE
- human bronchial epithelia.
REFERENCES
- 1. Boucher R. C. (2007) Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu. Rev. Med. 58, 157–170 [DOI] [PubMed] [Google Scholar]
- 2. Donaldson S. H., Boucher R. C. (2007) Sodium channels and cystic fibrosis. Chest 132, 1631–1636 [DOI] [PubMed] [Google Scholar]
- 3. Ratjen F. A. (2009) Cystic fibrosis: pathogenesis and future treatment strategies. Respir. Care 54, 595–605 [DOI] [PubMed] [Google Scholar]
- 4. Riordan J. R. (2008) CFTR function and prospects for therapy. Annu. Rev. Biochem. 77, 701–726 [DOI] [PubMed] [Google Scholar]
- 5. Riordan J. R. (2005) Assembly of functional CFTR chloride channels. Annu. Rev. Physiol. 67, 701–718 [DOI] [PubMed] [Google Scholar]
- 6. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 7. Quinton P. M. (2010) Role of epithelial HCO3 transport in mucin secretion: lessons from cystic fibrosis. Am. J. Physiol. Cell Physiol 299, C1222–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 9. Ratjen F., Döring G. (2003) Cystic fibrosis. Lancet 361, 681–689 [DOI] [PubMed] [Google Scholar]
- 10. Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 11. Denning G. M., Ostedgaard L. S., Cheng S. H., Smith A. E., Welsh M. J. (1992) Localization of cystic fibrosis transmembrane conductance regulator in chloride secretory epithelia. J. Clin. Invest. 89, 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du K., Sharma M., Lukacs G. L. (2005) The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 12, 17–25 [DOI] [PubMed] [Google Scholar]
- 13. Kopito R. R. (1999) Biosynthesis and degradation of CFTR. Physiol. Rev. 79, S167–173 [DOI] [PubMed] [Google Scholar]
- 14. Yang H., Shelat A. A., Guy R. K., Gopinath V. S., Ma T., Du K., Lukacs G. L., Taddei A., Folli C., Pedemonte N., Galietta L. J., Verkman A. S. (2003) Nanomolar affinity small molecule correctors of defective Delta F508-CFTR chloride channel gating. J. Biol. Chem. 278, 35079–35085 [DOI] [PubMed] [Google Scholar]
- 15. Pedemonte N., Lukacs G. L., Du K., Caci E., Zegarra-Moran O., Galietta L. J., Verkman A. S. (2005) Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Invest. 115, 2564–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carlile G. W., Robert R., Zhang D., Teske K. A., Luo Y., Hanrahan J. W., Thomas D. Y. (2007) Correctors of protein trafficking defects identified by a novel high-throughput screening assay. Chembiochem 8, 1012–1020 [DOI] [PubMed] [Google Scholar]
- 17. Robert R., Carlile G. W., Liao J., Balghi H., Lesimple P., Liu N., Kus B., Rotin D., Wilke M., de Jonge H. R., Scholte B. J., Thomas D. Y., Hanrahan J. W. (2010) Correction of the Delta phe508 cystic fibrosis transmembrane conductance regulator trafficking defect by the bioavailable compound glafenine. Mol. Pharm. 77, 922–930 [DOI] [PubMed] [Google Scholar]
- 18. Robert R., Carlile G. W., Pavel C., Liu N., Anjos S. M., Liao J., Luo Y., Zhang D., Thomas D. Y., Hanrahan J. W. (2008) Structural analog of sildenafil identified as a novel corrector of the F508del-CFTR trafficking defect. Mol. Pharm. 73, 478–489 [DOI] [PubMed] [Google Scholar]
- 19. Loo T. W., Bartlett M. C., Clarke D. M. (2005) Rescue of DeltaF508 and other misprocessed CFTR mutants by a novel quinazoline compound. Mol. Pharm 2, 407–413 [DOI] [PubMed] [Google Scholar]
- 20. Van Goor F., Straley K. S., Cao D., González J., Hadida S., Hazlewood A., Joubran J., Knapp T., Makings L. R., Miller M., Neuberger T., Olson E., Panchenko V., Rader J., Singh A., Stack J. H., Tung R., Grootenhuis P. D., Negulescu P. (2006) Rescue of DeltaF508-CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am. J. Physiol. 290, L1117–1130 [DOI] [PubMed] [Google Scholar]
- 21. Clancy J. P., Rowe S. M., Accurso F. J., Aitken M. L., Amin R. S., Ashlock M. A., Ballmann M., Boyle M. P., Bronsveld I., Campbell P. W., Deboeck K., Donaldson S. H., Dorkin H. L., Dunitz J. M., Durie P. R., Jain M., Leonard A., McCoy K. S., Moss R. B., Pilewski J. M., Rosenbluth D. B., Rubenstein R. C., Schechter M. S., Botfield M., Ordonez C. L., Spencer-Green G. T., Vernillet L., Wisseh S., Yen K., Konstan M. W. (2011) Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 67, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trzcinska-Daneluti A. M., Ly D., Huynh L., Jiang C., Fladd C., Rotin D. (2009) High-content functional screen to identify proteins that correct F508del-CFTR function. Mol. Cell. Proteomics 8, 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zabner J., Zeiher B. G., Friedman E., Welsh M. J. (1996) Adenovirus-mediated gene transfer to ciliated airway epithelia requires prolonged incubation time. J. Virol. 70, 6994–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Chiaw P., Huan L. J., Gagnon S., Ly D., Sweezey N., Rotin D., Deber C. M., Bear C. E. (2009) Functional rescue of DeltaF508-CFTR by peptides designed to mimic sorting motifs. Chem. Biol. 16, 520–530 [DOI] [PubMed] [Google Scholar]
- 25. Ostedgaard L. S., Zabner J., Vermeer D. W., Rokhlina T., Karp P. H., Stecenko A. A., Randak C., Welsh M. J. (2002) CFTR with a partially deleted R domain corrects the cystic fibrosis chloride transport defect in human airway epithelia in vitro and in mouse nasal mucosa in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 3093–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bebök Z., Venglarik C. J., Pánczél Z., Jilling T., Kirk K. L., Sorscher E. J. (1998) Activation of DeltaF508 CFTR in an epithelial monolayer. Am. J. Physiol. 275, C599–607 [DOI] [PubMed] [Google Scholar]
- 27. Shen Y., Boivin R., Yoneda N., Du H., Schiller S., Matsushima T., Goto M., Shirota H., Gusovsky F., Lemelin C., Jiang Y., Zhang Z., Pelletier R., Ikemori-Kawada M., Kawakami Y., Inoue A., Schnaderbeck M., Wang Y. (2010) Discovery of anti-inflammatory clinical candidate E6201, inspired from resorcylic lactone LL-Z1640–2, III. Bio. Med. Chem. Lett. 20, 3155–3157 [DOI] [PubMed] [Google Scholar]
- 28. Muramoto K., Goto M., Inoue Y., Ishii N., Chiba K., Kuboi Y., Omae T., Wang Y. J., Gusovsky F., Shirota H. (2010) E6201, a novel kinase inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase-1 and mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase-1: in vivo effects on cutaneous inflammatory responses by topical administration. J. Pharm. Exp. Therap. 335, 23–31 [DOI] [PubMed] [Google Scholar]
- 29. Johnson L. G., Olsen J. C., Sarkadi B., Moore K. L., Swanstrom R., Boucher R. C. (1992) Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat. Genet. 2, 21–25 [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Button B., Gabriel S. E., Burkett S., Yan Y., Skiadopoulos M. H., Dang Y. L., Vogel L. N., McKay T., Mengos A., Boucher R. C., Collins P. L., Pickles R. J. (2009) CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 7, e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas S. R., Jaffe A., Geddes D. M., Hodson M. E., Alton E. W. (1999) Pulmonary disease severity in men with deltaF508 cystic fibrosis and residual chloride secretion. Lancet 353, 984–985 [DOI] [PubMed] [Google Scholar]
- 32. Dorin J. R., Farley R., Webb S., Smith S. N., Farini E., Delaney S. J., Wainwright B. J., Alton E. W., Porteous D. J. (1996) A demonstration using mouse models that successful gene therapy for cystic fibrosis requires only partial gene correction. Gene Ther. 3, 797–801 [PubMed] [Google Scholar]
- 33. Fajac I., Hubert D., Bienvenu T., Richaud-Thiriez B., Matran R., Kaplan J. C., Dall'Ava-Santucci J., Dusser D. J. (1998) Relationships between nasal potential difference and respiratory function in adults with cystic fibrosis. Eur. Respir. J. 12, 1295–1300 [DOI] [PubMed] [Google Scholar]
- 34. Rabeh W. M., Bossard F., Xu H., Okiyoneda T., Bagdany M., Mulvihill C. M., Du K., di Bernardo S., Liu Y., Konermann L., Roldan A., Lukacs G. L. (2012) Correction of both NBD1 energetics and domain interface is required to restore DeltaF508 CFTR folding and function. Cell 148, 150–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laird A. D., Christensen J. G., Li G., Carver J., Smith K., Xin X., Moss K. G., Louie S. G., Mendel D. B., Cherrington J. M. (2002) SU6668 inhibits Flk-1/KDR and PDGFRbeta in vivo, resulting in rapid apoptosis of tumor vasculature and tumor regression in mice. FASEB J. 16, 681–690 [DOI] [PubMed] [Google Scholar]
- 36. Kuenen B. C., Giaccone G., Ruijter R., Kok A., Schalkwijk C., Hoekman K., Pinedo H. M. (2005) Dose-finding study of the multitargeted tyrosine kinase inhibitor SU6668 in patients with advanced malignancies. Clin. Cancer Res. 11, 6240–6246 [DOI] [PubMed] [Google Scholar]
- 37. Xiong H. Q., Herbst R., Faria S. C., Scholz C., Davis D., Jackson E. F., Madden T., McConkey D., Hicks M., Hess K., Charnsangavej C. A., Abbruzzese J. L. (2004) A phase I surrogate endpoint study of SU6668 in patients with solid tumors. Invest. New Drugs 22, 459–466 [DOI] [PubMed] [Google Scholar]
- 38. Porter J. R., Fritz C. C., Depew K. M. (2010) Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy. Curr. Opin. Chem. Biol. 14, 412–420 [DOI] [PubMed] [Google Scholar]
- 39. Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997) Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 [DOI] [PubMed] [Google Scholar]
- 40. Sun L., Tran N., Liang C., Tang F., Rice A., Schreck R., Waltz K., Shawver L. K., McMahon G., Tang C. (1999) Design, synthesis, and evaluations of substituted 3-[(3- or 4-carboxyethylpyrrol-2-yl)methylidenyl]indolin-2-ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. J. Med. Chem. 42, 5120–5130 [DOI] [PubMed] [Google Scholar]
- 41. Grand E. K., Chase A. J., Heath C., Rahemtulla A., Cross N. C. (2004) Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia 18, 962–966 [DOI] [PubMed] [Google Scholar]
- 42. Mohammadi M., Froum S., Hamby J. M., Schroeder M. C., Panek R. L., Lu G. H., Eliseenkova A. V., Green D., Schlessinger J., Hubbard S. R. (1998) Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 17, 5896–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kubo K., Shimizu T., Ohyama S., Murooka H., Iwai A., Nakamura K., Hasegawa K., Kobayashi Y., Takahashi N., Takahashi K., Kato S., Izawa T., Isoe T. (2005) Novel potent orally active selective VEGFR-2 tyrosine kinase inhibitors: synthesis, structure-activity relationships, and antitumor activities of N-phenyl-N′-{4-(4-quinolyloxy)phenyl}ureas. J. Med. Chem. 48, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 44. Sweeney W. E., Futey L., Frost P., Avner E. D. (1999) In vitro modulation of cyst formation by a novel tyrosine kinase inhibitor. Kidney Int. 56, 406–413 [DOI] [PubMed] [Google Scholar]
- 45. Iverson C., Larson G., Lai C., Yeh L. T., Dadson C., Weingarten P., Appleby T., Vo T., Maderna A., Vernier J. M., Hamatake R., Miner J. N., Quart B. (2009) RDEA119/BAY869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer. Cancer Res. 69, 6839–6847 [DOI] [PubMed] [Google Scholar]
- 46. Barrett S. D., Bridges A. J., Dudley D. T., Saltiel A. R., Fergus J. H., Flamme C. M., Delaney A. M., Kaufman M., LePage S., Leopold W. R., Przybranowski S. A., Sebolt-Leopold J., Van Becelaere K., Doherty A. M., Kennedy R. M., Marston D., Howard W. A., Jr., Smith Y., Warmus J. S., Tecle H. (2008) The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 18, 6501–6504 [DOI] [PubMed] [Google Scholar]
- 47. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 48. Shaul Y. D., Seger R. (2007) The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 49. Wang S., Koromilas A. E. (2009) Stat1 is an inhibitor of Ras-MAPK signaling and Rho small GTPase expression with implications in the transcriptional signature of Ras transformed cells. Cell Cycle 8, 2070–2079 [DOI] [PubMed] [Google Scholar]
- 50. Banerjee Mustafi S., Chakraborty P. K., Raha S. (2010) Modulation of Akt and ERK1/2 pathways by resveratrol in chronic myelogenous leukemia (CML) cells results in the downregulation of Hsp70. PLoS One 5, e8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chu B., Soncin F., Price B. D., Stevenson M. A., Calderwood S. K. (1996) Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J. Biol. Chem. 271, 30847–30857 [DOI] [PubMed] [Google Scholar]
- 52. Chu B., Zhong R., Soncin F., Stevenson M. A., Calderwood S. K. (1998) Transcriptional activity of heat shock factor 1 at 37 degrees C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Calpha and Czeta. J. Biol. Chem. 273, 18640–18646 [DOI] [PubMed] [Google Scholar]
- 53. Nunes S. L., Calderwood S. K. (1995) Heat shock factor-1 and the heat shock cognate 70 protein associate in high molecular weight complexes in the cytoplasm of NIH-3T3 cells. Biochem. Biophys. Res. Commun. 213, 1–6 [DOI] [PubMed] [Google Scholar]
- 54. Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R. (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 [DOI] [PubMed] [Google Scholar]
- 55. Jope R. S., Johnson G. V. (2004) The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 56. Leost M., Schultz C., Link A., Wu Y. Z., Biernat J., Mandelkow E. M., Bibb J. A., Snyder G. L., Greengard P., Zaharevitz D. W., Gussio R., Senderowicz A. M., Sausville E. A., Kunick C., Meijer L. (2000) Paullones are potent inhibitors of glycogen synthase kinase-3beta and cyclin-dependent kinase 5/p25. Eur. J. Biochem. 267, 5983–5994 [DOI] [PubMed] [Google Scholar]
- 57. Bain J., McLauchlan H., Elliott M., Cohen P. (2003) The specificities of protein kinase inhibitors: an update. Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xavier I. J., Mercier P. A., McLoughlin C. M., Ali A., Woodgett J. R., Ovsenek N. (2000) Glycogen synthase kinase 3beta negatively regulates both DNA-binding and transcriptional activities of heat shock factor 1. J. Biol. Chem. 275, 29147–29152 [DOI] [PubMed] [Google Scholar]
- 59. Pirkkala L., Nykänen P., Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 60. Bao X. Q., Liu G. T. (2009) Induction of overexpression of the 27- and 70-kDa heat shock proteins by bicyclol attenuates concanavalin A-Induced liver injury through suppression of nuclear factor-kappaB in mice. Mol. Pharm. 75, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 61. Murapa P., Gandhapudi S., Skaggs H. S., Sarge K. D., Woodward J. G. (2007) Physiological fever temperature induces a protective stress response in T lymphocytes mediated by heat shock factor-1 (HSF1). J. Immunol. 179, 8305–8312 [DOI] [PubMed] [Google Scholar]
- 62. Rada A., Merentes E., Rodríguez M., Anselmi G., Strauss M. (2010) Human hepatoma cell line (HepG2) cellular response to hypothermic stress with recovery. Induction of Hsp70, Hsp60 and Hsf1 expression. Invest. Clin. 51, 479–488 [PubMed] [Google Scholar]
- 63. Saito K., Dai Y., Ohtsuka K. (2005) Enhanced expression of heat shock proteins in gradually dying cells and their release from necrotically dead cells. Exp. Cell Res. 310, 229–236 [DOI] [PubMed] [Google Scholar]
- 64. Walsh D., Li Z., Wu Y., Nagata K. (1997) Heat shock and the role of the HSPs during neural plate induction in early mammalian CNS and brain development. Cell. Mol. Life Sci. 53, 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hayashida N., Fujimoto M., Tan K., Prakasam R., Shinkawa T., Li L., Ichikawa H., Takii R., Nakai A. (2010) Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT. EMBO J. 29, 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Parsell D. A., Lindquist S. (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496 [DOI] [PubMed] [Google Scholar]
- 67. Choo-Kang L. R., Zeitlin P. L. (2001) Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am. J. Physiol. 281, L58–68 [DOI] [PubMed] [Google Scholar]
- 68. Banz V. M., Medová M., Keogh A., Furer C., Zimmer Y., Candinas D., Stroka D. (2009) Hsp90 transcriptionally and post-translationally regulates the expression of NDRG1 and maintains the stability of its modifying kinase GSK3beta. Biochim. Biophys. Acta 1793, 1597–1603 [DOI] [PubMed] [Google Scholar]
- 69. Norez C., Bilan F., Kitzis A., Mettey Y., Becq F. (2008) Proteasome-dependent pharmacological rescue of cystic fibrosis transmembrane conductance regulator revealed by mutation of glycine 622. J. Pharmacol. Exp. Ther. 325, 89–99 [DOI] [PubMed] [Google Scholar]
- 70. Fuller W., Cuthbert A. W. (2000) Post-translational disruption of the delta F508 cystic fibrosis transmembrane conductance regulator (CFTR)-molecular chaperone complex with geldanamycin stabilizes delta F508 CFTR in the rabbit reticulocyte lysate. J. Biol. Chem. 275, 37462–37468 [DOI] [PubMed] [Google Scholar]
- 71. Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 72. Tuo B., Wen G., Zhang Y., Liu X., Wang X., Dong H. (2009) Involvement of phosphatidylinositol 3-kinase in cAMP- and cGMP-induced duodenal epithelial CFTR activation in mice. Am. J. Physiol. Cell Physiol. 297, C503–515 [DOI] [PubMed] [Google Scholar]
- 73. Jung H. J., Kwon T. H. (2010) Membrane Trafficking of Collecting Duct Water Channel Protein AQP2 Regulated by Akt/AS160. Electrolyte Blood Press. 8, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Robertson S. D., Matthies H. J., Owens W. A., Sathananthan V., Christianson N. S., Kennedy J. P., Lindsley C. W., Daws L. C., Galli A. (2010) Insulin reveals Akt signaling as a novel regulator of norepinephrine transporter trafficking and norepinephrine homeostasis. J. Neurosci. 30, 11305–11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.