Abstract
Hel308 is a superfamily 2 helicase conserved in eukaryotes and archaea. It is thought to function in the early stages of recombination following replication fork arrest, and has a specificity for removal of the lagging strand in model replication forks. A homologous helicase constitutes the N-terminal domain of human DNA polymerase Q. The Drosophila homologue mus301 is implicated in double strand break repair and meiotic recombination. We have solved the high-resolution crystal structure of Hel308 from the crenarchaeon Sulfolobus solfataricus, revealing a five-domain structure with a central pore lined with essential DNA binding residues. The fifth domain is shown to act as a molecular brake, clamping the ssDNA extruded through the central pore of the helicase structure to limit the enzyme’s helicase activity. This provides an elegant mechanism to tune the enzyme’s processivity to its functional role. Hel308 can displace streptavidin from a biotinylated DNA molecule, suggesting that one function of the enzyme may be in the removal of bound proteins at stalled replication forks and recombination intermediates.
DNA helicases unwind duplex DNA, and are essential components of the DNA replication, recombination and repair machinery in all cellular organisms and many viruses. DNA helicases utilize the energy released by ATP hydrolysis to undergo conformational cycling and translocate along DNA, displacing a DNA strand in the process. Many helicases belong to one of three superfamilies (SF1, 2 & 3), classified according to the conservation of specific sequence motifs 1. SF1 and SF2 helicases possess two motor domains with RecA-like folds, which couple ATP hydrolysis to DNA translocation 2. SF2 DNA helicases include RecG in bacteria, hepatitis C virus NS3 (HCV NS3) and the RecQ family helicases, which all translocate along ssDNA with a 3′ to 5′ polarity 3 (MIGHT BE MY ENDOTE BUT FORMAT IS WRONG). The RecQ helicases play a key role in maintaining genomic integrity by stabilising stalled replication forks and removing intermediates of DNA recombination 4. Previous studies have shown that RecQ proteins target specialised DNA structures, specifically branched substrates that mimic replication forks and Holliday junctions. In humans, RecQ family helicases include the BLM and WRN proteins, mutated in certain rare inherited diseases in humans 5.
The Hel308 family SF2 helicases, like RecQ, are implicated in DNA repair, recombination and genome stability. The founding member, Mus308 from Drosophila melanogaster, was identified in a screen for mutations conferring hypersensitivity to DNA cross-linking reagents 6. (MIGHT BE MY ENDOTE BUT FORMAT IS WRONG) Mus308 consists of an N-terminal SF2 helicase fused to a C-terminal DNA polymerase. The human orthologues, PolQ, has the same arrangement (7), and the polymerase domain has been shown to function efficiently in the bypass of damaged DNA templates, consistent with a role in DNA repair (8,9). In addition to this helicase-polymerase fusion protein, metazoans also encode an orthologue of the helicase alone. This protein, known as Hel308 in H. sapiens, has been characterized biochemically, and shown to function as a typical SF2, 3′ to 5′ DNA helicase with limited processivity (10). The orthologues from D. melanogaster, Mus301, has been shown to function in double strand break repair and meiotic recombination (11). The Hel308 family helicases therefore have RecQ-like properties.
Bacteria and fungi lack orthologues of the Hel308 family, but clear homologues are present in archaea. Hel308 proteins from Pyrococcus furiosus (also known as Hjm helicase) and Methanothermobacter thermautotrophicum have been cloned and studied biochemically (12,13). Both proteins have recQ-like activities in vitro, targeting branched DNA substrates that are models for stalled replication forks and unwinding lagging strands. 7,8. Hel308 from M. thermautotrophicum functions like RecQ in a genetic screen for synthetic lethality in Escherichia coli, reinforcing the impression that the two proteins may have related functions 7.
Here we report the high-resolution crystal structure of Hel308 from the crenarchaeote Sulfolobus solfataricus strain PBL2025. The structure reveals a five domain organization, with the first four domains, including the two motor domains, a winged-helix (WH) domain 3 and domain 4 forming a ring structure with a central cavity for ssDNA. The fifth domain adopts a Helix-Loop-Helix structure known to function in DNA binding in many other proteins. Site directed mutagenesis of conserved arginine residues confirms the path of ssDNA through the central cavity, and mutant helicases with domain 5 removed or mutated have significantly higher processivity than the wild-type enzyme. This suggests that domain 5 acts as an autoinhibitory domain to control the enzyme’s processivity in vivo. Hel308 displaces streptavidin from a biotinylated oligonucleotide efficiently, consistent with a role in the removal of bound proteins from stalled replication forks or recombination intermediates. Together with the recent report of the crystal structure of Hel308 from Archaeoglobus fulgidus bound to a DNA substrate 9, (MIGHT BE MY ENDOTE BUT FORMAT IS WRONG) these observations provide considerable new information on the molecular basis for the function of this important class of DNA helicases.
Experimental procedures
Overexpression and purification of recombinant Hel308
The S. solfataricus strain PBL2025 Hel308 gene was cloned into a pDest14 vector using the Gateway® cloning system and provided by Scottish Structural Proteomics Facility (SSPF), St Andrews University. The gene was sequenced and submitted to the EMBL database with accession number AM778123. Recombinant Hel308 was expressed with a His-tag in C43 cells; the cultures were grown at 37 °C to an OD600 of approximately 0.8 in LB broth containing ampicillin at a final concentration of 100 μg/ml. Protein expression was induced by addition of 0.1 mM IPTG, and the cells were incubated for a further 3 hours at 30 °C before being harvested. Seleno-labeled Hel308 protein was produced in BL21(DE3) cells using a published procedure that employs a simplified selenomethionine media (15). An overnight culture in 100 ml of Luria broth supplemented with 100 μg/ml ampicillin was harvested and the pellet washed gently three times in the simple selenomethionine media, resulting in a final volume of 10ml. This 10ml of washed cells were used to inoculate 1l of the same media supplemented with 100 μg/ml of ampicillin. The cells were grown in shaker flasks at 200 RPM and 37 °C to an optical density of 0.6, whereupon the temperature was reduced to 25 °C and expression was induced with 0.2 mM IPTG. The cells were harvested the following day.
For purification of native and seleno-labeled protein, cell pellets with overexpressed PBL2025 Hel308 protein were resuspended in lysis buffer (50 mM sodium phosphate pH 7.5, 500 mM NaCl, 1 mg ml−1 lysozyme) with appropriate protease inhibitors and lysed on ice using a Constant Systems cell disrupter at 207 MPa. The crude lysate was centrifuged (15 000 g, 30 min, 277 K) and the cleared lysate was filtered through a 0.22 μm filter. Protein was purified from cleared lysate by two-step nickel-affinity chromatography. The lysate was batch bound to nickel Sepharose 6 fast flow medium (GE Healthcare), poured into a column and washed with 50 column volumes of lysis buffer plus 20 mM imidazole. The His-tagged protein was eluted in ten column volumes of lysis buffer plus 500 mM imidazole and immediately desalted into 50 mM Tris, 500 mM NaCl pH 7.5 on a HiPrep 26/10 desalting column (GE Healthcare) to remove imidazole. His-tagged TEV protease was added to the protein at a 1:10 mass ratio to remove the N-terminal His tag, leaving the native protein. The TEV/target protein mixture was incubated at room temperature for 15 h. Cleaved Hel308 was separated from His-tagged TEV by difference purification on Ni resin and polished by a a hiload supderdex 16/60 S-200 gel filtration column (GE Healthcare) equilibrated and eluted with 10mM Tris-chloride, pH7.5, 150 mM sodium chloride. The purified protein was characterized by SDS–PAGE and mass spectrometry. For crystallization trials, the protein was concentrated to 10 mg ml−1.
Crystallization
Initial hits for crystallization were found by performing sitting drop experiments of 1μl of protein solution (10 mg.ml−1 in 10 mM Tris-chloride, pH7.5, 150 mM sodium chloride) plus 1 μl of well solution against the following four 96 condition crystallization screens: the Classics, Pegs and JCSG+ suites (Qiagen); and JMAC, a homemade peg-based screen (unpublished). The crystals used for data collection were grown from hanging drops containing 1 μl of the 10 mg.ml−1 protein solution mixed with 0.5 μl of the 0.5 ml well solution containing, for native protein crystals, 15.3 % PEG8000, 0.1 M sodium cacodylate, pH 6.5, 0.13 M ammonium sulfate and 0.03 M magnesium chloride, and for selenium methionine-labelled protein, 13.6 % PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.12 M ammonium sulfate and 0.03 M magnesium chloride. Crystals were prepared for data collection by soaking in a cryoprotecting solution containing 16 % PEG 8000 (w/v), 0.1 M sodium cacodylate, pH 6.5, 0.1 M ammonium sulfate, 0.05 M magnesium chloride and 20 % PEG 400 (w/v). The cryoprotected crystals were flash frozen in liquid nitrogen and stored at −80 °C for shipment to the European Synchrotron Radiation Facility (ESRF).
Data collection, structure solution and refinement
Data from the crystal of the native protein for refinement were collected to 2.3 Å in 0.5° slices at x-ray wavelength of 0.934 Å on beamline ID14-1 at the ESRF using a ADSC Q210 detector. The native data were processed using Mosflm and scaled using Scala from the CCP4 suite 10 (CCP no.4,1994; Table I). Data from the crystal of the selenomethionine-labeled protein were collected to 2.6 Å in 0.5° slices at the selenium peak (0.979 Å) on beamline BM14 at the ESRF using a MAR225 detector. The selenium peak data were processed using the HKL2000 interface 11 to Denzo and scaled using Scalepack with the ‘no merge original index’ flag set. Scaling statistics for Table I were recalculated using Scala.
Table I.
Crystallographic statistics for data collected on native and selenomethionine Hel308
| Native | Se-Peak | |
|---|---|---|
| Data collection | ||
| Space Group | P21 | P21 |
| Unit Cell | a=61.7 | a=61.8 |
| b=138.1 | b=138.0 | |
| c=107.6 | c=107.5 | |
| β=94.7 | β=95.2 | |
| Resolution (Å) | 30-2.3 (2.42-2.3) | 30-2.6 (2.74-2.6) |
| Wavelength (Å) | 0.934 | 0.979 |
| Unique reflections | 79222 (11421) | 54876 (7481) |
| Multiplicity | 6.0 (3.6) | 3.7 (3.7) |
| Completeness (%) | 99.8 (99.3) | 99.5 (98.3) |
| Rmerge (%) | 7.8 (30.9) | 8.3 (35.8) |
| I/σ(I) | 14.7 (4.2) | 11.2 (4.3) |
| Structure solution | ||
| Monomers | 2 | 2 |
| Selenium sites | 24 | |
| Refinement | ||
| Protein atoms | 11116 | |
| Sulfate ions | 7 | |
| Waters | 146 | |
| Average B-factors (Å2) | ||
| Chain A | 45.76 | |
| Chain B | 45.92 | |
| Sulfate ions | 48.75 | |
| Waters | 39.58 | |
| Monomer superposition rmsd(Å) | ||
| Domains 1 to 5 | 0.874 (690 residues) | |
| Domains 1 to 4 | 0.446 (630 residues) | |
| Domain 5 | 0.389 (61 residues) | |
| r.m.s.d. bond lengths(Å) | 0.011 | |
| Angles (°) | 1.18 | |
| Rfactor/Rfree (%) | 21.2 (25.9) | |
| Validation | ||
| Ramachandran angles | ||
| Favored(#,%) | 97.5 | |
| Disallowed(#,%) | 0.07 |
Molprobity definition (26).
The 24 selenium sites were readily located and confirmed using the ShelxC, ShelxD and ShelxE programs 12 as implemented in the HKL2MAP interface 11. Phases were improved by 2-fold averaging density improvement routines in the Solve/Resolve (19,20) software package. The structure was automatically partially built using Buccaneer 13 from the CCP4 suite and completed by building manually into the experimentally phased electron density map using Xfit 14.
The structure was refined against the 2.3Å native data using Refmac5 15 from the CCP4 suite and iteratively rebuilt using Xfit. Simulated annealing was performed 16 and analyzed by superposition using the program Sequoia (25) to help with the building of less ordered loops. TLS parameters were refined for each domain, giving ten in total. Seven large difference peaks near positively charged side chains were modeled as sulfate ions. The final structure has two monomers of Hel308 comprising 1388 amino acid residues, 7 sulfate ions and 146 water molecules. (Table I). Only two gaps occur, one of 14 residues (358 to 371) in monomer A and one of 13 residues (358 to 370) in monomer B, where should be found the ‘ploughshare’ motif that physically separates the two DNA strands 9. The structure was analyzed using the Molprobity software (26). Side chain rotamers, mainchain bond angles and Cα-Cβ bond distances were corrected and the structure refined in an interactive manner resulting in a final structure with overall geometry scores in the 97th percentile of structures with resolutions from 2.1 Å to 2.5 Å (Table I).
Site Directed Mutagenesis
Site directed mutant forms of the Hel308 protein were prepared using the Quikchange protocol (Stratagene) followed by sequencing of the entire gene to ensure that no spurious mutations had been introduced. The oligonucleotide sequences used for mutagenesis are available from the corresponding author on request.
Assembly and purification of DNA Substrates
Oligonucleotides were 5′ γ-32P-ATP end-labeled and the substrates assembled by slow cooling, from 95 °C to room temperature, overnight, followed by purification on 12 % native acrylamide: TBE gels, as described previously 17
Fluorescence Anisotropy
The DNA binding affinity of Hel308 was investigated using fluorescence anisotropy using a Cary Eclipse fluorimeter with automatic polarisers. All experiments were carried out under temperature control at 20 °C. For direct titration, a 15T oligonucleotide with a 5′ fluorescein label was used at a final concentration of 20 nM in 150 μl of anisotropy buffer (20 mM HEPES pH 7.6, 100 mM NaCl, 1 mM DTT, 0.01 % Triton-X100). The first measurement was taken prior to the addition of protein; this was subtracted from the data as a blank. The protein concentration was increased cumulatively and further readings were taken after each addition, with corrections made for dilution. Changes in fluorescence intensity were also recorded; in order to avoid anisotropy effects on fluorescence intensity, “magic angle” conditions were used 18. Decreases in fluorescence intensity of up to 30 % were observed, which is not unusual given the pH sensitive nature of the fluorophore 19. Measurements were also taken for a double stranded 15mer (15T strand annealed to a 15A complement). Data was fitted, using Kaleidagraph, to the following equation:
where A is the measured anisotropy; E is the total protein concentration; and D represents the total DNA concentration. Amin (minimum anisotropy) is the anisotropy of free DNA; Amax (maximum anisotropy) is the anisotropy of the DNA-protein complex; and KD is the dissociation constant 18.
For the competition assays, 1 μM protein was incubated with 100 nM fluorescently labeled 15T DNA in anisotropy buffer to a final volume of 150 μl, and unlabelled 15T oligonucleotide was added progressively, with measurements taken as described above. The titration curves were fitted as described in Reid, et al. assuming a 1:1 interaction between the protein and the DNA 20. The KD values calculated from direct and competitive titrations were in good agreement.
Helicase Assay
The helicase reactions were set up containing 1 × helicase buffer (100 mM MES pH 6.0, 5 mM DTT, 100 mM NaCl, 0.1 mg/ml BSA), 10 nM 32P-labelled DNA substrates, and 0.5 μM protein. The assay was carried out at either 60 °C or 45 °C, as stated, in a final volume of 150 μl. Reactions were equilibrated at this temperature for 5 min and initiated by the addition of 1 mM ATP-MgCl2. 10 μl samples were taken at relevant time points and added to chilled STOP Solution (1 × STOP solution buffer (100 mM Tris pH 8.0, 50 mM EDTA), 0.5% SDS, 1 mg/ml Proteinase K, 300 mM NaCl, and 5 μM of a competitor DNA designed to bind to the displaced strand to prevent reannealing). Gel loading dye was added and samples were analysed on 12 % native acrylamide: TBE gels. The gels were phosphor-imaged and quantified as described previously (31).
Streptavidin Displacement Assay
To monitor the displacement of streptavidin from a biotinylated oligonucleotide, a 32P-labelled 50mer DNA oligonucleotide (oligo J150B 17), with a biotin label on the 5′ end was incubated at a final concentration of 10 nM with 300 nM streptavidin (Sigma) in 1 × helicase buffer with 1 mM Mg.ATP at 45 °C for 5 min to allow the streptavidin to bind to the oligo. 6 μM free biotin was then added as a streptavidin trap and the reaction was initiated by the addition of 0.5 μM Hel308 (final volume 60 μl). At the indicated time points 10 μl aliquots of the reaction mixture were removed and added to 10 μl STOP solution (1 M NaCl, 200 mM EDTA pH 8.0) together with 10 μM non-biotinylated J150B oligonucleotide to bind to the protein and reduce band shifting). The samples were separated on a 12% native acrylamide: TBE gel as described for the helicase assays above.
RESULTS
Gene cloning, site directed mutagenesis and protein expression
The hel308 gene from the crenarchaeote Sulfolobus solfataricus strain PBL2025 (ssHel308) 21 was amplified by PCR, cloned into the expression vector pDEST14 and expressed in E. coli with an N-terminal his-tag. The recombinant protein was purified using immobilized metal affinity chromatography and gel filtration, as described in the methods. For crystallization, the tag was removed by cleavage with the Tev protease during the purification process. A K52A variant corresponding to a mutation in the canonical Walker A motif was constructed as a negative control lacking ATPase and helicase activity. Two conserved arginine residues expected to have a role in DNA binding were mutated to produce the R255A and R320A mutants. A further two mutants targeted the C-terminal domain 5 of ssHel308. The first arginine of the conserved RAR motif was mutated to alanine (R662A), and the entire domain 5 was removed by truncation of the protein in another mutant (K646-stop). Site-directed mutant versions of the protein were expressed and purified as for the wild-type.
Structure of Hel308
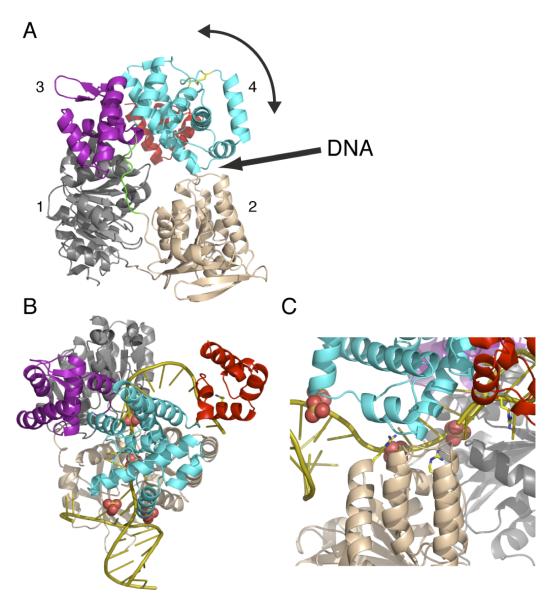
The asymmetric unit contains two monomers of the protein. Analysis using PISA 22 indicates the protein is a monomer, consistent with gel filtration results. The monomer can be decomposed into five domains, as reported previously 9 (figure 1A). Domain 1 (residues 1 – 197) and domain 2 (200 – 416) are the classical ATP binding motor domains seen in all SF1 and SF2 helicase structures. These two domains share the same core α/β fold, the root mean square deviation of 106 superimposable Cα atoms is 2.6 Å. The interface between these two domains forms the ATP binding site and the conformation changes induced during the complete cycle of ATP hydrolysis are thought to drive duplex unwinding (reviewed in 3). Domain 3 (426-501) is a winged helix domain commonly seen in nucleic acid binding proteins and closely matches Histone H5 (1.5 Å rmsd for 62 overlapping carbons). A simple search of structural similarity using SSM reveals over 400 such domains have been structurally characterized. Domain 4 (502-644) is a seven-helical bundle which is found relatively rarely in proteins, allowing little meaningful functional assignment of its role. Domain 5 (647-705) is a simple helix hairpin helix motif found in very many proteins and is commonly associated with single stranded nucleic acid binding. This domain contains the RAR motif (residues R660, R662) conserved in all Hel308 family helicases, in an α helix. The positively charged N-terminus of this α helix points towards to central pore formed by domains 1-4. Domain 5 hangs down like a cap behind the central hole. The ring is not covalently closed, and opening of a gap between domains 2 and 4 would be necessary to allow the helicase to load onto substrates lacking a 3′ ssDNA end, such as stalled replication forks. An analysis of the interface between domains 2 and 4 reveals only a weak interaction. The domain:domain contact buries a total of 650 Å2 and many of the interactions are salt bridges or polar contacts. Such a weak interaction would allow the two domains to separate to allow the passage of ssDNA into the central pore. The hinge for this movement has not been identified, although we note there is a long loop (416-426) connecting domain 2 and domain 3 which could act as a hinge (figure 1A, B).
Fig. 1.
Structure and domain organization of Hel308
A. Shown in ribbon is the structure of Hel308 from S. solfataricus. Domain 1 (1-199) is colored grey, domain 2 (200-416) wheat, domain 3 (426-501) purple, domain 4 (501-646) cyan and domain 5 (648-705) red. The extended linker (417-425) connecting domains 2 and 3 is shown in green. A disulfide bond linking the only two cysteines in the structure, C519 and C555, is shown in yellow.
B. Cartoon showing the domain arrangement of Hel308 and the proposed opening of the interface between domains 2 and 4 to allow loading onto DNA.
Overall the five domain arrangement is very similar to that reported for the structure of Hel308 from A. fulgidus 9, although the proteins share only 39 % identity. For 600 superimposable Cα atoms the rmsd deviation is 2.0 Å, the main differences are in the positions of some secondary structure elements and loops. Our apo structure superimposes equally well on the apo and DNA-bound forms of A. fulgidus Hel308, suggesting that this helicase adopts quite a rigid structure that is not perturbed significantly by DNA binding. This has allowed us to model the DNA from the A. fulgidus complex into our apo S. solfataricus structure (figure 2). The DNA threads through the central pore, forming interactions with domains 3 and 4, and engages domain 5 on the opposite side. The interactions with domain 3 and domain 4 are extensive. Loss of these domains uncouples ATP hydrolysis from helicase activity and domain 4 has been proposed as the ratchet powered by ATP hydrolysis which unwinds the DNA 9. Four sulfates present in our crystal structure coincide with the likely positions of phosphate residues in the DNA backbone. In the Hel308-DNA co-crystal structure, the DNA duplex is split by a loop almost at the entrance to the central hole which forms a small two stranded β sheet (residues 347-359). In our apo structure this loop is disordered, as was observed in the apo form of A. fulgidus Hel308 9. Three conserved arginine residues (R255, R320 and R662) mutated in this study clearly adopt positions consistent with a role in DNA binding (figure 2c).
Fig. 2.
Hel308 interaction with DNA
A. Ribbon representation of Hel308 coloured as for figure 1. The experimentally located sulfate ions are shown as space filling spheres, with oxygen colored in red, sulfur yellow. The side chains of R255, R320 (both domain 2) and R662 (domain 5), which have been mutated, are shown as sticks. Also shown is DNA double helix colored as gold taken from the structure PDB 2p6r 9.
B. The same as figure 2A but rotated approximately 90° around a horizontal axis in the plane of the paper. The ssDNA bends around in a “dog leg” from the motor domains to make contact with domain 5, the braking domain.
C. A close view of the likely path of ssDNA through the central pore, highlighting the 3 conserved arginine residues (left to right: R255, R320, R662) implicated in DNA binding.
DNA binding by wild-type and mutant Hel308 proteins
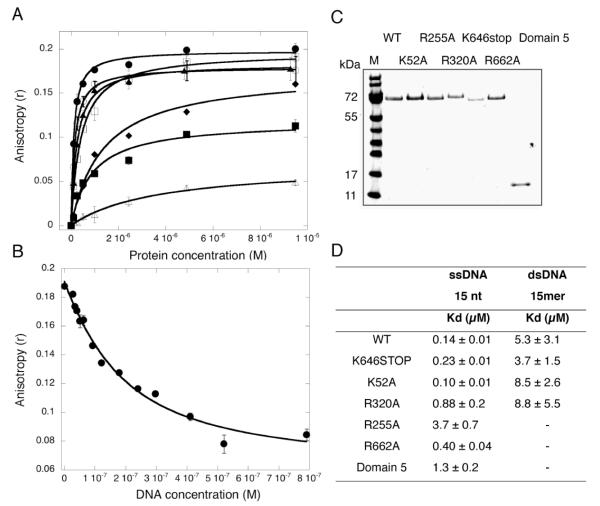
DNA binding by the wild-type and mutant versions of Hel308 was assessed by fluorescence anisotropy, using a 15mer oligonucleotide with a 5′-fluorescein dye as a reporter. A duplex DNA ligand was formed by annealing the fluorescent oligonucleotide to an oligonucleotide of complementary sequence. Equilibrium dissociation constants were obtained by plotting the change in fluorescence anisotropy in response to increasing concentrations of Hel308 (figure 3). Wild-type Hel308 bound the single-stranded and duplex DNA ligands with KD’s of 0.14 μM and 5.3 μM, respectively. The strong preference for binding to ssDNA is consistent with the function of the helicase, which must track along ssDNA and displace a duplex DNA strand. To ensure that the DNA binding affinity was not affected by protein-dye interactions, competition assays were performed by first forming a complex of Hel308 with the fluorescent oligo, and then titrating an unlabelled competitor ssDNA of the same sequence 18. The decrease in anisotropy observed in figure 3B yielded a KD of 0.13 μM, in good agreement with that calculated from the forward titration, suggesting that the influence of the fluorescein was minimal. As expected, the K52A mutant bound ssDNA and dsDNA with similar affinities to the wild type protein. Both the R255A and R320A mutants had significantly reduced ssDNA binding affinities, with the R255A mutant showing the largest effect – a 25-fold increase in the KD for ssDNA. These data confirm an important role in ssDNA binding for Arg-320 and particularly Arg-255, which line the central pore predicted as the path for ssDNA (figure 2). The truncated protein lacking domain 5 and the R662A mutant both had ssDNA binding affinities comparable or only slightly weaker than the wild-type enzyme for this short oligonucleotide. A longer oligonucleotide might be expected to engage more fully with domain 5, as this domain can be expressed independently and binds ssDNA in vitro (data not shown).
Fig. 3.
Affinity of DNA binding by wild-type and mutant Hel308
A. ssDNA binding affinities of wild-type and mutant forms of Hel308, measured by fluorescence anisotropy. Direct titration of increasing concentrations of Hel308 into a solution containing a 15T oligonucleotide with a 5′-fluorescein fluorescent reporter. Protein binding to the DNA leads to an increase in the fluorescence anisotropy, from which a dissociation constant can be calculated. The mean of triplicate measurements was plotted and the standard errors are shown. Wild-type Hel308, open circles; K646-stop, closed triangles; K52A, closed circles; R320A, closed squares; R255A, open triangles; R662A, open squares.
B. Competition analysis of Hel308 WT binding to ssDNA. Competitor DNA (unlabelled) was titrated into the assay containing 1 μM protein and 100 nM fluorescently labelled DNA. Assays were carried out at 20 °C. Each data set was carried out in triplicate and the standard errors are shown.
C. Table summarizing the dissociation constants measured for wild-type and mutant Hel308 binding to ssDNA and dsDNA. Standard errors derived from the curve fitting in Kaleidagraph are shown.
Helicase activity of wild-type and mutant Hel308 proteins
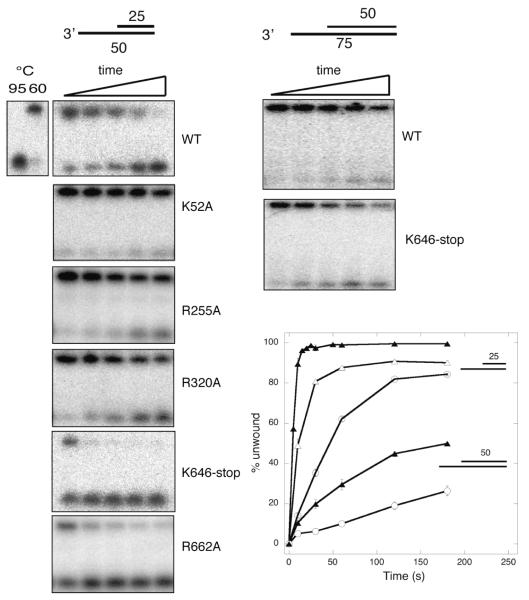
The 3′-5′ helicase activity of Hel308 was assayed using a minimal substrate: a 3′overhang with a 25 nt ssDNA region and a 25 bp duplex region. Helicase assays were carried out at 60 °C over a 3 min time course and analyzed by gel electrophoresis (figure 4). The wild-type protein displaced the duplex strand efficiently, as observed previously for the homologues from archaea and H. sapiens (10,13). As expected, the K52A mutant was unable to function as a helicase, and the R255A and R320A mutants had significantly reduced helicase activity, consistent with a defect in ssDNA binding. Surprisingly, both the K646-stop mutant lacking domain 5, and the R662A mutant lacking the first arginine of the RAR motif in domain 5 both showed significantly faster rates of DNA unwinding than the wild-type protein. To follow up this observation, we tested the helicase activity of the wild-type and truncated proteins using a DNA substrate with a longer duplex region of 50 bp (figure 5). Hel308 has been shown previously to have a limited processivity, and a limited ability to displace longer DNA duplex strands 7. Consistent with these observations, the wild-type ssHel308 protein showed only a very weak helicase activity against the larger substrate. However, significantly faster unwinding was observed with the mutant lacking domain 5, consistent with the effects observed for the shorter substrate.
Fig. 4.
Helicase activity of wild-type and mutant Hel308
The helicase activity was measured using 10 nM 32P-labeled 3′overhang substrate and 0.5 μM protein at 60 °C, as described in the methods. The plot (right) shows a quantification of the time course of unwinding for each protein, showing the means and standard errors obtained from triplicate experiments. Symbols are as indicated for figure 2.
Fig. 5.
Hel308 wild-type and truncated mutant activities with longer DNA duplexes.
The helicase activity of the wild-type and K646-stop mutants was assayed using DNA substrates with 25 bp and 50 bp duplex regions. Both forms of the enzyme unwound the longer duplexes significantly more slowly than the shorter ones. In both cases, the K646-stop truncated mutant lacking domain 5 exhibited faster helicase activity than the wild-type protein. The plot on the left shows the quantification of triplicate measurements of helicase activity. K646-stop (triangles); wild-type (circles); 25 bp and 50 bp duplex substrates are indicated by open and closed symbols, respectively. Standard errors are shown.
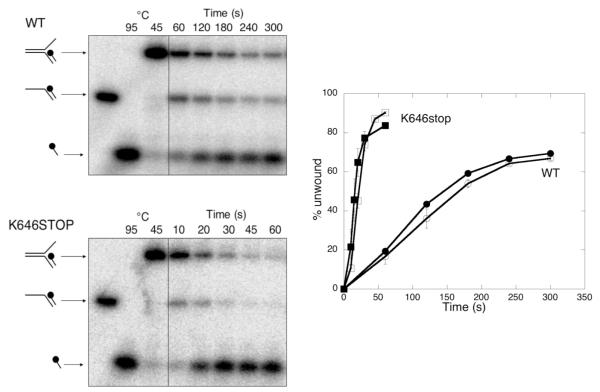
Previously biochemical studies have identified branched DNA structures, and specifically substrates resembling a stalled DNA replication fork, as the preferred substrates of Hel308 7. Accordingly, we compared the activities of the wild-type and truncation mutant using a model replication fork substrate with a 25 bp duplex region corresponding to the lagging strand of a replication fork (figure 6). These assays were carried out at the reduced temperature of 45 °C to allow more accurate determination of reaction progress. The transient production and then disappearance of the 3′-overhang intermediate at early time points suggested that unwinding may proceed via displacement of the top strand, yielding a 3′-overhang substrate intermediate that is then unwound to yield the final products, but this needs further study. With this substrate we observed the most dramatic effect of the removal of domain 5, with the truncation mutant displacing about 80 % of the lagging strand within 30 s, whereas the wild-type enzyme unwound only about 15 % of the substrate in the same period. Taken together, these observations suggest that domain 5, which is conserved in all Hel308 proteins, functions as an autoinhibitory domain, limiting the processivity of the helicase.
Fig. 6.
Wild-type and truncated Hel308 helicase activity with a stalled replication fork model substrate
The activity of wild-type and K646-stop mutant of Hel308 were compared using a model substrate resembling a stalled replication fork. The black circle indicates the 5′-32P-labelled DNA end. Quantification of the reaction (right) shows that he truncated mutant (triangles) unwinds this substrate much faster than the wild-type protein (circles). The means of triplicate experiments are shown along with standard errors.
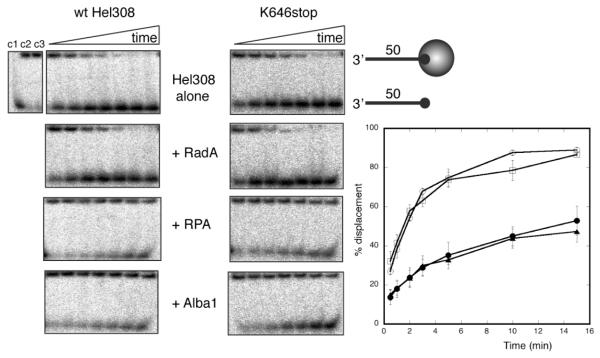
Hel308 displaces streptavidin from a biotinylated oligonucleotide
The translocation of Hel308 along ssDNA was investigated by its ability to displace streptavidin bound to a biotinylated oligonucleotide. Wild-type and mutant Hel308 proteins were incubated with the DNA substrate in the presence of ATP at 45 °C for the times indicated. Samples were run on acrylamide:TBE gels in order to separate the free biotinylated probes from those bound by streptavidin 23. Wild-type Hel308 was able to displace more than 80 % of the streptavidin in 10 min (figure 7). As expected, the inactive K52A mutant was unable to displace any streptavidin during the time course, and the R320A mutant displayed significantly lower activity, consistent with the weaker binding to ssDNA observed for this mutant. The K646-stop truncation mutant lacking domain 5 retained the ability to displace streptavidin, but was not as active as the wild-type protein. The ability of Hel308 to translocate along ssDNA with a force sufficient to dissociate a tightly bound biotin:streptavidin linkage suggests that the helicase could act to displace proteins bound to ssDNA in vivo 24.
Fig. 7.
Streptavidin displacement by Hel308
Wild-type and mutant versions of Hel308 were incubated with an oligonucleotide bound to streptavidin via a 5′ biotin modification. The wild-type and K646-stop mutant lacking domain 5 can displace streptavidin efficiently, whilst the Walker A box mutant K52A showed no activity and the R320A mutant, which has reduced ssDNA binding affinity and helicase activity, had severely reduced protein displacement activity. The plot shown on the right represents a quantification of triplicate experiments, with means and standard errors shown. Wild-type (open circles); K646-stop (triangles); K52A (closed circles); R320A (squares).
DISCUSSION
The high-resolution crystal structure of Hel308 presented here, together with the site directed mutagenesis and biochemical data, provide several new insights into the molecular basis of this conserved SF2 DNA helicase. The helicase is organized into 5 domains. The first four form a ring with a central pore large enough for the passage of single-stranded, but not double-stranded DNA.
In common with other DNA helicases, we predict that ssDNA is drawn across the top of domains 1 and 2, propelled by cyclic conformational changes in these two motor domains powered by ATP binding and hydrolysis. Conserved arginine residues lining the central pore have been shown to be important for DNA binding and helicase activity. The data are fully consistent with the recently published structure of Hel308 from A. fulgidus in the presence and absence of a DNA substrate, which shows the DNA duplex abutting a beta hairpin motif (disordered in the apo-structure) and ssDNA passing through the pore 9.
The crystal structure of Hel308 suggests an ancillary role for domain 5, which sits perpendicular to the ring formed by the first 4 domains, and which has been shown to engage the 3′ DNA end that extrudes through the central pore as the helicase translocates DNA. Hopfner and colleagues have suggested a role as a specificity domain for domain 5, predicting that it confers binding specificity for the branched DNA substrates preferred by the enzyme 9. However our data show clearly that either deletion of domain 5 or abrogation of its DNA binding affinity in the R662A mutant results in an increase in processivity of the helicase. Importantly, the truncated mutant retains a faster helicase activity than the wild-type enzyme, and can displace longer DNA strands more effectively. The effect is equally pronounced with the minimal 3′ overhang substrate and the preferred lagging strand substrate, suggesting that the function of domain 5 in vivo is to act as a molecular brake, limiting the extent of DNA unwinding by Hel308.
Autoinhibitory domains have been observed in other helicases. The SF1 helicase Rep, which functions in replication restart, in inhibited by its 2B subdomain, which is thought to block the Rep helicase from invading the DNA duplex (36). This inhibition is alleviated by dimerisation of the Rep protein. Recently, the C-terminal D7 domain of the transcription coupled repair protein Mfd from E. coli has also been shown to inhibit the helicase activity of Mfd in vitro 25. In this case, the interaction of Mfd with a stalled RNA polymerase molecule is thought to result in a conformational change in the D7 domain, alleviating the repression of the helicase. Finally, domain 4 of the bacterial DNA repair helicase UvrB has also been shown to function as an autoinhibitory domain in vitro, where it is proposed to regulate the repair activity of the UvrABC system and prevent spurious incision events (38). For all these examples, inhibition is thought to be “on” by default, and to be relieved by the formation of cognate protein:protein interactions. It remains to be determined whether the autoinhibitory domain of Hel308 has a similar role, or whether it functions as a constitutive brake on the helicase activity.
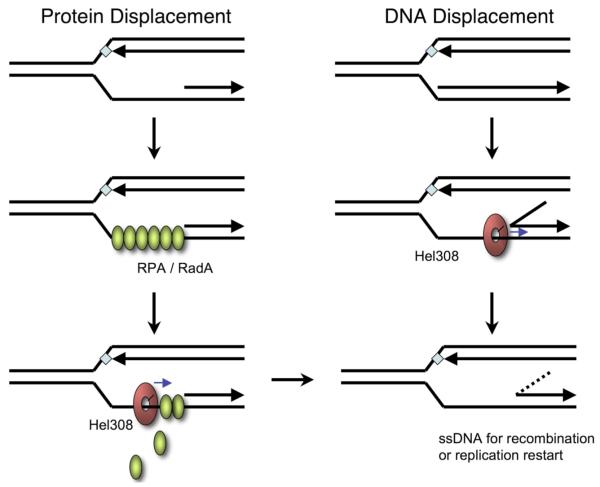
The limited helicase activity of Hel308 is consistent with the proposed function in vivo in the removal of short sections of the lagging strand next to a stalled replication fork to clear a ssDNA binding site for proteins that can restart replication or initiate recombination 7 (figure 8). In E. coli the 3′-5′ helicase activities of PriA or Rep carry out this role (reviewed in 26). However, as we have also shown that Hel308 can displace streptavidin from biotinylated ssDNA, a plausible additional function of the enzyme may be to displace RPA or other ssDNA binding proteins from the site of a stalled replication fork, allowing binding of Rad51 (RadA in archaea) and the initiation of recombination, or the binding of other proteins for DNA replication (Figure 7). This is analogous to the role proposed for RecFOR in the displacement of SSB from stalled forks to allow RecA-mediated recombination 27. Equally, Hel308 could function like bacterial UvrD, another 3′-5′ helicase that has been shown to disrupt RecA nucleoprotein filaments and thus limit recombination 28. In this context, it is intriguing to note that Ishino and colleagues report a direct interaction between Hel308 and RadA from P. furiosus 8.
Fig. 8.
Schematic model for possible roles of Hel308 at stalled replication forks
Hel308 can translocate in a 3′ to 5′ direction along ssDNA, and displace either bound protein or a duplex DNA strand. In vivo, the protein could function to generate an area of ssDNA on the lagging strand to allow replication restart or the initiation of recombination. This could involve the removal of bound proteins such as RPA or RadA (archaeal Rad51), as shown on the left, or the displacement of a short section of the lagging strand, as shown on the right.
Acknowledgments
Thanks to the BBSRC for financial support of the enzymology. Special thanks to Georg Lipps for the kind donation of PBL2025 chromosomal DNA. The native structure of Hel308 was determined by the Scottish Structural Proteomics Facility, which is funded by the BBSRC, Scottish Funding Council and The University of St Andrews.
Footnotes
Figure S1. Sequence alignment of hel308 homologues
Figure S2. SDS-PAGE analysis showing purified wild-type and mutant Hel308 proteins.
REFERENCES
- 1.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 2.Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol. 2002;184:1819–26. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Doherty KM, Brosh RM., Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–37. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007 doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd JB, Golino MD, Nguyen TD, Green MM. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics. 1976;84:485–506. doi: 10.1093/genetics/84.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guy CP, Bolt EL. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucl. Acids Res. 2005;33:3678–90. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujikane R, Shinagawa H, Ishino Y. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells. 2006;11:99–110. doi: 10.1111/j.1365-2443.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- 9.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 10.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–94. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 11.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- 12.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 13.Cowtan K. Fast Fourier feature recognition. Acta Crystallogr D Biol Crystallogr. 2001;57:1435–44. doi: 10.1107/s0907444901010812. [DOI] [PubMed] [Google Scholar]
- 14.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. Journal of structural biology. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.Kvaratskhelia M, White MF. An archaeal Holliday junction resolving enzyme with unique properties. J. Mol. Biol. 2000;295:193–202. doi: 10.1006/jmbi.1999.3363. [DOI] [PubMed] [Google Scholar]
- 18.Reid SL, Parry D, Liu HH, Connolly BA. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry. 2001;40:2484–94. doi: 10.1021/bi001956p. [DOI] [PubMed] [Google Scholar]
- 19.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–12. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 20.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–10. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 21.Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol. 2004;186:427–37. doi: 10.1128/JB.186.2.427-437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007 doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–71. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 24.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–8. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 25.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucl. Acids Res. 2007;35:1802–11. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell. Biol. 2006;7:932–43. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 27.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA. 2004;101:12783–8. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–9. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]