Summary
Maturation of all cytoplasmic mRNAs in trypanosomes involves trans-splicing of a short exon at the 5′ end. Inhibition of trans-splicing results in an accumulation of partially processed oligocistronic mRNAs. Here, we show that the accumulation of newly synthesised partially processed mRNAs results in the formation of foci around the periphery of the nucleus. These nuclear periphery granules (NPGs) contain the full complement of P-body proteins identified in trypanosomes to date, as well as poly(A)-binding protein 2 and the trypanosome homologue of the RNA helicase VASA. NPGs resemble perinuclear germ granules from metazoa more than P-bodies because they: (1) are localised around the nuclear periphery; (2) are dependent on active transcription; (3) are not dissipated by cycloheximide; (4) contain VASA; and (5) depend on nuclear integrity. In addition, NPGs can be induced in cells depleted of the P-body core component SCD6. The description of NPGs in trypanosomes provides evidence that there is a perinuclear compartment that can determine the fate of newly transcribed mRNAs and that germ granules could be a specialised derivative.
Key words: Pre-mRNA, Trans-splicing, RNA granules, VASA, P-bodies, Trypanosoma brucei
Introduction
The maturation of all nuclear-encoded mRNAs in trypanosomes requires trans-splicing. Polycistronic pre-mRNA is co-transcriptionally processed to monocistronic mRNAs by two coupled steps that trans-splice a 39-nucleotide leader from the spliced leader RNA (SL RNA) to the 5′ end of the mRNA, and then cleave and polyadenylate the 3′ end of the upstream mRNA (Campbell et al., 1984; LeBowitz et al., 1993; Ullu et al., 1993; Matthews et al., 1994). The SL RNA is transcribed by RNA polymerase II (Gilinger and Bellofatto, 2001) and is capped (Mair et al., 2000); trans-splicing provides a mechanism to add a 5′ cap to the mature mRNA. Cis-splicing is very rare and occurs in only two genes (Mair et al., 2000; Jaé et al., 2010; Kolev et al., 2010; Siegel et al., 2010). The mechanism of trans-splicing is analogous to the cis-splicing reaction (Murphy et al., 1986; Sutton and Boothroyd, 1988). The spliceosomes that catalyse cis- and trans-splicing are similar: five snRNAs each associate with seven core Sm proteins in ring-shaped structures – SmB, SmD1, SmD2, SmD3, SmE, SmF and SmG for U1, U2, U4 and U5 snRNPs, and Lsm2–Lsm8 for U6. In trans-splicing, the SL RNA replaces the U1 snRNA in the spliceosome (Thomas et al., 1988; Van Doren and Hirsh, 1988; Hannon et al., 1991; Palfi et al., 1991).
Inhibition of trans-splicing in trypanosomes by heat shock results in the accumulation of incompletely processed mRNAs that are capped and polyadenylated but contain two or more open reading frames in tandem (Muhich and Boothroyd, 1988; Muhich et al., 1989). A convenient assay for incompletely processed mRNAs takes advantage of the structure of the tubulin locus which contains 19 tandem repeats of α- and β-tubulin genes (..αβαβα..) (Seebeck et al., 1983; Thomashow et al., 1983; Ersfeld et al., 1998); trans-splicing of β-tubulin is more efficient than of α-tubulin (Muhich and Boothroyd, 1988; Muhich et al., 1989; Siegel et al., 2005) and upon inhibition of trans-splicing, there is an accumulation of β-α dicistronic RNA, β-α-β-α tetracistronic RNA, and so on, with a lesser accumulation of odd number cistronic RNAs. Low levels of dicistronic tubulin RNA can be detected in cells under normal growth conditions (Muhich and Boothroyd, 1988) and the accumulation of oligocistronic RNAs on heat shock thus probably represents an increase in a pre-existing pool rather than the appearance of novel RNAs. The incompletely processed tubulin mRNAs induced by heat shock are not processed into mature mRNAs during recovery from heat shock (Muhich and Boothroyd, 1988), but little is known of their fate and whether they are retained in the nucleus or exported to the cytoplasm.
After export to the cytoplasm, there is spatial regulation of mRNA metabolism with aggregation of mRNA and associated proteins into granules. Cytoplasmic mRNP granules are thought to regulate mRNA turn-over and translation by regulating access to translation, storage and decay pathways, including nonsense-mediated decay and microRNA-induced gene silencing (Anderson and Kedersha, 2009). P-bodies are a subset of these mRNP granules and contain enzymes involved in 5′ to 3′ decay: the decapping complex Dcp1–Dcp2 and the 5′ to 3′ exoribonuclease Xrn1, as well as proteins with a function in translational repression, such as Scd6 and the RNA helicase Dhh1. The behaviour of P-body proteins is consistent with an equilibrium existing between P-bodies and polysomes; cycloheximide prevents polysome disassembly and diminishes P-bodies, whereas puromycin causes polysome disassembly and increases P-bodies (Sheth and Parker, 2003; Kedersha et al., 2005; Anderson and Kedersha, 2006). mRNAs also move between P-bodies and polysomes (Brengues et al., 2005; Bhattacharyya et al., 2006). Although the components and dynamic behaviour of P-bodies have been characterised, the exact functions remain unclear.
Other mRNP granules are restricted to particular developmental stages or appear in response to stress. Germ granules, such as P-granules in Caenorhabditis elegans and polar granules in Drosophila melanogaster, store a subset of maternal mRNAs in developing embryos. In C. elegans, germ granules are cytoplasmic in the early embryo, but become perinuclear in germ line cells of later stages (Eddy and Ito, 1971; Mahowald, 1971; Mahowald and Hennen, 1971; Strome and Wood, 1982; Strome and Wood, 1983; Hay et al., 1988b). Such perinuclear germ granules are associated with nuclear pores, and this location suggests a possible function in the regulation of export and/or quality control and/or determination of fate (Pitt et al., 2000; Sheth et al., 2010). Germ granules interact with P-bodies and share some components, but are a distinct type of mRNP granule (Gallo et al., 2008); prominent germ granule proteins are Pgl-1 in C. elegans and the RNA helicase Vasa in Drosophila (Glh-1 in C. elegans) (Strome, 2005).
mRNP granules have been described in trypanosomes, and include starvation stress granules (Cassola et al., 2007), heat shock stress granules (Kramer et al., 2008) and P-bodies (Cassola et al., 2007; Holetz et al., 2007; Kramer et al., 2008). P-bodies are constitutively present and contain DHH1, SCD6 and XRNA, the Xrn1 homologue (Cassola et al., 2007; Holetz et al., 2007; Kramer et al., 2008), but the decapping reaction probably differs from yeast and mammals because the enzyme, Dcp1–Dcp2 and the regulators of decapping, Lsm1 and Pat1, are not apparent in the genome, although decapping activity has been identified (Milone et al., 2002).
Here, it is shown that the inhibition of trans-splicing and consequent accumulation of incompletely processed mRNA causes the redistribution of a set of proteins involved in mRNA metabolism in the cytoplasm to granules close to the periphery of the nucleus. The proteins included all P-body components characterised to date in trypanosomes and additionally poly(A) binding protein 2 and the homologue of Vasa. Proteins involved in mRNA maturation and export from the nucleus did not localise to the granules. The nuclear periphery granules differ from P-bodies because their presence is dependent on active transcription, and they are unaffected by cycloheximide. The inclusion of Vasa in particular suggests a similarity to perinuclear germ granules, and it is possible that the trypanosome nuclear periphery granules fulfil a similar function in sorting and/or quality control of newly transcribed mRNAs.
Results
Inhibition of trans-splicing causes P-body proteins to relocalise to granules at the nuclear periphery
Trans-splicing can be inhibited by a range of methods: degradation of U2, U4 or U6 snRNAs by the addition of complementary oligonucleotides and RNAse H to permeabilised cells (Tschudi and Ullu, 1990), inhibition of methylation of the SL RNA using sinefungin, an S-adenosylmethionine analogue (McNally and Agabian, 1992), and by RNAi depletion of the spliceosome components SmE and SmD (Mandelboim et al., 2003). Sinefungin was chosen for the initial experiments because it does not require permeabilised cells, and accumulation of partially processed mRNAs occurs within a few minutes. By contrast, the onset of phenotype after RNAi depletion of spliceosome components takes more than 24 hours. The addition of sinefungin has immediate and downstream effects. For mRNA, the immediate effect is to inhibit methylation of SL RNA, the consequence is an inhibition of trans-splicing, which, in turn, has the effect of decreasing the cytoplasmic mRNA pool through turn-over because the mean half-life, measured after inhibition of transcription and trans-splicing, is less than 30 minutes (Manful et al., 2011).
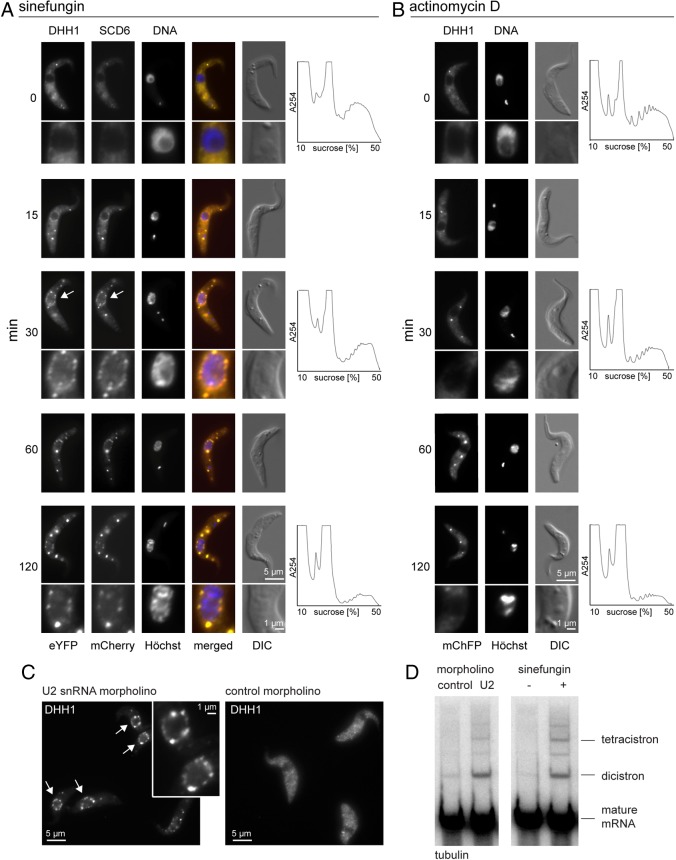
The starting procyclic trypanosome cell line expressed eYFP–DHH1 and SCD6–mChFP from modified endogenous loci and has been described previously (Kelly et al., 2007; Kramer et al., 2008). In untreated cells, both proteins are cytoplasmic, with an average of around three microscopically visible P-bodies (Kramer et al., 2008) (Fig. 1A). Upon addition of sinefungin, a fraction of each protein relocalised to >10 granules distributed around the nucleus within 15 minutes: these were named nuclear periphery granules (NPGs). The remainder of each protein continued to be localised in the cytoplasm and was concentrated in P-bodies distant from the nucleus. This distribution altered with time: 60 minutes after sinefungin addition, P-bodies had increased in size and were more prominent than the NPGs (Fig. 1A). The increase in number and size of P-bodies coincided with a decrease in polysomes (Fig. 1A).
Fig. 1.
Nuclear periphery granules. (A,B) Time course of the effect of (A) sinefungin or (B) actinomycin D on cells expressing eYFP–DHH1 and SCD6–mChFP. Images of typical cells and polysome profiles are shown, arrows indicate NPGs. (C,D) Inhibition of splicing using a morpholino oligo antisense to part of U2 snRNA in a cell line expressing eYFP–DHH1. (C) Cells were imaged 2 hours after electroporation, arrows indicate NPGs. (D) Northern blot analysis of incompletely processed tubulin mRNAs in untreated cells, cells treated with sinefungin for 30 minutes and cells harvested 2 hours after electroporation of the morpholino.
The event(s) necessary for relocation of eYFP–DHH1 and SCD6–mChFP to NPGs were investigated. Transcription was inhibited by actinomycin D; there was no formation of NPGs, whereas P-bodies increased in size, and polysomes decreased, indicating that NPGs do not form as a response to the absence of newly synthesised mRNAs or to the decrease in polysomes (Fig. 1B). To determine whether the formation of NPGs was caused by the inhibition of trans-splicing or some other effect of sinefungin, a morpholino oligo designed to base pair with bases 27–51 of the U2 snRNA was introduced into the cells by electroporation (Tschudi et al., 1990). The same approach has been used to inhibit cis-splicing in metazoa (Matter and König, 2005). Electroporation of the U2 morpholino, but not a control oligo, caused eYFP–DHH1 to localise to NPGs (Fig. 1C). To confirm that the morpholino inhibited splicing, RNA from electroporated cells was analysed for the presence of partially processed tubulin mRNAs. Cells electroporated with the U2 snRNA antisense morpholino contained tubulin dicistrons at approximately the same level as cells treated with sinefungin for 30 minutes (Fig. 1D). These data indicate that NPGs form as a consequence of the inhibition of trans-splicing and not from the absence of another methylation reaction inhibited by sinefungin.
A titration of sinefungin was performed to determine the minimum concentration required for nuclear periphery granule formation and the accumulation of incompletely processed tubulin mRNAs. Sinefungin was titrated from 5.2 µM (2 µg/ml) used in the experiments above, to 5.2 nM. Both NPG formation and the appearance of incompletely processed tubulin mRNAs had a similar threshold concentration of sinefungin, between 5.2 and 52 nM (supplementary material Fig. S1A,B). With increasing sinefungin concentration, the relative amounts of dicistronic tubulin mRNAs decreased and tetracistronic and polycistronic RNAs increased, reflecting a greater inhibition of trans-splicing (supplementary material Fig. S1B). The effect of sinefungin on nuclear periphery granule formation could be blocked by co-incubation with excess S-adenosyl methionine, as expected (supplementary material Fig. S1C).
A third method to inhibit trans-splicing is RNAi depletion of the Sm core protein SmE (Mandelboim et al., 2003). When this RNAi experiment was repeated in a cell line carrying an eYFP–DHH1 transgene, growth arrest occurred after roughly two population doubling times, but there was a reduced accumulation of incompletely processed tubulin mRNA compared with results obtained in the presence of sinefungin, possibly because of incomplete inhibition of trans-splicing (supplementary material Fig. S2). However, eYFP–DHH1 did not localise to NPGs, suggesting that almost complete inhibition of trans-splicing is necessary for NPG formation, a model reinforced by the observation that NPGs can form in the majority of SmE-depleted cells upon addition of sinefungin (supplementary material Fig. S2C). Taken together, the data provide strong evidence that NPGs form as a consequence of an accumulation of incompletely processed mRNAs above a threshold concentration.
NPGs are located outside the nuclear membrane and do not form if nuclear integrity is compromised by knockdown of NUP158
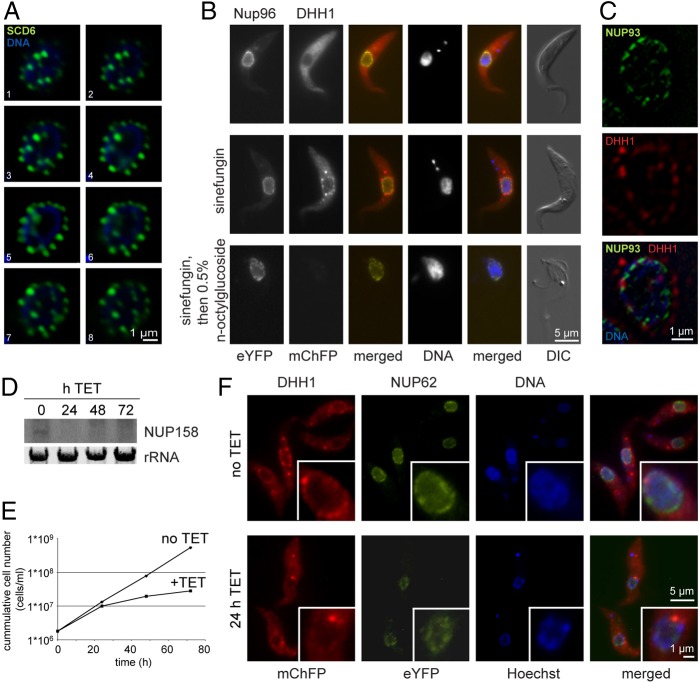
The subcellular localisation of the NPGs was characterised in more detail. A cell line expressing three times wild-type levels of SCD6–eYFP was treated with sinefungin and individual nuclei were imaged at high resolution. In serial optical sections, the granules are relatively uniform in size and are evenly distributed around the nucleus (Fig. 2A). To determine whether the NPGs were localised inside or outside the nuclear membrane, the nuclear periphery was visualised by expressing NUP96–eYFP in a cell line expressing mChFP–DHH1. Trypanosoma brucei NUP96 (Tb927.10.7060) was identified in the proteome of T. brucei nuclear pores and localises to a series of discrete spots at the nuclear periphery that probably coincide with nuclear pores (DeGrasse et al., 2009). NUP96–eYFP localised to small spots at the rim of the nucleus, and its localisation was unaffected by sinefungin (Fig. 2B). The NPGs containing mChFP–DHH1 were localised outside the NUP96–eYFP spots (Fig. 2B,C), suggesting that the granules formed in the cytoplasm and not in the nucleus. NPG stability after addition of 0.5% n-octylglucoside was determined, this concentration dissolves the plasma membrane but not the nuclear membrane. Upon addition of n-octylglucoside, the localisation of NUP96–eYFP was unaffected, whereas NPGs disappeared, providing further evidence that the NPGs are cytoplasmic (Fig. 2B).
Fig. 2.
NPGs are localised outside the nucleus and require nuclear integrity. (A) Deconvolved optical sections (0.24 µm intervals) of a typical cell expressing SCD6–eYFP after incubation for 120 minutes with sinefungin. (B) Stability of NPGs after detergent lysis of the plasma membrane of cells expressing NUP96–eYFP and mChFP–DHH1. Cells were not lysed (top), treated with sinefungin (middle) or treated with sinefungin followed by detergent lysis (bottom). (C) Localisation of NUP96–eYFP and mChFP–DHH1 after incubation with sinefungin for 60 minutes. A single optical section from a deconvolved Z-stack is shown. (D–F) Tetracycline-inducible RNAi depletion of NUP158 in cells expressing mChFP–DHH1 and NUP62–eYFP. (D) Northern blot probed for NUP158. (E) Growth after induction of NUP158 RNAi. (F) Images of cells after incubation with sinefungin for 60 minutes.
To test whether the formation of the NPGs was dependent on nuclear integrity, the nucleoporin NUP158 (Tb11.03.0140) (DeGrasse et al., 2009) was knocked down using tetracycline inducible RNAi in a cell line expressing both mChFP–DHH1 and NUP62–eYFP (Tb927.4.5200) as a marker of nuclear pores (DeGrasse et al., 2009). Both transgenes were expressed from modified endogenous loci. RNAi induction resulted in a decrease of the mRNA coding for NUP158 within 24 hours (Fig. 2D), a reduction in growth rate between 24 and 48 hours (Fig. 2E) and a change in localisation of NUP62–eYFP from predominantly at the edge of the nucleus to a more diffuse nucleoplasmic localisation within 24 hours (Fig. 2F). The redistribution of NUP62–eYFP indicated that nuclear structure was disrupted after induction of NUP158 knockdown. Addition of sinefungin 24 hours after RNAi induction resulted in a reduced formation of NPGs (Fig. 2F). This observation indicates that NPG formation requires nuclear integrity.
Proteins present in NPGs
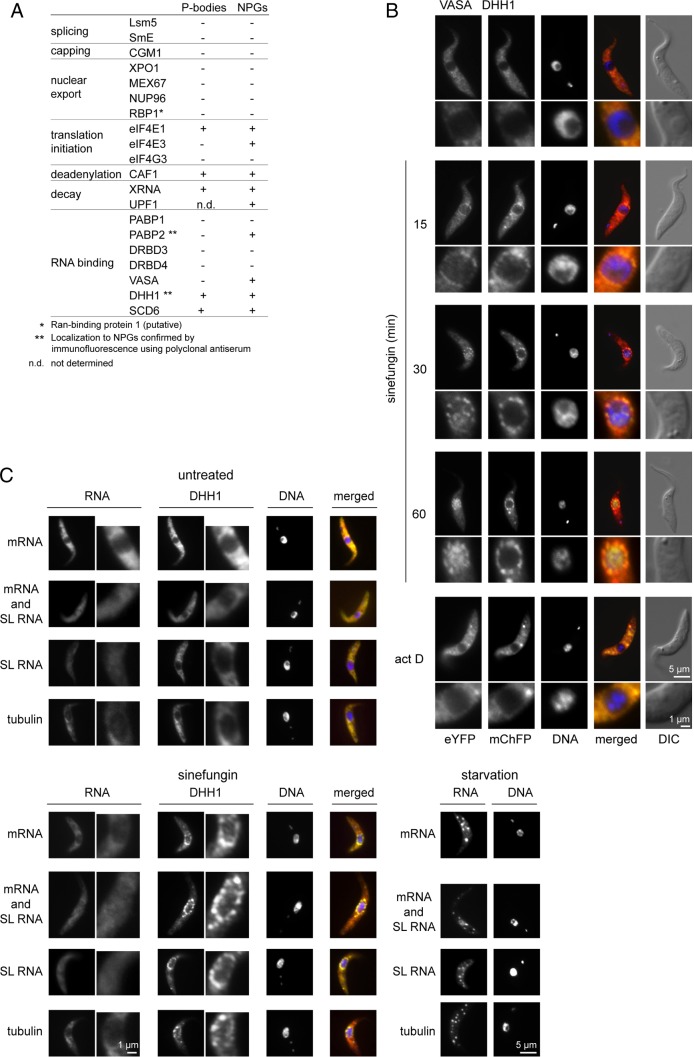
The re-localisation of proteins involved in various mRNA metabolism pathways to NPGs upon inhibition of trans-splicing was further investigated. Targets were expressed as eYFP fusion proteins in cell lines containing transgenes encoding mChFP–DHH1 or SCD6–mChFP or, in one case, PABP2–mChFP. All were expressed after modification of endogenous loci with the exception of LSM5–eYFP which was expressed from a derivative of pDEX377 (Kelly et al., 2007). The expression of transgenes was verified by western blotting with anti-GFP (supplementary material Fig. S3). Any localisation of the proteins to P-bodies in untreated cells or to NPGs after sinefungin addition was determined (Fig. 3A; supplementary material Fig. S4). The localisation of DHH1 and PABP2 to NPGs was confirmed by immunofluorescence (Fig. 3A,C). For other proteins, it is possible that the eYFP tag has altered the normal localisation and has prevented detection in P-bodies and/or NPGs. The proteins split into three groups: (1) proteins present in both P-bodies and NPGs; (2) proteins present in NPGs but not in P-bodies; and (3) proteins that localised to neither P-bodies nor NPGs.
Fig. 3.
Proteins and RNAs present in NPGs. (A) Summary of localisation of eYFP fusion proteins to P-bodies or NPGs. Designation as present in P-bodies indicates that the protein was readily visualised in P-bodies in untreated cells. However, all the proteins localised to NPGs also localised to P-body-like structures under a range of conditions that increased P-body size, including sinefungin treatment. (B) Subcellular localisation of VASA–eYFP and mChFP–DHH1 over a time course of incubation with sinefungin or actinomycin D. (C) Fluorescence in situ hybridisation of untreated cells, cells treated with sinefungin for 60 minutes or cells incubated in PBS for 3 hours (starvation). The RNAs detected are shown on the left: mRNA (dT44 probe); mRNA and SL RNA (antisense SL RNA exon probe); SL RNA (antisense SL RNA intron probe); and β-tubulin mRNA (antisense β-tubulin mRNA probe).
Proteins present in both P-bodies and NPGs included SCD6 and DHH1, the translation initiation factor eIF4E1 (Dhalia et al., 2005), as well as three components of the mRNA degradation pathway: the deadenylase CAF1 (Schwede et al., 2008), the 5′ to 3′ exonuclease XRNA (Li et al., 2006). The T. brucei homologue to the NMD factor UPF1 (Delhi et al., 2011) localised to NPGs, but the expression level of UPF1–eYFP was too low to come to a firm conclusion about whether it was present in P-bodies.
Proteins present in NPGs but not in P-bodies were poly(A) binding protein 2 (Pitula et al., 1998; Hotchkiss et al., 1999; da Costa Lima et al., 2010), an RNA helicase (Tb10.61.2130) that is the closest homologue to VASA in D. melanogaster and to GLH1-4 in C. elegans (supplementary material Fig. S5), and eIF4E3 (Dhalia et al., 2005); the latter was only weakly concentrated in NPGs.
Proteins localising to neither P-bodies nor NPGs included all tested proteins with a role in mRNA maturation and export: the splicing factors LSM5 and SmE (Mandelboim et al., 2003; Liu et al., 2004), the 5′ cap guanylyltransferase-methyltransferase CGM1 (Ruan et al., 2007; Takagi et al., 2007), the mRNA export factors XPO1/CRM1 (Zeiner et al., 2003; Cuevas et al., 2005; Biton et al., 2006), MEX67 (Schwede et al., 2009; Kramer et al., 2010a), the putative Ran-binding protein 1 (DeGrasse et al., 2009), as well as DRBD3 and DRBD4, RNA-binding proteins related to the polypyrimidine tract binding protein (De Gaudenzi et al., 2005; Estévez, 2008; Stern et al., 2009). Proteins that either bind the 5′ cap directly or are part of complexes that bind the cap were tested, but none of the following were present in either P-bodies or NPGs: the nuclear mRNA cap-binding protein CBP20 (Li and Tschudi, 2005), the translation initiation factors eIF4E2 (Dhalia et al., 2005) and eIF4G3 (Dhalia et al., 2005), and PABP1, the other poly(A)-binding protein (Bates et al., 2000; Milone et al., 2004; da Costa Lima et al., 2010). Three of these, CBP20 (Li and Tschudi, 2005), eIF4E2 and eIF4E4 (Dhalia et al., 2005), were expressed at low levels that might have prevented the detection of a small fraction in any granule (supplementary material Fig. S3). These proteins are not listed in Fig. 3A, but are shown in supplementary material Fig. S4B.
The data suggest that the NPGs are related to P-bodies in their composition. All proteins identified to be normally present in P-bodies also localise to NPGs. Proteins present in NPGs but not normally in P-bodies, PABP2, eIF4E3 and VASA, become visible in P-body derivatives under conditions that result in a dramatic reduction in protein synthesis, such as sinefungin, actinomycin D or carbon source starvation, indicating that they might be minor P-body components under reduced growth conditions. Further evidence for a similarity between P-bodies and NPGs is provided by DHH1 mutant proteins: DHH1 mutants impaired in either RNA-binding (R74A and K76A) (Cheng et al., 2005) or catalytic activity (E182Q) show impaired localisation to P-bodies (Kramer et al., 2010b) and also a reduced localisation to NPGs (supplementary material Fig. S6). Together, these results suggest a similarity between NPGs and P-bodies.
The addition of sinefungin had a second effect on the subcellular distribution of VASA (Fig. 3B) and DRBD3 (supplementary material Fig. S4B): both proteins concentrated in the nucleus. This behaviour is consistent with proteins that normally shuttle between the cytoplasm and the nucleus. The relocalisation to the nucleus upon addition of sinefungin could have been caused by a loss of co-export with a mature mRNA or sequestration by binding to the increased pool of incompletely processed mRNAs in the nucleus. To distinguish between these two possibilities, transcription was inhibited using actinomycin D, which prevents both export of newly synthesised mRNA and nuclear accumulation of incompletely processed mRNAs. After addition of actinomycin D, there was no readily observable change in the localisation of VASA or DRBD3 (Fig. 3; supplementary material Fig. S3). Furthermore, addition of actinomycin D 30 minutes before sinefungin prevented nuclear accumulation of VASA. These data suggest that the nuclear accumulation is caused by the presence of incompletely processed mRNAs and not by the absence of fully processed mRNAs.
RNA content of the NPGs
To investigate any RNA content of the NPGs, in-situ hybridisation experiments were performed using the following oligonucleotides: (1) dT44 to detect all polyadenylated mRNAs; (2) antisense SL RNA exon to detect mRNAs and SL RNA; (3) antisense SL RNA intron to detect only the SL RNA; and (4) antisense β-tubulin mRNA (Fig. 3C). Sense oligonucleotides were used as controls; all gave significantly weaker signals than the antisense probes (not shown). In untreated cells, mRNA detected by dT44 and antisense tubulin probes was distributed throughout the cytoplasm where it was more concentrated than in the nucleus. SL RNA was present at similar concentrations in the cytoplasm and nucleus. On inhibition of trans-splicing, no changes in the localisation of mRNA or SL RNA were observed in the majority of cells. Polyadenylated RNA was detected in the NPGs in the occasional cell but too few (<1%) to attach significance. DHH1 protein was readily detectable in NPGs by immunofluorescence indicating that the granules survived the in situ hybridisation procedure. The accumulation of mRNAs in starvation stress granules (Cassola et al., 2007) was used to test the efficacy of the in situ hybridisation procedure: all antisense probes detected the accumulation of mRNAs in starvation stress granules. These experiments suggested that neither mRNAs nor the SL RNA are enriched in NPGs, but it remains possible that any RNA in the granules was not accessible to the probes under the conditions used. The sensitivity of the ISH did not allow us to probe specifically for incompletely spliced mRNAs.
Dependency on translation and transcription
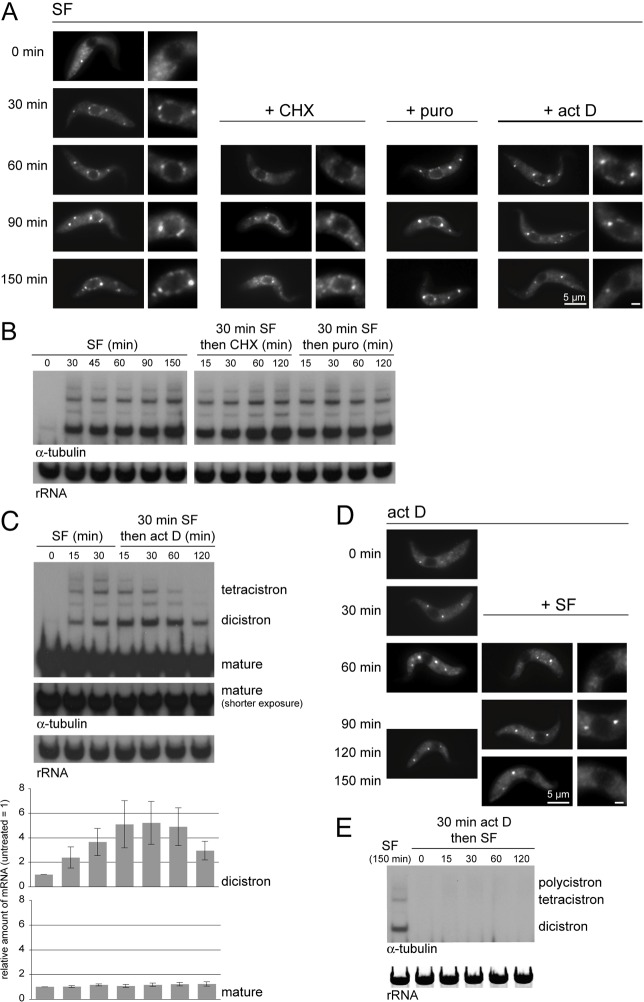
The number and size of P-bodies is often the inverse of the number of polysomes; puromycin releases ribosomes from polysomes and causes an increase in P-bodies, whereas cycloheximide traps ribosomes in polysomes and causes a decrease in P-bodies (Sheth and Parker, 2003; Anderson and Kedersha, 2006). This relationship is often interpreted as the physical movement of proteins and mRNAs between polysomes and P-bodies. By contrast, germ granules are not affected by translational inhibitors but, unlike P-bodies, are dependent on active transcription (Sheth et al., 2010). Cells expressing an eYFP–DHH1 transgene were used to examine any relationship between NPGs, translation and transcription. NPGs were induced by the addition of sinefungin for 30 minutes and the effect of cycloheximide, puromycin or actinomycin D was determined (Fig. 4A–C; supplementary material Fig. S7A,B). Neither cycloheximide nor puromycin dissolved NPGs, whereas both affected P-bodies within 30 minutes: cycloheximide caused visible P-bodies to disappear within 30 minutes and puromycin caused the expected P-body increase (Kramer et al., 2008). These observations suggest that the protein components localised to NPGs do not exchange with polysomes, or do so at a much slower rate than the exchange between P-bodies and polysomes. Neither cycloheximide nor puromycin had any effect on the oligocistronic tubulin RNAs (Fig. 4B).
Fig. 4.
Actinomycin D, but not cycloheximide, dissolves NPGs and prevents NPG formation. (A) Cells expressing eYFP–DHH1 were incubated with sinefungin for 30 minutes to induce NPG formation and then with other inhibitors as indicated. Images of representative cells are shown, in some cases with an enlargement of the nucleus (Scale bar: 1 µm). (B,C) Northern blot analysis of incompletely processed tubulin mRNAs from the cells in A. For actinomycin-treated cells, the amount of tubulin dicistrons and β-tubulin mature mRNAs was quantified. Error bars represent s.d. from three independent experiments. (D) Cells expressing eYFP–DHH1 were incubated with actinomycin D for 30 minutes and then with sinefungin as indicated. Images of representative cells are shown, in some cases with an enlargement of the nucleus (Scale bar: 1 µm). (E) Northern blot analysis of incompletely processed β-tubulin mRNAs from the cells in D. Representative results from three separate experiments are shown.
By contrast, actinomycin D caused the disappearance of NPGs and a decrease in tubulin oligocistrons within 120 minutes, providing evidence that NPGs are dependent on newly synthesised oligocistronic mRNAs (Fig. 4A,C). This model was reinforced when transcription was inhibited with actinomycin D for 30 minutes before sinefungin, when neither NPGs nor tubulin oligocistrons formed (Fig. 4D,E). Actinomycin D also caused P-bodies to increase in size, as previously reported (Marnef et al., 2012). The cause of this increase is not understood but might result from a feedback mechanism in which the absence of newly synthesised mRNAs causes a reduction in translation and thus an increase in P-bodies. The experiment also showed that the incompletely processed tubulin mRNAs have a shorter half-life than the mature mRNAs (Fig. 4C), which provides evidence for a selective turnover of aberrant mRNAs.
Depletion of SCD6 causes a reduction in P-bodies but not in NPGs
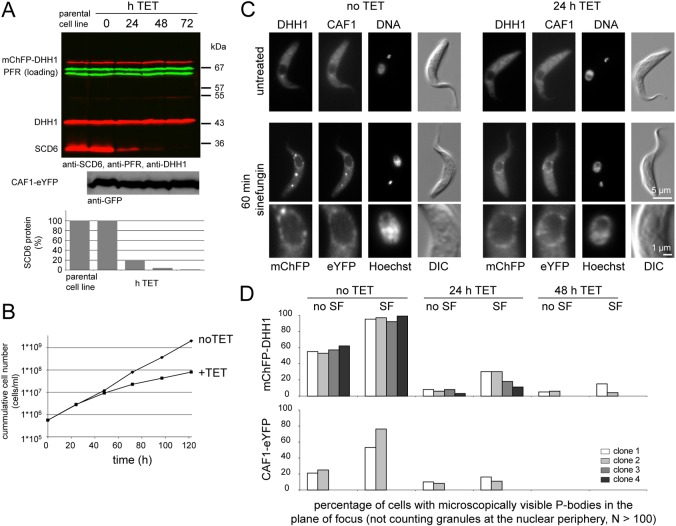
SCD6 is an RNA-binding protein necessary for the formation of membrane-stress induced P-bodies in S. cerevisiae (Kilchert et al., 2010) and its orthologue RAP55 is required for P-body formation in HeLa cells (Tanaka et al., 2006; Yang et al., 2006). However, SCD6 is not required for starvation-stress-induced P-body formation in S. cerevisiae (Teixeira and Parker, 2007). RNAi depletion of SCD6 was used to determine whether SCD6 is a core component of trypanosome P-bodies and whether it is required for NPG formation.
The experiments were performed in cell lines expressing mChFP–DHH1 and CAF1–eYFP through modification of endogenous loci (clones 1 and 2) or mChFP–DHH1 alone (clones 3 and 4). Induction of RNAi caused a reduction in SCD6 protein to <2% after 72 hours (Fig. 5A) and a decrease in the rate of growth after 48 hours (Fig. 5B) but had no effect on the amount of either mChFP–DHH1 or CAF1–eYFP protein over the time course (Fig. 5A). The localisation of both mChFP–DHH1 and CAF1–eYFP to P-bodies was greatly reduced after induction of SCD6 RNAi (Fig. 5C,D; supplementary material Fig. S8). The effect was quantified by determining the percentage of cells with mChFP–DHH1 localising to microscopically visible P-bodies in one plane of focus. This percentage was reduced from >50% to <10% within 24 hours of RNAi induction in all four clonal cell lines (Fig. 5D). In addition, the P-bodies still present after RNAi induction appeared smaller. Similarly, the localisation of CAF1–eYFP to P-bodies was reduced from 20% of CAF1–eYFP localising to visible P-bodies in one plane of focus, to 10% after 24 hours of SCD6 RNAi induction (Fig. 5D). Note that CAF1–eYFP was concentrated in P-bodies to a lesser extent than DHH1. The addition of sinefungin resulted in an increase in P-bodies (Fig. 1A); however, after depletion of SCD6 this effect was greatly reduced. Before the induction of SCD6 RNAi, 95% (mChFP–DHH1) or 50% (CAF1–eYFP) of cells contained visible P-bodies in one plane of focus (Fig. 5D). After 24 or 48 hours of SCD6 RNAi, these were reduced to <30% (DHH1) and <20% (CAF1). The reduction of localisation of the two proteins to P-bodies is probably a direct consequence of SCD6 knockdown, the effect is present at 24 hours when the growth rate is normal. As a control, growth arrest was induced by RNAi knockdown of either FLA1 (LaCount et al., 2002) or Polo-like kinase (Hammarton et al., 2007); in both cases, there was no decrease in P-bodies (supplementary material Fig. S9), indicating that the reduction in P-bodies following SCD6 RNAi is not caused by the reduction in growth or by the RNAi procedure. Thus, T. brucei SCD6 is required for correct P-body localisation of at least two P-body markers, consistent with a function as a core-protein of P-bodies.
Fig. 5.
The effect of SCD6 depletion on NPG formation. SCD6 was depleted in cell lines constitutively expressing either both mCHFP–DHH1 and CAF1–eYFP (clones 1 and 2) or just mChFP–DHH1 (clones 3 and 4). (A) Western blot analysis of the parental cell line (Lister 427 pSPR2.1) and a time course of SCD6 RNAi. The blots were probed for DHH1, SCD6 and PFR (loading control). The reduction in SCD6 protein was quantified. One representative blot from four experiments is shown. A non-quantitative western blot was probed for CAF1–eYFP using anti-GFP antibody. (B) Growth after induction of SCD6 RNAi. One representative growth analysis from four experiments is shown. (C) The effect of incubation with sinefungin on cells before and after induction of SCD6 RNAi. Images of typical cells before (no TET) and 24 hours after RNAi induction SCD6 RNAi (24 h TET) are shown. (D) Quantification of the sinefungin-induced relocalisation of mChFP–DHH1 and CAF1–eYFP before and after induction of SCD6 RNAi. The percentage of cells with mChFP–DHH1 or CAF1-eYFP localised to cytoplasmic granules other than NPGs in one plane of focus was quantified from >100 microscopy images.
By contrast, the localisation of mChFP–DHH1 and CAF1–eYFP to NPGs after addition of sinefungin was unaffected by SCD6 knockdown (Fig. 5C; supplementary material Fig. S8). A minor change in the abundance of the NPGs cannot be excluded, because the dissociation of the cytoplasmic P-bodies slightly increases the cytoplasmic fluorescence, making a direct comparison of microscopic images difficult. However, addition of sinefungin caused the appearance of clearly visible NPGs in every cell at 24, 48 and 72 hours after induction of SCD6 RNAi. These data indicate that SCD6 is required for P-body formation, but not for the formation of the NPGs. The independence of NPG formation from SCD6 prevented a readily accessible investigation of the effect of perturbing NPG formation on mRNAs.
Incompletely processed mRNAs are present in the cytoplasm
Inhibition of trans-splicing caused the formation of NPGs in the cytoplasm and accumulation of partially spliced mRNAs. To determine whether the incompletely processed mRNAs were confined to the nucleus, both control and sinefungin-treated cells were fractionated and protein and RNA prepared. Northern blots were probed for expression of tubulin and HSP83, which is also transcribed from tandemly arranged genes and therefore suitable for the detection of incompletely spliced mRNAs (Mottram et al., 1989), as well as for control RNAs: tRNAleu as a cytoplasmic marker, U3 snRNA as a nuclear marker. Western blots were probed for histone H3 as a nuclear marker, PFR (a marker of the cytoskeleton which co-fractionates with nuclei in the procedure used) and for BiP (an ER protein that was used as a cytoplasmic marker after detergent lysis) (Fig. 6). The majority of the U3 snRNA was in the nuclear fraction, suggesting little contamination of the cytoplasmic fraction with nuclear RNAs. All the tRNAleu was present in the cytoplasmic fraction indicating no contamination of the nuclear fraction with cytoplasm. More than half of dicistronic mRNA encoding β-tubulin and HSP83 were found in the cytoplasmic fraction. The β-tubulin dicistrons were quantified using phosphorimager analysis and the cytoplasmic fraction/nuclear fraction (C/N) ratio was 1.33, the C/N ratio for U3 snRNA was 0.30. These measurements indicate that the dicistrons present in the cytoplasmic fraction do not arise from nuclear contamination. Very low levels of tubulin dicistrons were detectable in the cytoplasmic fraction even in the absence of sinefungin (asterisk in Fig. 6), suggesting that low levels of incompletely spliced mRNAs in the cytoplasm occur normally. These data provide strong evidence for the presence of incompletely processed mRNAs in the cytoplasm. The larger incompletely processed mRNAs, equivalent to tetracistronic tubulin and larger, showed a greater retention in the nucleus. It is possible that a nuclear retention factor that binds the intron sequences had become limiting and the larger incompletely processed mRNAs with more introns are more likely to be retained. Another possible explanation is that the larger RNAs are retained in the nucleus simply because of their larger size.
Fig. 6.
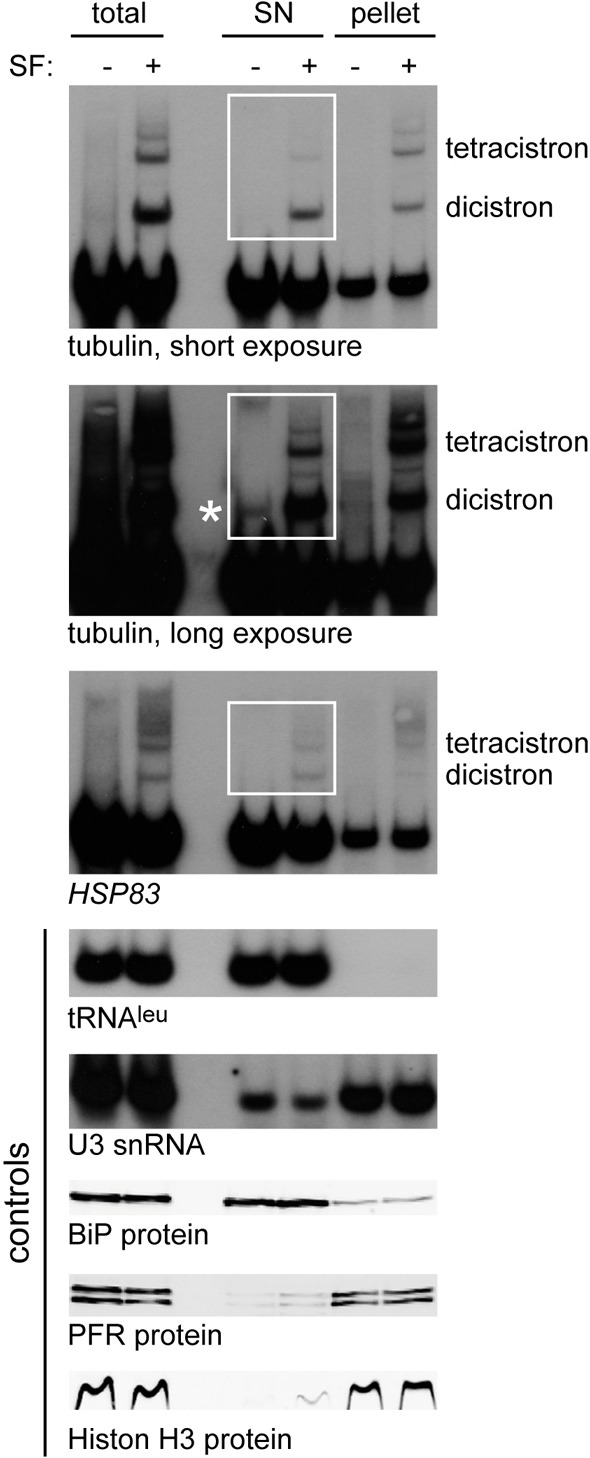
Partially processed mRNAs are present in the cytoplasm. RNA and protein from total, cytoplasmic (SN) and nuclear (pellet) fractions of a subcellular fractionation were analysed by Northern blotting for tubulin and HSP83 to detect partially processed mRNAs and for tRNAleu, U3 snRNA to control for fractionation. The asterisk indicates the dicistronic tubulin mRNA in the cytoplasmic fraction, visible in the absence of sinefungin. In parallel, protein samples were analysed by western blotting for histone H3, BiP and PFR. The same cell equivalents were loaded in each track.
Inhibition of splicing in HeLa cells does not cause the formation of NPGs
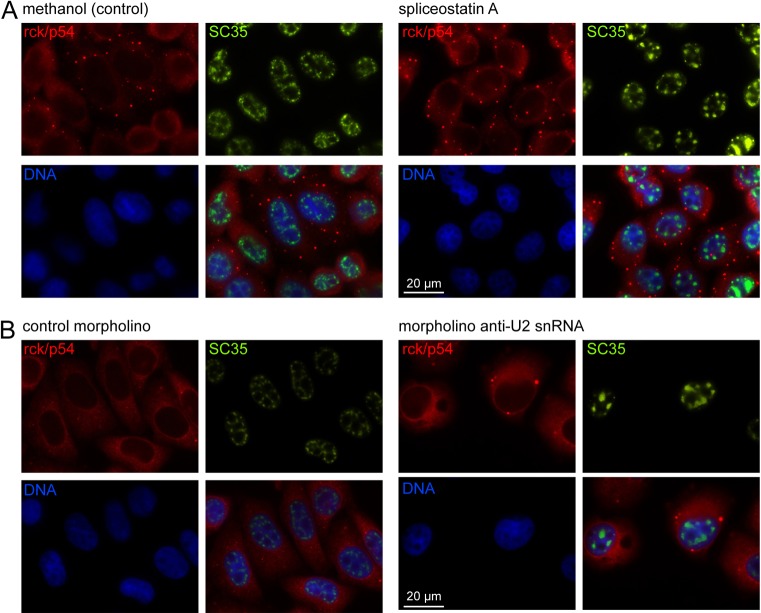
Trypanosomes represent a branch of eukaryotes that are as divergent from metazoa as any eukaryote (Walker et al., 2011). To investigate whether NPGs are a general phenomenon, the effect of inhibition of splicing in HeLa cells was investigated. Splicing was inhibited by the addition of spliceostatin A, previously shown to inhibit splicing in HeLa cells by binding to the splicing factor SF3B (Kaida et al., 2007; Lo et al., 2007). The inhibition of splicing was monitored by the reorganisation of splicing factors within the nucleus from 20–50 irregularly shaped foci into a smaller number of larger foci (O'Keefe et al., 1994; Tanackovic and Krämer, 2005; Kaida et al., 2007). Addition of spliceostatin A to HeLa cells caused re-localisation of SC35 into larger foci in the majority of the cells within 2 hours (Fig. 7A), as previously described (Kaida et al., 2007). However, rck (p54), the orthologue of DHH1, did not localise to granules at the nuclear periphery up to the endpoint of 5 hours of incubation with spliceostatin A (Fig. 7A). A second method to inhibit splicing was used: a morpholino oligonucleotide designed to pair with U2 snRNA, equivalent in sequence to the one used above with trypanosomes, was introduced into HeLa cells by electroporation (Matter and König, 2005). Again, SC35 relocalised to a smaller number of enlarged foci within the nucleus indicating successful inhibition of splicing, but no rck (p54) granules around the nuclear periphery were observed 24 hours after transfection (Fig. 7B). The results of these experiments indicate that it is unlikely that NPGs form in HeLa cells in response to inhibition of splicing, but it remains possible that inhibition was incomplete. The induction of NPGs in trypanosomes but not HeLa cells could result from a trivial difference in the effectiveness of inhibition: spliceostatin A only partially inhibits splicing of a reporter in HeLa cells, even at very high concentrations (Kaida et al., 2007). However, the U2 morpholinos target the same reaction and might be expected to have a similar effect. There could also be a mechanistic difference. It is worth noting that all trypanosome mRNAs are trans-spliced but ∼6% of human genes have a single exon (Wang et al., 2008) and the export of these would be expected to continue when splicing was inhibited, perhaps this is enough to stop visible NPGs forming.
Fig. 7.
Inhibition of splicing in HeLa cells does not result in NPG formation. Inhibition of splicing in HeLa cells by Spliceostatin A (100 ng/ml for 5 hours using a 100 µg/ml stock in methanol) (A) or a morpholino antisense to the U2 snRNA (B). Methanol (0.1%) or a morpholino sense to T. brucei β-tubulin served as controls. Rck (p54) and SC35 proteins were detected by immunofluorescence, using anti-rck/p54 (Bethyl Laboratories) or anti-SC35 (Sigma) antibodies.
Discussion
Trans-splicing is an essential step in mRNA processing in many eukaryotes; in trypanosomes, all mRNAs require trans-splicing, whereas only 2 of the >5000 genes contain a cis-intron. The trans-splicing reaction adds the capped 39 nucleotide exon from the SL RNA to the 5′ end of all nuclear encoded mRNAs (Sather and Agabian, 1985). The trypanosome 5′ cap is particularly ornate, with 5 to 7 methylations on the first 4 nucleotides of the SL-RNA-derived exon (Bangs et al., 1992), and methylation of the SL RNA is required before trans-splicing (Ullu and Tschudi, 1991). The extensive methylation of SL RNA probably causes the extreme sensitivity of trans-splicing to sinefungin, an S-adenosyl methionine analogue. The efficiency of trans-splicing varies with the sequence around the splice site (Siegel et al., 2005), yet only very low levels of incompletely spliced mRNAs are detected in normally growing cells. This infers that there is a quality control mechanism that normally recognises and degrades incompletely spliced mRNAs. Further evidence for such a pathway comes from experiments that investigated the fate of accumulated incompletely processed β-tubulin mRNAs produced during heat shock; on recovery from heat shock, these were degraded rather than processed to mature mRNAs (Muhich and Boothroyd, 1988), and in the experiments presented here, sinefungin-induced tubulin dicistrons decayed more rapidly than mature tubulin mRNA (Fig. 4).
Here, the cellular response to increased partially processed mRNAs has been investigated. The main findings are: (1) inhibition of trans-splicing by either sinefungin or an U2 snRNA antisense morpholino oligonucleotide causes the formation of nuclear periphery granules (NPGs) external to the nucleus. NPG formation is dependent on the integrity of the nucleus. (2) NPGs contain a set of proteins involved in regulating mRNA fate. (3) The presence of NPGs is dependent on newly synthesised but incompletely processed mRNAs, because they do not form when transcription is inhibited and, once formed, dissociate when transcription is inhibited. (4) Part of the incompletely spliced mRNAs are present in cytoplasmic fractions. (5) NPG formation does not require the P-body core protein SCD6.
NPGs have not been reported in response to heat shock, a stress that causes the accumulation of partially processed mRNAs (Kramer et al., 2008) and the question arises as to whether the NPGs represent a true compartment for mRNA metabolism. However, there is an important difference between heat shock and the inhibition of trans-splicing by sinefungin. The action of sinefungin is rapid and completely blocks mRNA maturation; whereas during stress, gene expression continues, although at a reduced rate, and thus inhibition of trans-splicing is not complete (Kramer et al., 2008). The evidence points towards the formation of visible NPGs requiring a fast and (almost) complete inhibition of trans-splicing, currently only achieved with sinefungin or the morpholino antisense to the U2 snRNA. It is likely that NPGs represent structures that are otherwise present in a smaller or very transient form.
Incomplete trans-splicing results in mRNAs that are capped and polyadenylated, contain more than one open reading frame and one or more introns. Export of such mRNAs into the cytoplasm is potentially deleterious because most dicistrons will contain mixed regulatory elements. The observation that partially processed β-tubulin mRNAs are more concentrated in the nucleus [∼50% is present in the cytoplasm but the volume is ∼eightfold larger than the nucleus (Grünfelder et al., 2002)], indicates that some retention system must be present. The finding that incompletely processed mRNA is readily detectable in the cytoplasm, even when trans-splicing is not inhibited, as well as the previously reported cytoplasmic location of a dicistronic mRNA (Jäger et al., 2007) argues that any quality control system may not be perfect. In other organisms, incompletely cis-spliced mRNAs encode aberrant, potentially harmful proteins. In yeast, control systems at the nuclear pore prevent the majority of incompletely processed mRNAs from entering the cytoplasm. Nuclear retention requires the assembly of the spliceosome on the pre-mRNA (Legrain and Rosbash, 1989; Rain and Legrain, 1997; Rutz and Séraphin, 2000), suggesting that the presence of the spliceosome prevents pre-mRNA export. The control system is not perfect, and small amounts of unspliced mRNAs do enter the cytoplasm; this was shown by measuring the activity of gene products arising from the translation of unspliced reporter mRNAs containing in-frame introns (Yost and Lindquist, 1988; Legrain and Rosbash, 1989). Cytoplasmic incompletely cis-spliced mRNAs are targets for nonsense-mediated decay (NMD) (He et al., 1993; Rutz and Séraphin, 2000; Hilleren and Parker, 2003; Jaillon et al., 2008; Sayani et al., 2008; Kawashima et al., 2009), although there might be exceptions (Kaida et al., 2007). Several yeast genes involved in retention of immature mRNAs have been identified including the perinuclear pore protein Mlp1 (Galy et al., 2004) and the nuclear components Pml1 (Dziembowski et al., 2004) and Pml39 (Palancade et al., 2005). Orthologues of Pml1 and Pml39 are not readily identified in the trypanosome genome and a putative orthologue of Mlp1 has been identified based on location and domain architecture rather than sequence (DeGrasse et al., 2009). However, the evolutionary distance between yeast and trypanosomes is such that the failure to identify clear orthologues does not indicate the absence of a conserved pathway.
NPGs have an overlapping protein composition with P-bodies, but have more similarities with germ granules present in metazoan adult gonads; they share: (1) a localisation at the nuclear periphery (Eddy and Ito, 1971; Mahowald, 1971; Mahowald and Hennen, 1971; Strome and Wood, 1982; Strome and Wood, 1983; Hay et al., 1988b), (2) the presence of VASA (Hay et al., 1988a), (3) stability in the presence of cycloheximide (Sheth et al., 2010), (4) a dependence on transcription (Sheth et al., 2010) and (5) a requirement for integrity of the nucleus (Voronina and Seydoux, 2010). At this stage, it has not been possible to determine whether NPGs are associated with clusters of nuclear pore complexes enriched for mRNA export factors, a feature of some germ granules. A further clear difference between NPGs and P-bodies is the independence of NPG formation from SCD6, a core protein in P-bodies. Perinuclear germ granules mark the site of mRNA exit from the nucleus and have been suggested to act as sorting compartments to determine the further fate of the mRNAs (Sheth et al., 2010); they might represent a cytoplasmic extension of nuclear pores (Updike et al., 2011). It is tempting to speculate that trypanosome NPGs might fulfil a similar function.
On addition of sinefungin, the pool of fully methylated SL RNA decreases over a few minutes and at a threshold concentration splicing stops with an accumulation of many times the normal concentration of partially processed mRNAs and the formation of NPGs. Inhibition of transcription does not result in NPG formation and inhibition of transcription once NPGs have formed causes them to disappear. Thus, it is the increase in newly synthesised but incompletely processed mRNAs that results in the formation of NPGs. The function of NPGs remains uncertain, but it can be speculated that NPGs are an enlarged form of a normal regulatory compartment that functions not only in quality control but also to label newly synthesised mRNAs for one of their possible fates elsewhere in the cytoplasm: translation, storage or degradation. Furthermore, the similarities between NPGs and the germ granules present in metazoa suggest that the compartment was present before the divergence of eukaryotes and so represents a basic step in mRNA metabolism.
Materials and Methods
Trypanosomes
Trypanosoma brucei Lister 427 procyclic cells (a kind gift from Keith Gull; Sir William Dunn School of Pathology, University of Oxford, UK) were used for most experiments. RNAi experiments were done in either Lister 427 pSPR2.1 cells (Sunter et al., 2012) (SCD6, SmE, NUP158) or PTT (Philippe Bastin, Institute Pasteur, Paris, France) (FlaI, Polo-like-kinase); over-expression in either Lister 427 SIMP (Bill Wickstead, Sir William Dunn School of Pathology, Oxford, UK) (Lsm5) or PTT (Philippe Bastin, Institute Pasteur, Paris, France) (DHH1 and dhh1 mutants) or Lister 427 pSPR2.1 cells (Sunter et al., 2012) (SCD6). Transgenic trypanosomes were generated using standard procedures (McCulloch et al., 2004). All experiments were performed with logarithmically growing trypanosomes at a cell density of less than 1×107 cells/ml.
Plasmids and cloning
Details of all plasmids are described in supplementary material Table S1. RNAi of Polo-like kinase was carried out with a previously described plasmid (Hammarton et al., 2007).
Inhibitor treatments
Puromycin and cycloheximide were used at 50 µg/ml, sinefungin at 2 µg/ml; all concentrated stocks were dissolved in SDM79 without serum and haemin. Actinomycin D was used at 10 µg/ml from a 100× stock dissolved in ethanol.
Trypanosome morpholino experiments
Morpholino oligos were synthesised by GeneTool: 5′-TGATAAGAACAGTTTAATAACTTGA-3′ (U2 snRNA, Tb927.2.5680); 5′-GAAAATAGTTCAAACGAATTATGCG-3′ (control, β-tubulin sense).
1.5×107 cells were washed once in cytomix (10 mM K2HPO4/KH2PO4, pH 7.6, 2 mM EGTA, 120 mM KCl, 150 µM CaCl2, 25 mM HEPES, 5 mM MgCl2, 0.5% glucose, 1 mM hypoxanthine, 100 µg/ml BSA), resuspended in 100 µl cytomix + 16 µl morpholino (at 500 µM) and electroporated using the Amaxa Nucleofector electroporator (programme: X-001). Cells were analysed 2 hours following electroporation by fluorescence microscopy or Northern blot.
RNA work
Quantitative Northern blots and sucrose density gradients were performed as described (Kramer et al., 2008). For better detection of β-tubulin mRNA precursors, an on-column DNAse treatment was performed during RNA isolation (Qiagen). To control for loading, blots were reprobed for rRNA, in some cases non-radioactively by using an oligo antisense to the 18S rRNA (5′-CCTTCGCTGTAGTTCGTCTTGGTGCGGTCTAAGAATTTC-3′) coupled to IRDye800. Hybridisation of blots probed with oligos was performed at 42°C and the blot was washed at room temperature in 4× SSC, 0.1%SDS and RNA was detected and quantified using the Odyssey system (Licor).
Nucleus cytoplasm separation
Subcellular fractionation of RNA was essentially done as described previously (Jäger et al., 2007). Preparation of RNA from the fractions was carried out using hot phenol extraction, followed by phenol–chloroform extraction, ethanol precipitation and treatment with RNase-free DNase (Qiagen).
Microscopic imaging and detergent lysis
Microscopic imaging of cells was performed as described (Kramer et al., 2008). For deconvolution of a Z-stack image, the theoretical point spread function and the iterative algorithm of the AxioVision software was used (default settings). For detergent lysis, 1×107 cells were resuspended in 285 µl SDM79 without serum and haemin, lysed by the addition of 15 µl 10% n-octylglycoside (final concentration 0.5%), Hoechst 33342 DNA stain was added and images were taken immediately.
Immunofluorescence and fluorescent in situ hybridisation for RNA detection
2×107 cells were washed once in SDM79 without serum and haemin, resuspended in 5 ml SDM79 without serum and haemin and fixed for 15 minutes by the addition of 5 ml 8% paraformaldehyde (dissolved in PBS) whilst rotating. 5 ml fixed cells were washed once in 45 ml PBS, resuspended in 2 ml PBS and allowed to settle on slides. Slides were washed in 25 mM NH4Cl for 10 minutes. Cells were permeabilised and blocked for 1 hour in blocking solution (taken from fluorescent antibody enhancer set for DIG detection, Roche) containing 0.5% saponin. Prehybridisation was done for two hours in hybridisation solution (1× blocking solution, 5× Denhardt, 4× SSC, 35% deionised formamide, 0.5 mg/ml tRNA). Hybridisation was performed overnight in a humid chamber at room temperature in hybridisation solution containing 0.5 ng/µl (poly A, spliced leader) or 5 ng/µl (SL RNA, tubulin) biotinylated oligo. Slides were washed twice in 4× SSC, 35% formamide, once in 4× SSC, once in 2× SSC and once in PBS. Detection was by blocking for 30 minutes in blocking solution, followed by 60 minutes incubation with Alexa-Fluor-568–streptavidin (1∶200) and DHH1 antiserum (1∶5000) in blocking solution, followed by four washing steps in PBS, followed by 60 minutes incubation with Alexa Fluor 488 goat anti-rabbit (1∶200) antibody in blocking solution, followed by four washing steps in PBS. Slides were mounted in FluorSave Reagent (Calbiochem), containing Hoechst 33342 DNA stain at 5 µg/ml. Oligo sequences were: 5′ Biotin (T)44 3′ Biotin (poly A), 5′-Biotin-CAATATAGTACAGAAACTGTTCTAATAATAGCGTT-3′-Biotin (spliced leader); 5′-Biotin-GGTCATCCGACCCCACCTTCCAGATTCCCGCAGTA-3′-Biotin (SL RNA); 5′-Biotin-CAGACGCTGTCAGGTAGCGGCCGTGACGAGGATCTGCAGC-3′ (β-tubulin).
Control oligos were antisense to the oligos above, used in the same concentrations and cells were imaged at the same exposure times. Immunofluorescence experiments were performed in an identical manner, but without the prehybridisation and hybridisation steps.
Western blots
Western blots were performed using standard protocols. Detection was either by ECL or using the Odyssey Infrared Imaging System (LI-COR). For quantification, the Odyssey software was used (background method: the average of a three pixel width line at the top and bottom of each band was subtracted from each pixel). Unequal loading was corrected by reprobing the blots for BiP or PFR (L13D6).
HeLa cell experiments
HeLa cells were routinely maintained in DMEM. For immunofluorescence, cells were grown on 13 mm glass coverslips for 24 hours. Splicing was inhibited by the addition of spliceostatin A to the cells at 100 ng/ml for up to 5 hours using a 100 µg/ml stock in methanol for a maximum of 5 hours (the same volume of methanol served as control). Cells were rinsed with PBS, fixed in 4% paraformaldehyde in PBS for 20 minutes, rinsed twice in PBS, permeabilised in 0.5% TritonX-100 in PBS for 4.5 minutes, rinsed twice in PBS, incubated with primary antibodies (rabbit anti-rck/p54 1∶1000 and mouse anti-SC35 1∶500 in PBS with 5% BSA) for 1 hour, washed twice in PBS, incubated with secondary antibody conjugated to Rhodamine (rabbit) or dy488 (mouse) (1∶1000 in PBS, 5% BSA) for 1 hour, washed twice in PBS. After rinsing three times with PBS, cells were stained with DAPI (1.25 µg/ml) for 10 seconds. Coverslips were mounted in Citifluor (Citifluor Labs, Birmingham, UK).
Splicing was also inhibited by the electroporation of a morpholino (TGATAAGAACAGATACTACACTTGA, antisense to U2 snRNA, 10 µl of 1 mM stock) into 2×105 cells of trypsinised HeLa cells resuspended in 100 µl supplemented nucleofector solution using the Amaxa Nucleofector (programme I-013). The control morpholino used was 5′-GAAAATAGTTCAAACGAATTATGCG-3′ (β-tubulin sense). After electroporation, cells were resuspended in 500 µl pre-warmed medium and transferred into a 24-well plate containing a 13 mm glass coverslip. Fixation and immunofluorescence were performed 24 hours after electroporation as above.
Supplementary Material
Acknowledgments
The authors would like to thank Jenny Reed for expert technical assistance; Antonio Estevez (Instituto de Parasitología y Biomedicina ‘Lopez-Neyra’, Granada, Spain) for discussions, plasmids and cell lines; Sophie Piper for preparing the XRNA RNAi plasmid; Steffanie Seisenberger for preparing the FlaI RNAi plasmid; Michael Boshart (LMU Munich, Germany) for sending the Polo-like kinase p2T7 plasmid and the Fla1 RNAi plasmid; Rachana Ramarao (this lab) and Ines Subota (Institut Pasteur, Paris, France) for helpful advice on in situ hybridisations; Chris Smith (Department of Biochemistry, Cambridge, UK) for suggesting the morpholino experiment; Minory Yoshida (Chemical Genetics Laboratory, RIKEN, Japan) for the Spliceostatin A; Osvaldo de Melo Neto (Instituto Osvaldo Cruz, Recife, Brasil) for anti-PABP2; Jay Bangs (University of Wisconsin-Madison, Madison, WI) for the BiP antibody; Keith Gull (Sir William Dunn School of Pathology, University of Oxford, UK) for the L13D6 PFR antibody; Deirdre Scadden (Department of Biochemistry, Cambridge, UK) for the anti-SC35; Christian Janzen (Biocenter, Würzburg, Germany) for anti-Histone H3; and Jack Sunter for discussions and proofreading.
Footnotes
Funding
This work was funded by the Wellcome Trust [grant number 085956/Z/08/Z to M.C. and S.K.]; and a Biotechnology and Biological Sciences Research Council (BBSRC) Research Studentship (to A.M. and N.S.). Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.099275/-/DC1
References
- Anderson P., Kedersha N. (2006). RNA granules. J. Cell Biol. 172, 803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Bangs J. D., Crain P. F., Hashizume T., McCloskey J. A., Boothroyd J. C. (1992). Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267, 9805–9815 [PubMed] [Google Scholar]
- Bates E. J., Knuepfer E., Smith D. F. (2000). Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 28, 1211–1220 10.1093/nar/28.5.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Biton M., Mandelboim M., Arvatz G., Michaeli S. (2006). RNAi interference of XPO1 and Sm genes and their effect on the spliced leader RNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 150, 132–143 10.1016/j.molbiopara.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. A., Thornton D. A., Boothroyd J. C. (1984). Apparent discontinuous transcription of Trypanosoma brucei variant surface antigen genes. Nature 311, 350–355 10.1038/311350a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassola A., De Gaudenzi J. G., Frasch A. C. (2007). Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol. Microbiol. 65, 655–670 10.1111/j.1365-2958.2007.05833.x [DOI] [PubMed] [Google Scholar]
- Cheng Z., Coller J., Parker R., Song H. (2005). Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA 11, 1258–1270 10.1261/rna.2920905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas I. C., Frasch A. C. C., D'Orso I. (2005). Insights into a CRM1-mediated RNA-nuclear export pathway in Trypanosoma cruzi. Mol. Biochem. Parasitol. 139, 15–24 10.1016/j.molbiopara.2004.11.002 [DOI] [PubMed] [Google Scholar]
- da Costa Lima T. D., Moura D. M. N., Reis C. R. S., Vasconcelos J. R. C., Ellis L., Carrington M., Figueiredo R. C. B. Q., de Melo Neto O. P. (2010). Functional characterization of three leishmania poly(a) binding protein homologues with distinct binding properties to RNA and protein partners. Eukaryot. Cell 9, 1484–1494 10.1128/EC.00148-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi J., Frasch A. C., Clayton C. (2005). RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot. Cell 4, 2106–2114 10.1128/EC.4.12.2106-2114.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse J. A., DuBois K. N., Devos D., Siegel T. N., Sali A., Field M. C., Rout M. P., Chait B. T. (2009). Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8, 2119–2130 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhi P., Queiroz R., Inchaustegui D., Carrington M., Clayton C. (2011). Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS ONE 6, e25112 10.1371/journal.pone.0025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalia R., Reis C. R., Freire E. R., Rocha P. O., Katz R., Muniz J. R., Standart N., de Melo Neto O. P. (2005). Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 140, 23–41 10.1016/j.molbiopara.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Ventura A. P., Rutz B., Caspary F., Faux C., Halgand F., Laprévote O., Séraphin B. (2004). Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23, 4847–4856 10.1038/sj.emboj.7600482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy E. M., Ito S. (1971). Fine structural and radioautographic observations on dense perinuclear cytoplasmic material in tadpole oocytes. J. Cell Biol. 49, 90–108 10.1083/jcb.49.1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K., Asbeck K., Gull K. (1998). Direct visualisation of individual gene organisation in Trypanosoma brucei by high-resolution in situ hybridisation. Chromosoma 107, 237–240 10.1007/s004120050302 [DOI] [PubMed] [Google Scholar]
- Estévez A. M. (2008). The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 36, 4573–4586 10.1093/nar/gkn406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C. M., Munro E., Rasoloson D., Merritt C., Seydoux G. (2008). Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 323, 76–87 10.1016/j.ydbio.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont–Racine M., Romano A., Jacquier A., Nehrbass U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- Gilinger G., Bellofatto V. (2001). Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29, 1556–1564 10.1093/nar/29.7.1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünfelder C. G., Engstler M., Weise F., Schwarz H., Stierhof Y-D., Boshart M., Overath P. (2002). Accumulation of a GPI-anchored protein at the cell surface requires sorting at multiple intracellular levels. Traffic 3, 547–559 10.1034/j.1600-0854.2002.30805.x [DOI] [PubMed] [Google Scholar]
- Hammarton T. C., Kramer S., Tetley L., Boshart M., Mottram J. C. (2007). Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 65, 1229–1248 10.1111/j.1365-2958.2007.05866.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. J., Maroney P. A., Nilsen T. W. (1991). U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J. Biol. Chem. 266, 22792–22795 [PubMed] [Google Scholar]
- Hay B., Jan L. Y., Jan Y. N. (1988a). A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55, 577–587 10.1016/0092-8674(88)90216-4 [DOI] [PubMed] [Google Scholar]
- Hay B., Ackerman L., Barbel S., Jan L. Y., Jan Y. N. (1988b). Identification of a component of Drosophila polar granules. Development 103, 625–640 [DOI] [PubMed] [Google Scholar]
- He F., Peltz S. W., Donahue J. L., Rosbash M., Jacobson A. (1993). Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 90, 7034–7038 10.1073/pnas.90.15.7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. J., Parker R. (2003). Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell 12, 1453–1465 10.1016/S1097-2765(03)00488-X [DOI] [PubMed] [Google Scholar]
- Holetz F. B., Correa A., Avila A. R., Nakamura C. V., Krieger M. A., Goldenberg S. (2007). Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356, 1062–1067 10.1016/j.bbrc.2007.03.104 [DOI] [PubMed] [Google Scholar]
- Hotchkiss T. L., Nerantzakis G. E., Dills S. C., Shang L., Read L. K. (1999). Trypanosoma brucei poly(A) binding protein I cDNA cloning expression, and binding to 5 untranslated region sequence elements. Mol. Biochem. Parasitol 98, 117–129 10.1016/S0166-6851(98)00156-X [DOI] [PubMed] [Google Scholar]
- Jaé N., Wang P., Gu T., Hühn M., Palfi Z., Urlaub H., Bindereif A. (2010). Essential role of a trypanosome U4-specific Sm core protein in small nuclear ribonucleoprotein assembly and splicing. Eukaryot. Cell 9, 379–386 10.1128/EC.00353-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger A. V., De Gaudenzi J. G., Cassola A., D'Orso I., Frasch A. C. (2007). mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes. Proc. Natl. Acad. Sci. USA 104, 2035–2042 10.1073/pnas.0611125104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Bouhouche K., Gout J. F., Aury J. M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B. M., Ségurens B., et al. (2008). Translational control of intron splicing in eukaryotes. Nature 451, 359–362 10.1038/nature06495 [DOI] [PubMed] [Google Scholar]
- Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H., et al. (2007). Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 10.1038/nchembio.2007.18 [DOI] [PubMed] [Google Scholar]
- Kawashima T., Pellegrini M., Chanfreau G. F. (2009). Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 15, 2236–2247 10.1261/rna.1736809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke–Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Reed J., Kramer S., Ellis L., Webb H., Sunter J., Salje J., Marinsek N., Gull K., Wickstead B., et al. (2007). Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 154, 103–109 10.1016/j.molbiopara.2007.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C., Weidner J., Prescianotto–Baschong C., Spang A. (2010). Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol. Biol. Cell 21, 2624–2638 10.1091/mbc.E10-02-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N. G., Franklin J. B., Carmi S., Shi H., Michaeli S., Tschudi C. (2010). The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 6, e1001090 10.1371/journal.ppat.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J. D., Clayton C., Carrington M. (2008). Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 121, 3002–3014 10.1242/jcs.031823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S., Kimblin N. C., Carrington M.2010a). Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major. BMC Genomics 11283 10.1186/1471-2164-11-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S., Queiroz R., Ellis L., Hoheisel J. D., Clayton C., Carrington M.2010b). The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J. Cell Sci. 123699–711 10.1242/jcs.058511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Barrett B., Donelson J. E. (2002). Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem 277, 17580–17588 10.1074/jbc.M200873200 [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., Smith H. Q., Rusche L., Beverley S. M. (1993). Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7, 996–1007 10.1101/gad.7.6.996 [DOI] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. (1989). Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57, 573–583 10.1016/0092-8674(89)90127-X [DOI] [PubMed] [Google Scholar]
- Li C-H., Irmer H., Gudjonsdottir–Planck D., Freese S., Salm H., Haile S., Estévez A. M., Clayton C. (2006). Roles of a Trypanosoma brucei 5′->3′ exoribonuclease homolog in mRNA degradation. RNA 12, 2171–2186 10.1261/rna.291506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tschudi C. (2005). Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol. Cell. Biol. 25, 2216–2226 10.1128/MCB.25.6.2216-2226.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liang X. H., Uliel S., Belahcen M., Unger R., Michaeli S. (2004). Identification and functional characterization of lsm proteins in Trypanosoma brucei. J. Biol. Chem. 279, 18210–18219 10.1074/jbc.M400678200 [DOI] [PubMed] [Google Scholar]
- Lo C-W., Kaida D., Nishimura S., Matsuyama A., Yashiroda Y., Taoka H., Ishigami K., Watanabe H., Nakajima H., Tani T., et al. (2007). Inhibition of splicing and nuclear retention of pre-mRNA by spliceostatin A in fission yeast. Biochem. Biophys. Res. Commun. 364, 573–577 10.1016/j.bbrc.2007.10.029 [DOI] [PubMed] [Google Scholar]
- Mahowald A. P. (1971). Polar granules of Drosophila. IV. Cytochemical studies showing loss of RNA from polar granules during early stages of embryogenesis. J. Exp. Zool. 176, 345–352 10.1002/jez.1401760309 [DOI] [PubMed] [Google Scholar]
- Mahowald A. P., Hennen S. (1971). Ultrastructure of the “germ plasm” in eggs and embryos of Rana pipiens. Dev. Biol. 24, 37–53 10.1016/0012-1606(71)90045-5 [DOI] [PubMed] [Google Scholar]
- Mair G., Ullu E., Tschudi C. (2000). Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275, 28994–28999 10.1074/jbc.M004193200 [DOI] [PubMed] [Google Scholar]
- Mandelboim M., Barth S., Biton M., Liang X. H., Michaeli S. (2003). Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 278, 51469–51478 10.1074/jbc.M308997200 [DOI] [PubMed] [Google Scholar]
- Manful T., Fadda A., Clayton C. (2011). The role of the 5′-3′ exoribonuclease XRNA in transcriptome-wide mRNA degradation. RNA 17, 2039–2047 10.1261/rna.2837311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A., Weil D., Standart N. (2012). RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol. Biol. Cell 23, 213–224 10.1091/mbc.E11-05-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N., König H. (2005). Targeted ‘knockdown’ of spliceosome function in mammalian cells. Nucleic Acids Res. 33, e41 10.1093/nar/gni041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. R., Tschudi C., Ullu E. (1994). A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8, 491–501 10.1101/gad.8.4.491 [DOI] [PubMed] [Google Scholar]
- McCulloch R., Vassella E., Burton P., Boshart M., Barry J. D. (2004). Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262, 53–86 [DOI] [PubMed] [Google Scholar]
- McNally K. P., Agabian N. (1992). Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol 12, 4844–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone J., Wilusz J., Bellofatto V. (2002). Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 30, 4040–4050 10.1093/nar/gkf521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone J., Wilusz J., Bellofatto V. (2004). Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10, 448–457 10.1261/rna.5180304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram J. C., Murphy W. J., Agabian N. (1989). A transcriptional analysis of the Trypanosoma brucei hsp83 gene cluster. Mol. Biochem. Parasitol. 37, 115–127 10.1016/0166-6851(89)90108-4 [DOI] [PubMed] [Google Scholar]
- Muhich M. L., Boothroyd J. C. (1988). Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol. Cell. Biol. 8, 3837–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhich M. L., Hsu M. P., Boothroyd J. C. (1989). Heat-shock disruption of trans-splicing in trypanosomes: effect on Hsp70, Hsp85 and tubulin mRNA synthesis. Gene 82, 169–175 10.1016/0378-1119(89)90042-5 [DOI] [PubMed] [Google Scholar]
- Murphy W. J., Watkins K. P., Agabian N. (1986). Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell 47, 517–525 10.1016/0092-8674(86)90616-1 [DOI] [PubMed] [Google Scholar]
- O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R., Spector D. L. (1994). Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249–260 10.1083/jcb.124.3.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., Doye V. (2005). Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell 16, 5258–5268 10.1091/mbc.E05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfi Z., Günzl A., Cross M., Bindereif A. (1991). Affinity purification of Trypanosoma brucei small nuclear ribonucleoproteins reveals common and specific protein components. Proc. Natl. Acad. Sci. USA 88, 9097–9101 10.1073/pnas.88.20.9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt J. N., Schisa J. A., Priess J. R. (2000). P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315–333 10.1006/dbio.2000.9607 [DOI] [PubMed] [Google Scholar]
- Pitula J., Ruyechan W. T., Williams N. (1998). Trypanosoma brucei: identification and purification of a poly(A)-binding protein. Exp. Parasitol 88, 157–160 10.1006/expr.1998.4211 [DOI] [PubMed] [Google Scholar]
- Rain J. C., Legrain P. (1997). In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J 16, 1759–1771 10.1093/emboj/16.7.1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J-P., Shen S., Ullu E., Tschudi C. (2007). Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol. Biochem. Parasitol. 156, 246–254 10.1016/j.molbiopara.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B., Séraphin B. (2000). A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19, 1873–1886 10.1093/emboj/19.8.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather S., Agabian N. (1985). A 5′ spliced leader is added in trans to both alpha- and beta-tubulin transcripts in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 82, 5695–5699 10.1073/pnas.82.17.5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S., Janis M., Lee C. Y., Toesca I., Chanfreau G. F. (2008). Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31, 360–370 10.1016/j.molcel.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A., Ellis L., Luther J., Carrington M., Stoecklin G., Clayton C. (2008). A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36, 3374–3388 10.1093/nar/gkn108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A., Manful T., Jha B. A., Helbig C., Bercovich N., Stewart M., Clayton C. (2009). The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 37, 5511–5528 10.1093/nar/gkp571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck T., Whittaker P. A., Imboden M. A., Hardman N., Braun R. (1983). Tubulin genes of Trypanosoma brucei: a tightly clustered family of alternating genes. Proc. Natl. Acad. Sci. USA 80, 4634–4638 10.1073/pnas.80.15.4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Pitt J., Dennis S., Priess J. R. (2010). Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137, 1305–1314 10.1242/dev.044255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel T. N., Tan K. S., Cross G. A. (2005). Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei. Mol. Cell. Biol. 25, 9586–9594 10.1128/MCB.25.21.9586-9594.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. M. (2010). Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38, 4946–4957 10.1093/nar/gkq237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. Z., Gupta S. K., Salmon–Divon M., Haham T., Barda O., Levi S., Wachtel C., Nilsen T. W., Michaeli S. (2009). Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA 15, 648–665 10.1261/rna.1230209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. (2005). Specification of the germ line. WormBook: The Online Review Of C. Elegans Biology (ed. The C. elegans Research Community), pp. 1-10 Pasadena, California: WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W. B. (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 79, 1558–1562 10.1073/pnas.79.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W. B. (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15–25 10.1016/0092-8674(83)90203-9 [DOI] [PubMed] [Google Scholar]
- Sunter J., Wickstead B., Gull K., Carrington M.2012). A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PloS One 7e35167 10.1371/journal.pone.0035167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. E., Boothroyd J. C. (1988). Trypanosome trans-splicing utilizes 2′-5′ branches and a corresponding debranching activity. EMBO J. 7, 1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y., Sindkar S., Ekonomidis D., Hall M. P., Ho C. K. (2007). Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J. Biol. Chem 282, 15995–16005 10.1074/jbc.M701569200 [DOI] [PubMed] [Google Scholar]
- Tanackovic G., Krämer A. (2005). Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell 16, 1366–1377 10.1091/mbc.E04-11-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. J., Ogawa K., Takagi M., Imamoto N., Matsumoto K., Tsujimoto M. (2006). RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 281, 40096–40106 10.1074/jbc.M609059200 [DOI] [PubMed] [Google Scholar]
- Teixeira D., Parker R. (2007). Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2274–2287 10.1091/mbc.E07-03-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. D., Conrad R. C., Blumenthal T. (1988). The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell 54, 533–539 10.1016/0092-8674(88)90075-X [DOI] [PubMed] [Google Scholar]
- Thomashow L. S., Milhausen M., Rutter W. J., Agabian N. (1983). Tubulin genes are tandemly linked and clustered in the genome of trypanosoma brucei. Cell 32, 35–43 10.1016/0092-8674(83)90494-4 [DOI] [PubMed] [Google Scholar]
- Tschudi C., Ullu E. (1990). Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell 61, 459–466 10.1016/0092-8674(90)90527-L [DOI] [PubMed] [Google Scholar]
- Tschudi C., Williams S. P., Ullu E. (1990). Conserved sequences in the U2 snRNA-encoding genes of Kinetoplastida do not include the putative branchpoint recognition region. Gene 91, 71–77 10.1016/0378-1119(90)90164-M [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. (1991). Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 88, 10074–10078 10.1073/pnas.88.22.10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Matthews K. R., Tschudi C. (1993). Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 13, 720–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D. L., Hachey S. J., Kreher J., Strome S. (2011). P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939–948 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren K., Hirsh D. (1988). Trans- spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature 335, 556–559 10.1038/335556a0 [DOI] [PubMed] [Google Scholar]
- Voronina E., Seydoux G. (2010). The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 137, 1441–1450 10.1242/dev.047654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G., Dorrell R. G., Schlacht A., Dacks J. B. (2011). Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 138, 1638–1663 10.1017/S0031182010001708 [DOI] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-H., Yu J. H., Gulick T., Bloch K. D., Bloch D. B. (2006). RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12, 547–554 10.1261/rna.2302706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. (1988). Translation of unspliced transcripts after heat shock. Science 242, 1544–1548 10.1126/science.3201243 [DOI] [PubMed] [Google Scholar]
- Zeiner G. M., Sturm N. R., Campbell D. A. (2003). Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2, 222–230 10.1128/EC.2.2.222-230.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.