Summary
Much of our understanding of the biological mechanisms that underlie cellular functions, such as migration, differentiation and force-sensing has been garnered from studying cells cultured on two-dimensional (2D) glass or plastic surfaces. However, more recently the cell biology field has come to appreciate the dissimilarity between these flat surfaces and the topographically complex, three-dimensional (3D) extracellular environments in which cells routinely operate in vivo. This has spurred substantial efforts towards the development of in vitro 3D biomimetic environments and has encouraged much cross-disciplinary work among biologists, material scientists and tissue engineers. As we move towards more-physiological culture systems for studying fundamental cellular processes, it is crucial to define exactly which factors are operative in 3D microenvironments. Thus, the focus of this Commentary will be on identifying and describing the fundamental features of 3D cell culture systems that influence cell structure, adhesion, mechanotransduction and signaling in response to soluble factors, which – in turn – regulate overall cellular function in ways that depart dramatically from traditional 2D culture formats. Additionally, we will describe experimental scenarios in which 3D culture is particularly relevant, highlight recent advances in materials engineering for studying cell biology, and discuss examples where studying cells in a 3D context provided insights that would not have been observed in traditional 2D systems.
Key words: 3D culture models, Cell adhesion, Dimensionality, Mechanotransduction, Microenvironment, Soluble factors
Introduction
Our current understanding of many biological processes is based largely on studies of homogenous populations of cells cultured on flat, two-dimensional (2D) plastic or glass substrates. However, in vivo, cells primarily exist embedded within a complex and information-rich environment that contains multiple extracellular matrix (ECM) components, mixed cell populations that interact heterotypically and a medley of cell-secreted factors. The striking disparity between traditional monolayer culture and the in vivo scenario has been a double-edged sword: the simplicity of 2D culture has enabled reductionist approaches to understanding individual cellular phenomena but these findings have come with the caveat that the 2D model might not faithfully capture the physiological behavior of cells in vivo.
Indeed, many cell types, when isolated from tissues and placed into planar cell culture, become progressively flatter, divide aberrantly and lose their differentiated phenotype (von der Mark et al., 1977; Petersen et al., 1992). Interestingly, some of these cell types can regain their physiological form and function when embedded in a three-dimensional (3D) culture environment. For instance, encapsulation of dedifferentiated chondrocytes restores their physiological phenotype, including cell shape and the expression of cartilaginous markers (Benya and Shaffer, 1982). Similarly, mammary epithelial cells embedded in a 3D environment halt uncontrolled division, assemble into acinar structures and establish a de novo basement membrane (Emerman and Pitelka, 1977; Lee et al., 1984; Petersen et al., 1992).
These observations have led to the notion that the dimension in which cells are cultured is a crucial fate determinant, and to the vague impression that culturing cells in monolayer drives abnormal cell function or dedifferentiation, whereas 3D culture elicits a more physiological state. However, we must be wary of oversimplifying these comparisons into a single difference between two states, i.e. three-dimensionality versus two-dimensionality. Presently, dimensionality has become a blanket statement for what entails many potential differences between traditional culture in a 2D monolayer, 3D culture systems and the physiological setting. Rather than the overall dimensional shape of the cell or culture, functional consequences instead originate from the finer features that are inherent to each of these contexts. Thus, rather than simply concluding that a dimensionality factor is at play, we must identify and understand the salient features of each experimental setting and strive to demystify exactly what 3D culture provides to the cells that differs from more traditional 2D settings.
With this goal in mind, this Commentary will examine the main avenues by which microenvironmental cues are known to impact cell function – cell adhesions, mechanical forces and diffusible factors – and how such cues may be presented in 3D versus 2D culture. Beyond providing appropriate physiological cues, 3D culture also facilitates biological responses that might not be observable on 2D substrates. For example, the collective cell migration, force generation and tissue folding that occurs during gastrulation, the angiogenic sprouting of blood vessels, and the migration of cancerous cells through stroma and into lymphatics during metastasis, are all cases of higher-order cell processes that are inherently 3D (Fig. 1). Deconstructing these 3D microenvironments and the associated processes into adhesive, mechanical and chemical components will aid us in understanding the underlying mechanisms that guide these processes. Furthermore, because the technologies for engineering the cellular environment are rapidly evolving, we also examine some of the methods that can be employed for studying these different cues in vitro (see Boxes 1 and 2). This Commentary is not intended to be an exhaustive compilation of the literature on cell biology in 3D but, rather, seeks to identify some salient features of 3D experimental systems that should be considered in the questions we pose and the studies we conduct.
Fig. 1.
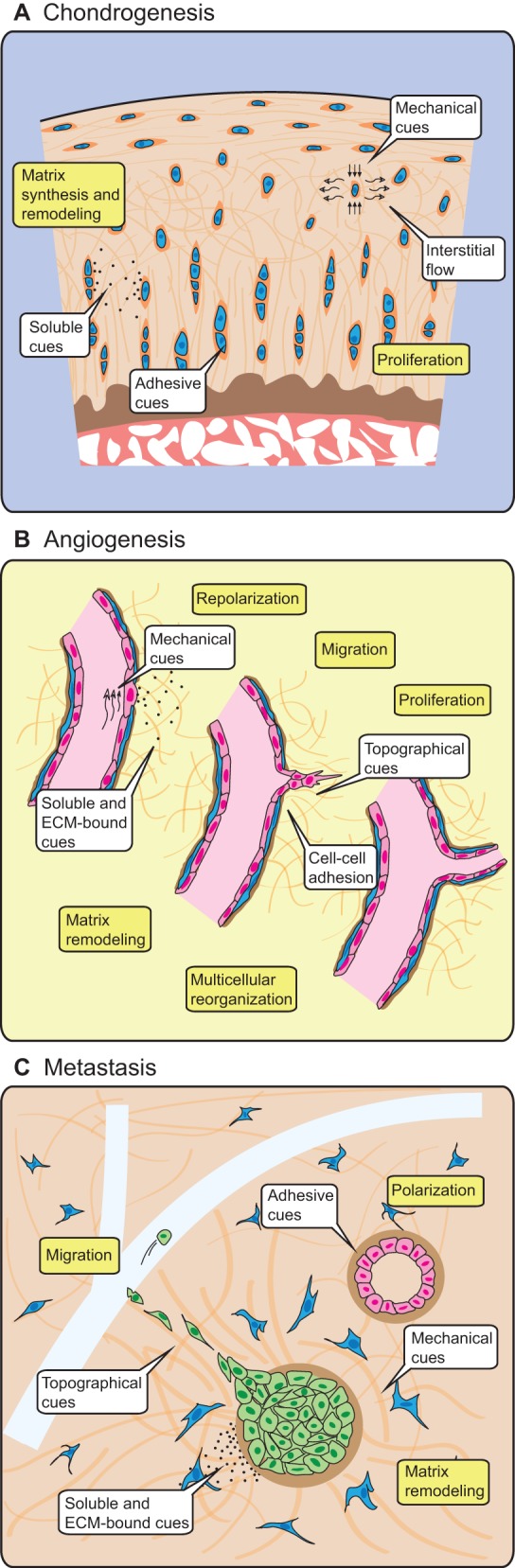
3D cellular phenomena in development, tissue homeostasis and disease are conducted by adhesive, mechanical and chemical cues originating from other cells and the extracellular environment. (A) Chondrocytes (blue) reside within a specialized pericellular ECM, where they are exposed to compressive forces, interstitial fluid flow, adhesive cues and soluble cues in the form of cytokines, which allow the cells to form and maintain the surrounding cartilage. (B) In response to soluble and matrix-bound growth factors and flow-induced mechanical forces on the blood vessel wall, endothelial cells (pink) alter their polarity and cell-cell contacts, and degrade the surrounding basement membrane (brown) and stromal ECM (orange) in order to collectively invade the surrounding tissue and form tubular sprouts. (C) The formation of normal epithelial structures (pink) requires adhesive and mechanical cues from neighboring cells and the basement membrane (brown) in order to tightly regulate proliferation and apoptosis. Misregulation of proliferation through genetic or extracellular changes initiates a cascade of soluble signals that activate fibroblasts (blue) in the surrounding stroma. Subsequent mechanical and structural changes in the stromal ECM enable transformed epithelial cells (green) to migrate towards neighboring vasculature (light blue) and, eventually, to metastasize. Drawings not to scale.
Box 1. Materials and systems for 3D culture.
An overwhelming number of biomaterials have been developed for studying, as well as directing, cellular interactions in 3D (Langer and Tirrell, 2004; Lutolf and Hubbell, 2005). In order for experiments to be logistically feasible, these systems typically begin with a liquid precursor containing suspended cells, which gels or solidifies in a cyto-compatible and hydrated manner. The ensuing gels are porous to enable nutrient and waste exchange, and possess sufficient mechanical properties to be self-supporting. The most basic biological functionality is achieved by addition of cell- adhesive ligands, commonly in the form of Arg–Gly–Asp peptides. In experiments where cell migration, matrix remodeling or multicellular organization is of interest, the embedded cells will need to overcome the steric constraints of their surroundings. For this to occur, the 3D ECM must contain structural entities that are susceptible to degradation, either through proteolytic cleavage or hydrolysis. Finally, specific biological activity and interactions can be facilitated through the addition of soluble or insoluble factors and biological domains. Such materials can be of natural or synthetic origin, but much of the engineering behind synthetic materials has been informed by our understanding of how natural ECMs function.
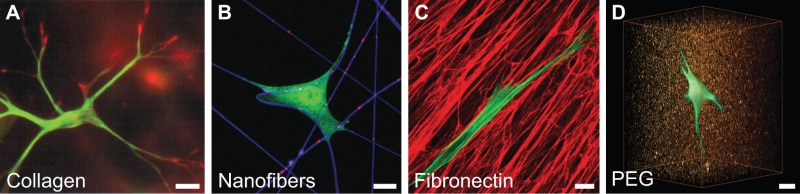
For several decades, ECM from natural sources has served as an important tool for biologists. These materials include purified collagen type I (A in Figure) (Grinnell, 2003), fibrin gels formed by thrombin cleavage of fibrinogen, reconstituted basement membrane (e.g. Matrigel) (Kleinman and Martin, 2005) and stromal ECM synthesized by fibroblasts (C in Figure) (Beacham et al., 2007). As a result of their cellular origin, these materials inherently possess adhesive ligands and other biological activity, and can readily be remodeled by cells. For this same reason, these systems can prove disadvantageous in isolating certain cell responses. For example, Matrigel comprises collagens, laminin and entactin, but also possesses an uncharacterized population of growth factors that varies substantially between batches (Hughes et al., 2010). Additional limitations of natural systems include the challenge of modifying these systems to incorporate additional functional moieties and the difficulty of tuning different features of the microenvironment independently. For example, in a collagen gel it is impossible to modulate stiffness without also altering the density of the adhesive ligand, pore size and porosity.
The development of synthetic gels has rapidly advanced during the past decade, and has been motivated by the desire to provide greater control over material and biological properties than can be achieved by using their natural counterparts (Lutolf and Hubbell, 2005). These materials typically possess a structural backbone, cell-binding ligands, and a ‘cell-friendly’ crosslinking mechanism. The most common of these include polyethylene glycol (PEG)-based hydrogels (D in Figure) (Mann et al., 2001; Burdick and Anseth, 2002; Raeber et al., 2005; Miller et al., 2010) and self-assembling peptides (Kisiday et al., 2002; Zhang, 2003; Mata et al., 2009). These gels are highly tunable and often modular in their inclusion of additional functionalities, such as matrix metalloprotease-cleavable domains or growth-factor- binding sites. The advantages of synthetic materials are best evidenced by the ever-expanding flexibility and diversity that is achievable in these systems. For example, gels in which dynamic changes in stiffness can be induced in response to light have been developed recently (Kloxin et al., 2009).
Images in the Figure were adapted with permission from Grinnell et al., 2003 (A), Doyle et al., 2009 (C) and Legant et al., 2010 (D) (Grinnell et al., 2003; Doyle et al., 2009; Legant et al., 2010). Scale bars: 20 µm.
Box 2. How to engineer microenvironments to capture the features of 3D culture.
There is a growing interest in isolating and harnessing the specific microenvironmental cues that 3D culture can provide. With these tools in hand, reconstituting a particular cellular phenotype might be possible without the requirement for 3D culture and the experimental drawbacks associated with it (e.g. non-optimal imaging and interdependent microenvironmental factors). Examples include methods to control cell adhesion and shape, matrix mechanics and topography, and the delivery of soluble factors.
Adhesion
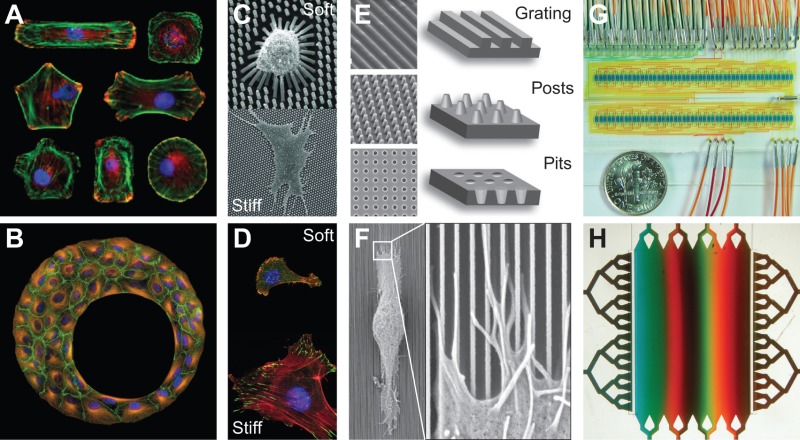
Micropatterned substrates contain defined arrangements of cell-adhesive and protein-adsorption-resistant regions that are presented on a flat surface. These substrates can precisely control cell adhesion and spreading, without altering physical or chemical attributes of the microenvironment (Whitesides et al., 2001; Théry, 2010). Depending on the geometry of the patterns, such substrates have been used to dictate the size and distribution of integrin-mediated adhesions, cell shape and cytoskeletal architecture (A in Figure) as well as multicellular organization (B in Figure), and have been instrumental in demonstrating the importance of these factors in regulating the tight coupling between cell structure, signaling, and function. Nanotopographic materials (E–F in Figure) are typically stiff substrates with nano-scale topological features that are designed to constrain the size and geometry of cell–matrix adhesions. These materials can be fabricated using several methods, including lithography, chemical and physical roughening, electrospinning or electrospray (Dalby et al., 2004; Anselme et al., 2010). As the scale of the features approaches nm scales, the local curvature and roughness can influence the positioning of the plasma membrane, surface receptors and structures associated with them. For example, Salaita and colleagues employed nm-scale gratings to restrict the movement and clustering of EPH receptor A2 (EPHA2) receptors and, thereby, alter signaling from these receptors (Salaita et al., 2010).
Mechanics
Nanometer-scale features can also be used to alter the mechanical properties of the cellular microenvironment when this is formed in softer materials. For example, we have generated arrays of silicone posts of sub-micrometer diameters and differing lengths in order to study the effects of ECM stiffness on cells (C in Figure, Fu et al., 2010). Silicones can also be prepared with different crosslinking densities to tune the stiffness of the resulting substrate (Prager-Khoutorsky et al., 2011). Similarly, numerous hydrogels, such as and a growing number of designer systems, can be differentially crosslinked to alter substrate stiffness (D in Figure). A salient feature that is intrinsic to all of these systems is the orthogonal control of stiffness and ligand density.
Soluble factors
High-throughput fluid-handling methods and microfluidic provide two means by which the soluble environment can be stringently defined. High-throughput screening approaches can be used to expose cells to libraries of soluble factors and examine the resulting biological response. Huang and co-workers have used a high-throughput approach to optimize the soluble factors that promote mesenchymal stem cell chondrogenesis (Huang et al., 2008). Microfluidic devices (G,H in Figure) that are based on soft lithography can be used to control the finer features of the soluble microenvironment, including the timing and spatial presentation of biological factors (Quake and Scherer, 2000), or can simply reduce the effective culture volume and, thus, amplify autocrine signals (Yu et al., 2007). These devices range in design from simple free diffusion systems to more complex flow-based gradient generators that provide greater control and flexibility in producing temporally and spatially dynamic profiles. Microfluidics are now used routinely to provide gradients of soluble growth factors to cells, primarily to study polarization and chemotaxis (Kim et al., 2010).
The images in the Figure were adapted with permission from Kilian et al., 2010 (A), Desai et al., 2009 (B), Fu et al., 2010 (C), Bettinger et al., 2009 (E), Teixeira et al., 2003 (F), G�mez-Sj�berg et al., 2007 (G) (Kilian et al., 2010; Desai et al., 2009; Fu et al., 2010; Bettinger et al., 2009; Teixeira et al., 2003; G�mez-Sj�berg et al., 2007). Images in D, courtesy of Colin Choi and Christopher Chen. Image H, courtsey of Albert Folch, (University of Washington, Seattle, WA).
Cell adhesion and structure
For anchorage-dependent cells, adhesive interactions with the surrounding ECM and neighboring cells define cell shape and organization. The organization, composition and number of adhesions are among the better understood signals that are integrated by the cell in order to regulate many fundamental cellular behaviors, including survival, differentiation, proliferation and migration (Wozniak et al., 2004; Weber et al., 2011; Orr et al., 2006; Geiger et al., 2009). Unavoidably, studying cell biology in vitro necessitates stripping cells of these native cell–cell and cell–ECM interactions, and introducing them into a foreign adhesive environment that is defined by the culture system.
One of the most striking differences observed when comparing cells in 2D and 3D is the dissimilarity in morphology (Fig. 2). Cells grown in a monolayer are flat, and can adhere and spread freely in the horizontal plane but have no support for spreading in the vertical dimension. One consequence of this is that cells that are cultured on 2D surfaces have a forced apical–basal polarity. This polarity is arguably relevant for some cell types, such as epithelial cells, but is unnatural for most mesenchymal cells, which – when embedded in a 3D ECM – assume a stellate morphology and only polarize from front to rear during migration (Mseka et al., 2007). These changes in cell geometry and organization can directly impact cell function. For instance, apical-basal polarity has been shown to modulate the sensitivity of cells to apoptosis (Weaver et al., 2002). It also has been suggested that the flattening of cells can alter the effective surface-to-volume (i.e. membrane-to-cytoplasm) ratio, such that signaling from the cell surface is better propagated into the cell (Meyers et al., 2006). By using micropatterned adhesive islands on 2D substrates, we and others have demonstrated that altering the degree of cell spreading can impact cell proliferation, apoptosis and differentiation (Singhvi et al., 1994; Chen et al., 1997; Thomas et al., 2002; McBeath et al., 2004). In addition to the total area of spreading, the geometric shape (e.g. circular versus star-shaped, cuboidal versus elongated) that is assumed by the cell can impact its function (Brock et al., 2003; Théry et al., 2006; Théry et al., 2007; Mahmud et al., 2009; Kilian et al., 2010). Despite the apparent role of shape and area on cell function in 2D culture, it remains unclear how these insights map to 3D settings. Methods to control cell adhesion in 3D are just beginning to be established (Lee et al., 2008; DeForest et al., 2009; Khetan and Burdick, 2010; Klein et al., 2011). Indeed, the process of spreading or extending into a 3D matrix might be quite different from the processes that occur on a planar surface. For example, on 2D substrates, integrin-mediated adhesion is followed by lamellipodial extension, myosin-mediated cytoskeletal tension and the reinforcement of focal adhesions (Hynes, 1987; Burridge et al., 1988; Lauffenburger and Horwitz, 1996; Reinhart-King et al., 2005). Overall this process takes a short period of time on these restraint-free substrates. In 3D settings, however, cells must often negotiate or proteolytically cleave the physical scaffold in order to extend. Thus, cell spreading occurs over hours, and, in some instances, days rather than minutes (Khetan and Burdick, 2010).
Fig. 2.
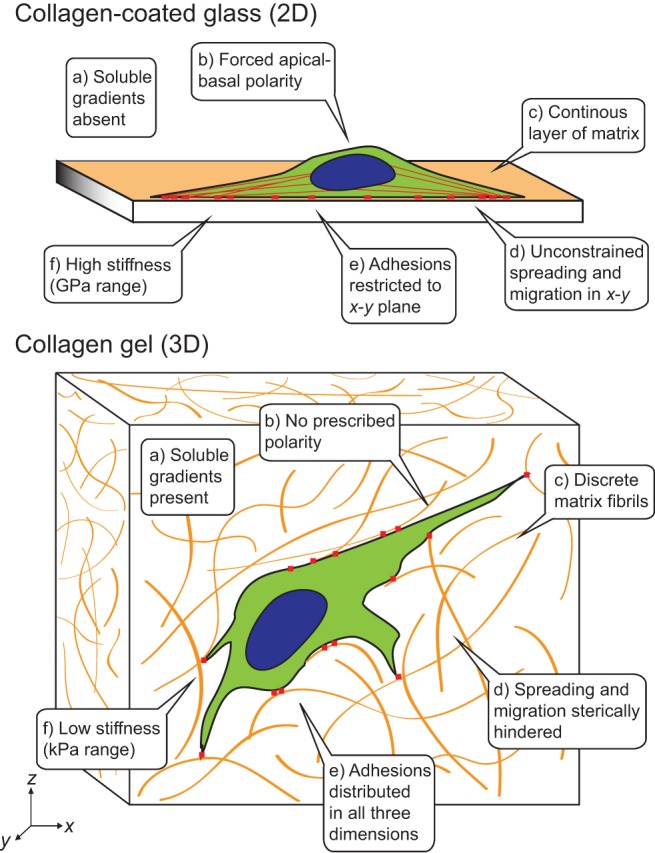
Adhesive, topographical, mechanical, and soluble cues in 2D and 3D. The cues encountered by a cell are strikingly different between an ECM-coated glass or plastic surface (2D) and a typical 3D ECM, such as collagen.
Cells assume 2D or 3D geometries largely on the basis of whether integrin-mediated adhesions to the extracellular matrix form on one face of the cell or all around the cell surface (Fig. 2). Different cellular responses in 2D versus 3D culture could arise from these variations in the spatial distribution of adhesions. For example, Beningo et al. ‘sandwiched’ fibroblasts between two ECM-coated polyacrylamide gels to simultaneously engage both dorsal and ventral integrins (Beningo et al., 2004). With integrin binding now occurring on two opposing planes, lamellipodial formation diminished in favor of a stellate morphology with long actin-rich extensions, akin to fibroblast morphology observed in vivo (Langevin et al., 2005). Similar ‘sandwich’ cultures have been shown to maintain the differentiated function of hepatocytes (Dunn et al., 1989). How spatial distributions of adhesions in 3D could impact cell signaling and function remains largely speculative but, given the body of knowledge we have acquired from controlling adhesion distributions in 2D, such pursuits are well motivated.
Advances in our ability to control ECM presentation on 2D surfaces have facilitated studies that begin to explain how spatial distributions of ligand impact integrin-mediated adhesion and function (Théry, 2010; Geiger et al., 2009). For example, the distribution of adhesions underlying a cell can bias the axis of planar polarity and the mitotic spindle (Théry et al., 2006; Théry et al., 2007). Using 8-nm gold particles coated with Arg-Gly-Asp (RGD) peptides that permit the binding of only a single integrin, Arnold and colleagues determined the minimum ligand spacing required for integrin-clustering-induced signaling to occur (Arnold et al., 2004). The technology for spatially patterning adhesive ligands in 3D is still in its infancy, efforts to develop approaches analogous to these 2D technologies will be crucial in moving our understanding of cell adhesion into a 3D context. Nonetheless, these results suggest that the nanometer-scale architecture of the ECM impacts the structure, function and, possibly, composition of integrin-mediated adhesions. This question of architecture is particularly important given the wide variety of nanometer-scale fibrillar structures of native ECMs that can change dynamically during the progression of disease (Levental et al., 2009; Amatangelo et al., 2005; Provenzano et al., 2006; Räsänen and Vaheri, 2010; Beacham and Cukierman, 2005).
Given the structural diversity of the extracellular environment, it is not surprising that adhesions in 3D are highly variable. Factors such as matrix stiffness and topography are likely to contribute to the recruitment of proteins to adhesion sites and might explain some of the variations observed in different 3D contexts (Harunaga and Yamada, 2011; Fraley et al., 2010; Kubow and Horwitz, 2011). Approaches to quantitatively describe these variations have not yet emerged, owing to the additional hurdle that imaging focal adhesions in 3D contexts is challenging because of the decreased size or intensity of adhesions, or non-optimal optics in a non-planar sample. Despite these challenges, 3D matrix adhesions on cell-derived matrices have been described, and it has been suggested that the type of integrin employed by the cell is differentially specified by 3D versus 2D microenvironments (Cukierman et al., 2001). Interestingly, follow-up work has suggested that these 3D adhesions can be reproduced by culturing cells on flat surfaces that present narrow strips of micropatterned ECM (Doyle et al., 2009). In other words, the fibrillar nature of the ECM might be responsible for modulating adhesion structure and signaling, and one important mechanism by which 3D culture might impact cell function is through such nanoscale features. Looking forward, whereas a few natural ECMs are already accessible as models for studying these topographical effects (e.g. collagen I and fibrin), recent progress in synthetic matrices might afford more tunable control over the structural and mechanical features of the 3D environment.
Mechanotransduction
It has become widely appreciated that mechanical forces are ever-present between cells and their surroundings, and that these forces provide a crucial set of signals that can control cell structure and function (Eyckmans et al., 2011; Hoffman et al., 2011). Mechanical stresses generated or experienced by cells as they adhere to the ECM and to their neighbors represent a central component of how cells transduce adhesion-mediated signaling and processes (Orr et al., 2006). However, in contrast to adhesions, where much of our understanding has been derived from studies on 2D surfaces, some of the earliest evidence suggesting a role for ECM mechanics arose from comparing cells cultured in gels that remained bound to the dish (attached) with those cultured in gels that were detached and hence were allowed to contract (floating). For instance, mammary epithelial cell acini and tubule formation were found to occur only in floating gels (Emerman and Pitelka, 1977; Parry et al., 1985; Keely et al., 1995). Here, we consider the possibility that 3D culture impacts mechanical forces and their transduction in cells.
Traditional 2D culture on glass or plastic substrates places cells in a static mechanical environment that is supraphysiological in terms of stiffness (Fig. 2). Recognizing the disparity between these artificial conditions and the markedly more-compliant microenvironment of most tissues, recent work using soft 2D gels has confirmed that ECM stiffness can influence adhesions, morphogenesis, and stem cell differentiation and maintenance (Engler et al., 2006; Paszek et al., 2005; Gilbert et al., 2010). Low stiffness is not necessarily an intrinsic property of 3D environments; however, it is a feature common to most 3D systems and a factor that should be taken into consideration when differences are noticed between cell behavior in 2D and 3D. Our understanding of how cells sense stiffness or rigididty is still developing, and most of the current efforts are seeking to identify the mechanisms by using 2D gels. However, it is possible that the mechanisms cells use to sense stiffness differ between 2D surfaces and 3D microenvironments. For example, Huebsch and co-authors examined the influence of gel stiffness on mesenchymal stem cell (MSC) differentiation in 3D RGD-modified alginate gels (Huebsch et al., 2010). Interestingly, MSC differentiation demonstrated a bimodal response, with osteogenesis occurring maximally at an intermediate stiffness, whereas 2D studies suggested a plateau response for which both glass and plastic substrates efficiently induce osteogenesis. Probing cell–ECM interactions by using FRET-imaging, the authors also found that osteogenesis in a 3D context requires integrin clustering by cell-exerted traction forces, and that this clustering is abrogated in an overly stiff 3D matrix. For unknown reasons, this dependency on integrin clustering was not observed on 2D substrates (Huebsch et al., 2010).
In addition to sensing passive mechanical cues, such as the compliance of the surrounding matrix, tissues commonly experience a variety of active loads, which result in force transduction and deformation at the cellular level (Hoffman et al., 2011). Although it remains unclear whether exogenous and internally generated forces are sensed through common mechanotransduction mechanisms (Chen, 2008), there are clear differences in how forces are experienced by cells in 2D versus 3D contexts. For example, in typical 2D experiments, the effect of tensile deformations is investigated by stretching cells that adhere to flat, ECM-coated silicone membranes. The resulting strain fields are smooth and homogeneous, and cell deformation occurs in a predictable, affine manner. By contrast, most 3D tissues are fibrous and, therefore, structurally heterogeneous and anisotropic (Pathak and Kumar, 2011). The way in which force is transmitted to the cell depends on the scale and organization of matrix fibers relative to that of the cell, as well as whether the cell is directly bound and/or physically constrained by the material. For example, neighboring cells located within fibrocartilage experience considerably different amounts of stretch depending on their proximity and adhesions to collagen fibrils (Upton et al., 2008). Adding to the complexity, cell morphology and orientation with respect to the direction of applied forces and matrix architecture can have a profound effect on the cellular response (Kurpinski et al., 2006; Nathan et al., 2011). Heo and colleagues observed that the divergence between the orientation of applied forces and structural anisotropy in a fibrous material alters cellular deformation and differentially influences gene expression (Heo et al., 2011). Given these numerous intricacies, making sense of the effects of exogenous loads on cells embedded within structurally complex materials in 3D will require a combination of experimental and computational modeling approaches (Niklason et al., 2010), thus engendering the need for cross-disciplinary collaborations between biologists and engineers.
Even putting the complexities of heterogeneous, anisotropic fibrous materials aside, 3D culture gives rise to important distinctions with regards to the mechanical condition. In 2D culture, the cell is allowed to deform in the out-of-plane axis and free of mechanical or physical constraint. In 3D culture, this is not the case. For example, discrepancies between how the cell and surrounding matrix narrow as they elongate under stretch, could generate stresses in the transverse plane of the cell. Similarly, the orientation of stress with respect to focal adhesions is tangential to the surface of the cell on a 2D substrate (i.e. it is causing shearing at the adhesions) and leads to force transmission along the basally located stress fibers (Dembo and Wang, 1999; Tan et al., 2003) (Fig. 2). By contrast, stretching an embedded cell will lead to stresses that lie perpendicular to the membrane and result in forces that travel through the midline of the cells. How cells and adhesions transduce mechanical forces could very well change depending on the orientation of those forces. Although we used applied forces to illustrate the differences between the stresses experienced in 2D and 3D environments, cell-generated forces might also be experienced quite differently in 2D versus 3D contexts. Recent developments in measuring cellular traction forces in 2D and 3D settings have allowed initial characterization of these differences and will be instrumental in achieving a better understanding of cell–ECM force transduction (Maskarinec et al., 2009; Legant et al., 2010).
Effector transport
Beyond serving as an adhesive and mechanical support for cells, the ECM also has an integral role in regulating the spatial distribution of nutrients, gases (such as oxygen and nitric oxide) and soluble effector molecules (including morphogens, growth factors, hormones and cytokines). These gradients are essential for the regulation of fundamental cell processes, such as cell migration and homing (Kay et al., 2008), angiogenic sprouting (Adams and Alitalo, 2007; Otrock et al., 2007) and tissue patterning during early development (Gurdon and Bourillot, 2001; Saha and Schaffer, 2006; Manjón et al., 2007; Bollenbach et al., 2008) (Fig. 1). The spatial presentation of diffusible factors in tissues is complex and is dictated by the structure and porosity of the surrounding ECM, and the presence and distribution of vasculature and surrounding cells (both of which can serve as sources or sinks of these factors). The ensuing compartmentalization of our tissues during development is a result of, as well as a subsequent contributor to, the formation of signaling gradients, and this feedback loop between ECM structure and effector transport is essential to the maintenance of differentiated structures in the adult.
Under traditional 2D monolayer culture conditions, cell-secreted or exogenously added soluble factors convectively mix and diffuse freely throughout the medium, thereby rapidly equilibrating. Temporary gradients can be created in these contexts to study short-lived chemotactic events such as cell polarization and migration on 2D surfaces (see Box 2), but longer-term morphogenetic events require gradients that are sustained for the duration of hours to days. The mere presence of ECM, whether created by using 3D culture or by simply overlaying matrices on a 2D culture, slows down the transport and equilibration of soluble factors, and can support such sustained gradients. A pelleted cluster of cells that lacks exogenous ECM is a relatively simple 3D model that can also capture diffusion-mediated radial gradients. For example, tumor-cell spheroids of sufficient size possess a hypoxic necrotic core that is reminiscent of actual tumors, and have proved useful for studying the role of hypoxia in the production of tumorigenic factors and drug resistance (Hirschhaeuser et al., 2010; Mueller-Klieser, 1997).
When cells are encapsulated in a 3D ECM, structural features of the ECM, such as pore size, interconnectivity and gel dimensions – in addition to cell density, solute size and charge – all modulate the diffusion of soluble factors (Ramanujan et al., 2002). In fact, because cells are typically embedded within diffusion-limited mm-scale gels, it is often difficult to distinguish the direct effects of encapsulation within the 3D ECM from differences in the timing of soluble signals diffusing through the material. To address this, we generated µm-sized 3D gels that, owing to their small scale, lack diffusion barriers. Comparing the response of MDCK cells to soluble hepatocyte growth factor (HGF) in µm- and traditional mm-scale gels, we confirmed that the timing and level of signaling was highly sensitive to diffusion rates of HGF rather than dimensionality. Strikingly, HGF-induced ERK phosphorylation was robustly elevated in µm-scale gels but nearly absent in larger-scale gels, even over the course of hours (Raghavan et al., 2010). Thus, although the effects of diffusion through in vitro 3D tissues are often ignored, limitations in the diffusion of soluble factors might account for some of the differences frequently observed between 2D and 3D settings. These diffusion effects can lead to surprisingly local responses. For example, Nelson and co-workers identified a role for inhibitory morphogens that act in an autocrine manner to dictate the precise spatial location of sprouting in 3D-patterned µm-scale epithelial tubes (Nelson et al., 2006). Similarly, oxygen gradients in 3D tissues – owing to the low solubility of oxygen in aqueous media – can sharply define boundaries of metabolically active cells and areas of hypoxia or cell death (Volkmer et al., 2008).
Control over the spatial presentation of soluble factors throughout the 3D ECM is not only governed by the laws of diffusion. Pressure gradients and macroscale tissue deformations result in directed interstitial fluid flow and convective transport. Indeed, the coupling between mechanical forces and solute transport might be another important means by which mechanical signals can be transduced into cell signaling in three dimensions (Griffith and Swartz, 2006). Furthermore, the ECM not only limits the diffusive and convective transport of soluble factors, but can also actively sequester soluble factors. For example, heparin sulfate proteoglycans have long been known to actively bind growth factors in the ECM and transforming growth factor β (TGF-β) has been shown to have an affinity towards type IV collagen (Vlodavsky et al., 1987; Park et al., 1993; Paralkar et al., 1991). The storage of factors within the ECM is likely to be an essential mechanism by which the timing and spatial presentation of factors to cells is tightly orchestrated. Such factors can then be released through a variety of mechanisms, such as proteolytic degradation of the matrix (Bashkin et al., 1989) or active release through mechanical tugging by cells (Wipff et al., 2007), illustrating not only a means by which cells can control the consumption of ECM-bound factors, but another mechanism in which mechanical and chemical signaling are closely entwined.
Concluding remarks
The field of cell biology is continuously moving towards the dimension, striving to validate 2D findings in a more physiological setting, to develop more robust drug screens and organotypic models, and to keep tissue engineering or regenerative purposes in mind. In all these endeavors, a critical examination of the microenvironment, rather than attributing observed discrepancies to nonspecific effects of dimensionality, will be essential to revealing the specific cues and mechanisms behind a desired effect. Depending on the circumstances, we have learned that there can be dramatically different and often multiple reasons why cell behavior differs between 2D and 3D contexts. Fruitful topics to be examined in the future include how the structure, composition and 3D distribution of adhesive domains alter cell shape and cytoskeletal organization and, in turn, influence cell signaling and function, how passive and active mechanical force transduction in 3D ECMs differs greatly from the static and stiff or linearly elastic mechanics of tissue culture plastic or flat gels, respectively, and how the chemical and physical structure of matrix alters the transport and availability of soluble and bound effectors. Although the factors involved might be numerous, complex and interdependent, as scientists it is our nature as well as our task to bring to light the hidden mechanisms that govern cell behavior.
Applying this scrutinizing outlook to the examples of chondrocyte and mammary epithelial cell culture mentioned above, we see that there are substantial differences in the 3D culture systems that are employed to maintain the differentiated phenotype of chondrocytes compared with mammary epithelial cells, suggesting that distinct underlying mechanisms are involved. Chondrocytes maintain their functionality when embedded within biologically inert agarose gels (Benya and Shaffer, 1982), whereas encapsulation in 3D type I collagen gel accelerates dedifferentiation towards a fibroblastic phenotype (van Susante et al., 1995). The operative feature of encapsulation in 3D was not, as initially thought, the factor that exerted these effects on cell shape (Benya and Shaffer, 1982). Instead, it was the removal of adhesive ligands that resulted in phenotypic maintenance (Mallein-Gerin et al., 1991; Langelier et al., 2000; Woods et al., 2005; Connelly et al., 2007). Thus, monolayer culture is non-physiological for chondrocytes because it promotes an overabundance of adhesion. Moreover, reducing the level of adhesion (whether on a 2D or 3D substrate) is sufficient to restore chondrocyte morphology and function (Glowacki et al., 1983; Woods and Beier, 2006; Gao et al., 2010). By contrast, the formation of mammary acini is highly dependent on the amount of laminin and collagen IV within the matrix (Weaver et al., 1997), the presence of a low-stiffness substrate (Paszek et al., 2005) and apoptotic signals that trigger lumen formation (Debnath et al., 2002). In fact, encapsulating mammary epithelial cells in 3D is not essential for this response, because plating cells on top of basement membrane results in similar morphogenetic events (Debnath et al., 2003). Thus, the vital changes needed to restore normal cell function arise not directly from 3D culture but from important underlying features of select microenvironments that, perhaps initially, were found by happenstance in a particular 3D setting.
In addition to modulating the symphony of adhesive, mechanical and chemical cues that regulate cell function, the 3D context also allows for higher-order phenomena to occur, where the cellular responses themselves are structural in nature. Morphogenesis, tissue remodeling, and cancer cell migration alter the 3D organization of the microenvironment itself and it is difficult, if not impossible, to recapitulate these processes in 2D culture systems (Fig. 1). Even simple processes can assume dramatically different forms in 2D and 3D settings. For instance, migration on 2D surfaces iteratively progresses through several steps: extension of the leading edge, adhesion formation, traction generation and subsequent retraction of the trailing edge (Lauffenburger and Horwitz, 1996; Ridley et al., 2003). In contrast to this rather defined sequence, the way in which cells migrate in 3D depends on the topography, steric hindrance and anisotropic mechanics specific to the fibrous ECM the cells are embedded in, as well as the activation of proteolytic machinery to bore into the matrix or activation of mechanisms needed to squeeze through pores (Doyle et al., 2009; Zaman et al., 2006; Raeber et al., 2005; Wolf et al., 2003). It is, therefore, not surprising that a large variety of migration modes and mechanisms have been described for cells in various 3D environments.
In conclusion, there is a great risk of oversimplifying the differences between cell culture and native tissue environments as being simply a matter of dimensionality. Instead, it is more productive to think of the classic microenvironmental cues that drive cell function – cell adhesions, mechanical forces and diffusible factors – and how they are modulated by different 2D and 3D cultures as compared with in vivo. Through such an understanding, we will be better suited to engineer microenvironments (Box 2) that more accurately capture the in vivo scenario, or even those that can dictate cell function and harness the cell for regenerative purposes. Lastly, a discussion on the cellular microenvironment and its regulation of cell behavior would be incomplete without coming full circle and acknowledging that the origin of the ECM is the cell itself. This two-way interaction between matrix and cells, termed ‘dynamic reciprocity’ (Bissell et al., 1982), leaves us with a humble reminder that our cultures, whether 2D or 3D, are only controlled in terms of the initial condition. The rest is left to the cells themselves.
Supplementary Material
Acknowledgments
The authors thank Sarah Stapleton, Britta Trappmann and Michele Wozniak (University of Pennsylvania, PA) for thoughtful discussions and suggestions.
Footnotes
Funding
This work was supported in part from grants from the NIH [grant numbers EB00262, EB08396, HL73305, GM74048] and Center for Engineering Cells and Regeneration of the University of Pennsylvania. B.M.B. acknowledges financial support from a Ruth L. Kirschstein National Research Service Award. Deposited in PMC for release after 12 months.
This article is part of a Minifocus on Mechanotransduction. For further reading, please see related articles: ‘Finding the weakest link – exploring integrin-mediated mechanical molecular pathways’ by Pere Roca-Cusachs et al. (J. Cell Sci. 125, 3025-3038). ‘Signalling through mechanical inputs – a coordinated process’ by Huimin Zhang and Michel Labouesse (J. Cell Sci. 125, 3039-3049). ‘United we stand – integrating the actin cytoskeleton and cell–matrix adhesions in cellular mechanotransduction’ by Ulrich S. Schwarz and Margaret L. Gardel (J. Cell Sci. 125, 3051-3060). ‘Mechanosensitive mechanisms in transcriptional regulation’ by Akiko Mammoto et al. (J. Cell Sci. 125, 3061-3073). ‘Molecular force transduction by ion channels – diversity and unifying principles’ by Sergei Sukharev and Frederick Sachs (J. Cell Sci. 125, 3075-3083).
References
- Adams R. H., Alitalo K. (2007). Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 8, 464–478 10.1038/nrm2183 [DOI] [PubMed] [Google Scholar]
- Amatangelo M. D., Bassi D. E., Klein–Szanto A. J. P., Cukierman E. (2005). Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am. J. Pathol. 167, 475–488 10.1016/S0002-9440(10)62991-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselme K., Davidson P., Popa A. M., Giazzon M., Liley M., Ploux L. (2010). The interaction of cells and bacteria with surfaces structured at the nanometre scale. Acta Biomater. 6, 3824–3846 10.1016/j.actbio.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Arnold M., Cavalcanti–Adam E. A., Glass R., Blümmel J., Eck W., Kantlehner M., Kessler H., Spatz J. P. (2004). Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem 5, 383–388 10.1002/cphc.200301014 [DOI] [PubMed] [Google Scholar]
- Bashkin P., Doctrow S., Klagsbrun M., Svahn C. M., Folkman J., Vlodavsky I. (1989). Basic fibroblast growth factor binds to subendothelial extracellular matrix and is released by heparitinase and heparin-like molecules. Biochemistry 28, 1737–1743 10.1021/bi00430a047 [DOI] [PubMed] [Google Scholar]
- Beacham D. A., Cukierman E. (2005). Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin. Cancer Biol. 15, 329–341 10.1016/j.semcancer.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Beacham D. A., Amatangelo M. D., Cukierman E. (2007). Preparation of Extracellular Matrices Produced by Cultured and Primary Fibroblasts. Curr. Protoc. Cell Biol. 33, 10.9.1–10.9.21 [DOI] [PubMed] [Google Scholar]
- Beningo K. A., Dembo M., Wang Y. L. (2004). Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc. Natl. Acad. Sci. USA 101, 18024–18029 10.1073/pnas.0405747102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benya P. D., Shaffer J. D. (1982). Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 30, 215–224 10.1016/0092-8674(82)90027-7 [DOI] [PubMed] [Google Scholar]
- Bettinger C. J., Langer R., Borenstein J. T. (2009). Engineering substrate topography at the micro- and nanoscale to control cell function. Angew. Chem. Int. Ed. Engl. 48, 5406–5415 10.1002/anie.200805179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M. J., Hall H. G., Parry G. (1982). How does the extracellular matrix direct gene expression? J. Theor. Biol. 99, 31–68 10.1016/0022-5193(82)90388-5 [DOI] [PubMed] [Google Scholar]
- Bollenbach T., Pantazis P., Kicheva A., Bökel C., González–Gaitán M., Jülicher F. (2008). Precision of the Dpp gradient. Development 135, 1137–1146 10.1242/dev.012062 [DOI] [PubMed] [Google Scholar]
- Brock A., Chang E., Ho C-C., LeDuc P., Jiang X., Whitesides G. M., Ingber D. E. (2003). Geometric determinants of directional cell motility revealed using microcontact printing. Langmuir 19, 1611–1617 10.1021/la026394k [DOI] [PubMed] [Google Scholar]
- Burdick J. A., Anseth K. S. (2002). Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials 23, 4315–4323 10.1016/S0142-9612(02)00176-X [DOI] [PubMed] [Google Scholar]
- Burridge K., Fath K., Kelly T., Nuckolls G., Turner C. (1988). Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu. Rev. Cell Biol. 4, 487–525 10.1146/annurev.cb.04.110188.002415 [DOI] [PubMed] [Google Scholar]
- Chen C. S. (2008). Mechanotransduction - a field pulling together? J. Cell Sci. 121, 3285–3292 10.1242/jcs.023507 [DOI] [PubMed] [Google Scholar]
- Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E. (1997). Geometric control of cell life and death. Science 276, 1425–1428 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- Connelly J. T., García A. J., Levenston M. E. (2007). Inhibition of in vitro chondrogenesis in RGD-modified three-dimensional alginate gels. Biomaterials 28, 1071–1083 10.1016/j.biomaterials.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D. R., Yamada K. M. (2001). Taking cell-matrix adhesions to the third dimension. Science 294, 1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Dalby M. J., Riehle M. O., Sutherland D. S., Agheli H., Curtis A. S. G. (2004). Use of nanotopography to study mechanotransduction in fibroblasts-methods and perspectives. Eur. J. Cell Biol. 83, 159–169 10.1078/0171-9335-00369 [DOI] [PubMed] [Google Scholar]
- Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., Brugge J. S. (2002). The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 10.1016/S0092-8674(02)01001-2 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K., Brugge J. S. (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- DeForest C. A., Polizzotti B. D., Anseth K. S. (2009). Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 8, 659–664 10.1038/nmat2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M., Wang Y-L. (1999). Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76, 2307–2316 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R. A., Gao L., Raghavan S., Liu W. F., Chen C. S. (2009). Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 122, 905–911 10.1242/jcs.028183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A. D., Wang F. W., Matsumoto K., Yamada K. M. (2009). One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 184, 481–490 10.1083/jcb.200810041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. C., Yarmush M. L., Koebe H. G., Tompkins R. G. (1989). Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. FASEB J. 3, 174–177 [DOI] [PubMed] [Google Scholar]
- Emerman J. T., Pitelka D. R. (1977). Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro 13, 316–328 10.1007/BF02616178 [DOI] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L., Discher D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Eyckmans J., Boudou T., Yu X., Chen C. S. (2011). A hitchhiker's guide to mechanobiology. Dev. Cell 21, 35–47 10.1016/j.devcel.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley S. I., Feng Y., Krishnamurthy R., Kim D-H., Celedon A., Longmore G. D., Wirtz D. (2010). A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 10.1038/ncb2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wang Y-K., Yang M. T., Desai R. A., Yu X., Liu Z., Chen C. S. (2010). Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 7, 733–736 10.1038/nmeth.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., McBeath R., Chen C. S. (2010). Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells 28, 564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P., Bershadsky A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Gilbert P. M., Havenstrite K. L., Magnusson K. E. G., Sacco A., Leonardi N. A., Kraft P., Nguyen N. K., Thrun S., Lutolf M. P., Blau H. M. (2010). Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 10.1126/science.1191035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacki J., Trepman E., Folkman J. (1983). Cell shape and phenotypic expression in chondrocytes. Proc. Soc. Exp. Biol. Med. 172, 93–98 [DOI] [PubMed] [Google Scholar]
- Gómez–Sjöberg R., Leyrat A. A., Pirone D. M., Chen C. S., Quake S. R. (2007). Versatile, fully automated, microfluidic cell culture system. Anal. Chem. 79, 8557–8563 10.1021/ac071311w [DOI] [PubMed] [Google Scholar]
- Griffith L. G., Swartz M. A. (2006). Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 7, 211–224 10.1038/nrm1858 [DOI] [PubMed] [Google Scholar]
- Grinnell F. (2003). Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 13, 264–269 10.1016/S0962-8924(03)00057-6 [DOI] [PubMed] [Google Scholar]
- Grinnell F., Ho C-H., Tamariz E., Lee D. J., Skuta G. (2003). Dendritic fibroblasts in three-dimensional collagen matrices. Mol. Biol. Cell 14, 384–395 10.1091/mbc.E02-08-0493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J. B., Bourillot P. Y. (2001). Morphogen gradient interpretation. Nature 413, 797–803 10.1038/35101500 [DOI] [PubMed] [Google Scholar]
- Harunaga J. S., Yamada K. M. (2011). Cell-matrix adhesions in 3D. Matrix Biol. 30, 363–368 10.1016/j.matbio.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S-J., Nerurkar N. L., Baker B. M., Shin J-W., Elliott D. M., Mauck R. L. (2011). Fiber stretch and reorientation modulates mesenchymal stem cell morphology and fibrous gene expression on oriented nanofibrous microenvironments. Ann. Biomed. Eng. 39, 2780–2790 10.1007/s10439-011-0365-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller–Klieser W., Kunz–Schughart L. A. (2010). Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 148, 3–15 10.1016/j.jbiotec.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Hoffman B. D., Grashoff C., Schwartz M. A. (2011). Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. H., Motlekar N. A., Stein A., Diamond S. L., Shore E. M., Mauck R. L. (2008). High-throughput screening for modulators of mesenchymal stem cell chondrogenesis. Ann. Biomed. Eng. 36, 1909–1921 10.1007/s10439-008-9562-4 [DOI] [PubMed] [Google Scholar]
- Huebsch N., Arany P. R., Mao A. S., Shvartsman D., Ali O. A., Bencherif S. A., Rivera–Feliciano J., Mooney D. J. (2010). Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 9, 518–526 10.1038/nmat2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C. S., Postovit L. M., Lajoie G. A. (2010). Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890 10.1002/pmic.200900758 [DOI] [PubMed] [Google Scholar]
- Kay R. R., Langridge P., Traynor D., Hoeller O. (2008). Changing directions in the study of chemotaxis. Nat. Rev. Mol. Cell Biol. 9, 455–463 10.1038/nrm2419-c2 [DOI] [PubMed] [Google Scholar]
- Keely P. J., Fong A. M., Zutter M. M., Santoro S. A. (1995). Alteration of collagen-dependent adhesion, motility, and morphogenesis by the expression of antisense alpha 2 integrin mRNA in mammary cells. J. Cell Sci. 108, 595–607 [DOI] [PubMed] [Google Scholar]
- Khetan S., Burdick J. A. (2010). Patterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogels. Biomaterials 31, 8228–8234 10.1016/j.biomaterials.2010.07.035 [DOI] [PubMed] [Google Scholar]
- Kilian K. A., Bugarija B., Lahn B. T., Mrksich M. (2010). Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 107, 4872–4877 10.1073/pnas.0903269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H. J., Jeon N. L. (2010). Biological applications of microfluidic gradient devices. Integr Biol (Camb) 2, 584–603 10.1039/c0ib00055h [DOI] [PubMed] [Google Scholar]
- Kisiday J., Jin M., Kurz B., Hung H., Semino C., Zhang S., Grodzinsky A. J. (2002). Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc. Natl. Acad. Sci. USA 99, 9996–10001 10.1073/pnas.142309999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Richter B., Striebel T., Franz C. M., von Freymann G., Wegener M., Bastmeyer M. (2011). Two-component polymer scaffolds for controlled three-dimensional cell culture. Adv. Mater. (Deerfield Beach Fla.) 23, 1341–1345 10.1002/adma.201004060 [DOI] [PubMed] [Google Scholar]
- Kleinman H. K., Martin G. R. (2005). Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15, 378–386 10.1016/j.semcancer.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Kloxin A. M., Kasko A. M., Salinas C. N., Anseth K. S. (2009). Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59–63 10.1126/science.1169494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow K. E., Horwitz A. R. (2011). Reducing background fluorescence reveals adhesions in 3D matrices. Nat. Cell Biol. 13, 3–5, author reply 5–7 10.1038/ncb0111-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpinski K., Chu J., Hashi C., Li S. (2006). Anisotropic mechanosensing by mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 103, 16095–16100 10.1073/pnas.0604182103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier E., Suetterlin R., Hoemann C. D., Aebi U., Buschmann M. D. (2000). The chondrocyte cytoskeleton in mature articular cartilage: structure and distribution of actin, tubulin, and vimentin filaments. J. Histochem. Cytochem. 48, 1307–1320 10.1177/002215540004801002 [DOI] [PubMed] [Google Scholar]
- Langer R., Tirrell D. A. (2004). Designing materials for biology and medicine. Nature 428, 487–492 10.1038/nature02388 [DOI] [PubMed] [Google Scholar]
- Langevin H. M., Bouffard N. A., Badger G. J., Iatridis J. C., Howe A. K. (2005). Dynamic fibroblast cytoskeletal response to subcutaneous tissue stretch ex vivo and in vivo. Am. J. Physiol. Cell Physiol. 288, C747–C756 10.1152/ajpcell.00420.2004 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F. (1996). Cell migration: a physically integrated molecular process. Cell 84, 359–369 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Lee E. Y., Parry G., Bissell M. J. (1984). Modulation of secreted proteins of mouse mammary epithelial cells by the collagenous substrata. J. Cell Biol. 98, 146–155 10.1083/jcb.98.1.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H., Moon J. J., West J. L. (2008). Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials 29, 2962–2968 10.1016/j.biomaterials.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant W. R., Miller J. S., Blakely B. L., Cohen D. M., Genin G. M., Chen C. S. (2010). Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 7, 969–971 10.1038/nmeth.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F. T., Csiszar K., Giaccia A., Weninger W.et al. (2009). Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P., Hubbell J. A. (2005). Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55 10.1038/nbt1055 [DOI] [PubMed] [Google Scholar]
- Mahmud G., Campbell C. J., Bishop K. J. M., Komarova Y. A., Chaga O., Soh S., Huda S., Kandere–Grzybowska K., Grzybowski B. A. (2009). Directing cell motions on micropatterned ratchets. Nat. Phys. 5, 606–612 10.1038/nphys1306 [DOI] [Google Scholar]
- Mallein–Gerin F., Garrone R., van der Rest M. (1991). Proteoglycan and collagen synthesis are correlated with actin organization in dedifferentiating chondrocytes. Eur. J. Cell Biol. 56, 364–373 [PubMed] [Google Scholar]
- Manjón C., Sánchez–Herrero E., Suzanne M. (2007). Sharp boundaries of Dpp signalling trigger local cell death required for Drosophila leg morphogenesis. Nat. Cell Biol. 9, 57–63 10.1038/ncb1518 [DOI] [PubMed] [Google Scholar]
- Mann B. K., Gobin A. S., Tsai A. T., Schmedlen R. H., West J. L. (2001). Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials 22, 3045–3051 10.1016/S0142-9612(01)00051-5 [DOI] [PubMed] [Google Scholar]
- Maskarinec S. A., Franck C., Tirrell D. A., Ravichandran G. (2009). Quantifying cellular traction forces in three dimensions. Proc. Natl. Acad. Sci. USA 106, 22108–22113 10.1073/pnas.0904565106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata A., Hsu L., Capito R., Aparicio C., Henrikson K., Stupp S. I. (2009). Micropatterning of bioactive self-assembling gels. Soft Matter 5, 1228–1236 10.1039/b819002j [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- Meyers J., Craig J., Odde D. J. (2006). Potential for control of signaling pathways via cell size and shape. Curr. Biol. 16, 1685–1693 10.1016/j.cub.2006.07.056 [DOI] [PubMed] [Google Scholar]
- Miller J. S., Shen C. J., Legant W. R., Baranski J. D., Blakely B. L., Chen C. S. (2010). Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials 31, 3736–3743 10.1016/j.biomaterials.2010.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mseka T., Bamburg J. R., Cramer L. P. (2007). ADF/cofilin family proteins control formation of oriented actin-filament bundles in the cell body to trigger fibroblast polarization. J. Cell Sci. 120, 4332–4344 10.1242/jcs.017640 [DOI] [PubMed] [Google Scholar]
- Mueller–Klieser W. (1997). Three-dimensional cell cultures: from molecular mechanisms to clinical applications. Am. J. Physiol. 273, C1109–C1123 [DOI] [PubMed] [Google Scholar]
- Nathan A. S., Baker B. M., Nerurkar N. L., Mauck R. L. (2011). Mechano-topographic modulation of stem cell nuclear shape on nanofibrous scaffolds. Acta Biomater. 7, 57–66 10.1016/j.actbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M., Vanduijn M. M., Inman J. L., Fletcher D. A., Bissell M. J. (2006). Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 314, 298–300 10.1126/science.1131000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklason L. E., Yeh A. T., Calle E. A., Bai Y., Valentín A., Humphrey J. D. (2010). Enabling tools for engineering collagenous tissues integrating bioreactors, intravital imaging, and biomechanical modeling. Proc. Natl. Acad. Sci. USA 107, 3335–3339 10.1073/pnas.0907813106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A. W., Helmke B. P., Blackman B. R., Schwartz M. A. (2006). Mechanisms of mechanotransduction. Dev. Cell 10, 11–20 10.1016/j.devcel.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Otrock Z. K., Mahfouz R. A. R., Makarem J. A., Shamseddine A. I. (2007). Understanding the biology of angiogenesis: review of the most important molecular mechanisms. Blood Cells Mol. Dis. 39, 212–220 10.1016/j.bcmd.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Paralkar V. M., Vukicevic S., Reddi A. H. (1991). Transforming growth factor beta type 1 binds to collagen IV of basement membrane matrix: implications for development. Dev. Biol. 143, 303–308 10.1016/0012-1606(91)90081-D [DOI] [PubMed] [Google Scholar]
- Park J. E., Keller G. A., Ferrara N. (1993). The vascular endothelial growth factor (VEGF) isoforms: differential deposition into the subepithelial extracellular matrix and bioactivity of extracellular matrix-bound VEGF. Mol. Biol. Cell 4, 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Lee E. Y-H., Farson D., Koval M., Bissell M. J. (1985). Collagenous substrata regulate the nature and distribution of glycosaminoglycans produced by differentiated cultures of mouse mammary epithelial cells. Exp. Cell Res. 156, 487–499 10.1016/0014-4827(85)90556-7 [DOI] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart–King C. A., Margulies S. S., Dembo M., Boettiger D.et al. (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pathak A., Kumar S. (2011). Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 3, 267–278 10.1039/c0ib00095g [DOI] [PubMed] [Google Scholar]
- Petersen O. W., Rønnov–Jessen L., Howlett A. R., Bissell M. J. (1992). Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA 89, 9064–9068 10.1073/pnas.89.19.9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager–Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A. D. (2011). Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 10.1038/ncb2370 [DOI] [PubMed] [Google Scholar]
- Provenzano P. P., Eliceiri K. W., Campbell J. M., Inman D. R., White J. G., Keely P. J. (2006). Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quake S. R., Scherer A. (2000). From micro- to nanofabrication with soft materials. Science 290, 1536–1540 10.1126/science.290.5496.1536 [DOI] [PubMed] [Google Scholar]
- Raeber G. P., Lutolf M. P., Hubbell J. A. (2005). Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys. J. 89, 1374–1388 10.1529/biophysj.104.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S., Shen C. J., Desai R. A., Sniadecki N. J., Nelson C. M., Chen C. S. (2010). Decoupling diffusional from dimensional control of signaling in 3D culture reveals a role for myosin in tubulogenesis. J. Cell Sci. 123, 2877–2883 10.1242/jcs.055079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujan S., Pluen A., McKee T. D., Brown E. B., Boucher Y., Jain R. K. (2002). Diffusion and convection in collagen gels: implications for transport in the tumor interstitium. Biophys. J. 83, 1650–1660 10.1016/S0006-3495(02)73933-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räsänen K., Vaheri A. (2010). Activation of fibroblasts in cancer stroma. Exp. Cell Res. 316, 2713–2722 10.1016/j.yexcr.2010.04.032 [DOI] [PubMed] [Google Scholar]
- Reinhart–King C. A., Dembo M., Hammer D. A. (2005). The dynamics and mechanics of endothelial cell spreading. Biophys. J. 89, 676–689 10.1529/biophysj.104.054320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (1987). Integrins: a family of cell surface receptors. Cell 48, 549–554 10.1016/0092-8674(87)90233-9 [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Saha K., Schaffer D. V. (2006). Signal dynamics in Sonic hedgehog tissue patterning. Development 133, 889–900 10.1242/dev.02254 [DOI] [PubMed] [Google Scholar]
- Salaita K., Nair P. M., Petit R. S., Neve R. M., Das D., Gray J. W., Groves J. T. (2010). Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science 327, 1380–1385 10.1126/science.1181729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi R., Kumar A., Lopez G. P., Stephanopoulos G. N., Wang D. I., Whitesides G. M., Ingber D. E. (1994). Engineering cell shape and function. Science 264, 696–698 10.1126/science.8171320 [DOI] [PubMed] [Google Scholar]
- van Susante J. L., Buma P., van Osch G. J., Versleyen D., van der Kraan P. M., van der Berg W. B., Homminga G. N. (1995). Culture of chondrocytes in alginate and collagen carrier gels. Acta Orthop. Scand. 66, 549–556 10.3109/17453679509002314 [DOI] [PubMed] [Google Scholar]
- Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., Chen C. S. (2003). Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA 100, 1484–1489 10.1073/pnas.0235407100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A. I., Abrams G. A., Bertics P. J., Murphy C. J., Nealey P. F. (2003). Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 116, 1881–1892 10.1242/jcs.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M. (2010). Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 123, 4201–4213 10.1242/jcs.075150 [DOI] [PubMed] [Google Scholar]
- Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J-B., Bornens M. (2006). Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 103, 19771–19776 10.1073/pnas.0609267103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Jiménez–Dalmaroni A., Racine V., Bornens M., Jülicher F. (2007). Experimental and theoretical study of mitotic spindle orientation. Nature 447, 493–496 10.1038/nature05786 [DOI] [PubMed] [Google Scholar]
- Thomas C. H., Collier J. H., Sfeir C. S., Healy K. E. (2002). Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA 99, 1972–1977 10.1073/pnas.032668799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton M. L., Gilchrist C. L., Guilak F., Setton L. A. (2008). Transfer of macroscale tissue strain to microscale cell regions in the deformed meniscus. Biophys. J. 95, 2116–2124 10.1529/biophysj.107.126938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I., Folkman J., Sullivan R., Fridman R., Ishai–Michaeli R., Sasse J., Klagsbrun M. (1987). Endothelial cell-derived basic fibroblast growth factor: synthesis and deposition into subendothelial extracellular matrix. Proc. Natl. Acad. Sci. USA 84, 2292–2296 10.1073/pnas.84.8.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E., Drosse I., Otto S., Stangelmayer A., Stengele M., Kallukalam B. C., Mutschler W., Schieker M. (2008). Hypoxia in static and dynamic 3D culture systems for tissue engineering of bone. Tissue Eng. Part A 14, 1331–1340 10.1089/ten.tea.2007.0231 [DOI] [PubMed] [Google Scholar]
- von der Mark K., Gauss V., von der Mark H., Müller P. (1977). Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature 267, 531–532 10.1038/267531a0 [DOI] [PubMed] [Google Scholar]
- Weaver V. M., Petersen O. W., Wang F., Larabell C. A., Briand P., Damsky C., Bissell M. J. (1997). Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 137, 231–245 10.1083/jcb.137.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver V. M., Lelièvre S., Lakins J. N., Chrenek M. A., Jones J. C., Giancotti F., Werb Z., Bissell M. J. (2002). β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216 10.1016/S1535-6108(02)00125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. F., Bjerke M. A., DeSimone D. W. (2011). Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 124, 1183–1193 10.1242/jcs.064618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M., Ostuni E., Takayama S., Jiang X., Ingber D. E. (2001). Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 3, 335–373 10.1146/annurev.bioeng.3.1.335 [DOI] [PubMed] [Google Scholar]
- Wipff P-J., Rifkin D. B., Meister J-J., Hinz B. (2007). Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J. Cell Biol. 179, 1311–1323 10.1083/jcb.200704042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U. H., Deryugina E. I., Strongin A. Y., Bröcker E-B., Friedl P. (2003). Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160, 267–277 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Beier F. (2006). RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J. Biol. Chem. 281, 13134–13140 10.1074/jbc.M509433200 [DOI] [PubMed] [Google Scholar]
- Woods A., Wang G., Beier F. (2005). RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem. 280, 11626–11634 10.1074/jbc.M409158200 [DOI] [PubMed] [Google Scholar]
- Wozniak M. A., Modzelewska K., Kwong L., Keely Patricia. J. (2004). Focal adhesion regulation of cell behavior. BBA Mol. Cell Res., 1692, 103–119 10.1016/j.bbamcr.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Yu H., Alexander C. M., Beebe D. J. (2007). Understanding microchannel culture: parameters involved in soluble factor signaling. Lab Chip 7, 726–730 10.1039/b618793e [DOI] [PubMed] [Google Scholar]
- Zaman M. H., Trapani L. M., Sieminski A. L., Mackellar D., Gong H., Kamm R. D., Wells A., Lauffenburger D. A., Matsudaira P. (2006). Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 103, 10889–10894 10.1073/pnas.0604460103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. (2003). Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 21, 1171–1178 10.1038/nbt874 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.