Summary
PINCH, integrin-linked kinase (ILK) and Ras suppressor-1 (RSU-1) are molecular scaffolding proteins that form a physical complex downstream of integrins, and have overlapping roles in cellular adhesion. In Drosophila, PINCH and ILK colocalize in cells and have indistinguishable functions in maintaining wing adhesion and integrin to actin linkage in the muscle. We sought to determine whether the direct physical interaction between PINCH and ILK was essential for their functions using transgenic flies expressing a version of PINCH with a point mutation that disrupts ILK binding (PINCHQ38A). We demonstrate that the PINCH-ILK interaction is not required for viability, for integrin-mediated adhesion of the wing or muscle, or for maintaining appropriate localization or levels of either PINCH or ILK. These results suggest alternative modes for PINCH localization, stabilization and linkage to the actin cytoskeleton that are independent of a direct interaction with ILK. Furthermore, we identified a synthetic lethality in flies carrying both the PINCHQ38A mutation and a null mutation in the gene encoding RSU-1. This lethality does not result from PINCH mislocalization or destabilization, and illustrates a novel compensatory role for RSU-1 in maintaining viability in flies with compromised PINCH-ILK binding. Taken together, this work highlights the existence of redundant mechanisms in adhesion complex assembly that support integrin function in vivo.
Key words: Drosophila, Integrin-linked kinase, PINCH, Ras suppressor-1, Adhesion, Integrins
Introduction
Integrin-mediated adhesion is essential for proper development and for the maintenance of adult tissues (Bökel and Brown, 2002; Brown et al., 2002; Brower, 2003). Integrin receptors regulate cellular adhesion by mediating communication between the extracellular matrix and the cell interior (Hynes, 2002). The association of the intracellular domains of integrins with cytoplasmic protein partners influences signal transduction cascades and the actin cytoskeleton to control cell adhesion (Brakebusch and Fässler, 2003; Delon and Brown, 2007). Understanding the functions and contributions of the protein complexes that link integrins to the actin cytoskeleton is fundamental to understanding how cellular adhesion is regulated.
In Drosophila, loss of integrin function results in perturbation of cell adhesion in vitro and in vivo (Jani and Schöck, 2007; Pines et al., 2011). Genetic analysis has revealed that several cytoplasmic factors, including PINCH (particularly interesting cysteine-histidine-rich protein) and ILK (integrin-linked kinase), are required to support integrin-mediated adhesion in vivo (Prout et al., 1997; Walsh and Brown, 1998; Zervas et al., 2001; Löer et al., 2008). Null mutations that eliminate either PINCH (encoded by the steamer duck gene, stck) or ILK (encoded by the ilk gene) result in late embryonic or early larval lethality, and homozygous mutant clones of either gene in the wing result in wing blisters similar to what is observed when integrin function is compromised (Brower and Jaffe, 1989; Brabant et al., 1996; Zervas et al., 2001; Clark et al., 2003). PINCH and ILK colocalize at muscle attachment sites in the Drosophila embryo and both proteins are required to maintain the attachment of actin filaments to integrin-rich membrane domains (Zervas et al., 2001; Clark et al., 2003). In contrast, other integrin effectors, including rhea (Talin) and wech, are required for integrin-dependent anchorage of the cell membrane to the extracellular matrix (Brown et al., 2002; Löer et al., 2008). In the mouse, targeted gene disruption of either Pinch or Ilk results in embryonic lethality at the peri-implantation stage. Embryoid bodies derived from Pinch and Ilk null embryos display abnormal epiblast polarity, impaired cavitation, and detachment of the endoderm and epiblast from the basement membrane (Sakai et al., 2003; Li et al., 2005).
Several lines of investigation have led to the idea that PINCH and ILK operate as a functional unit to tether actin filaments to integrin-rich membranes domains and support adhesion. First, the N-terminal LIM domain of PINCH interacts directly with the N-terminal ankyrin repeat domain (ANKR) of ILK (Li et al., 1999; Tu et al., 1999). Second, depletion of either ILK or PINCH results in reduction in the levels of the other, indicating a mutual stabilization of these two proteins (Fukuda et al., 2003; Stanchi et al., 2009; Meder et al., 2011). Third, targeted disruption of the interaction between PINCH and ILK in mammalian cell culture experiments by a point mutation in LIM1 of PINCH results in disturbances in cell spreading and survival, as well as reduced stability of both PINCH and ILK (Velyvis et al., 2001; Zhang et al., 2002; Xu et al., 2005). Fourth, ILK is required for localizing PINCH at integrin-rich sites (Zervas et al., 2011). Parvin, which binds both to ILK and to Actin, is often included in this functional complex. The ILK-PINCH-Parvin complex may provide a mechanism for direct coupling of integrins to the actin cytoskeleton (Tu et al., 2001).
Ras suppressor-1 (RSU-1) is also recovered in a complex with PINCH, ILK, and Parvin (Kadrmas et al., 2004). RSU-1 was first identified in a screen for genes whose expression suppressed Ras-dependent oncogenic transformation in mammalian cells (Cutler et al., 1992). RSU-1 interacts directly with LIM5 of PINCH (Kadrmas et al., 2004; Dougherty et al., 2005) and cooperates with PINCH to regulate JNK signaling in Drosophila (Kadrmas et al., 2004). RSU-1 is encoded by the icarus (ics) gene in Drosophila (Kadrmas et al., 2004). ics null flies are viable and fertile, but display wing blisters characteristic of a failure of integrin-dependent adhesion, illustrating a role for RSU-1 in adhesion processes that also depend upon PINCH and ILK (Kadrmas et al., 2004).
Although the data from both vertebrate and invertebrate systems largely support the idea that PINCH-ILK complexes are critical for cell adhesion, protein localization, and protein stability, some recent findings suggest independent roles for PINCH and ILK. First, while the phenotypes of mice with targeted gene disruptions in Pinch or Ilk are similar, they are not identical. The Pinch null mouse survives slightly longer (E6.5–E7.5) than the Ilk null mouse (E5.5–E6.5). Furthermore, Pinch null embryoid bodies display additional defects in cell-cell adhesion of the endoderm and the epiblast and contain apoptotic cells within the endodermal layer that are not seen in embryoid bodies derived from Ilk null embryos (Sakai et al., 2003; Li et al., 2005). Genetic studies in Drosophila also support the view that ILK and PINCH, though performing many common functions, have some unique and independent roles. For example, ILK accumulation at muscle attachment sites is compromised in wech mutants whereas PINCH localization is reported to be undisturbed, raising the possibility that PINCH is not completely dependent on ILK for its appropriate subcellular localization (Löer et al., 2008). Consistent with this view, a PINCH variant that lacks LIM1 and cannot bind ILK (or perform any other putative LIM1-dependent functions), retains some capacity to localize to muscle attachment sites (Zervas et al., 2011).
Despite the work by many labs demonstrating that PINCH and ILK are required for integrin-mediated adhesion, controversy exists regarding how they contribute to this critical cell behavior. In particular, as highlighted above, the literature contains conflicting conclusions regarding the question of whether or not a direct association of PINCH and ILK is required to carry out their functions. Here, we have endeavored to resolve this controversy and extend our understanding of the key requirements for PINCH-ILK function in integrin-dependent adhesion by testing directly the importance of the PINCH-ILK interaction in Drosophila. We report that a direct association between PINCH and ILK is not required for viability or integrin-dependent adhesion in the muscle or wing. However, in ics mutants that lack RSU-1, the inability of PINCH to bind ILK results in synthetic lethality. Our work suggests that redundancy exists within adhesion complexes to preserve integrin-mediated adhesion in vivo.
Results
A conserved glutamine residue in LIM1 of PINCH is required for ILK binding
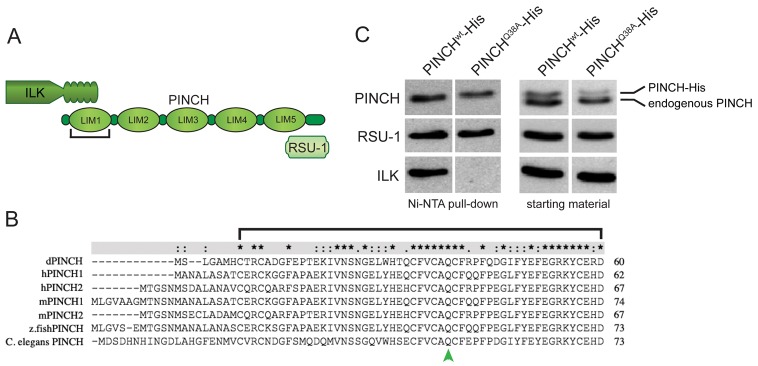
PINCH-ILK complexes are essential for integrin-dependent cell adhesion of cultured cells, however this has not been demonstrated definitively within an intact organism. To test whether the PINCH-ILK interaction is required for integrin-mediated cell adhesion in vivo, we established a system for specific disruption of PINCH-ILK binding in Drosophila. PINCH is comprised of five tandemly arrayed LIM domains, with a short N-terminus and a C-terminal tail (Fig. 1A). The ILK N-terminal ANKR domain interacts with LIM1 of PINCH. Sequence alignment of PINCH proteins from Drosophila, human, mouse, zebrafish, and C. elegans reveals the high degree of sequence conservation of the N-terminal LIM domain of PINCH (LIM1) (Fig. 1B). In mammalian cells, a variant of PINCH in which a universally conserved amino acid (glutamine 40) is mutated to alanine (Q40A) does not bind to ILK, fails to localize at focal adhesions, and causes defects in cell spreading and adhesion (Zhang et al., 2002; Xu et al., 2005). We engineered and expressed Drosophila PINCHwt-His or PINCHQ38A-His (the fly mutation corresponding to PINCHQ40A) in Drosophila S2 cells, captured His-tagged proteins with their partners under native conditions using Ni-NTA agarose, and performed western blots for known binding partners. PINCHwt-His was associated with the LIM5 partner, RSU-1, and the LIM1 partner ILK (Fig. 1A,C). In contrast, PINCHQ38A-His was associated with RSU-1, but not ILK (Fig. 1C). The demonstration that PINCHQ38A-His retains the capacity to interact with RSU-1 provides evidence for conformational integrity of PINCHQ38A and illustrates a critical role of glutamine 38 in specifying a binding interface for ILK.
Fig. 1.
Drosophila PINCHQ38A is a mutation that disrupts the interaction with ILK. (A) PINCH schematic demonstrating the five LIM domain structure with a short N-terminus and a C-terminal tail. LIM1 binds to the N-terminal ANKR domain of ILK, and LIM5 binds to RSU-1. (B) Sequence alignment of the N-terminus through LIM1 of various PINCH sequences. The conserved glutamine is marked by a green arrowhead. Asterisks indicate positions which have a single, fully conserved residue, whereas colon (:) and full point (.) indicate strong and weaker conservation defined by ClustalX. (C) Ni-NTA pull-down of PINCHwt-His and PINCHQ38A-His expressed in S2 cells. Western blots of purified proteins were probed with antibodies raised against PINCH, RSU-1 and ILK. PINCHQ38A-His does not show any detectible binding to ILK, but is still able to bind to RSU-1.
The interaction between PINCH and ILK is not required for viability in Drosophila
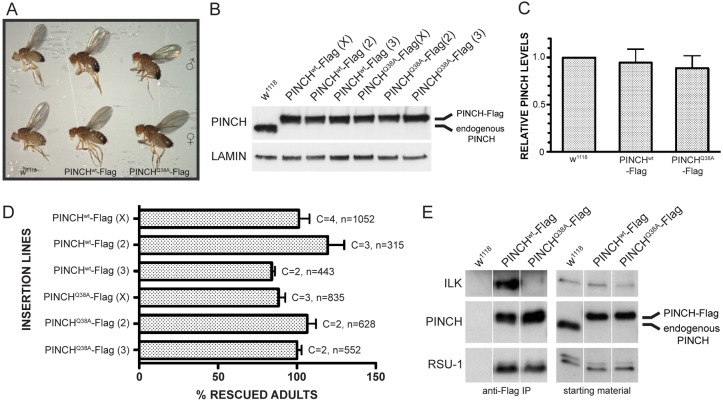
In order to evaluate the functional consequences of disrupting the PINCH-ILK interaction in vivo, we generated transgenic flies carrying either PINCHwt-Flag or PINCHQ38A-Flag under control of the endogenous stck promoter. No dominant phenotypes were observed when either PINCHwt-Flag or PINCHQ38A-Flag was expressed in otherwise wild-type flies (data not shown). We introduced either the PINCHwt-Flag or PINCHQ38A-Flag transgenes into embryos that lack zygotic PINCH expression to determine whether they could rescue the defects observed in stck null mutants. As expected, the PINCHwt-Flag transgene rescues the stck null mutant phenotypes, including both the embryonic/early larval lethality observed with complete loss of PINCH and the adult wing blisters caused by specific loss of PINCH in the wing (Fig. 2A). Expression of the PINCHQ38A-Flag transgene also rescues the lethality of the stck null mutant, indicating that the interaction between PINCH and ILK is not required for viability (Fig. 2A). Given that loss of either PINCH or ILK specifically in the wing results in blisters, it was possible that this phenotype would be observed in flies where the PINCH-ILK interaction was disrupted. However, we did not observe wing blisters in adult rescued PINCHQ38A flies, indicating that the PINCH-ILK interaction is not required to maintain adhesion of the adult wing (Fig. 2A). PINCHQ38A-Flag rescued flies are fertile, and stocks were established and maintained for future experiments, indicating that the maternal contribution of endogenous PINCH was not responsible for the observed rescue.
Fig. 2.
PINCHwt-Flag and PINCHQ38A-Flag transgenes rescue the lethality of the stck null mutant. (A) Adult rescued flies do not have wing blisters or any other overt phenotypes. PINCHwt-Flag and PINCHQ38A-Flag adult flies appear similar to each other, and to w1118 controls. (B) Western blots of adult rescued flies probed with anti-PINCH antibody show a lack of endogenous PINCH and only show the higher migrating PINCH-Flag species. Three different insertion lines are shown for both PINCHwt-Flag and PINCHQ38A-Flag on the X, 2nd and 3rd chromosomes. Anti-Lamin antibody is used as a loading control. (C) PINCHwt-Flag and PINCHQ38A-Flag transgenic protein levels are similar to levels of endogenous PINCH in wild-type flies. Bars represent the median ± range of transgenic PINCH from three different chromosomal insertions for both PINCHwt-Flag and PINCHQ38A-Flag rescued flies. (D) PINCHwt-Flag transgenes rescue 84–120% and PINCHQ38A-Flag transgenes rescue 88–107%. Bars represent the mean ± s.e.m. of the percentage rescued flies counted for the number of crosses set. C denotes the number of times the rescue cross was set. n is the total number of flies counted across all crosses. (E) Anti-Flag immunoprecipitations confirm disruption of the PINCH-ILK interaction in adult PINCHQ38A-Flag rescued flies. PINCHwt-Flag complexes contain both ILK and RSU-1, whereas PINCHQ38A-Flag complexes contain only RSU-1. w1118 lysate was used as a control to assess nonspecific binding. All proteins were present in the starting material.
We confirmed rescue of the stck null mutant in three independent insertion lines for both PINCHwt-Flag and PINCHQ38A-Flag transgenes by western blot and observed in each case the loss of endogenous PINCH expression and the presence of the Flag-tagged PINCH variant, which migrated at a higher molecular mass in adult rescued flies (Fig. 2B). Furthermore, PINCHwt-Flag and PINCHQ38A-Flag transgenes are expressed at comparable levels to endogenous PINCH (Fig. 2C). To evaluate whether there were subtle differences in developmental fitness between rescue with PINCHwt-Flag and PINCHQ38A-Flag transgenes, a quantitative analysis was performed to assess the numbers of viable progeny resulting from each rescue cross. The mean percent rescue was calculated for three different insertion lines of PINCHwt-Flag and PINCHQ38A-Flag, where each rescue cross was repeated two-four times (Fig. 2D). We observe rescue between 84–120% of expected values for all lines tested and no statistically significant difference was observed between rescue with the PINCHwt-Flag and PINCHQ38A-Flag transgenes. The lack of interaction between PINCHQ38A-Flag and ILK was confirmed by native immunoprecipitations using lysates derived from rescued transgenic fly lines (Fig. 2E). Importantly, although PINCHQ38A-Flag fails to recruit ILK in vivo, it does recruit the LIM5 binding partner, RSU-1, providing biochemical confirmation that PINCH protein structure is preserved.
The PINCH-ILK interaction is not required for establishment or maintenance of muscle cytoarchitecture
Because both PINCH and ILK have been shown to be required for muscle structure and function in Drosophila (Zervas et al., 2001; Clark et al., 2003), we performed high-resolution fluorescence microscopy to evaluate the integrity of actin-membrane linkages in PINCHQ38A-Flag rescued flies. The Drosophila embryonic somatic muscle consists of segments of muscle fibers, where two muscle segments meet specialized tendon cells that are part of the epidermis (Bökel and Brown, 2002; Brower, 2003; Schweitzer et al., 2010). The junction of these two cell types is maintained by β-integrin dependent adhesion to extracellular matrix that is deposited between muscle and tendon cells. PINCHwt-Flag and PINCHQ38A-Flag rescued embryos were stained with anti-β-integrin antibody to label adhesion sites at the muscle ends and with Phalloidin to label the actin cytoskeleton (Fig. 3). We observe strong and coherent localization of β-integrin at the myotendinous junction (Fig. 3A,C,D,F), with a normal distribution of actin fibers extending between these sites (Fig. 3B,C,E,F) in both PINCHwt-Flag and PINCHQ38A-Flag rescued embryos, indicating that disruption of the PINCH-ILK interaction is not sufficient to alter muscle structure or stability of the actin cytoskeleton in the Drosophila embryo.
Fig. 3.
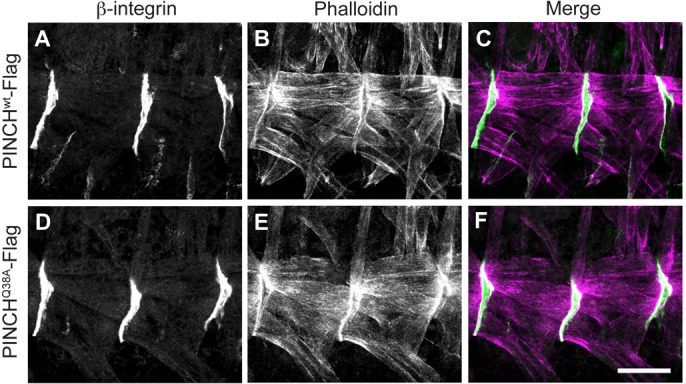
Muscle structure is preserved in PINCHQ38A-Flag rescued embryos. Stage 16–17 embryos were stained with anti-β-integrin antibody (A,C,D,F) and Phalloidin (B,C,E,F) to mark the muscle attachment site (green) and the actin cytoskeleton (magenta). β-integrin localizes normally to the muscle cell membrane and Phalloidin staining indicates that the actin cytoskeleton is stable and that the muscle fibers are properly distributed between muscle attachment sites in both PINCHwt-Flag rescued embryos (A–C) and in PINCHQ38A-Flag rescued embryos (D–F). Scale bar: 20 µm.
PINCH and ILK have an independent capacity to localize at muscle attachment sites in vivo
Prior work in cultured mammalian cells suggested that disruption of the PINCH-ILK interaction compromises the ability of PINCH to accumulate at integrin rich adhesion sites (Zhang et al., 2002). Therefore, we tested whether the PINCHQ38A-Flag rescue of the stck null mutant was occurring independent of the ability of PINCHQ38A-Flag to localize at muscle attachment sites. PINCHwt-Flag and PINCHQ38A-Flag rescued embryos were double labeled with anti-Flag antibody to detect transgenic PINCH and anti-myosin heavy chain (MHC) antibody to label body wall muscle. Both PINCHwt-Flag and PINCHQ38A-Flag display robust localization at muscle termini in the Drosophila muscle (Fig. 4A,B). At higher resolution, it is clear that both PINCHwt-Flag and PINCHQ38A-Flag colocalize with the LIM5 binding partner RSU-1 at muscle attachment sites (Fig. 4C–H). These findings illustrate that the PINCH-ILK interaction is not required for appropriate subcellular localization of PINCH or RSU-1 in vivo, and highlight the importance of identifying other factors that contribute to the targeting of PINCH to sites of adhesion.
Fig. 4.
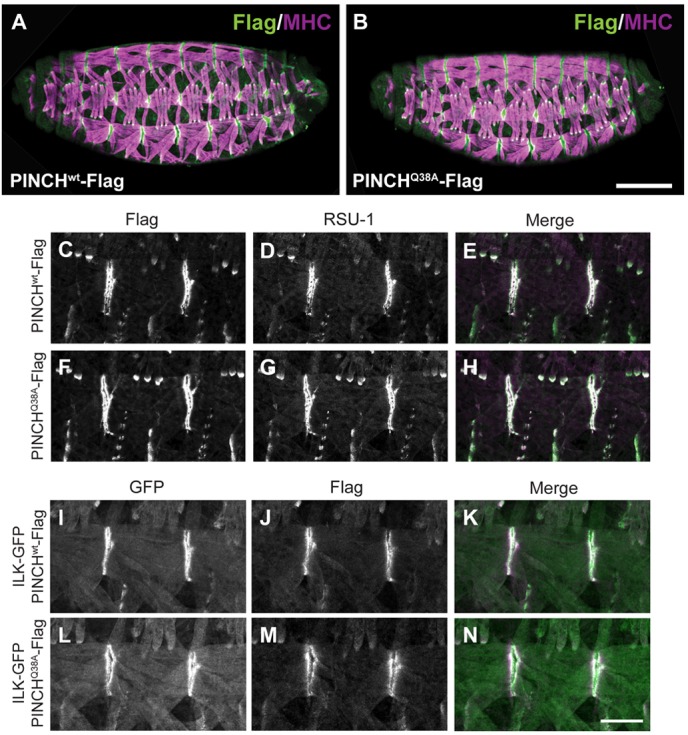
PINCH, RSU-1, and ILK are properly localized in PINCHQ38A-Flag rescued embryos. (A,B) Stage 16-17 embryos were stained with anti-Flag antibody to label PINCH transgenic protein (green) and anti-Myosin Heavy Chain (MHC) to label the embryonic body wall muscle (magenta). PINCHwt-Flag and PINCHQ38A-Flag localization and muscle structure are normal throughout the embryo. Scale bar: 50 µm. (C–H) Stage 16-17 embryos stained with anti-Flag antibody (green) to label PINCH transgenic protein (C,E,F,H) and anti-RSU-1 antibody (magenta) (D,E,G,H) show that both proteins localize in the absence of a PINCH-ILK interaction. Colocalization appears white in the merge (E,H). (I–N) Stage 16-17 embryos stained with anti-GFP antibody to label ILK-GFP (green) (I,K,L,N) and anti-Flag antibody to label PINCH transgenic protein (magenta) (J,K,M,N) show that PINCH and ILK colocalize in the absence of their direct interaction. Colocalization appears white in the merge (K,N). Scale bar: 20 µm.
Given that PINCHQ38A-Flag localizes to muscle attachment sites independently of an interaction with ILK, we conducted a complementary experiment to evaluate whether proper ILK localization occurs in PINCHQ38A-Flag rescued embryos. We introduced an ILK-GFP transgene into PINCHwt-Flag and PINCHQ38A-Flag rescued flies and observed that ILK-GFP does indeed accumulate at muscle attachment sites (Fig. 4I,K,L,N), colocalizing with both PINCHwt-Flag and PINCHQ38A-Flag (Fig. 4J,K,M,N). These results confirm that ILK localization is not dependent on direct binding to PINCH in Drosophila muscle.
The PINCH-ILK interaction is not required to stabilize PINCH or ILK protein levels
In cell culture, reductions in both ILK and PINCH protein levels are observed when either ILK or PINCH levels are depleted either by RNA interference or targeted gene disruption (Fukuda et al., 2003; Xu et al., 2005; Stanchi et al., 2009). A recent publication has also demonstrated mutual stabilization of PINCH and ILK in vivo using antisense morpholinos in zebrafish (Meder et al., 2011). We have tested whether this stabilization phenomenon occurs in Drosophila by examining the levels of PINCH-ILK complex constituents in late stage stck and ilk null embryos. We found that PINCH protein levels are reduced to 34% of wild-type levels in ilk1 mutant embryos and that ILK protein levels are reduced to 12% of wild-type levels in stck17/18 m/z mutant embryos (Fig. 5A,B). In agreement with other studies, our findings demonstrate that PINCH and ILK are required for their mutual stabilization in vivo and across species. We extended the analysis of protein stability to include the PINCH binding partner, RSU-1, as well as βPS-integrin. In agreement with previously published work in mid-stage Drosophila embryos (Kadrmas et al., 2004), we demonstrate that PINCH and RSU-1 mutually stabilize each other (Fig. 5A,B). We observe a reduction of PINCH protein in ics null embryos (67% of wild-type levels) compared with a more dramatic reduction of PINCH protein in ilk null embryos (34% of wild-type levels). Notably, we observe a similarly dramatic reduction of ILK protein levels in both stck and ics null embryos (12% and 24% of wild-type levels, respectively), indicating that RSU-1 also has the capacity to stabilize ILK protein levels (Fig. 5A,B). We conclude that within the ILK–PINCH–RSU-1 complex, loss of any individual protein can affect the levels of the other two members to varying degrees. By contrast, the levels of βPS-integrin in all mutants examined are comparable to the levels observed in wild-type embryos, indicating that the stability of this protein complex is independent of the total levels of the receptor.
Fig. 5.
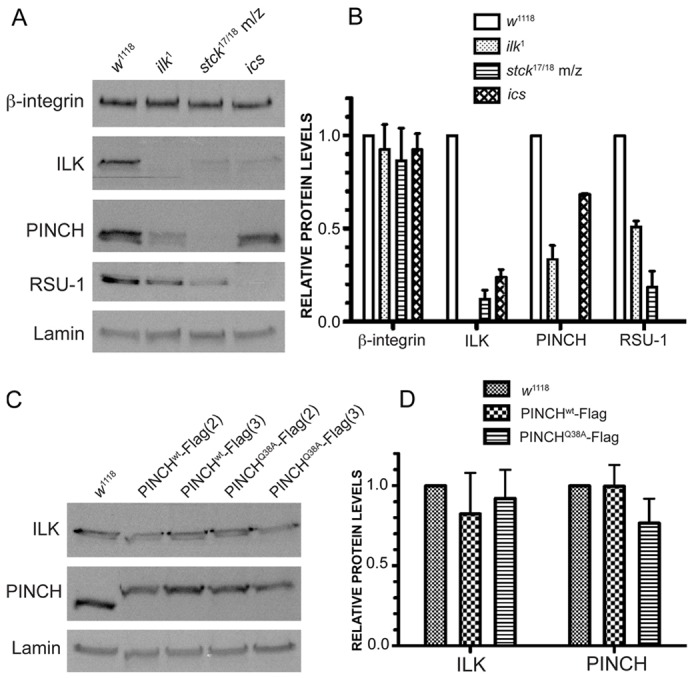
PINCHQ38A-Flag rescued embryos do not exhibit the same decreases in adhesion complex protein stability as null mutant embryos. (A) Stage 16–17 null embryos of the genotypes indicated were collected and Western blots were probed with antibodies raised against β-integrin, ILK, PINCH, RSU-1 and Lamin. (B) Protein levels for the genotypes indicated were quantified and are represented as a fraction of wild-type expression. Bars represent the mean ± range for two replicate experiments. (C) Stage 16–17 embryos were collected for two insertion lines each (2nd and 3rd chromosome) from PINCHwt-Flag and PINCHQ38A-Flag rescued stocks. western blots were probed with antibodies raised against PINCH, ILK and Lamin. (D) Levels of PINCH and ILK in PINCHwt-Flag and PINCHQ38A-Flag rescued embryos are similar to each other and to wild-type levels. Bars represent the median ± range of transgenic PINCH from three different chromosomal insertions for both PINCHwt-Flag and PINCHQ38A-Flag rescued flies.
It has been demonstrated that the physical interaction of PINCH with ILK is critical for their mutual stability in cell culture (Xu et al., 2005). To test directly whether the ability of PINCH to bind ILK is required for their stability in vivo, we performed western blots of PINCHwt-Flag and PINCHQ38A-Flag rescued embryos and observed that levels of PINCH and ILK are comparable in multiple transgenic lines (Fig. 5C,D). This result indicates that loss of the interaction between PINCH and ILK is not sufficient to destabilize the proteins and promote turnover in an intact organism.
RSU-1 is crucial for viability when PINCH-ILK binding is compromised
The ability of PINCH to localize appropriately and support integrin-dependent adhesion in the absence of a direct association with ILK suggested the involvement of additional factors. RSU-1 is an attractive candidate to contribute to ILK independent PINCH function for several reasons: (1) RSU-1 binds directly to PINCH. (2) ics null mutants are viable but display wing blisters indicating a defect in integrin function. (3) RSU-1 localizes to sites of adhesion in the Drosophila embryonic muscle (Kadrmas et al., 2004). We performed rescue crosses in which PINCHwt-Flag or PINCHQ38A-Flag transgenes were introduced into a stck and ics null background (ics; PINCHwt-Flag and ics; PINCHQ38A-Flag). Strikingly, we observe a profound reduction of viable progeny in ics; PINCHQ38A-Flag rescued animals (34% and 6%), whereas we observe full rescue of viability in ics; PINCHwt-Flag rescued animals (83% and 85%) (Fig. 6A). Although there is a range in viable adult ics; PINCHQ38A-Flag rescued progeny, all values are greatly reduced from both PINCHwt-Flag and PINCHQ38A-Flag rescue in the presence of RSU-1 (Fig. 2D; Fig. 6A). These data suggest that in the absence of a PINCH-ILK interaction, RSU-1 plays an important role in maintaining PINCH function, and highlight a deficit in PINCHQ38A-Flag rescued flies that can be discerned in the context of an ics null background.
Fig. 6.
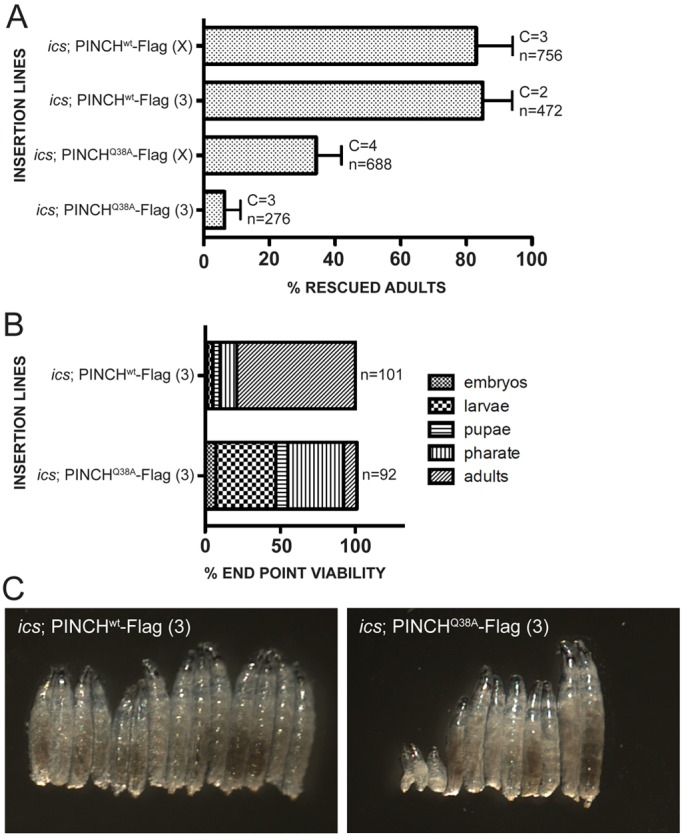
Loss of RSU-1 reduces the viability of PINCHQ38A-Flag rescued flies. (A) PINCHwt-Flag transgenic flies lacking RSU-1 (ics; PINCHwt-Flag) rescue at 83% and 85% whereas PINCHQ38A-Flag transgenic flies lacking RSU-1 (ics; PINCHQ38A-Flag) rescue at 34% and 6%. Bars represent the mean ± s.e.m. of the percentage rescued flies counted for the number of crosses set. C denotes the number of times the rescue cross was set. n is the total number of flies counted across all crosses. (B) End stage analysis of ics; PINCHwt-Flag and ics; PINCHQ38A-Flag rescued animals demonstrates that the majority of lethality in ics; PINCHQ38A-Flag animals occurs during the larval and pharate stages. (C) Heat fixed larvae at 66 hours AEL demonstrate a growth defect in ics; PINCHQ38A-Flag larvae compared with ics; PINCHwt-Flag.
To define the lethal phase of ics; PINCHQ38A-Flag animals, we collected late stage rescued embryos and monitored them over time (Fig. 6B). The majority of lethality in ics; PINCHQ38A-Flag rescued animals occurs during the larval (4% in ics; PINCHwt-Flag vs 40% in ics; PINCHQ38A-Flag) and pharate (11% in ics; PINCHwt-Flag vs 37% in ics; PINCHQ38A-Flag) stages. In this analysis, only 9% of ics; PINCHQ38A-Flag rescued animals survived to adulthood compared with 79% in ics; PINCHwt-Flag rescued animals (Fig. 6B). The increased lethality observed in ics; PINCHQ38A-Flag rescued larvae is associated with a growth defect that manifests during larval development. Approximately 66 hours after egg lay (AEL), ics; PINCHwt-Flag rescued larvae appear relatively uniform in size, while ics; PINCHQ38A-Flag rescued larvae display a range of sizes, including presumptive L1s that appear stumpy, exhibit sluggish movement, and do not progress in development (Fig. 6C).
Elimination of RSU-1 does not affect viability on its own, but can impact PINCH protein levels (Fig. 5A) (Kadrmas et al., 2004). Therefore, we explored the possibility that compared to PINCHwt-Flag, PINCHQ38A-Flag might be differentially destabilized in an ics null background, an outcome that could explain the synthetic lethality observed in ics; PINCHQ38A-Flag animals. Western blots were performed using lysates prepared from 66 hours AEL larvae, spanning the size range demonstrated (Fig. 6C, 7A). We observe a modest, but statistically significant decrease in the levels of PINCH (90% and 82% of wild-type levels) in larvae lacking RSU-1. Levels of ILK are also reduced in animals lacking RSU-1 (82% and 77% of wild-type levels) (Fig. 7A,B). This trend of complex destabilization is most pronounced in embryos, but is reliably observed at other developmental stages. Importantly, we did not observe a significant reduction in PINCH or ILK levels in ics; PINCHQ38A-Flag rescued larvae compared to ics; PINCHwt-Flag rescued larvae, indicating that the loss of viability observed in these flies is not due to altered levels of PINCH or ILK at this stage (Fig. 7A,B).
Fig. 7.
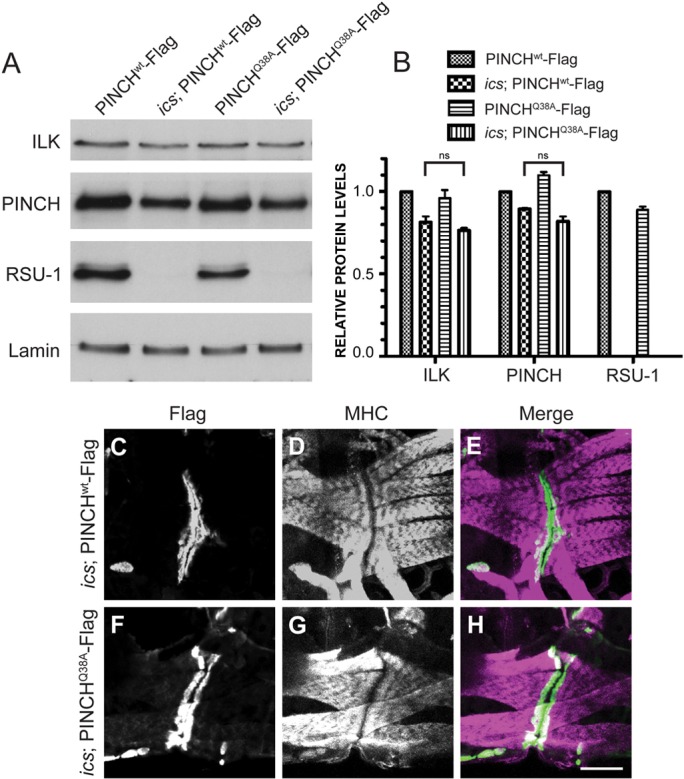
PINCH protein levels and localization are normal in ics; PINCHQ38A-Flag rescued larvae. (A) Western blots of pooled larvae at 66 hours AEL were probed with antibodies raised against ILK, PINCH, RSU-1 and Lamin. (B) Protein levels for the genotypes indicated were quantified and are represented as a fraction of PINCHwt-Flag expression. There is no statistical difference in PINCH or ILK levels between ics; PINCHwt-Flag and ics; PINCHQ38A-Flag rescued larvae. Bars represent the mean ± range for two replicate experiments. (C–H) 66 hour larval fillets were immunostained with anti-Flag antibody to label transgenic PINCH (green) (B,D,E,G) and anti-myosin heavy chain (MHC) to label the body wall muscle (magenta) (C,D,F,G). PINCH transgenes localize at muscle attachment sites in both ics; PINCHwt-Flag and ics; PINCHQ38A-Flag rescued larvae. Scale bar: 50 µm.
As described above, mutation of the ILK-binding domain of PINCH does not compromise the ability of PINCH to localize to integrin adhesion sites, illustrating that the PINCH-ILK interaction is not absolutely required for maintaining the proper subcellular distribution of PINCH in vivo. To test whether RSU-1 is required to support the appropriate localization of PINCH under conditions in which PINCH-ILK binding is compromised, we double-labeled 66 hour AEL L2 larval fillets to capture animals during the first major wave of lethality. Using anti-Flag antibody to visualize PINCHwt-Flag or PINCHQ38A-Flag and anti-MHC antibody to label body wall muscle, we observe PINCH localization at muscle attachment sites in both ics; PINCHwt-Flag and ics; PINCHQ38A-Flag animals (Fig. 7C,E,F,H), as well as normal muscle organization and intact sarcomeric structure (Fig.7D,E,G,H). These findings were recapitulated in both late stage embryos and L3 larval fillets. Moreover, β-integrin localizes to the muscle attachment sites of L3 animals of both genotypes (data not shown). These results indicate that the loss of viability observed in ics; PINCHQ38A-Flag larvae is not due to a complete failure of PINCH to localize in vivo. Loss of RSU-1 in flies where the interaction between PINCH and ILK has been disrupted results in synthetic lethality that is not explained by altered levels or localization of PINCH. These results are suggestive of a novel function for RSU-1 that is discernable in the absence of a PINCH-ILK interaction.
Discussion
Disrupting the PINCH-ILK interaction does not compromise cell adhesion or viability in Drosophila
In this study, we have tested the importance of the PINCH-ILK interaction in vivo via the generation of a genetically engineered PINCH variant that lacks ILK-binding capacity. We have shown that disrupting the PINCH-ILK interaction does not compromise viability, wing adhesion, or muscle structure or function. This was surprising given the prevailing idea that PINCH-ILK complexes form a functional unit, and from data in cell culture demonstrating loss of adhesion upon disruption of the PINCH-ILK interaction (Xu et al., 2005). Despite their capacity for physical association, tight co-localization at integrin-rich sites, and strikingly similar null phenotypes, the direct interaction between PINCH and ILK is not required in Drosophila. We conclude that PINCH and ILK can support integrin-dependent adhesion by mechanisms that are independent of their direct association.
A crucial role for RSU-1 in PINCH-ILK complex function
On their own, neither the elimination of RSU-1 function nor the rescue of stck null animals with the PINCHQ38A-Flag transgene reveals any lethality. Here we have identified a synthetic lethal condition in which the viability of PINCHQ38A-Flag transgenic flies is seriously compromised by elimination of RSU-1. These data suggest a critical function for RSU-1 that is made evident under conditions in which the PINCH-ILK association is compromised. Our analyses during the larval stages of development suggest that the lethality of ics; PINCHQ38A-Flag rescued animals is not due to a disruption of integrin function. However, we have not completely ruled out a role for integrin complex function or downstream signaling at other stages or in the 66 hour AEL larvae displaying a growth defect, leaving this an important unresolved issue that will require further study. Based on the results presented here, disruption of mechanisms that involve growth and survival signaling could also contribute to the lethality observed in ics; PINCHQ38A-Flag rescued animals. ILK and PINCH have both been implicated in survival signaling via AKT, and it will be important to determine the contributions of RSU-1 (Fukuda et al., 2003; Xu et al., 2005; Meder et al., 2011). Many defects observed from disruption of integrin adhesion complexes are attributed to improper ILK-PINCH-Parvin complex formation, function, and downstream signaling (Zhang et al., 2002; Legate et al., 2006; Stanchi et al., 2009; Wickström et al., 2010; Zervas et al., 2011). Here, we propose that the components of this integrin effector complex be expanded to include RSU-1.
The lack of complete lethality in ics; PINCHQ38A-Flag rescued flies suggests that RSU-1 is not the sole protein maintaining viability in the absence of PINCH-ILK binding, and highlights that multiple interactions within integrin complexes function together to maintain viability. Animals in a genetically sensitized background, like PINCHQ38A-Flag rescued flies, may serve as a model for assessing the contributions of other non-essential adhesion complex components, like Tensin, Vinculin and FAK, whose functions have been somewhat elusive (Alatortsev et al., 1997; Lee et al., 2003; Grabbe et al., 2004; Torgler et al., 2004). Like RSU-1, these proteins might support integrin function and viability in combination with other adhesion complex components.
Requirements for PINCH and ILK localization at muscle attachment sites
In Drosophila, ilk null mutational analyses have shown that ILK is required for the appropriate targeting of PINCH to integrin-rich adhesion sites (Zervas et al., 2011). In contrast, we have found that PINCHQ38A-Flag, which lacks the ability to bind ILK, assembles properly at muscle attachment sites, supporting the retention of cellular and muscle architecture, as well as viability of the organism. These data demonstrate the severity of consequence upon complete loss of ILK and indicate that in the fly there is some additional ILK-dependent mechanism that aids in localizing PINCH to muscle attachment sites beyond the interaction mediated by PINCHQ38. In cell culture, the interaction between PINCH and ILK is required for appropriate PINCH localization (Zhang et al., 2002). The contrast between the data in cell culture and our observations that PINCH and ILK localize appropriately in the PINCHQ38A-Flag mutants suggests that multiple redundant interactions within adhesion complexes contribute to localization in vivo and that perhaps these interactions are altered or lacking in cell culture. We tested whether RSU-1 could be functioning to preserve PINCH localization in the absence of an interaction with ILK. Although loss of RSU-1 in the PINCHQ38A-Flag rescued animals results in lethality, we demonstrate that RSU-1 is not required to localize PINCH in larvae.
Several lines of evidence support our finding that PINCH-ILK binding is not solely responsible for localizing PINCH, and suggest that both PINCH and ILK have additional protein interactions that contribute to targeting PINCH to adhesion sites. In mammalian cell culture, deletion of the C-terminal tail of PINCH results in a similar disruption of localization and function as seen with the PINCHQ40A mutant. This indicates that the PINCH C-terminal tail also participates in localization, even in the context of normal binding to ILK (Xu et al., 2005). A recent study, that is complementary to this report, has probed how various ILK deletion mutants affect protein assembly at muscle attachment sites (Zervas et al., 2011). This work demonstrates that expression and proper localization of the ANKR domain of ILK in an ilk null mutant is insufficient to localize PINCH despite the capacity of PINCH to bind this portion of ILK (Zervas et al., 2011). This suggests that either another portion of ILK or another protein interaction is required to localize PINCH. In the same study, a version of PINCH in which LIM1 is deleted (PINCHΔLIM1) demonstrates a reduced yet retained capacity to localize to muscle attachment sites (Zervas et al., 2011) (our unpublished results). The limited capacity for localization of PINCHΔLIM1 as compared to PINCHQ38A-Flag suggests that LIM1 has additional functions beyond ILK binding. Consistent with this perspective, a recently published crystal structure of LIM1 of PINCH bound to the ANKR domain of ILK confirms that human PINCHQ40 is a key residue in maintaining the PINCH-ILK interaction, and highlights that LIM1 of PINCH undergoes a conformational ‘twist’ upon binding to ILK that could create docking sites for additional partners (Chiswell et al., 2008; Yang et al., 2009). Several studies have identified novel binding partners for LIM1 of PINCH in C. elegans and in human cultured podocytes (Mercer et al., 2003; Qadota et al., 2007; Wang et al., 2011). However, additional research is required to assess the conservation of these PINCH-LIM1 binding partners in other organisms, as well as any potential roles they may have in PINCH localization and integrin effector function.
Contributions of PINCH, ILK and RSU-1 to adhesion complex stability
Reports that PINCH and ILK influence the stability of each other have made it difficult to assign distinct functions to the individual proteins. In agreement with data from mammalian cell culture and in zebrafish, we show that loss of PINCH affects ILK protein levels and vice versa, indicating that this is an evolutionarily conserved characteristic of proteins in this adhesion complex (Fukuda et al., 2003; Stanchi et al., 2009; Meder et al., 2011). Contrary to these data, a recent publication has claimed that ILK stabilizes PINCH, but that PINCH does not stabilize ILK, basing this conclusion on qualitative immunofluorescence images, where consistent with other published data ILK localization is preserved in a stck null mutant (Clark et al., 2003; Zervas et al., 2011). Here, we show via semi-quantitative western analysis that PINCH is indeed responsible for the stabilization of ILK protein levels, and conclude that a reduction in the level of ILK in stck mutants does not preclude the ability of the remaining ILK to localize properly.
Results presented here provide further evidence for the mutual protein stability of additional adhesion complex components. We demonstrate that RSU-1 stabilizes PINCH and ILK and vice versa. Stabilization within this complex is modulated not only by proteins with direct or primary interactions (PINCH–RSU-1 or PINCH-ILK), but by secondary protein interactions as well (ILK–RSU-1). This secondary regulation is also observed in other studies where loss of PINCH reduces Parvin and Tensin protein levels, which are both direct partners of ILK (Fukuda et al., 2003; Stanchi et al., 2009). Further work will be required to reveal the extent of this stabilization phenomenon within adhesion complexes in Drosophila. We conclude that PINCH, ILK and RSU-1 function together as a stabilized complex and postulate that stck and ilk null phenotypes do not arise from loss of a single protein, but that reductions in or loss of other complex components may also contribute to the defects observed.
Consistent with this view, the PINCH loss of function phenotype might be due in part to the destabilization of ILK. However, we demonstrate that the reduction in ILK that is associated with stck null mutants is comparable to the reduction in ILK observed in ics mutants, which display no deficits in muscle adhesion or viability. Furthermore, a recent study in zebrafish has shown that overexpresion of ILK in a PINCH morphant does not rescue the defects caused by loss of PINCH (Meder et al., 2011). Taken together, the phenotypes associated with loss of PINCH function cannot be fully attributed to a coordinate loss of ILK. In contrast, it remains possible that a reduction of PINCH protein in ilk mutants contributes to ILK loss-of-function phenotypes.
Levels of PINCH and ILK are not affected in PINCHQ38A-Flag rescued embryos, demonstrating a fundamental difference between disruption of the PINCH-ILK interaction and complete loss of either PINCH or ILK in vivo. The wild-type protein levels observed in PINCHQ38A-Flag rescued embryos may be attributed to other proteins that aid in the localization and/or stabilization of PINCH.
The role of PINCH in stabilizing actin-membrane interactions
In Drosophila, stck and ilk null mutants clearly exhibit a defect in anchoring the actin cytoskeleton to sites of integrin-mediated adhesion (Zervas et al., 2001; Clark et al., 2003). Because neither PINCH nor ILK directly binds Actin, the detachment phenotype must be mediated indirectly through an actin-binding protein within adhesion complexes. One clear candidate for this role is Parvin, which binds directly to ILK and Actin. The data presented here demonstrate that PINCHQ38A-Flag stabilizes the actin cytoskeleton independent of a direct link to ILK and, by association with ILK, to Parvin. This result suggests two alternatives for a PINCH-actin link. First, it is possible that a direct interaction exists between PINCH and Parvin. However, yeast 3-hybrid data from C. elegans refute this hypothesis, as PINCH and Parvin are only able to interact when ILK is present (Norman et al., 2007). Moreover, we have been unable to detect a direct biochemical interaction between PINCH and Parvin (our unpublished observations). Second, additional PINCH binding partners independent of ILK and Parvin may be required to maintain a connection to the actin cytoskeleton. Based upon the normal actin structure in ics mutants, and in ics; PINCHQ38A-Flag larvae, we show that RSU-1 is not required to maintain PINCH-Actin linkages. Thus, elucidating the mechanism of physical linkage of PINCH to the actin cytoskeleton will likely require the identification of additional complex components.
A model for ILK–PINCH–RSU-1 complex function
Integrin adhesion complexes are composed of many proteins that aid in maintaining linkage of integrin receptors to both the extracellular matrix and the actin cytoskeleton. Many proteins that assemble at the cytoplasmic face of integrins have overlapping and/or partially characterized functions and are the current focus of investigation of many labs. A subset of these proteins is known to aid in the assembly of ILK, PINCH and RSU-1 at integrin adhesion sites (Fig. 8A). Data generated from the in vivo disruption of the PINCH-ILK interaction have provided evidence to suggest that PINCH makes additional contacts that anchor it to adhesion sites and aid in stabilization of the actin cytoskeleton (Fig. 8B). Furthermore, loss of RSU-1 in the absence of a PINCH-ILK interaction demonstrates a novel function for RSU-1 in maintaining viability independent of PINCH localization and stability (Fig. 8B). Taken together, we demonstrate that disruption of a single protein-protein interaction within the integrin effector complex is not sufficient to compromise adhesion or viability, but that deficits are observed once a second disruption is introduced. Currently uncharacterized partners for both PINCH and ILK likely aid in maintaining their common or individual functions. Our future work will focus on understanding which domains of PINCH are required for targeting to integrin rich adhesion sites and for maintaining actin linkage, and identifying novel PINCH binding partners to further elucidate ILK–PINCH–RSU-1 complex function.
Fig. 8.
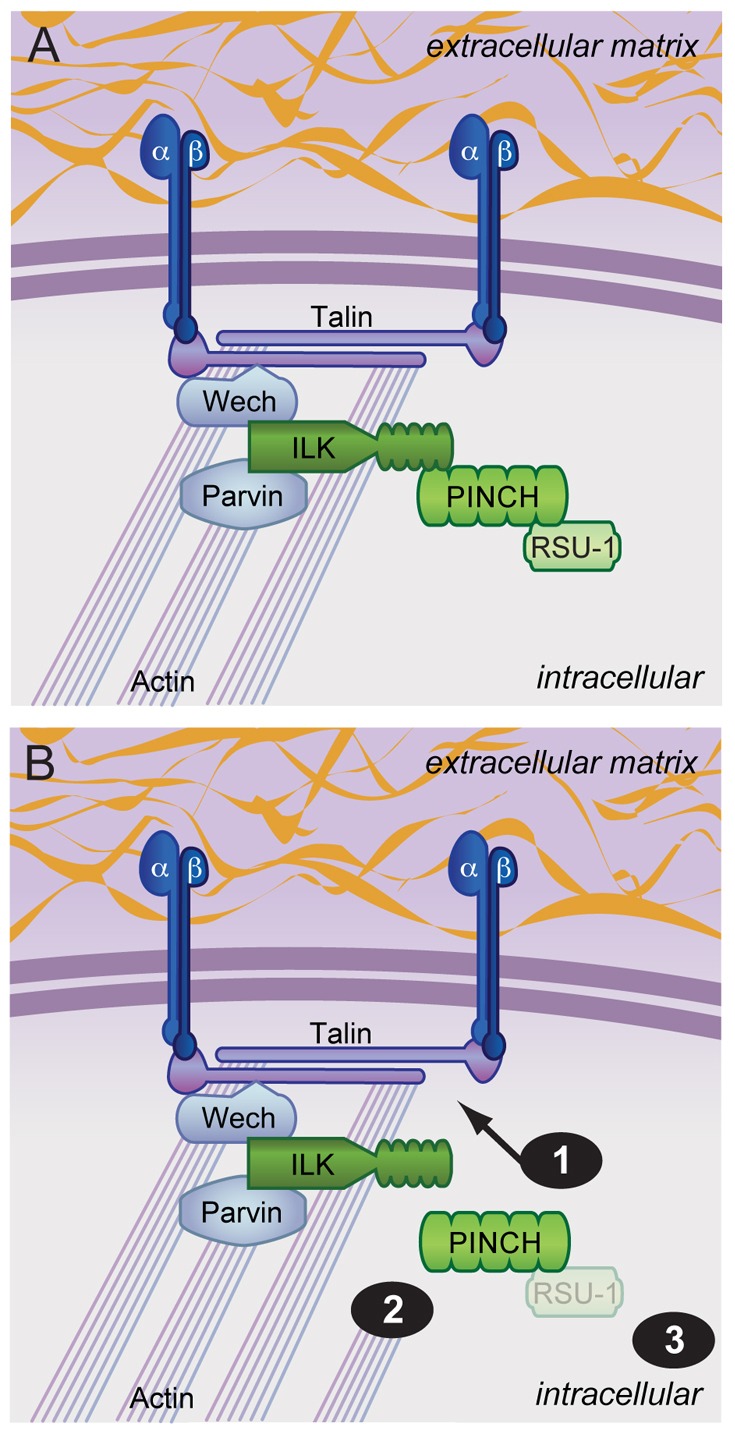
A model for ILK–PINCH–RSU-1 function. (A) Schematic of wild-type ILK, PINCH and RSU-1 within integrin adhesion complexes. A subset of protein interactions are highlighted demonstrating connection of the complex to integrins and the actin cytoskeleton. PINCH and ILK loss of function phenotypes are probably not due to the loss of a single protein, but to disruption of additional protein partners. (B) Specific disruption of the PINCH-ILK interaction does not affect viability or integrin function, and suggests that PINCH makes additional contacts that (1) anchor it to integrin rich adhesion sites and (2) connect it to the actin cytoskeleton. Loss of RSU-1 in PINCHQ38A rescued flies (3) results in lethality that is not due to mislocalization or destabilization of PINCH, suggesting a novel function for RSU-1.
Materials and Methods
Fly stocks
w1118 were used as wild-type controls in all experiments. ilk1 and icsBG02577 stocks were obtained from the Bloomington Stock Center. FRT82B stck17, FRT82B stck18 and ILK-GFP flies were previously described (Zervas et al., 2001; Clark et al., 2003). stck17 germ line clone females were generated as previously described and were crossed to stck18/TM3, Twist-GFP to generate stck17/18 maternal/zygotic (m/z) null embryos (Clark et al., 2003).
Sequence alignment and accession numbers
Sequence alignment was performed with ClustalX using the following PINCH protein sequences (NCBI accession numbers): human PINCH1 (AAH05341), human PINCH2 (AAH65816), mouse PINCH1 (AAH05621), mouse PINCH2 (NP_659111), zebrafish PINCH (NP_001019560), Drosophila PINCH isoform A (NP_524278), C. elegans unc-97 (NP_508943).
Generation of PINCHQ38A reagents
A glutamine to alanine mutation at position 38 in the dPINCHa cDNA was introduced by PCR mutagenesis into a previously described pMT-PINCHwt-His construct (Kadrmas et al., 2004). pMT-PINCHwt-His and pMT-PINCHQ38A-His were transfected into S2 cells using standard methods. Transgenic flies carrying the PINCHQ38A mutation were generated by p-element transposition using a previously described pCasper construct containing genomic PINCHwt-GFP (Clark et al., 2003; Kadrmas et al., 2004). A C-terminal 3×Flag tag replaced the GFP in both PINCHwt and PINCHQ38A constructs (referred to throughout the text as PINCHwt-Flag and PINCHQ38A-Flag). DNA constructs were injected into embryos by Genetic Services (Cambridge, MA).
PINCHwt and PINCHQ38A rescue experiments
To test for rescue, crosses were set to introduce one or two copies of the transgene and two copies of different null stck alleles into the progeny. For example: PINCHwt-Flag; stck17/TM3 × PINCHwt-Flag; stck18/TM3. It is expected that full rescue would produce a ratio of 1/3 rescued adult progeny to 2/3 adult TM3 balanced progeny. Rescue was calculated as the observed rescued flies over 1/2 of the observed balanced progeny as predicted by the cross. To assess rescued progeny during development, late stage embryos were collected at two hour intervals from established rescued stocks or from crosses that were set using TM3, Twist-GFP balancers and were sorted for lack of GFP. Embryos were aged on grape juice agar plates at 25°C and followed through development, or were collected 48 hours after sorting for larval analyses. Graphpad Prism was used for all graphical analyses.
Pull downs, immunoprecipitations and western blots
S2 cell lysates or adult fly lysates were prepared in lysis buffer (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.1% Triton X-100) and protease inhibitors, and were incubated with either Ni-NTA agarose (Qiagen) or anti-Flag M2 agarose (Sigma), and boiled in 2× Laemmli sample buffer followed by western blotting. For other western blots, equal numbers of adult flies or staged and sorted embryos or larvae were homogenized in 2× Laemmli sample buffer. Protein samples were run on SDS-PAGE gels and transferred to nitrocellulose. Antibodies used were: anti-PINCH (1∶5000) (Clark et al., 2003), anti-RSU-1 (1∶5000) (Kadrmas et al., 2004), anti-ILK (1∶500, BD #611802), anti-lamin (1∶5000, DSHB), anti-β-integrin (DX.468.1, 1∶1, DSHB). ImageJ was used for densitometry analyses of western blots. All values were normalized to the Lamin signal followed by normalization to the wild-type control. Graphpad Prism was used for all graphical analyses. All graphs represent two replicate experiments or three replicate samples within one experiment.
Immunofluorescence and imaging
Staged embryos were fixed in 4% paraformaldehyde or heat fixed according to previously published protocols (Clark et al., 2003). To preserve GFP or for staining with Phalloidin, embryos were devitellinized in 80% ethanol. Antibodies used were anti-Flag (M2) preabsorbed against w1118 embryos (1∶2000, Sigma), anti-RSU-1 preabsorbed against w1118 embryos (1∶1000), anti-β-integrin (CF.6G11, 1∶5, DSHB), anti-GFP (1∶1000, Clontech), anti-myosin heavy chain (MHC) (1∶500, kindly provided by Dan Kiehart), Phalloidin 568 or 647 (1∶100, Invitrogen). Secondary antibodies were Alexa Fluor anti-rabbit or anti-mouse 488 or 568 (1∶250, Invitrogen). All images were obtained on an Olympus Fluoview 300 using a 20× or 60× objective (University of Utah School of Medicine Cell Imaging Facility), and are presented as a merged Z-stack. Figures were assembled using Adobe CS4 programs.
Acknowledgments
The Developmental Studies Hybridoma Bank is acknowledged for providing antibodies used in this study, and we thank Dan Kiehart for providing the anti-myosin heavy chain antibody. We would like to acknowledge Kathleen Clark, members of the Beckerle Lab, Diana Lim and Oscar Ruiz for intellectual contributions and other support of the project.
Footnotes
Funding
The authors are supported by the National Institutes of Health [grant number R01 GM084103 to J.L.K.]; an NIH Genetics Training Grant [grant number T32 GM07464 to M.C.E.]; the Huntsman Cancer Institute; the Cancer Center [grant number P30CA042014] is acknowledged for critical support of shared resources. Deposited in PMC for release after 12 months.
References
- Alatortsev V. E., Kramerova I. A., Frolov M. V., Lavrov S. A., Westphal E. D. (1997). Vinculin gene is non-essential in Drosophila melanogaster. FEBS Lett. 413, 197–201 10.1016/S0014-5793(97)00901-0 [DOI] [PubMed] [Google Scholar]
- Bökel C., Brown N. H. (2002). Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell 3, 311–321 10.1016/S1534-5807(02)00265-4 [DOI] [PubMed] [Google Scholar]
- Brabant M. C., Fristrom D., Bunch T. A., Brower D. L. (1996). Distinct spatial and temporal functions for PS integrins during Drosophila wing morphogenesis. Development 122, 3307–3317 [DOI] [PubMed] [Google Scholar]
- Brakebusch C., Fässler R. (2003). The integrin-actin connection, an eternal love affair. EMBO J. 22, 2324–2333 10.1093/emboj/cdg245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower D. L. (2003). Platelets with wings: the maturation of Drosophila integrin biology. Curr. Opin. Cell Biol. 15, 607–613 10.1016/S0955-0674(03)00102-9 [DOI] [PubMed] [Google Scholar]
- Brower D. L., Jaffe S. M. (1989). Requirement for integrins during Drosophila wing development. Nature 342, 285–287 10.1038/342285a0 [DOI] [PubMed] [Google Scholar]
- Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. (2002). Talin is essential for integrin function in Drosophila. Dev. Cell 3, 569–579 10.1016/S1534-5807(02)00290-3 [DOI] [PubMed] [Google Scholar]
- Chiswell B. P., Zhang R., Murphy J. W., Boggon T. J., Calderwood D. A. (2008). The structural basis of integrin-linked kinase-PINCH interactions. Proc. Natl. Acad. Sci. USA 105, 20677–20682 10.1073/pnas.0811415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., McGrail M., Beckerle M. C. (2003). Analysis of PINCH function in Drosophila demonstrates its requirement in integrin-dependent cellular processes. Development 130, 2611–2621 10.1242/dev.00492 [DOI] [PubMed] [Google Scholar]
- Cutler M. L., Bassin R. H., Zanoni L., Talbot N. (1992). Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol. Cell. Biol. 12, 3750–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon I., Brown N. H. (2007). Integrins and the actin cytoskeleton. Curr. Opin. Cell Biol. 19, 43–50 10.1016/j.ceb.2006.12.013 [DOI] [PubMed] [Google Scholar]
- Dougherty G. W., Chopp T., Qi S. M., Cutler M. L. (2005). The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp. Cell Res. 306, 168–179 10.1016/j.yexcr.2005.01.025 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Chen K., Shi X., Wu C. (2003). PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J. Biol. Chem. 278, 51324–51333 10.1074/jbc.M309122200 [DOI] [PubMed] [Google Scholar]
- Grabbe C., Zervas C. G., Hunter T., Brown N. H., Palmer R. H. (2004). Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development 131, 5795–5805 10.1242/dev.01462 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Jani K., Schöck F. (2007). Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 179, 1583–1597 10.1083/jcb.200707045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J. L., Smith M. A., Clark K. A., Pronovost S. M., Muster N., Yates J. R., 3rd, Beckerle M. C. (2004). The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J. Cell Biol. 167, 1019–1024 10.1083/jcb.200408090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Cho K. S., Kim E., Chung J. (2003). blistery encodes Drosophila tensin protein and interacts with integrin and the JNK signaling pathway during wing development. Development 130, 4001–4010 10.1242/dev.00595 [DOI] [PubMed] [Google Scholar]
- Legate K. R., Montañez E., Kudlacek O., Fässler R. (2006). ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7, 20–31 10.1038/nrm1789 [DOI] [PubMed] [Google Scholar]
- Li F., Zhang Y., Wu C. (1999). Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J. Cell Sci. 112, 4589–4599 [DOI] [PubMed] [Google Scholar]
- Li S., Bordoy R., Stanchi F., Moser M., Braun A., Kudlacek O., Wewer U. M., Yurchenco P. D., Fässler R. (2005). PINCH1 regulates cell-matrix and cell-cell adhesions, cell polarity and cell survival during the peri-implantation stage. J. Cell Sci. 118, 2913–2921 10.1242/jcs.02422 [DOI] [PubMed] [Google Scholar]
- Löer B., Bauer R., Bornheim R., Grell J., Kremmer E., Kolanus W., Hoch M. (2008). The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat. Cell Biol. 10, 422–428 10.1038/ncb1704 [DOI] [PubMed] [Google Scholar]
- Meder B., Huttner I. G., Sedaghat–Hamedani F., Just S., Dahme T., Frese K. S., Vogel B., Köhler D., Kloos W., Rudloff J.et al. (2011). PINCH proteins regulate cardiac contractility by modulating integrin-linked kinase-protein kinase B signaling. Mol. Cell. Biol. 31, 3424–3435 10.1128/MCB.05269-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K. B., Flaherty D. B., Miller R. K., Qadota H., Tinley T. L., Moerman D. G., Benian G. M. (2003). Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell 14, 2492–2507 10.1091/mbc.E02-10-0676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman K. R., Cordes S., Qadota H., Rahmani P., Moerman D. G. (2007). UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev. Biol. 309, 45–55 10.1016/j.ydbio.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Pines M., Fairchild M. J., Tanentzapf G. (2011). Distinct regulatory mechanisms control integrin adhesive processes during tissue morphogenesis. Dev. Dyn. 240, 36–51 10.1002/dvdy.22488 [DOI] [PubMed] [Google Scholar]
- Prout M., Damania Z., Soong J., Fristrom D., Fristrom J. W. (1997). Autosomal mutations affecting adhesion between wing surfaces in Drosophila melanogaster. Genetics 146, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H., Mercer K. B., Miller R. K., Kaibuchi K., Benian G. M. (2007). Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell 18, 4317–4326 10.1091/mbc.E07-03-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Li S., Docheva D., Grashoff C., Sakai K., Kostka G., Braun A., Pfeifer A., Yurchenco P. D., Fässler R. (2003). Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 17, 926–940 10.1101/gad.255603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R., Zelzer E., Volk T. (2010). Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137, 2807–2817 10.1242/dev.047498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanchi F., Grashoff C., Nguemeni Yonga C. F., Grall D., Fässler R., Van Obberghen–Schilling E. (2009). Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J. Cell Sci. 122, 1800–1811 10.1242/jcs.044602 [DOI] [PubMed] [Google Scholar]
- Torgler C. N., Narasimha M., Knox A. L., Zervas C. G., Vernon M. C., Brown N. H. (2004). Tensin stabilizes integrin adhesive contacts in Drosophila. Dev. Cell 6, 357–369 10.1016/S1534-5807(04)00055-3 [DOI] [PubMed] [Google Scholar]
- Tu Y., Li F., Goicoechea S., Wu C. (1999). The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 19, 2425–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Huang Y., Zhang Y., Hua Y., Wu C. (2001). A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J. Cell Biol. 153, 585–598 10.1083/jcb.153.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velyvis A., Yang Y., Wu C., Qin J. (2001). Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain. J. Biol. Chem. 276, 4932–4939 10.1074/jbc.M007632200 [DOI] [PubMed] [Google Scholar]
- Walsh E. P., Brown N. H. (1998). A screen to identify Drosophila genes required for integrin-mediated adhesion. Genetics 150, 791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Li Y., Wu C., Liu Y. (2011). PINCH1 is transcriptional regulator in podocytes that interacts with WT1 and represses podocalyxin expression. PLoS ONE 6, e17048 10.1371/journal.pone.0017048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström S. A., Lange A., Montanez E., Fässler R. (2010). The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J. 29, 281–291 10.1038/emboj.2009.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Fukuda T., Li Y., Zha X., Qin J., Wu C. (2005). Molecular dissection of PINCH-1 reveals a mechanism of coupling and uncoupling of cell shape modulation and survival. J. Biol. Chem. 280, 27631–27637 10.1074/jbc.M504189200 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wang X., Hawkins C. A., Chen K., Vaynberg J., Mao X., Tu Y., Zuo X., Wang J., Wang Y. X.et al. (2009). Structural basis of focal adhesion localization of LIM-only adaptor PINCH by integrin-linked kinase. J. Biol. Chem. 284, 5836–5844 10.1074/jbc.M805319200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas C. G., Gregory S. L., Brown N. H. (2001). Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J. Cell Biol 152, 1007–1018 10.1083/jcb.152.5.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas C. G., Psarra E., Williams V., Solomon E., Vakaloglou K. M., Brown N. H. (2011). A central multifunctional role of integrin-linked kinase at muscle attachment sites. J. Cell Sci. 124, 1316–1327 10.1242/jcs.081422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Tu Y., Velyvis A., Yang Y., Qin J., Wu C. (2002). Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J. Cell Sci. 115, 4777–4786 10.1242/jcs.00166 [DOI] [PubMed] [Google Scholar]