Abstract
Agrobacterium species are plant-associated relatives of the rhizobia. Several species cause plant diseases such as crown gall and hairy root, although there are also avirulent species. A. tumefaciens is the most intensively studied species and causes crown gall, a neoplastic disease that occurs on a variety of plants. Virulence is specified by large plasmids, and in the case of A. tumefaciens this is called the Ti (tumor-inducing) plasmid. During pathogenesis virulent agrobacteria copy a segment of the Ti plasmid and transfer it to the plant, where it subsequently integrates into the plant genome, and expresses genes that result in the disease symptoms. A. tumefaciens has been used extensively as a plant genetic engineering tool and is also a model microorganism that has been well studied for host-microbe associations, horizontal gene transfer, cell-cell communication, and biofilm formation. This unit describes standard protocols for genetic manipulation of A. tumefaciens.
Keywords: Agrobacterium, Growth media, Genetic analyses, Taxonomy, Opines, Plant association, Virulence, Plasmids, Attachment, transposon
The mechanism of A. tumefaciens infection and the plant-microbe interaction has been thoroughly studied because of its importance for plant genetic engineering. However, it has also been intensively investigated as a model system for microbial cell-cell communication, plasmid conjugation, and biofilm formation. Several Agrobacterium genomes have been sequenced (available at the Virginia Bioinformatics Institute website: http://agro.vbi.vt.edu/public/) allowing for facile and robust genetic analysis of the species.
Basic Protocols 1 and 2, describe methods for plasmid introduction by electroporation and conjugation, respectively. Basic Protocol 3 describes a method for generating a mutant library using transposon mutagenesis. Basic Protocol 4 describes a method for mapping the insertion site of a transposon using Touchdown PCR. Basic Protocol 5 describes a highly effective method for generating markerless genetic mutants using a strategy of allelic replacement. Basic Protocol 6 outlines a method for the curing of Agrobacterium tumefaciens native plasmids, and related to this, Basic Protocol 7 describes a method for visualizing plasmid composition known as an Eckhardt gel.
BASIC PROTOCOL 1: TRANSFORMATION OF AGROBACTERIUM BY ELECTROPORATION (adapted from Mersereau et al. 1990)
In order to study the genetics of bacteria, it is necessary to manipulate their genetic composition. For Agrobacterium, one effective way to introduce foreign DNA, whether it is an allelic replacement construct, an expression vector, or a curing plasmid, is by electroporation. This procedure involves the application of an electrical field around the bacterial cells that leads to an increase in the electrical conductivity and permeability of the plasma membrane so that plasmid DNA is introduced into the cell.
Materials
8 ml LB broth
2 ml and 5 ml culture tubes
spectrophotometer
microcentrifuge
30% glycerol
1.6 ml microfuge tubes
electroporation cuvettes (e.g. Harvard apparatus, cat# 45-0124)
electroporator (eg. Bio-Rad E. coli Pulser)
ice
28°C incubator with shaking capabilities
ATGN plates supplemented with appropriate antibiotics (LB plates can also be used)
- Prepare electrocompetent cells
- Inoculate the strain of interest in 2 ml LB supplemented with the appropriate antibiotics.
- Incubate culture overnight to full turbidity (stationary phase).
- Subculture 50 µl of starter culture into 5 ml LB supplemented with the appropriate antibiotics.
- Incubate with optimal aeration until culture reaches OD600=0.6–0.7.
- Harvest all 5 ml of cells by sequential centrifugation in a sterile tube. Centrifuge 2 min at no greater than 9,000 × g in microfuge.
- Resuspend cells in 1 ml ice-cold, sterile water.
- Centrifuge, as in step 5.
- Repeat steps 6 and 7 four times.
- Resuspend cells in ~250 µl sterile ice cold 30% glycerol.
- Store at −70°C
Combine 40 µl electrocompetent cells and 2–10 µl DNA (this amount will vary depending on the concentration of the DNA preparation). For negative controls the same volume of TE buffer should be added to cells in a sterile 1.6 ml tube.
Incubate on ice for 5–10 min.
Add 60 µl sterile water, transfer entire mixture to an electroporation cuvette.
Electroporate at 25 volts/cm for a time constant of approximately 4.5–5.5msec.
Immediately add 400–800 µl of cold LB media, and return to 1.6 ml tube.
Incubate cells for 90 min at 28°C with aeration.
Centrifuge cells in a microfuge, resuspend in 100 µl of media and spread onto plates supplemented with appropriate antibiotics.
Incubate 2–4 days at 28°C until colonies appear.
BASIC PROTOCOL 2: PLASMID INTRODUCTION BY CONJUGATION
This method of plasmid introduction is more complicated and more laborious to perform than electroporation, but works very well for very large plasmids or introducing plasmid DNA that must recombine into the genome in order to be maintained. The protocol below describes the strategy for moving a plasmid from E. coli to A. tumefaciens, but it is worth noting that plasmids can be moved between Agrobacterium strains as well. Conjugation of the Ti plasmid is tightly regulated and requires the presence of conjugal opines or genetic manipulation. The At plasmid of strain C58 however will conjugate constitutively.
Materials
LB plates (no antibiotics), one per mating reaction plus two for controls
Sterile hydrophilic polyethersulfone filters: 25 mm, 0.2 µm
Donor strain of E. coli carrying plasmid of interest (plasmid must be self-conjugal or donor strain must be encode compatible conjugal machinery, tra+)
28°C incubator
37°C incubator
ATGN plates supplemented with appropriate antibiotics
- Grow overnight, fully turbid cultures of donor and recipient cultures.
- Required culture volume per mating:
- 2 ml culture of donor (E. coli)
- 2 ml culture of recipient (A. tumefaciens)
Collect cells by centrifugation (3 min @ 9000 × g).
Resuspend each pellet in 50 µl LB.
- Prepare mating mixtures as indicated below
Mating Donor Recipient Donor Control 50 µl cells ----------------------- Recipient Control ----------------------- 50 µl cells Experimental 50 µl cells 50 µl cells Spot entire mating on a filter on an LB plate without antibiotics. Allow fluid to soak into filter.
-
Incubate plates overnight at 28°C.
(E. coli grows optimally at 37°C but A. tumefaciens does not grow above 30°C).
Remove cells from filter by folding filter in half using sterile forceps (making a bacterial ‘taco’ with the cells inside) and transferring the folded filter to a 1.5 ml tube, adding 1 ml ATGN, and vortexing vigorously to dislodge the cells.
Make serial dilutions in ATGN of suspended cells in 1:10 increments.
Plate 100 µl of each dilution (from 100 to 10−3) on ATGN plates containing the appropriate antibiotic (only plate 100 µl of the undiluted donor-only and recipient-only controls). Incubate for 2–4 days at 28°C until colonies appear on donor plus recipient plates, make sure that there is no growth on donor only or recipient only controls.
BASIC PROTOCOL 3: TRANSPOSON MUTAGENESIS OF AGROBACTERIUM
This strategy allows you to generate a mutant library, by introduction of a transposon at random within the genome. Mariner is a very sequence specific transposon that inserts into sites in the genome containing an AT dinucleotide sequence; virtually every gene has at least one AT sequence. For this version of Mariner, the transposase is supplied in trans from a plasmid in the donor and cannot transpose or move within A. tumefaciens, thus the insertion(s) that occur at the time of transposon delivery are stable. We utilize the Mariner carried on the plasmid pFD1. (Lampe et al., 1999)
Materials
See ‘Plasmid Introduction by Conjugation’ protocol for all materials.
However, for this protocol the donor strain is E. coli carrying pFD1, and the recipient strain (to be mutated) must be sensitive to kanamycin.
Follow the protocol for ‘Plasmid introduction by Conjugation” Steps 1 – 9. Freeze down remaining cell resuspension (from Step 7) in 30% glycerol.
Perform a pilot experiment in which a dilution series is plated to determine transposition frequency. Incubate 2–4 days until A. tumefaciens colonies appear. The goal is to have isolated colonies (transposon mutants) on your plates so aim to have 50 – 150 colonies per plate. Inoculate multiple plates of frozen mutant library using the dilution factor that resulted in the optimal number of isolated colonies in the pilot experiment.
Patch colonies onto ATGN plates supplemented with Kanamycin.
Screen for phenotype of interest.
Identify insertion site using Touchdown PCR (see Basic Protocol 4).
BASIC PROTOCOL 4: TOUCHDOWN PCR (Levano-Garcia et al., 2005)
This technique is used to determine the insertion site of the transposon (or any gene or product with a known sequence). This strategy requires the use of two types of primers, a specific primer (sequence matching that of the transposon) and an arbitrary primer (sequence varies, to amplify from various sites throughout the genome). Because the specific primer anneals to the transposon and the arbitrary primer anneals to sites throughout the genome, this type of PCR typically yields multiple products of varying length that all originate from the same site within the transposon. The maximum length of product is determined by the limits of the DNA polymerase and the extension time in the reaction. Sequencing the resulting products using the specific primer provides information about the sequence adjacent to the site of insertion.
Materials
Genomic DNA from transposon mutant(s)
10 mM dNTPs
Taq polymerase and buffer
TD-PCR primers (specific and arbitrary)
20 mM MgCl2
0.2 ml tubes
thermocycler
gel electrophoresis apparatus and supplies (agarose, EtBr, UV imaging)
PCR purification kit
-
Set up the following PCR reaction mixture in 0.2 ml tubes:
Template (your genomic DNA solution 30–120 ng) 1.5 µl Primer 1 (specific primer, 20 pmol/µl) MarLseq 1.0 µl Primer 2 (arbitrary primer, 100 pmol/µl) MarTDL2 1.0 µl 10 mM dNTP mix 1.0 µl 10× Tricine Buffer 5.0 µl Taq polymerase 0.5 µl 20 mM MgCl2 (final concentration in reaction is 2.5 mM) 6.3 µl ddH2O 33.7 µl Total Volume 50 µl - Set 1: Amplifies from within the left side of the transposon:
- MarLseq (specific primer) 5’GGG AAT CAT TTG AAG GTT GGT 3’
- MarTDL2 (arbitrary primer) 5’GACACGGGCCTCGANGNNNCNTNGG 3’
- Set 2: Amplifies from within the right side of the transposon:
- MarRseq (specific primer) 5’ CGG GTA TCG CTC TTG AAG GGA 3’
- MarTDR1 (arbitrary primer) 5’ CAACCGTGGCGGGGNTNCNNGNCNCG 3’
If your TD-PCR reaction does not work with the MarLseq/MarTDL2, try the MarRseq/MarTDR1 set.
PCR program :Step Temperature Time 1 95°C 5 min 2 95°C 45 sec 3 60°C 45 sec; program to decrease by 0.5 °C per cycle 4 72°C 2 min 5 Go to Step 2 24 times 6 95°C 45 sec 7 50°C 45 sec 8 72°C 2 min 9 Go to Step 6 24 times 10 4°C Forever Run 5 µl of your PCR reaction mix on a 1.0 % agarose gel.
Compare the products of the reactions of putative mutants with the product from the negative control (wild-type DNA). Any reaction that yielded a product that is essentially the same as the negative control should be discarded. PCR reactions that yield one or more strong bands should be sequenced. Reactions that result in a smear should also be discarded.
-
PCR purify the products of the reactions that yielded appropriate products. Elute the purified product in sterile 30 µl mqH2O.
This removes unwanted reaction components such as free nucleotides, primers, salts, and polymerase.
Sequence purified PCR products.
BASIC PROTOCOL 5: CREATION OF MARKERLESS DELETIONS BY ALLELIC REPLACEMENT
This method relies on homologous recombination, and a counter-selectable plasmid marker to generate allelic replacement mutants. There are two vectors that work very well for this process, pKNG101 (SmR, Kaniga et. al. 1991) and pNPTS138 (KmR, Hibbing et al., 2011, Sarker, 1997 and Alley, unpublished). Neither can replicate in A. tumefaciens and both contain an antibiotic resistance marker in addition to the counter-selectable marker, sacB.
Materials
Plasmid to generate primary integrants (see Support Protocols 1 and 2)
28°C incubator
ATGN plates supplemented with appropriate antibiotics
ATSN plates with and without antibiotics
1.6 ml microcentrifuge tubes
5 ml glass culture tubes
Introduce the plasmid to generate primary integrants by one of two methods: A. electroporation, or B. conjugation with a donor strain of E. coli (See Basic Protocols I and 2, respectively)
Patch cells from antibiotic-resistant colonies onto ATGN plates (plus antibiotics), and onto 5% ATSN plates (plus antibiotics). Incubate at 28°C overnight, and the following day check for those clones that are antibiotic-resistant AND sucrose-sensitive. These primary integrants have the whole plasmid containing the antibiotic-resistance gene and sacB, conferring sucrose-sensitivity.
Streak purify several primary integrants on selective media (ATGN plus antibiotics) to eliminate non-resistant strains of A. tumefaciens, and in the case of conjugation, streak purify two successive times to remove any remaining E. coli cells.
Incubate several (3–4) primary integrants overnight, each in 2 ml ATGN liquid media without selection for the plasmid antibiotic resistance marker to allow for a secondary recombination event (excision of integrated plasmid DNA).
Plate 100 µl of overnight exponential phase cultures at a range of dilutions (this can vary somewhat depending on the genetic context) onto 5% ATSN (no antibiotics) and, incubate at 28°C to allow for secondary recombinant colonies to grow.
-
Patch sucrose-resistant colonies onto 5% ATSN plates, both with and without antibiotics. Select and streak purify all recombinants that are sucrose-resistant AND antibiotic-sensitive.
In C58, the hyperactive IS element, IS426, frequently inactivates sacB directly (Blair et. al, in prep). As a result, there can be a fair number of sucrose-resistant clones that retain resistance to the antibiotic. These clones still have the fully integrated plasmid and should be eliminated from further screening.
Screen by PCR to determine which secondary recombinants are wild-type and which are mutants (this should occur at a 50/50 ratio, but very often diverges from this frequency).
Support Protocol 1: Generating an Allelic Replacement Construct
This approach allows you to either i.) delete an existing gene, ii.) replace an existing gene with another gene of interest, or iii.) to site-specifically insert a novel gene into the genome. During the process above, you will select and screen for specific recombinants along the way, whereby you ultimately isolate a derivative containing the mutant sequence of DNA (carried on your allelic replacement construct).
To generate a plasmid for recombinational mutagenesis, it must include DNA sequence that is identical to the genomic DNA to allow for recombination.
Below are some examples of the sequence that should be included in an allelic replacement construct according to the intended purpose.
If the goal is to delete bases 600–1200 from a region of the genome, your plasmid should contain a sequence of DNA matching bases 1–599 and 1201–1700 in which base 599 is directly adjacent to base 1201.
If the goal is to insert a gene of interest into this region so that recombination would effectively replace bases 600–1200 with your gene of choice (e.g. antibiotic resistance or fluorescent protein gene). To do this, the gene of interest should be cloned into the allelic replacement construct, flanked on each side by fragments that will allow homologous recombination into the desired location (bases 1–599 and 1201–1700). It is however not mandatory that this new gene replaces an existing sequence in the genome (see iii.).
For a site-specific insertion, the gene of interest should be cloned directly between bases 1–599 and 600–1200.
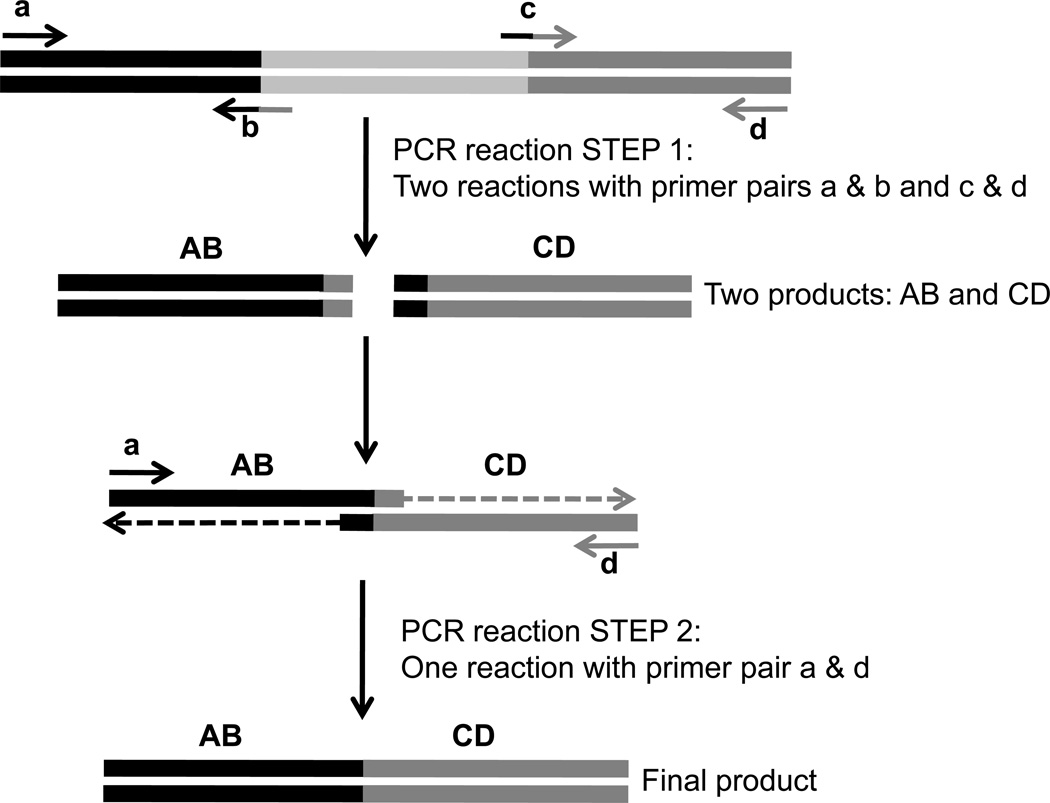
One highly effective way to generate a mutant sequence of nearly any of these types, is to take advantage of a PCR technique called SOEing (Splicing by Overlapping Extension) (See Figure 1). Below is a specific example of allelic replacement using the tetRA locus, but this method is an effective way to generate mutations in nearly any non-essential gene. To take advantage of this approach, four primers should be generated to amplify the regions flanking the section of DNA to be deleted. For SOEing to work, primers ‘b’ and ‘c’ should each contain sequences that are complimentary at the 3’ and 5’ regions, respectively. For example, primer ‘b’ should have 3’ sequence that is complimentary to the 5’ region of the downstream fragment (CD) that will be amplified (in Figure 1, this is represented by dark gray). In the example below, the primers are slightly different because they have been designed to include two restriction enzyme sites to allow for easy cloning of any other gene into the site.
Figure 1.
Schematic of PCR splicing by overlapping extension (SOEing). Light gray bars are representative of DNA sequence to be removed, whereas the black and dark gray bars represent sequence of the upstream and downstream flanking regions, respectively. Lower case letters denote primers, whereas upper case letters represent stretches of DNA.
To allow for efficient recombination to occur, it is recommended that the distance between primers ‘a’ and ‘b’ and primers ‘c’ and ‘d’ is at least 500 bp.
If the goal is to generate a non-polar deletion of a gene in an operon, the 3’ region of primer ‘b’ and the 5’ region of primer ‘c’ must be in frame with the coding sequence.
If the gene that is being deleted is at the beginning of an operon, sufficient sequence should be left (in fragment AB) to ensure maintenance of important regulatory elements and potential translational coupling between genes.
Support Protocol 2: Introducing a gene into the tetRA locus
One problem with using tetracycline in A. tumefaciens C58 is that spontaneous resistance to the antibiotic occurs at a high frequency via the activity of a normally cryptic tetracycline resistance locus, tetRA encoded on the C58 linear chromosome (Luo, 1998). Normally, the tetracycline efflux pump encoded by tetA is repressed by the adjacent repressor gene tetR. However, in A. tumefaciens there is an IS element, IS426 that will insert into the tetR gene rendering it inactive and leading to constitutive expression of tetA – these mutants are tetracycline resistant. One strategy that can be used to circumvent this issue is to remove this locus completely by allelic replacement as described above. In addition to making tetracycline a usable antibiotic, this provides an ideal location for introducing any gene of interest into the linear chromosome, such as a fluorescent protein or antibiotic resistance gene.
Primers for generating the allelic replacement mutation plasmid:
-
a.
tetRA-5'-5'-SpeI: ACT AGT CCT TCA TTT CGG CTG TTC AC
-
b.
tetRA -5'-3'-Link: CCA TGG TCT AGA GCT AGC CGG TAT GAA GGA GAA GCT GC
-
c.
tetRA -3'-5'-Link: GCT AGC TCT AGA CCA TGG CAC TTT ATC TTC TCG CCT TGC C
-
d.
tetRA -3'-3'-SalI: GTC GAC GGT ATT GGC GGC GGT ATT ATC
XbaI site = TCTAGA
NheI site = GCTAGC
-
STEP 1:
Use primer pairs a & b and c & d to amplify flanking regions of the tetRA locus.
-
STEP 2:
Use ab product and cd product as template with primer pair a & d to SOE the STEP 1 products together
-
STEP 3:
Ligate STEP 2 product into vector
Primers for mutation diagnostics:
-
e.
tetRA -ED-5': CTA CGT CGA AAT CAG GTG GT
-
f.
tetRA -ED-3': CTT CGT TTA CCT GAT TCT GTC CG
* ED = external diagnostic
BASIC PROTOCOL 6: PLASMID CURING
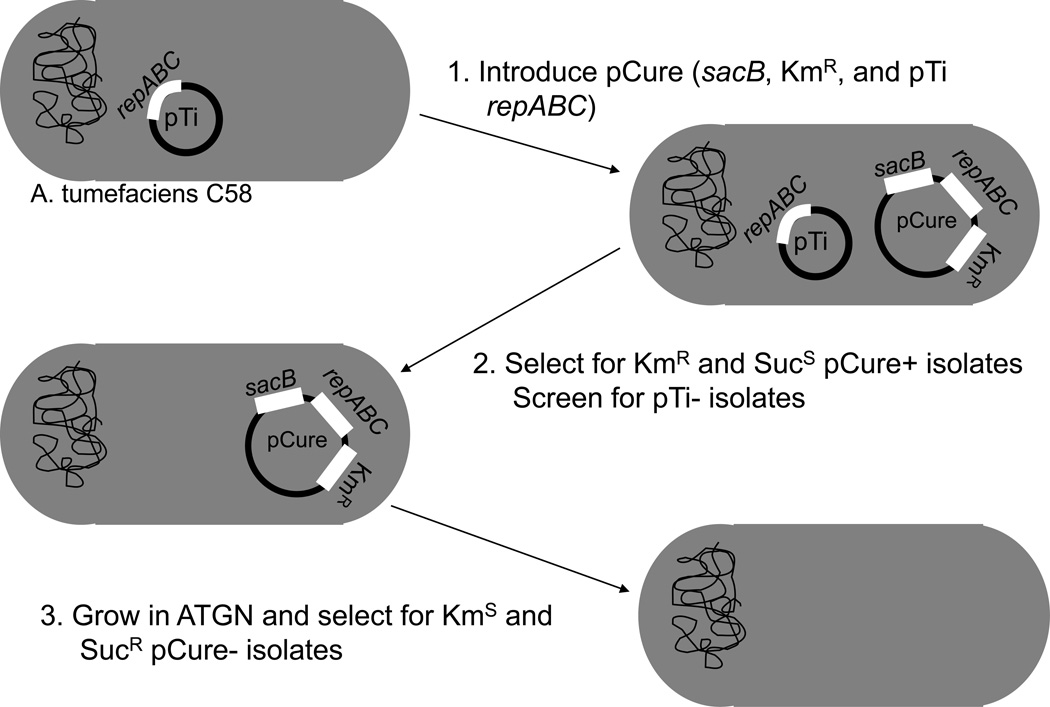
Plasmid incompatibility is largely determined by the sequences of the rep genes, and for agrobacterial plasmids these are of the repABC variety (Hynes, 1985 and Soberon et al., 2004). The effect is that only a specific number of plasmid copies containing the same incompatibility sequence can be maintained in a cell. Taking advantage of this phenomenon, plasmids can be ‘cured’ by generating a curing vector containing the same incompatibility region as the native plasmid (See Figure 2). This curing vector should possess an antibiotic resistance gene, as well as a counter-selectable marker such as sacB (conferring sucrose-sensitivity). Incompatibility conferred by repABC combined with antibiotic selection, will ensure that only those cells harboring the curing plasmid should persist. Subsequent growth on sucrose containing media will select for segregants that have lost the curing plasmid (by mis-segregation), thereby generating a plasmid-free strain.
Figure 2.
Diagram outlining strategy for plasmid curing by incompatibility. Gray ovals denote cells and black squiggly line represents chromosomal DNA. Plasmids are represented as black circles and associated genes as white rectangles.
Materials
Primers to amplify repABC (incompatibility) region of plasmid to be cured
A vector that lacks the replication genes suitable for multiplication in Agrobacterium, contains a selectable marker such as an antibiotic resistance gene, and a counter-selectable marker such as sacB (i.e. pNPTS138)
28°C incubator with shaking capablities
ATGN liquid and solid media (+/− antibiotics)
5% ATSN solid media (+/− antibiotics)
Petri plates
Grid for patching cells onto 2 types of media
Primers to amplify non-repABC sequences of the plasmid to be cured (for diagnostics)
Generate pCure containing repABC sequence of plasmid to be cured.
Introduce the pCure derivative into A. tumefaciens strain by electroporation or conjugation.
Select for cells containing the curing vector by plating onto selective ATGN media (plus antibiotic).
-
Patch antibiotic-resistant clones onto ATGN and 5% ATSN plates (both containing antibiotics) and incubate at 28°C overnight.
Note: Streak purify antibiotic-resistant clones three times if introducing plasmid by mating from E. coli
Check plates from Step 4 for clones that are both antibiotic-resistant AND sucrose-sensitive (these clones should have the curing plasmid). Inoculate several of these clones into liquid ATGN media containing antibiotics and incubate at 28°C with proper aeration.
Plate 10−7 dilutions of turbid cultures onto ATGN plus antibiotics and wait for colonies to form. Some or most of these clones should have lost the native plasmid. Diagnostics for this include a. PCR of a native plasmid sequence(s), b. phenotypic screen for loss of functions encoded on plasmids (eg. AHL production and degradation or opine catabolism), or c. Eckhardt gel (see Protocol 7).
Inoculate several liquid cultures of ATGN with plasmid-cured strains and incubate overnight with aeration.
Plate 100 µl of various dilutions (try 100 – 10−2) turbid cultures onto 5% ATSN plates (no antibiotics) to select for clones that have lost the curing vector. Incubate at 28°C and wait for colonies to form. Ideally, one would have at least 50 colonies to screen for sucrose-resistance, but this number can vary significantly and one colony could be sufficient.
Patch sucrose-resistant colonies onto 5% ATSN plates, both with and without antibiotics to screen for antibiotic-sensitive pCure clones.
Check for loss of both the native plasmid and the curing vector by performing PCR with repABC primers.
-
Freeze down plasmid-cured strains in 30% sterile glycerol at −80 C.
Genomic rearrangements during the curing process are possible and difficult to control. It is always a good idea to reintroduce the native plasmid back into the plasmid-cured strain and check for maintenance of wild-type phenotypes (growth, biofilm formation, motility, etc.)
PROTOCOL 7: ECKHARDT GEL FOR DETERMINING PLASMID COMPOSITION (Hynes et al., 1985)
This method is used to determine the plasmid composition of a strain. Large replicons such as pTi and pAt plasmids can be visualized. Our protocol is adapted from that used by Dr. Joel Griffitts (Brigham Young University) and colleagues for use with rhizobial plasmids.
Materials
28°C incubator with shaking capabilities
ATGN liquid media
5 ml culture tubes
spectrophotometer
electrophoresis apparatus and supplies
microcentrifuge
the following reagents:
5 × TBE Stock
0.5 × TBE
10% SDS in 0.5 × TBE
0.3% Sarkosyl in 0.5 × TBE
E1 solution
Eckhardt Lysis Solution (prepared fresh prior to use)
-
1
Grow overnight cultures of all strains of interest in ATGN growth media. Other media can be used, however a nutrient poor medium in which EPS production is low is preferable.
-
2Prepare 0.7% agarose, 1.0% SDS, 0.5 × TBE gel (for 100 ml gel):
- First dissolve 0.7 g agarose in 90 ml 0.5 × TBE by microwaving
- Allow to cool, and then add 10 ml 10% SDS in 0.5 × TBE
- Swirl to mix, and pour
- DO NOT add EtBr to gel or buffer
- Prep gel box with 0.5 × TBE (NO SDS)
-
3
When each culture has OD600 between 0.3 between 0.5, transfer 100 µl of culture to a microcentrifuge tube (on ice).
* Culture density can have an effect on results so try to harvest all strains at a similar optical density. It is important that the cells are actively growing, so if the cells have grown to a density that is too high, subculture and allow to grow for several hours before harvesting cells.
-
4
Add 500 µl of cold 0.3 % Sarkosyl in 0.5 × TBE. Spin for 3 min at 10,000 × g to pellet cells and then resuspend in 20 µl of freshly prepared Eckhardt lysis solution.
-
5
Load cell lysates into wells of gel. Prior to loading you may wish to flush wells of any SDS. Optionally, prior to loading you may add loading dye to your lysed cells to allow for visibility during well loading, but this is not necessary.
-
6
Electrophorese at 10 volts, 15 min to ensure that the cells are lysed.
-
7
Electrophorese at 70–80 volts, 6–8 h (depending on plasmids and separation desired). Alternatively you may run the gel for 12 hours/overnight at 40–50 volts. Either way, it is important that you leave the lid off the gel box because otherwise it gets too hot!
-
8
Post-stain gel for 30–45 min in a 0.5 µg/ml solution of EtBr (5 µl of EtBr Stock + 100 ml MQH2O). Note: EtBr cannot be added directly to gel and buffer in this protocol as it alters the migration of the plasmids and will differentially label DNA during the lysis procedure.
-
9
Optional: De-stain gel in MQH2O for 20 minutes. This helps detect faint bands.
-
12
Visualize with UV and photograph gel. Be careful because at this point the gel is more brittle than usual. Agrobacterium strains tend to have a more streaky background than those with Rhizobium and Sinorhizobium strains. The Ti plasmid typically migrates at approximately 3 cm in this protocol and sometimes the circular chromosome is visible above the plasmids.
REAGENTS AND SOLUTIONS
- 5 × TBE Stock
- 54 g Tris Base
- 27.5 g boric acid
- 20 ml of 0.5 M EDTA (pH 8.0)
- To 1L total using MQH2O
- 0.5 × TBE
- 1 part 5 × TBE Stock to 9 parts MQH2O
- 10% SDS in 0.5 × TBE
- 10 g SDS
- To 100 ml total using 0.5 × TBE
- 0.3% Sarkosyl in 0.5 × TBE
- 0.3g Sarkosyl
- To 100ml total using 0.5 × TBE
- Store at 4°C
- E1 solution
- 1g sucrose
- 5 µl RNase A solution (20 mg/ml)
- To 10 ml total 0.5 × TBE
- Filter sterilize
- Store at 4°C
- Eckhardt Lysis Solution (prepared fresh prior to use)
- Will use 20 µl per lane; prepare accordingly
- 1 part 10 mg/ml Lysozyme to 99 parts E1 solution
- Keep on ice
COMMENTARY
Background Information
Plasmid Introduction by Electroporation and Conjugation
Both of these strategies are highly effective ways of introducing foreign DNA into A. tumefaciens cells. Whether it is an expression plasmid, a curing plasmid, or an allelic replacement construct, they can all be delivered using either of these methods.
Transposon Mutagenesis
This strategy is primarily used for linking a phenotype of interest with one or multiple genes. It is most useful if you have an easily screenable phenotype, such as swimming motility or biofilm formation. Once the mutant library has been generated, individual clones can be assessed for the gain or loss of a particular function, which can subsequently be linked to a particular gene.
Touchdown PCR
This method for Touchdown PCR has been specifically designed for mapping the insertion site of the Mariner transposon Himar-1 but the basic concept can be used for any transposon. The protocol requires both a Mariner-specific and an arbitrary primer for amplifying the transposon and the flanking sequence. The strategy takes advantage of differential annealing based on a gradient of temperature changes, starting with a higher temperature (more specific annealing) and incrementally decreasing with each subsequent PCR cycle (less specific annealing). The result is multiple PCR products of varying lengths, all originating within the transposon.
Generation of a Markerless Deletion Strain
This approach uses PCR to generate a mutagenic deletion construct that can be used to drive homologous recombination to allow for highly efficient site-specific gene deletion. Alternatively, it provides a means for the introduction of another gene of interest, either directly into the genome, or by replacing an existing gene.
Plasmid Curing
Plasmids, particularly those associated with A. tumefaciens, although non-essential, carry many important genes. For example, crown gall disease for which A. tumefaciens is primarily known, in addition to its invaluable role as a tool for plant genetic engineering, are both characteristics of the species that are almost entirely conferred by the Ti plasmid. Alternative methods include exposing cells to stress such as heat shock or anitbiotics, or growing cells in the presence of DNA intercalating agents such as acridine orange or ethidium bromide (Trevors, 1986). These alternative methods however are likely to induce additional mutations to genomic DNA.
Just as with any specific isolated gene, to effectively study the role(s) of these plasmids in their entirety, it is imperative to be able to study derivatives that lack them. The method described in the protocol is considered to be one of the most effective, least problematic methods for curing a plasmid of this type.
Eckhardt Gel
This type of gel electrophoresis allows for the visualization of plasmid DNA. This can be informative in characterizing the plasmid profiles of a particular strain. For example, it can be an effective diagnostic tool in the above procedure to determine if a particular plasmid has been cured.
Critical Parameters and Troubleshooting
Plasmid Introduction by Electroporation
It is imperative that electrocompetent cells are in exponential phase at the time of harvest, are kept cold (on ice) at all times, and are washed thoroughly in sterile cold water to remove all residual salts from the media. If the salt concentration is high and ‘arcing’ occurs, a loud popping sound during the pulse application, it is because there is too much salt remaining in the cells or DNA preparation and they should be washed more thoroughly. Higher concentrations of glycerol can decrease transformation efficiency, so for storage of electrocompetent cells it is best to keep glycerol at 25–30%.
Plasmid Introduction by Conjugation/Transposon Mutagenesis
If there are difficulties obtaining transconjugants and/or transposon mutants it is advisable to check whether donor strain of E. coli is tra+ and that the plasmid can be mobilized. It is also never a bad idea to perform diagnostics (miniprep and restriction digest, or PCR) to confirm that the delivery strain harbors the correct plasmid. It is critical when transferring a plasmid from E. coli to A. tumefaciens that the mating cells are incubated at 28°C and that selection for transconjugants is performed using AT minimal media (on which, most laboratory strains of E. coli are auxotrophic mutants and cannot grow) and the antibiotic concentrations appropriate for A. tumefaciens. When transferring a plasmid from one A. tumefaciens strain to another, it is imperative that the recipient strain possesses some unique type of resistance relative to the donor for counterselection. If there is growth on the negative control plates (donor or recipient only), it is worth preparing fresh antibiotic solutions. Ampicillin for example, has been demonstrated to be pH and heat sensitive, and will degrade over time.
Touchdown PCR
If sequences that are obtained are not clean, one potential explanation is that the specific primer is binding elsewhere within the genome of the mutant, in addition to the site of transposon insertion. If this is the case, it may be necessary to design a new specific primer, that is unique to the transposon and will not amplify any product from the negative (unmutated strain) control. If a PCR product is not obtained, it is possible that antibiotic-resistant clones are spontaneous resistance mutants. This is more of an issue with certain species. A. radiobacter K84 for example will form spontaneous kanamycin-resistant mutants at a frequency that is similar to the rate of transposon insertion. Using the antibiotics kanamycin and neomycin in combination can help to circumvent this issue.
Generating Markerless Deletions by Allelic Replacement
The critical parameters for this protocol are more thoroughly outlined in the protocol itself. It is however worth noting that in A. tumefaciens C58, if there are difficulties obtaining primary integrants that are antibiotic-resistant and sucrose-sensitive, this could be due to the presence of the insertion element IS426, which inserts at high frequency into the sacB gene, rendering it non-functional. If this is an issue, it is recommended to screen a larger number of primary integrants. If the roadblock occurs at the second recombination step (generating antibiotic-sensitive and sucrose-resistant mutants), it could be that the mutation results in lethality for one reason or another (e.g. essentiality). Expression of the targeted gene from a plasmid using a controllable exogenous promoter before attempting to generate the mutant in the native gene is one way to potentially solve the issue of essentiality. If the mutant still cannot be isolated, it is possible that the gene is essential AND requires cis-acting regulatory elements in order to function properly.
Plasmid Curing
If difficulties are encountered in obtaining plasmid-cured derivatives, it can be helpful to try one of several additional measures. Growing A. tumefaciens cells at higher temperatures results in increased divisional errors and segregational loss. This is not an ideal method to employ because it puts stress on the cells that is likely to result in other mutations throughout the genome. The At plasmid of C58 for example, is very difficult to cure (Morton et al., in preparation). This may be attributed to one or more toxin-antitoxin systems carried on the plasmid. When growing pCure+ cells in the presence of antibiotic (the resistance to which is conferred by the curing vector), if the growth inhibition imparted by such toxin-antoxin systems is high enough, the frequency of recombination between the repABC regions of the two plasmids may be more likely than curing event. One way to limit the likeliness of recombination is to generate a pCure plasmid that only contains the region of incompatibility (rather than the entire rep region). This however will vary from plasmid to plasmid.
Eckardt Gel
If particular strains do not resolve well, omitting the Sarkosyl step may help, and in some cases this improves the success rate. Experimenting with different numbers of cells, different incubation times or slightly different media (e.g. lower CaCl2, more glucose) can also help.
Anticipated Results
The purpose of the methods and protocols outlined in this unit is to obtain cells and genetic derivatives that will be used for phenotypic analyses.
Time Considerations
All of the basic protocols described are only limited by the time it takes A. tumefeciens cells to grow. It is worth noting that plasmid introduction by electroporation (as opposed to conjugation) leads to longer recovery times (2–3 days as opposed to 1–2). Touchdown PCR is a basic protocol that can be completed in a matter of hours. Generation of allelic replacement mutants is a multi-step process that can take 1–2 weeks due to the time it takes to select, purify, and screen for mutants. Plasmid curing can be accomplished in a week, although this varies with the plasmid (the At plasmid for example, is very difficult to cure and may require multiple attempts and longer passaging of pCure+ isolates). Running an Eckhardt gel is a two day process, with the second day being the longest due to the time it takes to run the gel.
Acknowledgement
Studies of Agrobacterium in the Fuqua Lab are supported by the National Institutes of Health (GM092660 and GM080546).
Footnotes
INTERNET RESOURCES:
Genome Sequencing of Agrobacterium Biovar Type Strains, Virginia Bioinformatics Institute http://agro.vbi.vt.edu/public/
LITERATURE CITED
- Hibbing ME, Fuqua C. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J. Bacteriol. 2011;193(14):3461–3472. doi: 10.1128/JB.00317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes MF, Simon R, et al. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985;13(2):99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Delor I, et al. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, et al. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci., USA. 1999;96(20):11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levano-Garcia J, Verjovski-Almeida S, da Silva ACR. Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. Biotechniques. 2005;38:225–229. doi: 10.2144/05382ST03. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, Farrand SK. Cloning and characterization of a tetracyline resistance determinant present in Agrobacterium tumefaciens C58. J Bacteriol. 1999;181:618–626. doi: 10.1128/jb.181.2.618-626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersereau M, Pazour GJ, Das A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene. 1990;90(1):149–151. doi: 10.1016/0378-1119(90)90452-w. [DOI] [PubMed] [Google Scholar]
- Sarker M, Cornelis GR. An improved version of suicide vector pKNG101 for gene replacement in Gram negative bacteria. Molecular Microbiology. 1997;23:409–411. doi: 10.1046/j.1365-2958.1997.t01-1-00190.x. [DOI] [PubMed] [Google Scholar]
- Soberon N, et al. Incompatibility and the partitioning site of the repABC basic replicon of the symbiotic plasmid from Rhizobium etli. Plasmid. 2004;51(3):203–216. doi: 10.1016/j.plasmid.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Tomlinson AD, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr. Opin. Microbiol. 2009;12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors JT. Plasmid curing in bacteria. Fems Micr Rev. 1986;32(3–4):149–157. [Google Scholar]