Abstract
Viruses are the most abundant life forms on Earth, with an estimated 1031 total viruses globally. The majority of these viruses infect microbes, whether bacteria, archaea or microeukaryotes. Given the importance of microbes in driving global biogeochemical cycles, it would seem, based on numerical abundances alone, that viruses also play an important role in the global cycling of carbon and nutrients. However, the importance of viruses in controlling host populations and ecosystem functions, such as the regeneration, storage and export of carbon and other nutrients, remains unresolved. Here, we report on advances in the study of ecological effects of viruses of microbes. In doing so, we focus on an area of increasing importance: the role that ocean viruses play in shaping microbial population sizes as well as in regenerating carbon and other nutrients.
Introduction
The study of viruses of microbes has undergone a revolution in the past 20 years. What has been termed “The Third Age of Phage” [1] can be viewed more broadly as a renewal of interest in environmental viruses, and in particular in viruses of microbes [2-11]. This renewal has been spurred by advances in direct-imaging methods, sequencing technologies and bioinformatics that have revealed a previously unknown world of viral diversity in natural environments [12]. The study of viruses in the oceans has been key to many discoveries: from the finding that viruses have evolved novel lineages of key photosynthetic genes (as in cyanophages infecting Synechococcus and Prochlorococcus [13-17]) to the identification of many novel viral families that defy much of the conventional wisdom about viral life history (e.g. the discovery of “giant” algal-infecting viruses [18,19] and even viruses that exploit other viruses [20]). Altogether, it is well established that a diverse reservoir of viruses infect and lyse bacteria, archaea and microeukaryotes at the base of the ocean's food web [2,21]. Despite the growing literature on viruses of microbes, we remain relatively poorly informed concerning the dynamic, ecological effects of virus infection of marine microbes. In this report, we focus on the functional role of viruses within oceanic waters. In doing so, we highlight knowns, unknowns and challenges in three areas: (i) the role of ocean viruses in shaping microbial community composition; (ii) the role of ocean viruses in determining carbon and nutrient availability; (iii) modelling efforts to link viral infection of microbes with ecosystem-scale consequences.
Viral infection and microbial community composition
Viruses, in general, and viruses of microbes, in particular, infect a subset of available hosts in any environment. The subset of hosts that a virus typically infects is narrower than the range of microbes that are typically consumed by a grazer species (e.g. a protist or zooplankton). However, there is evidence that viruses of microbes can infect hosts from different species or even different genera. For example, some cyanophages can infect different strains within the same cyanobacteria species, either Synechococcus or Prochlorococcus, as well as strains of different cyanobacterial genera [13]. The quantitative study of the host-range of viruses of microbes is in its infancy [22-26], an issue we return to later. Successful viral infections can lead to lysis of hosts as well as the conversion of hosts into lysogens where the viral genome is integrated into that of the host, subsequently altering host physiology [2,21,27]. Lytic viral infection of microbes and its effects are our primary concerns here.
Viral lysis of microbes is thought to have direct effects on microbial community composition. Ostensibly, viral lysis should decrease the abundance of specific microbial lineages that are targeted by viral infection [28]. Estimates of bacterial-induced mortality suggest that viruses are, in some cases, as important as grazers in selectively killing microbes [29,30], whereas, in other cases, they may be the dominant source of microbial mortality [31,32]. The depletion of susceptible bacteria leads to the possibility of dynamic fluctuations in viral and microbial populations, a result predicted by simple population models [33]. Nonetheless, direct evidence for coupled oscillations between virus-microbe systems in the oceans is limited. The reasons are complex, but likely due to the fact that the emergence of new (or previously rare) viral subtypes occurs frequently and rapidly [34-37]. The changing identities of strains and their populations can make it difficult to infer the consequences of virus-microbe interactions. Hence, paradoxically, the seemingly most apparent consequences of viral-induced lysis of microbes may be hard to observe in practice. We anticipate that observations of virus and microbial abundances in situ via genomic and metagenomic methods, with improved time-scale resolution, will provide direct evidence for specific functional relationships between viruses and microbial hosts.
Viral infection and the availability of carbon and nutrients
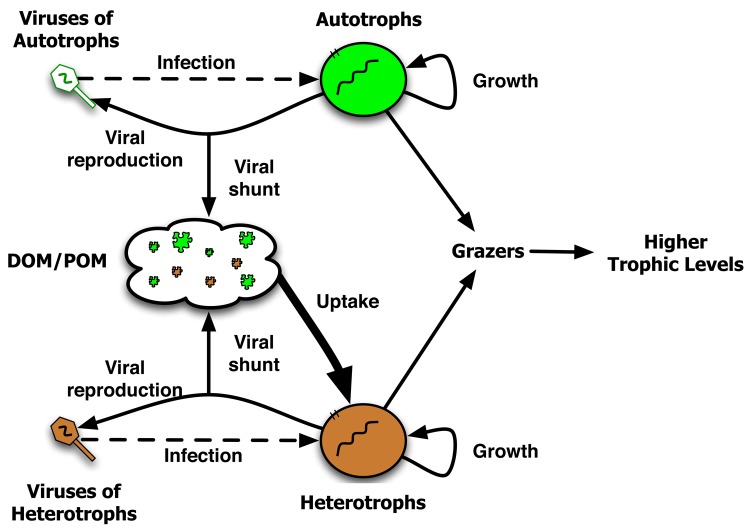
Virus-induced mortality of microbes has direct effects on ecosystem function. Lysis of microbes involves the release of organic carbon and other nutrients back into the environment. This redirection is known as the viral shunt [8]. The viral shunt denotes the fact that cellular materials released as particular or dissolved organic material are not directly available for utilization by organisms from higher trophic levels (e.g. plankton and fish) (see Figure 1) but are primarily utilized by predominantly heterotrophic bacteria, although some efforts have shown nutrients released in this manner to be rapidly assimilated by eukaryotic plankton [38].
Figure 1. Schematic of the viral shunt.
The lysis of microbes by viruses releases cellular material into the environment. Some of this cellular material can be utilized by microbes for subsequent metabolic processes. Here, we note that available dissolved organic material (DOM) and particulate organic material (POM) is utilized primarily by heterotrophs (the thicker arrow leading downward from the DOM/POM pool). Note that many other processes are not included in this schematic, e.g. sinking out of the system, exudation during growth, light input, or the influx of inorganic carbon and nutrients. Abbreviations: DOM, dissolved organic material; POM, particulate organic material.
Direct efforts to estimate the viral shunt are rare, with most indirectly determining virus turnover rates and from this inferring virus-mediated elemental release. While several approaches exist to estimate virus production [39], currently favoured approaches all depend on the same steps: dilution of samples and estimates of virus reoccurrence [31]. Application of these approaches in recent years has begun to provide a broad overview of the variability of virus effects across different oceanic realms [40-43], with all observations pointing to differing constraints on virus activities in different systems.
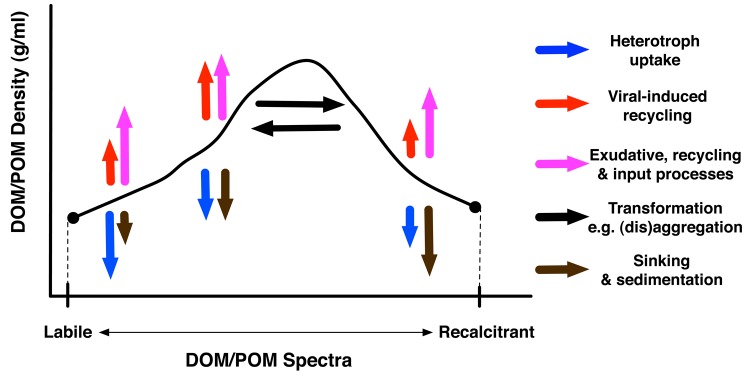
In addition, recent evidence suggests that viral lysis of microbes changes the relative distribution of dissolved organic matter with many indirect effects in ocean ecosystems. For example, viral lysis of microbes shifts organic matter from cells into dissolved and particular organic pools. The type of organic material released in viral lysis includes a spectrum of molecules ranging from bio-available (i.e. “labile”) to recalcitrant (see Figure 2) and may be dependent as much on the location of the lytic event as the players involved [44]. In the deep ocean, the fate of virus-released organic matter remains a mystery, but this process may drive the generation of the ancient organic carbon measured by marine chemists [45]. Moreover, it is known that virus activity may drive the formation of marine snow by releasing “sticky” components from within cells [46], while at the same time disaggregating particles through cell lysis [44]. Overall, we still know little about how virus activity changes the character of dissolved organic matter, the effect of viruses on carbon distribution (i.e. fixation, respiration, mineralization and export) in marine systems, despite the potential impacts on global budgets on both short-term and geological time scales.
Figure 2. Schematic of the role of viruses in the differential regeneration of organic matter.
The release of cellular materials by viral lysis depicted in Figure 1 increases the dissolved organic matter (DOM) and particulate organic matter (POM) in the environment. Lysis also changes the abundance of distinct biomolecules. Here, for purposes of illustration, we simplify the high-dimensional configuration space of biomolecules into a one-dimensional axis. The x-axis denotes the degree to which organic material is easy to utilize by organisms (i.e. labile) vs. hard to utilize by organisms (i.e. recalcitrant). The y-axis denotes the amount of organic material across this accessibility spectrum. The vertical arrows denote how a particular process can change the availability of different organic materials, potentially in non-uniform ways. The horizontal arrows denote that some processes may convert material from one state to another without changing the total amount of material. The size of the arrows represents an estimated magnitude of a given process. We note that characterizing the space of biomolecules and the role of viruses in modifying the relative abundance of biomolecules is an ongoing challenge. There are data to substantiate the existence of horizontal and vertical arrows (e.g. [41]) but less data to substantiate the correct size of horizontal and vertical arrows. Abbreviations: DOM, dissolved organic material; POM, particulate organic material.
In addition to direct effects, the differential conversion of cellular material by viruses into a spectrum of dissolved organic material may indirectly affect the growth of microbial populations. An intriguing hypothesis is that heterotrophs and autotrophs may be “primed” (i.e. stimulated) by the lysis of microbes [38,45,47-51]. This includes the release of lysis byproducts that may stimulate the growth of a subset of heterotrophs [3] as well as eukaryotic auxotrophs [52]. Characterizing priming and its consequences constitutes an important area of future research, as determining the fate of carbon released from lysed cells is critical to understand how it is recycled or removed (i.e. rendered recalcitrant) from marine carbon budgets [44,45,53]. The study of nutrients may also be critical to this goal, as it has been demonstrated that the release of bioavailable nutrient elements, including N and Fe [38,54], may maintain ecosystem productivity under conditions where nutrient availability limits carbon production.
Finally, the infection of microbes also alters host metabolism, often in significant ways. For example, cyanophage-infecting cyanobacteria, such as Prochlorococcus sp. and Synecochoccus sp., increase the overall photosynthetic rate of microbes, presumably changing the fixation rate of carbon from the environment before lysis. Cellular-based studies have confirmed that pathways within cells can be altered at the metabolic level [55-57]. Similarly, persistent infections, such as occurs during lysogeny, can change the rate and type of utilization processes. Hence, infection can alter the behaviour of microbes, as seen from an ecosystem perspective. The net effect of such differential metabolism at the ecosystem level, however, remains largely unknown. Estimates of the contribution of lysogeny production to virus activity in marine systems remain highly variable [21,58,59]. Future work should likely consider changes in cellular biochemistry as well as differences in cellular processes (e.g. growth efficiency, respiration and photosynthesis, etc.) between lysogens and uninfected cells. Quantifying these differences is likely to be an important component of efforts to census the overall rates at which viruses affect ecosystem functions.
Modelling efforts to link viral lysis with carbon and nutrient availability
Dynamic models of viral-host interactions in marine environments have, at their core, the same set of assumptions as were made for dynamics of Escherichia coli and associated phage in chemostats [33]. These now-classic “box” models consider how abundances of hosts, viruses and resources change with time due to the effects of resource uptake, cell division, viral-induced mortality of hosts, and viral reproduction via lysis. Extensions have taken into account a greater complexity of factors, including the possible covariation of bacterial life history traits and viral life history traits [34,35,60]. The most prominent extension to ocean systems is termed “Kill-the-Winner” [28,61], in which viruses specialize in distinct hosts and contribute to the successive decline of host strains that rise to dominance. Kill-the-Winner assumes one-to-one relationships between individual viruses and hosts. The cross-infection of viruses and hosts is more complex. For example, empirical assays of viral-host cross-infectivity reveal that viruses can infect many microbial hosts, and, similarly, microbes can be infected by many viruses [13,24-26]. A reanalysis of phage-bacteria cross-infection assays found a characteristic nested pattern [23]. The nested pattern implies there is a hierarchy of infection ability (amongst viruses) and susceptibility to infection (amongst bacteria) within the re-examined studies. Hence, a major question moving forward is: do the predictions of Kill-the-Winner and other population ecology models change given other cross-infection assumptions? In the context of the first direct effect outlined earlier, it would be important to quantify the relationship between who infects whom and the relative abundance and survival of host and viral strains.
Perhaps the biggest impediment to predictive modelling of direct and indirect effect of viruses of microbes on microbial community composition and ecosystem function is the empirical observation that rare genotypes can rise to observable levels rapidly and frequently [62-66]. These changes may be the result of de novo mutation of successful genotypes as well as emergence of pre-existing, yet rare, microbes that experience changes in conditions that dramatically affect fitness. Unifying ecological and evolutionary dynamics in a single model is difficult, and most models of host-viral dynamics neglect evolution entirely. A few models are available that have begun to assess how such evolutionary details matter. First, it has been proposed that evolutionary changes in allocation of phytoplankton to virus resistance vs. growth would lead to the evolution of co-limitation by hosts rather than the maximization of host productivity in a given environment [67]. However, the cost of resistance to viral infection remains largely uncharacterized in natural communities (see [68] for a laboratory study using marine cyanobacteria). Second, an evolutionary Kill-the-Winner model predicts that constant total diversity can emerge at the community level even if the identities of strains are in constant flux [69]. This model predicts that at the community scale there may be invariants for which fine-scale evolutionary details do not matter. Some empirical support for this idea comes from one study in which changes in system nutrient levels were buffered despite rapid changes in strains coevolving over a multi-month experiment [70]. Third, multiple coevolutionary models of viral-host interactions mediated by receptor resistance [71,72] and CRISPR immune defense [73] demonstrate principles under which coevolutionary-induced diversification may arise. Linking models to data and understanding when and which details matter is an issue relevant to the study of viruses of microbes and to problems in microbial ecology more generally [74].
Viruses are everywhere, but what are they doing and how important are they?
The potential role of viruses in marine biogeochemical cycles [75,76] and the viral shunt [8] has been discussed for nearly two decades now, yet the quantitative impact viruses have on regional and global scale processes remains generally unresolved apart from estimates from a few marine virus production surveys [e.g. 32,38,40,41,77-81]. This uncertainty with respect to quantifying viral effects manifests itself in two critical ways. First, there are a number of global earth systems and climate models that integrate geophysical processes with the biology of microbes to metazoans to predict the dynamics of carbon nutrients and biodiversity [82-84]. However, the smallest biotic agents on the planet – viruses – are rarely, if ever, included in such models. Second, the most recent Intergovernmental Panel on Climate Change released its last report in 2007 noting: “The overall reaction of marine biological carbon cycling (including processes such as nutrient cycling as well as ecosystem changes including the role of bacteria and viruses) to a warm and high-CO2 world is not yet well understood. Several small feedback mechanisms may add up to a significant one” [85]. The emphasis is ours. The assessment of quantitative effects of viruses on populations, communities and ecosystems in a form that can be integrated into large-scale models and climate change scenarios should be a major research goal.
There are many challenges to reach this goal. First, data on the quantitative effects of viruses on ecosystem processes are highly variable, and may reflect differences in collection and estimation methods in addition to natural biological variation. There is a need for increased standardization of methods (e.g. [86]) as well as the integration of existing data to relate viral interaction and effect data to environmental and geographic drivers. Second, new methods are being developed to characterize the genomes of viruses of microbes and the microbes they infect (e.g. [87]). Yet, how much do these details matter? That is, are there particular host-viral interactions, and even particular lineages or ecotypes, that are key to the flux of particular types of organic and inorganic pools? Multi-scale mathematical methods can help infer viral effects at large scale based on cellular level interactions. Finally, there is a growing emphasis on the quantification of genomic and transcriptomic diversity within ocean microbes. However, predicting ecosystem-level effects of viruses of microbes requires quantitative estimates of rates and interactions coinciding with genomic and transcriptomic surveys. We hope that future work on viruses of microbes in the oceans includes efforts to combine rates, -omics data and mathematical models in the service of answering a fundamental question: what effects do viruses have on the global earth system?
Acknowledgments
Joshua S. Weitz holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and acknowledges the support of an award from the James S. McDonnell Foundation. Steven W. Wilhelm acknowledges support from the National Science Foundation, Division of Biological and Chemical Oceanography and the Division of Environmental Biology. This work was conducted as a part of the Ocean Viral Dynamics Working Group at the National Institute for Mathematical and Biological Synthesis, sponsored by the National Science Foundation, the U.S. Department of Homeland Security, and the U.S. Department of Agriculture through NSF Award #EF-0832858, with additional support from The University of Tennessee, Knoxville
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/4/17
References
- 1.Mann NH. The third age of phage. PLoS biology. 2005;3(5):753–5. doi: 10.1371/journal.pbio.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiology and Molecular Biology Reviews. 2000;64(1):69–113. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, Kimmance SA, Middelboe M, Nagasaki K, Paul JH, Schroeder DC, Suttle CA, Vaqué D, Wommack KE. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME Journal. 2008;2(6):575–8. doi: 10.1038/ismej.2008.31. [DOI] [PubMed] [Google Scholar]
- 4.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459(7244):207–12. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 5.Suttle CA. Viruses in the sea. Nature. 2005;437(7057):356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 6.Suttle CA. Marine viruses - major players in the global ecosystem. Nature Reviews Microbiology. 2007;5(10):801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 7.Thurber RV. Current insights into phage biodiversity and biogeography. Current Opinion in Microbiology. 2009;12(5):582–7. doi: 10.1016/j.mib.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea - viruses play critical roles in the structure and function of aquatic food webs. Bioscience. 1999;49(10):781–8. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 9.Viruses of Microbes. http://events.embo.org/12-virus-microbe/ [DOI]
- 10.Aquatic Viral Workshop. http://www.avw6.org/avw6 [DOI]
- 11.Danovaro R, Corinaldesi C, Dell'Anno A, Fuhrman JA, Middelburg JJ, Noble RT, Suttle CA. Marine viruses and global climate change. FEMS Microbiology Reviews. 2011;35(6):993–1034. doi: 10.1111/j.1574-6976.2010.00258.x. [DOI] [PubMed] [Google Scholar]
- 12.Edwards RA, Rohwer F. Viral metagenomics. Nature Reviews Microbiology. 2005;3(6):504–10. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424(6952):1047–51. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 14.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438(7064):86–9. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]; F1000 Factor 15Evaluated by Tracy Palmer 08 Nov 2005, Daniel Chamovitz 16 Dec 2005, Oded Beja 06 Aug 2012
- 15.Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(30):11013–8. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Joshua Weitz 16 Aug 2012
- 16.Clokie MRJ, Shan JY, Bailey S, Jia Y, Krisch HM, West S, Mann NH. Transcription of a 'photosynthetic' T4-type phage during infection of a marine cyanobacterium. Environmental Microbiology. 2006;8(5):827–35. doi: 10.1111/j.1462-2920.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- 17.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature. 2003;424(6950):741–1. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 18.Derelle E, Ferraz C, Escande ML, Eychenie S, Cooke R, Piganeau G, Desdevises Y, Bellec L, Moreau H, Grimsley N. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One. 2008;3(5):e2250. doi: 10.1371/journal.pone.0002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson WH, Schroeder DC, Allen MJ, Holden MTG, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309(5737):1090–2. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 20.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455(7209):100–U165. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Chantal Abergel 13 Aug 2008, James Conway 01 Sep 2008, Joshua Weitz 16 Aug 2012
- 21.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiology Reviews. 2004;28(2):127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Comeau AM, Buenaventura E, Suttle CA. A persistent, productive, and seasonally dynamic vibriophage population within Pacific oysters (Crassostrea gigas) Applied and Environmental Microbiology. 2005;71(9):5324–4. doi: 10.1128/AEM.71.9.5324-5331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Statistical structure of host-phage interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):E288–97. doi: 10.1073/pnas.1101595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmfeldt K, Middelboe M, Nybroe O, Riemann L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Applied and Environmental microbiology. 2007;73(21):6730–9. doi: 10.1128/AEM.01399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poisot T, Lepennetier G, Martinez E, Ramsayer J, Hochberg ME. Resource availability affects the structure of a natural bacteria-bacteriophage community. Biology Letters. 2011;7(2):201–4. doi: 10.1098/rsbl.2010.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schütt C. Bacteriophage diversity in the North Sea. Applied and Environmental Microbiology. 1998;64(11):4128–33. doi: 10.1128/aem.64.11.4128-4133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson SJ, Paul JH. Environmental factors that influence the transition from lysogenic to lytic existence in the ϕHSIC/Listonella pelagia marine phage-host system. Microbial Ecology. 2006;52(2):217–25. doi: 10.1007/s00248-006-9113-1. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Joshua Weitz 16 Aug 2012
- 28.Thingstad TF, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquatic Microbial Ecology. 1997;13(1):19–27. doi: 10.3354/ame013019. [DOI] [Google Scholar]
- 29.Steward GF, Smith DC, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Marine Ecology Progress Series. 1996;131:287–300. doi: 10.3354/meps131287. [DOI] [Google Scholar]
- 30.Tuomi P, Kuuppo P. Viral lysis and grazing loss of bacteria in nutrient- and carbon-manipulated brackish water enclosures. Journal of Plankton Research. 1999;21(5):923–37. doi: 10.1093/plankt/21.5.923. [DOI] [Google Scholar]
- 31.Wilhelm SW, Brigden SM, Suttle CA. A dilution technique for the direct measurement of viral production: a comparison in stratified and tidally mixed coastal waters. Microbial Ecology. 2002;43(1):168–73. doi: 10.1007/s00248-001-1021-9. [DOI] [PubMed] [Google Scholar]
- 32.Winget DM, Wommack KE. Diel and daily fluctuations in virioplankton production in coastal ecosystems. Environmental Microbiology. 2009;11(11):2904–14. doi: 10.1111/j.1462-2920.2009.02038.x. [DOI] [PubMed] [Google Scholar]
- 33.Levin BR, Stewart FM, Chao L. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. American Naturalist. 1977;111:3–24. doi: 10.1086/283134. [DOI] [Google Scholar]
- 34.Middelboe M. Bacterial growth rate and marine virus-host dynamics. Microbial Ecology. 2000;40(2):114–24. doi: 10.1007/s002480000050. [DOI] [PubMed] [Google Scholar]
- 35.Middelboe M, Hagstrom A, Blackburn N, Sinn B, Fischer U, Borch NH, Pinhassi J, Simu K, Lorenz MG. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microbial Ecology. 2001;42(3):395–406. doi: 10.1007/s00248-001-0012-1. [DOI] [PubMed] [Google Scholar]
- 36.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320(5879):1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 37.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environmental Microbiology. 2009;11(2):457–66. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Anna-Louise Reysenbach 13 Mar 2009
- 38.Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. Viral release of iron and its bioavailability to marine plankton. Limnology and Oceanography. 2004;49(5):1734–41. doi: 10.4319/lo.2004.49.5.1734. [DOI] [Google Scholar]
- 39.Weinbauer MG, Rowe JM, Wilhelm SW. Determining rates of virus production in aquatic systems by the virus reduction approach. In: Wilhelm SW, Weinbauer MG, Suttle CA, Waco TX, editors. Manual of Aquatic Viral Ecology. ASLO; 2010. pp. 1–8. [DOI] [Google Scholar]
- 40.Matteson AR, Loar SN, Pickmere S, DeBruyn JM, Ellwood MJ, Boyd PW, Hutchins DA, Wilhelm SW. Production of viruses during a spring phytoplankton bloom in the South Pacific Ocean near of New Zealand. FEMS Microbiology Ecology. 2012;79(3):709–19. doi: 10.1111/j.1574-6941.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- 41.Rowe JM, DeBruyn JM, Poorvin L, LeCleir GR, Johnson ZI, Zinser ER, Wilhelm SW. Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. FEMS Microbiology Ecology. 2012;79(2):359–70. doi: 10.1111/j.1574-6941.2011.01223.x. [DOI] [PubMed] [Google Scholar]
- 42.Rowe JM, Saxton MA, Cottrell MT, DeBruyn JM, Berg GM, Kirchman DL, Hutchins DA, Wilhelm SW. Constraints on viral production in the Sargasso Sea and North Atlantic. Aquatic Microbial Ecology. 2008;52(3):233–44. doi: 10.3354/ame01231. [DOI] [Google Scholar]
- 43.Winter C, Herndl GJ, Weinbauer MG. Diel cycles in viral infection of bacterioplankton in the North Sea. Aquatic Microbial Ecology. 2004;35(3):207–16. doi: 10.3354/ame035207. [DOI] [Google Scholar]
- 44.Weinbauer MG, Chen F, Wilhelm SW. Virus-mediated redistribution and partitioning of carbon in the global oceans. In: Jiao N, Azam F, Sanders S, editors. Microbial carbon pump in the ocean. Washington DC: Science/AAAS; 2011. pp. 54–6. [Google Scholar]
- 45.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo TW, Chen F, Azam F. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nature Reviews Microbiology. 2010;8(8):593–9. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 46.Peduzzi P, Weinbauer MG. Impact of virus abundance on the formation of algal flocs (marine snow) in the sea. Nature. 1992 [Google Scholar]
- 47.Weinbauer MG, Bonilla-Findji O, Chan AM, Dolan JR, Short SM, Simek K, Wilhelm SW, Suttle CA. Synechococcus growth in the ocean may depend on the lysis of heterotrophic bacteria. Journal of Plankton Research. 2011;33(10):1465–76. doi: 10.1093/plankt/fbr041. [DOI] [Google Scholar]
- 48.Middelboe M, Jorgensen NOG. Viral lysis of bacteria: an important source of dissolved amino acids and cell wall compounds. Journal of the Marine Biological Association of the United Kingdom. 2006;86(3):605–12. doi: 10.1017/S0025315406013518. [DOI] [Google Scholar]
- 49.Middelboe M, Jorgensen NOG, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Applied and Environmental Microbiology. 1996;62(6):1991–7. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Middelboe M, Lyck PG. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquatic Microbial Ecology. 2002;27(2):187–94. doi: 10.3354/ame027187. [DOI] [Google Scholar]
- 51.Corinaldesi C, Dell'Anno A, Danovaro R. Viral infections stimulate the metabolism and shape prokaryotic assemblages in submarine mud volcanoes. ISME Journal. 2011;6(6):1250–9. doi: 10.1038/ismej.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Sãnudo-Wilhelmy SA. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnology and Oceanography. 1997;42(7):1492–504. doi: 10.4319/lo.1997.42.7.1492. [DOI] [Google Scholar]
- 53.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. The microbial carbon pump and the oceanic recalcitrant dissolved organic matter pool. Nature Reviews Microbiology. 2011;9:555–6. doi: 10.1038/nrmicro2386-c5. [DOI] [PubMed] [Google Scholar]
- 54.Shelford EJ, Middelboe M, Møller EF, Suttle CA. Virus-driven nitrogen cycling enhances phytoplankton growth. Aquatic Microbial Ecology. 2012;66:41–6. doi: 10.3354/ame01553. [DOI] [Google Scholar]
- 55.Llewellyn CA, Evans C, Airs RL, Cook I, Bale N, Wilson WH. The response of carotenoids and chlorophylls during virus infection of Emiliania huxleyi (Prymnesiophyceae) Journal of Experimental Marine Biology and Ecology. 2007;344(1):101–12. doi: 10.1016/j.jembe.2006.12.013. [DOI] [Google Scholar]
- 56.Evans C, Pond DW, Wilson WH. Changes in Emiliania huxleyi fatty acid profiles during infection with E. huxleyi virus 86: physiological and ecological implications. Aquatic Microbial Ecology. 2008;55(3):219–28. [Google Scholar]
- 57.Ankrah NYD, Middleton J, Jones D, Gooding J, Szanyi, Budinoff CR, LeCleir GR, Wilhelm SW, Campagna SR, Buchan A. Aquatic Virus Workshop VI: November 2011. Texel, NL: 2011. Metabolomics of phage infection. [Google Scholar]
- 58.McDaniel L, Houchin LA, Williamson SJ, Paul JH. Plankton blooms - lysogeny in marine Synechococcus. Nature. 2002;415(6871):496. doi: 10.1038/415496a. [DOI] [PubMed] [Google Scholar]
- 59.Wilson WH, Mann NH. Lysogenic and lytic viral production in marine microbial communities. Aquatic Microbial Ecology. 1997;13(1):95–100. doi: 10.3354/ame013095. [DOI] [Google Scholar]
- 60.Weitz JS, Dushoff J. Alternative stable states in host-phage dynamics. Theoretical Ecology. 2008;1(1):13–9. doi: 10.1007/s12080-007-0001-1. [DOI] [Google Scholar]
- 61.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnology and Oceanography. 2000;45(6):1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 62.Buckling A, Maclean RC, Brockhurst MA, Colegrave N. The Beagle in a bottle. Nature. 2009;457(7231):824–9. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- 63.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proceedings of the Royal Society of London B. 2002;269(1494):931–6. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, Lenski RE. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution. 2010;64(10):3024–34. doi: 10.1111/j.1558-5646.2010.01049.x. [DOI] [PubMed] [Google Scholar]
- 65.Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in phage Lambda. Science. 2012;335(6067):428–32. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Paul Rainey and Gayle Ferguson 17 Mar 2012
- 66.Mizoguchi K, Morita M, Fischer CR, Yoichi M, Tanji Y, Unno H. Coevolution of bacteriophage PP01 and Escherichia coli O157: H7 in continuous culture. Applied and Environmental Microbiology. 2003;69(1):170. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menge DNL, Weitz JS. Dangerous nutrients: evolution of phytoplankton resource uptake subject to virus attack. Journal of Theoretical Biology. 2009;257(1):104–15. doi: 10.1016/j.jtbi.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 68.Lennon JT, Khatana SAM, Marston MF, Martiny JBH. Is there a cost of virus resistance in marine cyanobacteria? ISME Journal. 2007;1(4):300–12. doi: 10.1038/ismej.2007.37. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nature Reviews Microbiology. 2009;7(11):828–36. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 70.Lennon JT, Martiny JBH. Rapid evolution buffers ecosystem impacts of viruses in a microbial food web. Ecology Letters. 2008;11(11):1178–88. doi: 10.1111/j.1461-0248.2008.01225.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Joshua Weitz 16 Aug 2012
- 71.Forde SE, Beardmore RE, Gudelj I, Arkin SS, Thompson JN, Hurst LD. Understanding the limits to generalizability of experimental evolutionary models. Nature. 2008;455(7210):220–3. doi: 10.1038/nature07152. [DOI] [PubMed] [Google Scholar]
- 72.Weitz JS, Hartman H, Levin SA. Coevolutionary arms races between bacteria and bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9535. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Childs LM, Held NL, Young MJ, Whitaker RJ, Weitz JS. Multi-scale model of CRISPR-induced coevolutionary dynamics: Diversification at the interface of Lamarck and Darwin. Evolution. 2012;66(7):2015–29. doi: 10.1111/j.1558-5646.2012.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gudelj I, Weitz JS, Ferenci T, Horner-Devine MC, Marx CJ, Meyer JR, Forde SE. An integrative approach to understanding microbial diversity: from intracellular mechanisms to community structure. Ecology Letters. 2010;13(9):1073–84. doi: 10.1111/j.1461-0248.2010.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Joshua Weitz 16 Aug 2012
- 76.Fuhrman JA, Wilcox RM, Noble RT, Law NC. Viruses in marine food webs. 1993. [Google Scholar]
- 77.Rowe JM, Dunlap JR, Gobler CJ, Anderson OR, Gastrich MD, Wilhelm SW. Isolation of a non-phage-like lytic virus infecting Aureococcus anophagefferens. Journal of Phycology. 2008;44(1):71–6. doi: 10.1111/j.1529-8817.2007.00453.x. [DOI] [PubMed] [Google Scholar]
- 78.Winget DM, Williamson KE, Helton RR, Wommack KE. Tangential flow diafiltration: an improved technique for estimation of virioplankton production. Aquatic Microbial Ecology. 2005;41:221–32. doi: 10.3354/ame041221. [DOI] [Google Scholar]
- 79.Winter C, Smit A, Szoek-Denes T, Herndl GJWMG. Viral production and virus-mediated bacterial mortality in the North Sea. American Society of Limnology and Oceanography. 2001 [Google Scholar]
- 80.Holmfeldt K, Titelman J, Riemann L. Virus production and lysate recycling in different sub-basins of the Northern Baltic Sea. Microbial Ecology. 2010;60(3):572–80. doi: 10.1007/s00248-010-9668-8. [DOI] [PubMed] [Google Scholar]
- 81.Weinbauer MG, Arrieta JM, Griebler C, Herndl GJ. Enhanced virus production and infection of bacterioplankton during an iron-induced phytoplankton bloom in the Southern Ocean. Limnology and Oceanography. 2009;54:774–84. doi: 10.4319/lo.2009.54.3.0774. [DOI] [Google Scholar]
- 82.Barton AD, Dutkiewicz S, Flierl G, Bragg J, Follows MJ. Patterns of Diversity in Marine Phytoplankton. Science. 2010;327(5972):1509–11. doi: 10.1126/science.1184961. [DOI] [PubMed] [Google Scholar]
- 83.Follows MJ, Dutkiewicz S, Grant S, Chisholm SW. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315(5820):1843–6. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]; F1000 Factor 19Evaluated by John Cullen 05 Apr 2007, Scott Doney 16 Apr 2007, John Marra 23 Apr 2007, Michael Pace 25 Apr 2007, Anand Gnanadesikan 10 May 2007, Helen Yap 14 Sep 2007, Joshua Weitz 16 Aug 2012
- 84.Stock CA, Alexander MA, Bond NA, Brander KM, Cheung WWL, Curchitser EN, Delworth TL, Dunne JP, Griffies SM, Haltuch MA, Hare JA, Hollowed AB, Lehodey P, Levin SA, Link JS, Rose KA, Rykaczewski RR, Sarmiento JL, Stouffer RJ, Schwing FB, Vecchi GA, Werner FE. On the use of IPCC-class models to assess the impact of climate on Living Marine Resources. Progress in Oceanography. 2011;88(1-4):1–27. doi: 10.1016/j.pocean.2010.09.001. [DOI] [Google Scholar]
- 85.Solomon S, Qin D., Manning M., Chen Z., Marquis M., Averyt K.B., Tignor M., Miller H.L., editors. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, 2007. Cambridge, UK and New York, NY: Cambridge University Press; 2007. [Google Scholar]
- 86.Wilhelm SW, Weinbauer MG, Suttle CA, editors. Manual of Aquatic Viral Ecology. Waco, TX:: American Society of Limnology and Oceanography; 2010. [DOI] [Google Scholar]
- 87.Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333(6038):58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Oded Beja 14 Jul 2012