Abstract
The human cytomegalovirus DNA polymerase subunit UL44 is a phosphoprotein, but its sites and roles of phosphorylation have not been investigated. We compared sites of phosphorylation of UL44 in vitro by the viral protein kinase UL97 and cyclin-dependent kinase 1 with those in infected cells. Transient treatment of infected cells with a UL97 inhibitor greatly reduced labeling of two minor UL44 phosphopeptides. Viruses containing alanine substitutions of most UL44 residues that are phosphorylated in infected cells exhibited at most modest effects on viral DNA synthesis and yield. However, substitution of highly phosphorylated sites adjacent to the nuclear localization signal abolished viral replication. The results taken together are consistent with UL44 being phosphorylated directly by UL97 during infection, and a crucial role for phosphorylation-mediated nuclear localization of UL44 for viral replication, but lend little support to the widely held hypothesis that UL97-mediated phosphorylation of UL44 is crucial for viral DNA synthesis.
Keywords: Human cytomegalovirus, UL44, UL97, phosphorylation, nuclear localization
INTRODUCTION
Human cytomegalovirus (HCMV) is an important human pathogen that can cause life-threatening disease in immunocompromised individuals and congenital defects in newborns (Mocarski et al., 2007). Although there are several antiviral drugs available for the treatment of HCMV disease, these drugs have disadvantages, such as toxicities and drug resistance (Biron, 2006). Thus, novel anti-HCMV compounds that exhibit a distinct mechanism of action, and are safe, potent and orally bioavailable, are needed. A new drug, maribavir, which showed promise in early clinical trials (Trofe et al., 2008), specifically inhibits the enzymatic activity of the HCMV-encoded protein kinase, UL97 (Biron et al., 2002). In addition, UL97 is also important for its ability to phosphorylate, and thus activate ganciclovir, the leading antiviral compound used to treat HCMV infections (Littler et al., 1992; Sullivan et al., 1992; Talarico et al., 1999).
UL97 is a member of the conserved herpesviral protein kinase (CHPK) family [reviewed in (Gershburg and Pagano, 2008)]. A subset of these viral kinases can phosphorylate cyclin dependent kinase (cdk) consensus sites within a number of cellular proteins (Baek et al., 2004; Hamirally et al., 2009; Hume et al., 2008; Kawaguchi and Kato, 2003; Kawaguchi et al., 2003; Kawaguchi et al., 1999; Kuny et al., 2010; Romaker et al., 2006), suggesting that these viral kinases may function as cdk mimics (Baek et al., 2004; Hamirally et al., 2009; Hume et al., 2008; Kawaguchi and Kato, 2003; Kawaguchi et al., 2003; Kawaguchi et al., 1999; Kuny et al., 2010; Romaker et al., 2006), Some CHPKs are essential for viral replication while others are dispensable, often depending on the cell type studied (Coulter et al., 1993; de Wind et al., 1992; Gershburg et al., 2007; Heineman and Cohen, 1995; Kamil and Coen, 2007; Kamil et al., 2009; Krosky et al., 2003; Moffat et al., 1998; Prichard et al., 2005; Prichard et al., 1999; Tanaka et al., 2005; Wolf et al., 2001).
Although UL97 is not an essential viral protein, it plays an important role in productive virus replication. Ablation of UL97, either genetically or pharmacologically, has been reported variously to cause defects in viral DNA synthesis, DNA packaging, nuclear egress of viral capsids, and cytoplasmic secondary envelopment and release (Azzeh et al., 2006; Biron et al., 2002; Goldberg et al., 2011; Kamil and Coen, 2007; Kamil et al., 2009; Krosky et al., 2003; Prichard et al., 2005; Prichard et al., 1999; Wolf et al., 2001). However, its role appears to be more important in non-dividing cells than in dividing cells (Kamil et al., 2009; Prichard et al., 2005), suggesting that cellular kinases active in dividing cells may partially substitute for UL97 kinase activity. This notion is consistent with increased antiviral activity when maribavir is added to cellular kinase inhibitors, including cdk inhibitors (Chou et al., 2006; Hertel et al., 2007). Moreover, the activities of cdks, mainly cdk-1, -2, -, and -9, are required for several stages of HCMV replication (Bresnahan et al., 1997a; Hertel et al., 2007; Kapasi and Spector, 2008; Rechter et al., 2009; Sanchez et al., 2004; Sanchez and Spector, 2006; Tamrakar et al., 2005; Zydek et al., 2010). It is thus possible that proteins identified as substrates of UL97 may also serve as substrates for cellular cdks.
One candidate substrate of UL97 is the HCMV DNA polymerase accessory subunit UL44, a phosphoprotein that is essential for viral replication (Alvisi et al., 2009; Appleton et al., 2006; Ertl and Powell, 1992; Gibson, 1983; Loregian et al., 2004a, b; Weiland et al., 1994). UL44 can be phosphorylated by purified UL97 in vitro (Krosky et al., 2003), and during infection, UL97 activity is important for the normal phosphorylation pattern of UL44 (Krosky et al., 2003). These data are consistent with UL44 being a substrate of UL97 during infection, and led to the widely promulgated hypothesis that UL97-mediated phosphorylation of UL44 is important for viral DNA synthesis (Jacob et al., 2011; Krosky et al., 2003; Marschall et al., 2003; Mercorelli et al., 2008; Michel and Mertens, 2004; Wolf et al., 2001). A corollary hypothesis is that inhibition of UL97 kinase activity by maribavir affects viral DNA replication by preventing phosphorylation of UL44 (Krosky et al., 2003; Marschall et al., 2003; Mercorelli et al., 2008; Michel and Mertens, 2004; Wolf et al., 2001).
Aside from phosphorylation by UL97, UL44 is very likely to be phosphorylated by cellular kinases in the infected cell (Krosky et al., 2003). Indeed, phosphorylation on serine 413, likely by casein kinase (CK2), has been reported to enhance the activity of the UL44 nuclear localization signal (NLS) in transfected cells (Alvisi et al., 2005). However, the phosphorylation sites of this indispensable viral protein during infection have not been determined, and no functional significance of the phosphorylation of UL44 for viral replication has been demonstrated. In this study, we determined sites of phosphorylation of UL44 both from in vitro kinase reactions with UL97 and cdk1, and from infected cells. We also investigated whether transient inhibition of UL97 or cdks reduces UL44 phosphorylation during infection. Finally, we constructed virus mutants with altered sites of phosphorylation on UL44, and investigated whether these mutations resulted in changes in viral replication and viral DNA synthesis.
RESULTS
Sites of UL44 Phosphorylated by GST-UL97 In Vitro
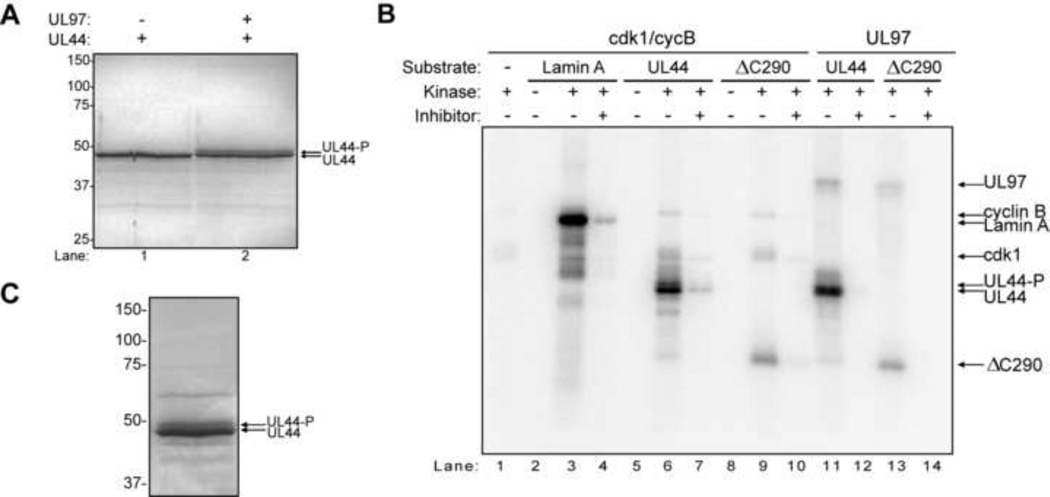
To identify the sites on UL44 phosphorylated by glutathione-S-transferase (GST)-UL97 in vitro, we first optimized our protocols for purifying UL44 and for phosphorylating it with GST-UL97 (Supplementary Fig. 1). We then performed a large-scale phosphorylation reaction in the presence or absence of GST-UL97. The proteins were resolved by SDS-PAGE and Coomassie stained. The gel from a representative experiment is shown in Fig. 1A. When UL44 was incubated alone, a single major UL44 band was observed (Fig. 1A, lane 1). When UL44 was incubated with GST-UL97, a second, slower migrating, presumably hyperphosphorylated form of UL44 was also observed (Fig. 1A, lane 2, and Supplementary Fig. 1B). Both species of UL44, as well as UL44 in control reactions, were excised and analyzed by mass spectrometry (MS) as described in Material and Methods.
Figure 1.
In vitro phosphorylation of UL44 by UL97 and cdk1/cyclin B. (A) A large-scale phosphorylation reaction was performed with UL44 alone (lane 1) or UL44 incubated with GST-UL97 (lane 2), and the products were resolved by SDS-PAGE and Coomassie-stained. (B) Radiolabeled phosphorylation reactions were performed with either cdk1/cyclin B (cycB) or UL97 and the substrates lamin A, UL44, or UL44ΔC290 (500 nM), and the products were resolved by SDS-PAGE. Reactions were assembled as indicated. The kinase inhibitors used were CGP74514A (cdk1 inhibitor) or maribavir (UL97 inhibitor). The positions of the various proteins are indicated on the right with arrows. (C) A large scale phosphorylation reaction assay was performed with UL44 and cdk1/cycB, and the products were resolved by SDS-PAGE. For all panels, the positions of UL44 and hyperphosphorylated UL44 (UL44-P) are indicated by arrows. The molecular weights of protein markers are indicated on the left.
Data from three independent experiments using two different MS facilities were compiled (Table 1, column 2). Coverage of the amino acid sequence of UL44 ranged from 83–95% in the three experiments. A total of 12 residues in UL44 were positively identified as sites of phosphorylation by GST-UL97 (Table 1, column 3; listed as confirmed). For six phosphopeptides, the phosphorylated residues could not be positively identified. Among these six phosphopeptides, ten additional sites are candidates for phosphorylation. These ten sites and the ones that are most likely to be phosphorylated are listed in Table 1. No phosphopeptides were recovered in control reactions lacking GST-UL97 (coverage >84%). A more detailed analysis of these phosphorylation sites can be found in Supplementary Table 1.
Table 1.
Compilation of UL44 Phosphorylation Residues identified by MS
| Residue | UL97 in vitroa |
Cdk1/cycB in vitrob |
HCMV infected cellsc |
|---|---|---|---|
| S8 | Confirmed | Confirmed | Confirmed |
| T12 | Confirmed | ||
| T45 | Confirmed | ||
| S47 | Confirmed | Possible | |
| T52 | Possible | ||
| S83 | Possible | ||
| T84 | Possible | Confirmed | |
| T164 | Confirmed | Confirmed | |
| T268 | Confirmed | ||
| Y282 | Possible | ||
| S286 | Confirmed | Confirmed | Confirmed |
| S289 | Possible | Confirmed | |
| S348 | Confirmed | ||
| S354 | Likely | Likely | Likely |
| S355 | Confirmed | Possible | Likely |
| S367 | Likely | Likely | Confirmed |
| S368 | Possible | Possible | |
| T378 | Possible | Likely | |
| S379 | Confirmed | Confirmed | |
| Y380 | Possible | Possible | |
| T382 | Possible | Possible | |
| S383 | Confirmed | Likely | |
| S387 | Confirmed | Confirmed | Confirmed |
| S402 | Confirmed | Confirmed | |
| S413 | Confirmed | Possible | Likely |
| S415 | Likely | Likely | |
| S418 | Confirmed | Possible | Likely |
| T420 | Possible | Possible | |
| T427 | Confirmed |
Residues were either positively confirmed as a phosphorylation site or the phosphorylation site could not be identified among two or more probable sites in a single peptide. In the latter case, the most likely sites and other possible sites are indicated. See supplementary information for more detailed analysis.
Data are compiled from three experiments and two MS facilities.
Data are compiled from one experiment and one MS facility.
Data are compiled from two experiments and two MS facilities.
Most of the sites of phosphorylation by GST-UL97 were located in the C-terminal third (aa 291–433) of the protein. Of the sites identified, only S379 has a basic residue at position +5, similar to the preferred sites of phosphorylation of histone H2B (Baek et al., 2002), while only S8 and S289 are predicted to be cdk sites (using the NetPhosK 1.0 software, scores >0.45) (Blom et al., 2004). Since the majority of the phosphorylation sites were located in the C-terminus, we tested whether the N-terminal two-thirds of the protein (UL44ΔC290) could be phosphorylated by GST-UL97 in vitro. We found that UL44ΔC290 was phosphorylated by GST-UL97 (Fig. 1A, lane 13 in a maribavir-sensitive manner (lane 14), but to a lesser extent than full-length UL44 (Fig. 1A, compare lanes 13 and 11), which is consistent with our MS data.
UL44 is a Substrate for Cdk1/Cyclin B In Vitro
Several lines of evidence indicate that UL97 and its homologs phosphorylate certain proteins on sites that are also phosphorylated by cdks (Baek et al., 2004; Hamirally et al., 2009; Hume et al., 2008; Kawaguchi and Kato, 2003; Kawaguchi et al., 2003; Kawaguchi et al., 1999; Kuny et al., 2010; Prichard et al., 2008; Romaker et al., 2006), and that cdk activities are required for HCMV replication (Bresnahan et al., 1997a; Bresnahan et al., 1997b; Feichtinger et al., 2011; Hertel et al., 2007; Sanchez et al., 2007; Sanchez et al., 2003; Sanchez et al., 2004; Sanchez and Spector, 2006). In addition, the HSV-1 homolog of UL44, UL42, has been reported to interact with and be phosphorylated by cdk1 (Advani et al., 2001, 2003). Thus, we tested whether UL44 can serve as a substrate of cdk1/cyclinB in vitro.
We incubated recombinant GST-cdk1 (cdk1) and GST-cyclin B (cycB) with either UL44 or UL44ΔC290 and [32P]-γATP. Lamin A, which is known to be a substrate of cdk1/cycB (Heald and McKeon, 1990; Peter et al., 1990; Ward and Kirschner, 1990), was used a positive control. When lamin A or UL44 were incubated alone, no 32P incorporation was observed (Fig. 1B, lanes 2 and 5), however, a robust signal was observed when these proteins were incubated with cdk1/cycB (lanes 3 and 6). Addition of an inhibitor of cdk1, CGP74514A, greatly decreased the level of phosphorylated lamin A and UL44 (lanes 4 and 7). UL44ΔC290 was also phosphorylated by cdk1/cycB (lane 9) in a CGP74514A-sensitive manner (lane 10), but as observed with UL97, to a lesser extent than full-length UL44 (Fig. 1B, compare lanes 6 and 9). In addition, the level of phosphorylation of UL44 or UL44ΔC290 by cdk1/cycB was comparable with the level of phosphorylation of these proteins by UL97 (Fig. 1B, compare lanes 6 and 11, and 9 and 14, respectively). Taken together, these data indicate that UL44 is phosphorylated by cdk1/cycB in vitro.
We then performed a large-scale phosphorylation of UL44 by cdk1/cycB in vitro. The reaction was resolved by SDS-PAGE and stained with colloidal blue (Fig. 1C). As with GST-UL97, we observed a slower migrating form of UL44 after incubation with cdk1/cycB (Fig. 1C). Both species of phosphorylated UL44 were excised together and subjected to MS. Coverage of the amino acid sequence of UL44 in this experiment was approximately 85%. Ten residues were positively identified as sites of phosphorylation by cdk1/cyclin B (Table 1, column 3, listed as confirmed). Both cdk minimal sites (S/T-P) present in UL44, T45 and T84, were definitively phosphorylated by cdk1/cycB. For six phosphopeptides, we were unable to positively identify phosphorylation sites, yielding a maximum of 14 additional possible sites of phosphorylation, with five of these residues identified as likely sites of phosphorylation (Table 1, column 3). A more detailed analysis of the phosphopeptides can be found in Supplementary Table 2.
Half of the sites that were confirmed to be phosphorylated by cdk1 corresponded to ones positively identified as being phosphorylated by UL97 (Table 1, compare columns 2 and 3). When the likely sites of phosphorylation were included, the overlap in sites phosphorylated by both enzymes was even more extensive (e.g. T84, S354, S383). Additionally, there were some UL44 residues that were uniquely phosphorylated by each kinase in vitro. The residues that we can identify definitively as being phosphorylated by one kinase but not the other in vitro are T12 and S402 (UL97) and T45, T268 and T427 (cdk1/cycB) (Table 1, columns 2 and 3). Interestingly, T427 is predicted to be a consensus site for cdk-mediated phosphorylation, and phosphorylation of UL44 residue T427 or its substitution with aspartate has been found to negatively regulate the activity of the UL44 NLS in transfection assays [(Fulcher et al., 2009) and Alvisi, G., et al., accompanying manuscript].
Sites of UL44 Phosphorylation in Infected Cells
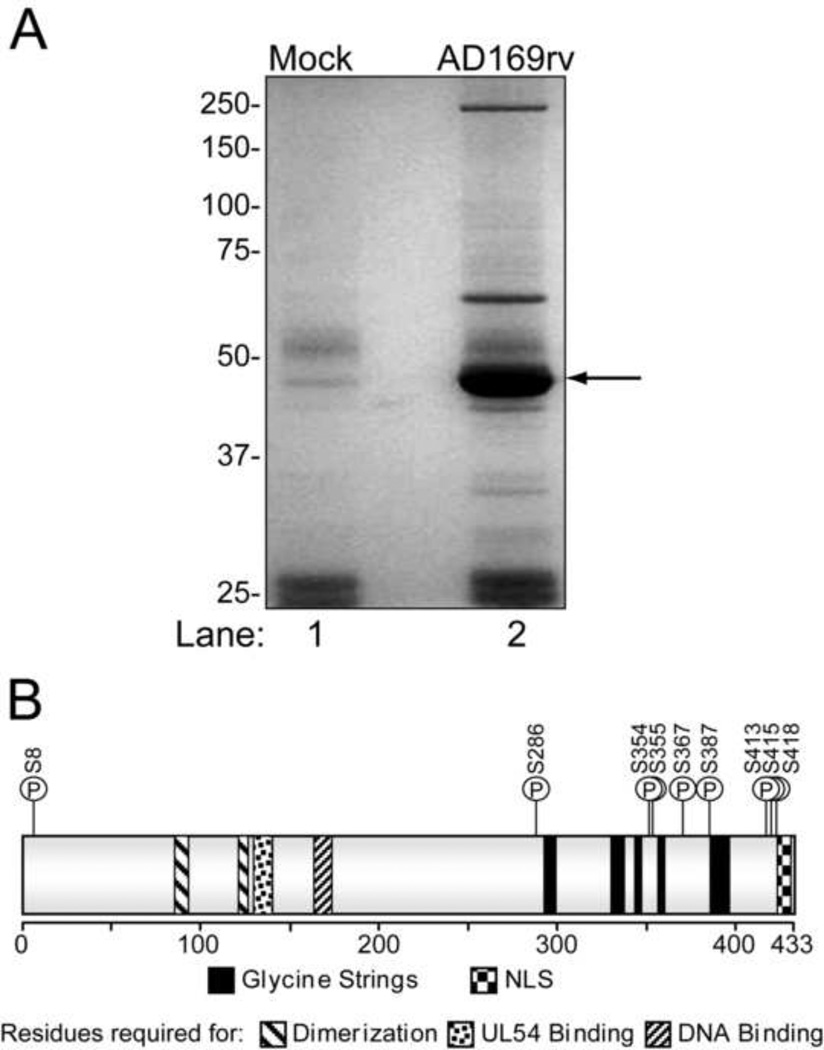
To identify UL44 phosphorylation sites in infected cells, HFF cells were either mock-infected or infected with strain AD169rv at an MOI of 3 PFU/cell. Cell lysates were harvested at 72 hpi, when viral DNA synthesis is robust, and incubated with anti-UL44 antibody. The resulting immunoprecipitates were resolved by SDS-PAGE and subjected to colloidal blue staining. One of two independent experiments is shown in Fig. 2A. UL44 was excised and subjected to MS analysis. Peptide fragments of UL44 of various sizes were identified with amino acid sequence coverage of 93% and 83% in the two experiments, respectively. For six peptides, we positively identified a single phosphorylation site (Table 1, column 4; listed as confirmed). We also recovered phosphopeptides in which phosphorylated residues could not be identified definitively. In these cases, the possible and most likely residues are listed (Table 1, column 4). A more detailed analysis of the UL44 phosphopeptides identified from AD169rv-infected cells can be found in Supplementary Table 3.
Figure 2.
Analysis of phosphorylation sites on UL44 from infected cells. (A) HFF cells (4.5 × 106) were either mock-infected (lane 1) or infected with AD169rv at an MOI of 3 PFU/cell (lane 2), and immunoprecipitates of UL44 were prepared and resolved by SDS-PAGE. Proteins were visualized by colloidal blue stain. The position of UL44 is indicated by an arrow. (B) Cartoon depiction of UL44. The amino acids required, but not necessarily sufficient for the known biochemical functions of the protein are shown as indicated. The positions of glycine-rich segments (glycine strings) and the NLS are also depicted. The locations of eleven residues of UL44 phosphorylated in cells infected with wild-type HCMV are indicated by a P in a circle with the amino acid denoted.
Taking the positively identified and most likely sites together, we found 11 sites of phosphorylation on UL44 from wt HCMV infected cells. All sites were serine residues, which correlates with Western blot data using phospho-specific antibodies (Supplementary Fig. 2). The locations of these 11 sites within UL44 are depicted in Fig. 2B. Interestingly, 10 of the 11 sites are located in the C-terminal third of the protein, which has been shown to be important for viral replication (Kim and Ahn, 2010; Silva et al., 2010). Importantly, many of the sites on UL44 that we identified as being phosphorylated during infection were also phosphorylated by UL97 or cdk1 in vitro (Table 1). In particular, S402, which was phosphorylated by UL97, but not by cdk1, in vitro, was also phosphorylated during infection, consistent with UL97 phosphorylating UL44 during infection.
In one of the two MS analyses of UL44 from infected cells, the intensities of the peaks of the non-phosphorylated and phosphorylated peptides were measured as an estimate of the extent of phosphorylation at given sites (Supplementary Table 3). The peptide containing the three sites (S413, S415, and S418) directly upstream of the NLS was essentially always phosphorylated on at least one of these three sites, with a monophosphorylated form estimated to account for about half of the peptides and the more highly phosphorylated forms the other half. Two phosphopeptides, containing S354/355 and S402, were estimated to be phosphorylated about half of the time. S8, S286, and S387 were estimated to be phosphorylated less than 20% of the time. S348 and S367 were not included in this analysis. Thus, the major sites of UL44 phosphorylation during infection are likely to be S413, S415, and S418.
The Effects of Transient Inhibition of UL97 and Cdks on UL44 Phosphorylation
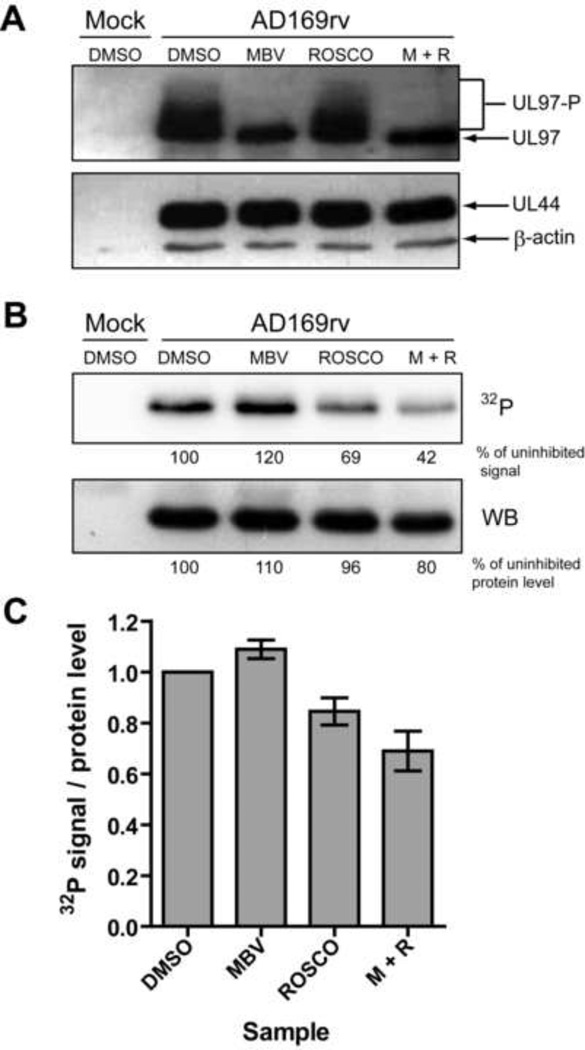
To further investigate whether UL44 is phosphorylated by UL97 or cdks during infection, we performed [32P]-orthophosphate metabolic labeling experiments of wt infected HFF cells in the presence or absence of maribavir, which specifically inhibits UL97 (Biron et al., 2002) or a cdk inhibitor, roscovitine, which inhibits cdks 1, 2, 5, 7 and 9 (Meijer et al., 1997; Meijer and Raymond, 2003). To limit the activity of cdks that might substitute for UL97 kinase activity, we performed the experiment with quiescent HFF cells, produced by serum-starvation. Furthermore, to ensure that the kinase inhibitors did not affect UL44 gene expression and to limit secondary effects of these compounds (Bresnahan et al., 1997a; Krosky et al., 2003; Sanchez et al., 2004; Sanchez and Spector, 2006), we allowed infections to proceed until 68 hpi before treating infected cells with 1 µM maribavir, 15 µM roscovitine, both drugs, or vehicle (DMSO) for 2 hours. At 70 hpi, cells were labeled with [32P]-orthophosphate for 2 h in the presence or absence of kinase inhibitors. At 72 hpi, cells were harvested and lysed. Levels of UL44, β-actin, and UL97 in the lysates, as assessed by Western blot analysis, were present at similar levels in all infected samples (Fig. 3A). However, in mock-treated and roscovitine-treated samples, slower migrating forms of UL97, presumably representing hyperphosphorylated forms of the protein, were detected (Fig. 3A), while only the fastest migrating form of UL97 was detected in samples treated with maribavir (Fig. 3A). These data are consistent with autophosphorylation, and thus the kinase activity, of UL97 being effectively inhibited by transient treatment with maribavir.
Figure 3.
Phosphorylation of UL44 during infection in the presence of UL97 and cdk inhibitors. Mock-infected or HCMV-infected (AD169rv) cells were pulse-labeled for 2 hours with [32P]-orthophosphate in the presence of 1 µM maribavir (MBV), 15 µM roscovitine (ROSCO), both drugs (M+R) or vehicle (DMSO), and whole cell lysates were prepared at 72 hpi. (A) UL97 (upper panel), UL44, and β-actin (lower panel) protein levels in lysates were assessed by Western blot. Phosphorylated forms of UL97 are indicated (UL97-P). (B) UL44 immunoprecipitations were prepared from cell lysates, resolved by SDS-PAGE, transferred to a PVDF membrane and then exposed to a phosphorscreen to detect 32P incorporation (upper panel). The 32P signal was quantified for each UL44 band and the values for 32P incorporation into UL44 are shown below the phosphorimage as percent of the DMSO-treated sample. The membrane was also subjected to Western blot analysis to detect UL44 protein levels (lower panel). The signal for UL44 was quantified, and the values are shown below the Western blot as percent of DMSO-treated sample. (C) Three independent immunoprecipitations of UL44 from lysates of the same orthophosphate labeling experiment were performed. The average normalized phosphate incorporation into UL44 relative to the untreated control is depicted. Error bars are the standard deviation of the average ratio of the three immunoprecipitations.
To evaluate whether UL44 phosphorylation was affected by the kinase inhibitors, we immunoprecipitated UL44 from each lysate, resolved immunoprecipitates by SDS-PAGE, transferred the proteins to a membrane, and measured 32P incorporation into UL44 by autoradiography (Fig. 3B, upper panel). We also probed the membrane with anti-UL44 antibody to quantify the amount of UL44 immunoprecipitated from the samples (Fig. 3B, lower panel). We calculated the ratio of the amount of phosphate incorporation into UL44 to the UL44 protein level for each sample (Fig. 3C). The normalized level of phosphorylation of UL44 was, if anything, slightly increased (~9%) in the presence of maribavir and modestly decreased (~16%) in the presence of roscovitine. Treatment with both drugs resulted in a reduction in the relative level of phosphorylation that was even greater (~31%) than the decrease observed with roscovitine alone. Nevertheless, more than half of the phosphorylation remained after treatment with both drugs. The remaining phosphorylation of UL44 is likely due to other cellular kinases (e.g. CK2).
The trends observed in the representative experiment (Fig. 3B and C) held true for multiple experiments in both quiescent cells and actively dividing cells [data not shown and (Hamirally et al., 2009)]. These data indicate that 1) the overall phosphorylation of UL44 can proceed efficiently in the absence of UL97 activity, 2) cdks likely contribute to the phosphorylation of UL44 in infected cells, and 3) other cellular kinases are responsible for the large majority of the phosphorylation of UL44 during infection, at least at 72 hpi. We suggest that CK2 is one such kinase, given the heavy phosphorylation of serines adjacent to the UL44 NLS (Supplementary Table 3), and work implicating CK2 in phosphorylation of this segment [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript]. In addition, our results suggest that UL97 may be more important for phosphorylating UL44 when cdks are inhibited (see Discussion).
Transient Inhibition by Maribavir Affects Two UL44 Phosphopeptides
We wished to investigate the possibility that, although overall phosphorylation of UL44 can proceed in the absence of UL97 activity in infected cells, this kinase might be responsible for phosphorylation of specific sites on UL44. Towards that end, the phosphorylated samples shown in Fig. 3B, as well as a sample of UL44 from an in vitro phosphorylation reaction with GST-UL97, were excised from the membrane and extensively digested with trypsin. The resulting peptides were resolved on an alkaline 40% polyacrylamide gel and visualized by autoradiography. Multiple tryptic phosphopeptide species of UL44 were observed following digestion of UL44 from mock-treated cells (Fig. 4, lane 2), as well as with samples from extracts of cells treated with maribavir, roscovitine or both drugs (Fig. 4, lanes 3, 4, and 5, respectively). As indicated by the arrows on the left of Fig. 4, a substantial reduction in the signal of two particular peptides was evident in samples that had been treated with maribavir, either alone (lane 3) or in the presence of roscovitine (lane 5), as compared to the mock-treated control (lane 2). The reduction in labeling of these two peptides was specific, as maribavir did not reduce the labeling of other peptides (Fig. 4). The two peptides and the reduction in labeling are more evident in a darker exposure of this region of the gel (Fig. 4, lower panel). Notably, these phosphopeptides migrated with the same mobility as phosphopeptides of UL44 from an in vitro phosphorylation reaction with UL97 (lane 7). Although we did not observe any major decrease in any specific phosphopeptide in samples treated with roscovitine, the phosphorylation of the majority of the peptides decreased (lanes 4 and 5). Similar results were observed in another independent experiment. Several attempts were made to identify the two peptides whose phosphorylation was diminished with maribavir treatment using MS; however, these efforts failed likely due to the low abundance of the peptides in the sample. Regardless, these results provide further evidence that UL44 is directly phosphorylated by UL97 during infection, albeit to a limited extent. The results also make it highly unlikely that UL97 is responsible for the heavy phosphorylation of the serines adjacent to the UL44 NLS.
Figure 4.
One-dimensional tryptic phosphopeptide analysis of UL44 from infected cells treated with kinase inhibitors. 32P-labeled UL44 from cells either mock-infected (DMSO, lane 1) or infected (AD169rv) and mock treated (DMSO, lane 2), or treated with 1 µM maribavir (MBV, lane 3), 15 µM roscovitine (ROSCO, lane 4) or both drugs (M+R, lane 5), was isolated from the membrane shown in Fig. 3B and extensively digested with trypsin. An unlabeled UL44 immunoprecipitation from an untreated AD169rv infection (Cold IP, lane 6) as well as UL44 from an in vitro kinase reaction with GST-UL97 (in vitro, lane 7) were also digested with trypsin. The resulting peptides were resolved on an alkaline 40% acrylamide gel and visualized using phosphorimager. Arrows point to the UL44 phosphopeptides that were absent when UL97 acitivity was inhibited by maribavir. A darker exposure of the region containing these peptides is shown in the lower panel.
Construction and Characterization of UL44 Phosphorylation Mutants
To determine whether particular UL44 phosphorylation sites are important for viral replication, we used site-directed mutagenesis to construct HCMV mutants that express UL44 that cannot be phosphorylated at certain sites (Phos mutants). For our initial set of mutants, eight of the eleven serine residues (all but those adjacent to the NLS) positively or most likely identified as sites of phosphorylation during infection were substituted with alanine in four different groupings (Table 2): Phos-1 contained a single alanine substitution at the only N-terminal site (serine 8); Phos-2 contained seven alanine substitutions at sites located in the C-terminus of the protein (serines 286, 348, 354/355, 367, 387 and 402); Phos-3 contained seven alanine substitutions at sites that were found to be phosphorylated during infection and also by GST-UL97 in vitro (serines 8, 286, 354/355, 367, 387, and 402); Phos-1/2 contained alanine substitutions at eight sites and is a combination of Phos-1 and Phos-2 mutants (serines 8, 286, 348, 354/355, 367, 387 and 402). These four Phos mutants were constructed as BACs as described in Materials and Methods and verified by sequencing. Each mutant BAC yielded infectious virus when electroporated into HFF cells, which indicates that phosphorylation of UL44 of these residues is not essential for viral replication. Additionally, Western blot experiments indicated that the mutations did not meaningfully alter the accumulation of UL44 (data not shown).
Table 2.
Alanine substitutions of phosphorylation sites in UL44 Phos mutants
| Mutation | Phos-1 | Phos-2 | Phos-3 | Phos-1/2 | Phos-4 |
|---|---|---|---|---|---|
| S8A | + | + | + | ||
| S286A | + | + | + | ||
| S348A | + | + | |||
| S354A | + | + | + | ||
| S355A | + | + | + | ||
| S367A | + | + | + | ||
| S387A | + | + | + | ||
| S402A | + | + | + | ||
| S413A | + | ||||
| S415A | + | ||||
| S418A | + |
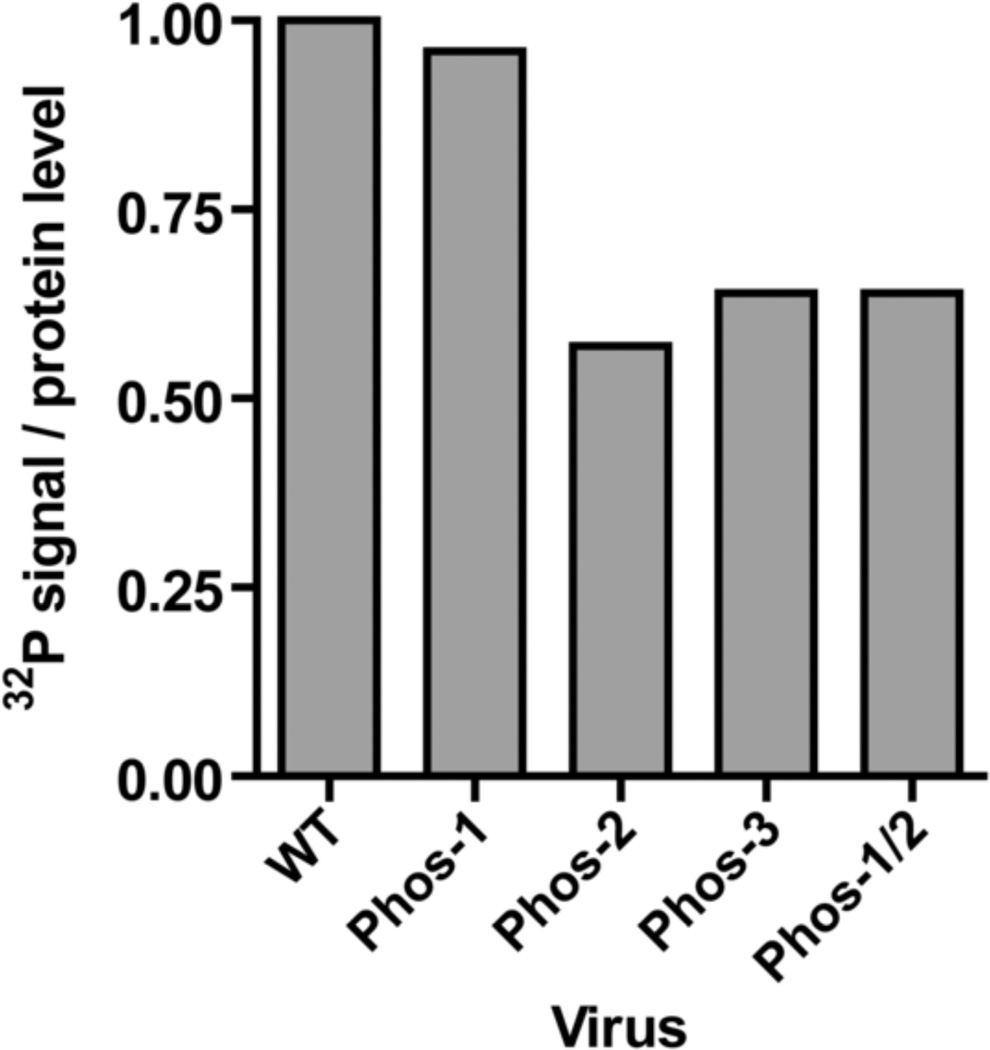
To investigate whether the alanine substitutions in UL44 affected phosphorylation, we performed [32P]-orthophosphate metabolic labeling. In this experiment, dividing HFF cells were infected with strain AD169rv or each mutant virus (MOI=1 PFU/cell), labeled with [32P]-orthophosphate at 70 hpi for 2 h, and whole cell lysates were harvested at 72 hpi. UL44 was immunoprecipitated from cell lysates, resolved by SDS-PAGE and transferred to a membrane. Phosphate incorporation into UL44 expressed from each virus was assessed by autoradiography and quantified (Supplementary Figure 3). The PVDF membrane was also probed with anti-UL44 antibody to determine UL44 levels (Supplementary Figure 3). We calculated the ratio of phosphate incorporation into UL44 to UL44 levels to obtain the normalized level of UL44 phosphorylation for each mutant virus as compared to AD169rv (Fig. 5, WT). We observed at most a slight decrease in the normalized level of UL44 phosphorylation from Phos-1, which contains only 1 alanine substitution. We observed larger decreases (~40%) in the levels of relative phosphorylation for the Phos mutants that contained 7 or 8 alanine substitutions (Phos-2, Phos-3 and Phos-1/2). Thus, substitutions at these sites lead to a decrease in the level of UL44 phosphorylation, as expected. However, these data indicate that a substantial amount of relative phosphorylation occurs at other possible sites of phosphorylation, most likely S413, S415, and S418.
Figure 5.
Relative phosphorylation levels of UL44 Phos mutants. HFF cells were either mock-infected or infected with parental (AD169rv) or Phos mutant viruses at an MOI of 1 PFU/cell and pulse-labeled for 2 hours with [32P]-orthophosphate at 70 hpi. UL44 immunoprecipitations were prepared from cell lysates harvested at 72 hpi, resolved by SDS-PAGE, transferred to a PVDF membrane. The normalized level of phosphorylation of UL44 from each virus was calculated as the ratio of phosphate incorporation (relative to the AD169rv control) to protein level (relative to the AD169rv control) as described for Fig. 3. The phosphorimage and Western blot of the membrane are shown in Supplementary Figure 3.
Replication and DNA Synthesis of Viable UL44 Phos Mutants
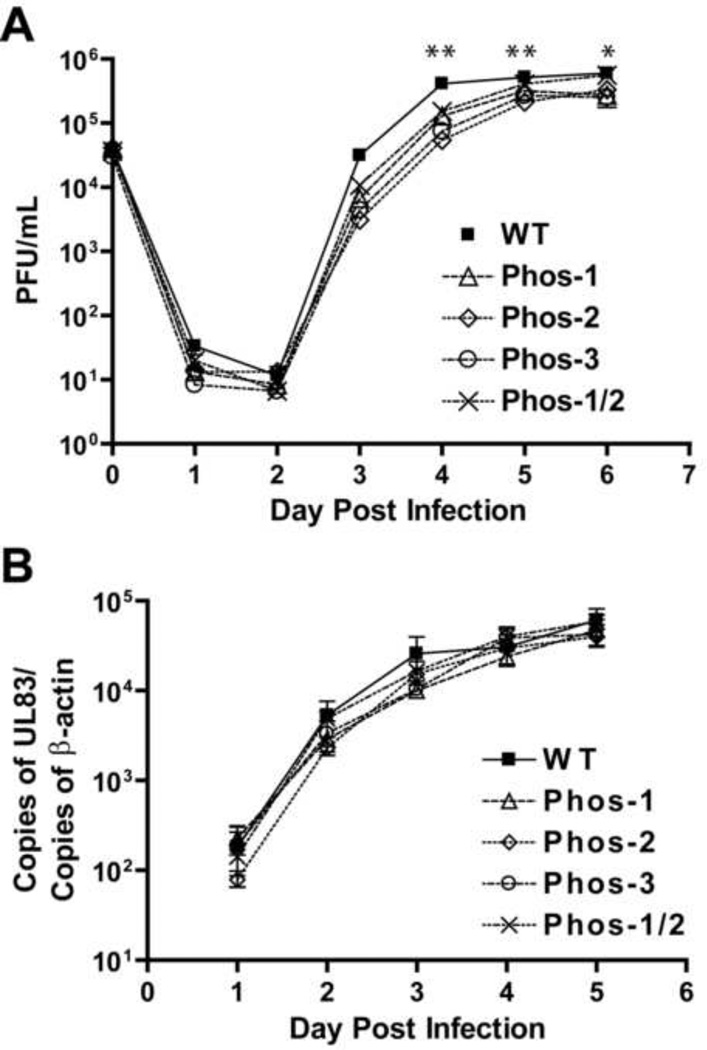
To test whether phosphorylation of UL44 at these sites affects viral replication, dividing HFF cells were infected with strain AD169rv or Phos mutants at an MOI of 1 PFU/cell. Virus produced from infected cultures was harvested every 24 h for six days following infection and titrated (Fig. 6A). Replication of the four Phos mutants was slightly delayed compared to the parental virus, AD169rv in each of three independent experiments (averages presented in Fig. 6A). At days 3 and 4 pi, we observed modest decrease (5- to 10- fold) in viral titers of the mutant viruses compared to AD169rv. These decreases became even less apparent (<5-fold) at days 5 and 6 pi (Fig. 6A). Nevertheless, the decrease in viral titers of all the Phos mutants was statistically significant on days 4 and 5 (P < 0.001 as determined by 2-way ANOVA followed by Bonferroni’s post-test; Fig. 6A, two asterisks), and the decrease in viral titers for Phos-1, Phos-2 and Phos-3 was statistically significant on day 6 as compared to the parental virus' titers (P < 0.001; Fig. 8A, one asterisk). We also assessed viral titers of the Phos mutants at 3 dpi in non-dividing cells, when mutant titers were lowest as compared to parental virus in dividing cells. HFF cells were serum-starved for three days and then infected with wt or mutant viruses (MOI=1 PFU/cell). The titers of progeny virus from the four Phos mutants at 3 dpi were again rather modest (within 3-fold of wt levels), but the slight decrease in titers was statistically significant (P<0.01) (Supplementary Fig. 4).
Figure 6.
Viral replication kinetics and viral DNA synthesis kinetics of Phos mutant viruses. (A) HFF cells were infected with parental (AD169rv), Phos-1, Phos-2, Phos-3 or Phos-1/2 mutant viruses at an MOI of 1 PFU/cell. Supernatant was harvested at the times indicated. Virus yields were determined by plaque assays. Error bars represent the standard deviation from the average titers of three independent experiments. Error bars shorter than the point symbols were omitted. *P<0.001 as determined by two-way ANOVA followed by Bonferroni multiple comparison post-tests. One asterisk indicates that Phos-1, Phos-2 and Phos-3 exhibited a statistically significant decrease in viral titers. Two asterisks indicate that all Phos mutants exhibited a statistically significant decrease in viral titers. (B) HFF cells were infected with AD169rv (WT), Phos-1, Phos-2, Phos-3 or Phos-1/2 mutant viruses at an MOI of 1 PFU/cell. Total DNA was harvested at the times indicated. Viral DNA accumulation was assayed by real-time PCR measuring the copies of the UL83 gene normalized to copies of the cellular β-actin gene. Error bars represent the standard deviation from the average of three independent experiments, except in the case of Phos-1 and Phos1/2 at day 5 when only duplicates are present and standard deviations were not calculated. Error bars shorter than the point symbols were omitted. P>0.05 for all samples on all days as determined by two-way ANOVA followed by Bonferroni multiple comparison post-tests.
Figure 8.
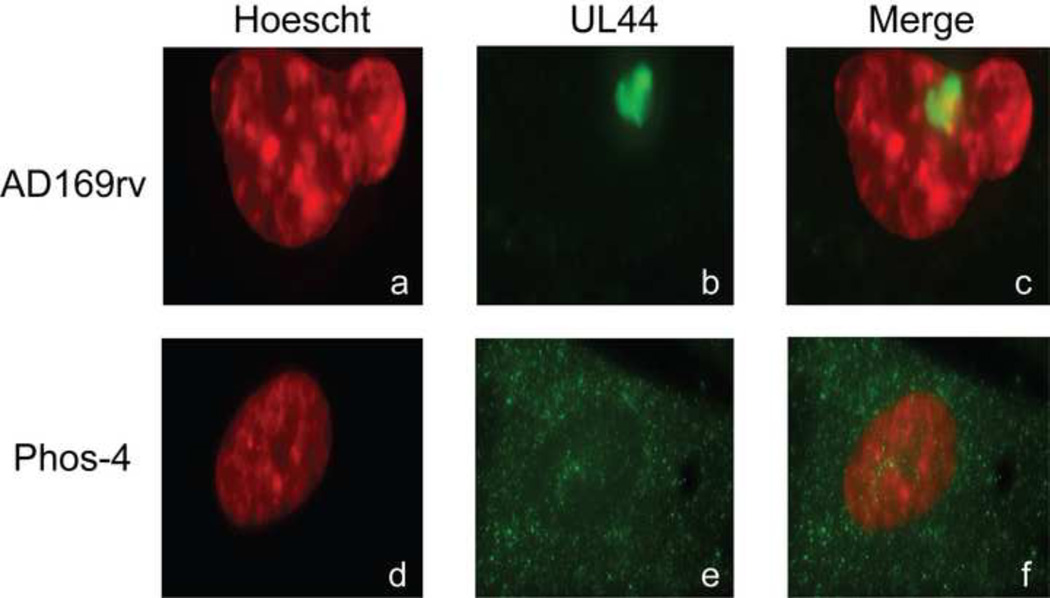
Subcellular localization of UL44. HFF cells electroporated with either AD169rv-BAC or BAC-Phos4 were seeded 72 hours after electroporation and stained with MAb recognizing UL44, secondary antibody conjugated to Alexa 488 and Hoechst reagent. Cells electroporated with AD169rv-BAC are shown in panels a-c, and cells electroporated with BAC-Phos4 are shown in panels d-f. Panels a and d in the left column show Hoechst staining and panels b and e in the middle column show the localization of UL44. Panels c and f in the right column show the merged images from the left and middle columns. Magnification: 63×
We also evaluated kinetics of viral DNA synthesis of the Phos mutants using quantitative real-time PCR, measuring copies of the UL83 gene normalized to copies of the cellular β-actin gene. With the parental virus, levels of viral DNA increased each day until day 4 pi when the levels of viral DNA began to plateau (Fig. 6B). The four UL44 phosphorylation mutants replicated their DNA with similar kinetics as the parental virus (Fig. 6B). Levels of viral DNA from cells infected with mutant viruses were within 3-fold of levels of the parental virus at each dpi, but these differences in DNA accumulation were not statistically significant (P>0.05 as determined by 2-way ANOVA followed by Bonferroni’s post-test). We obtained similar results in non-dividing, serum-starved cells (data not shown). The data indicate that these Phos mutants exhibit a modest defect in viral replication, and little or no defect in viral DNA synthesis.
Substitution of Serines at Positions 413, 415 and 418 in UL44 is Lethal to HCMV
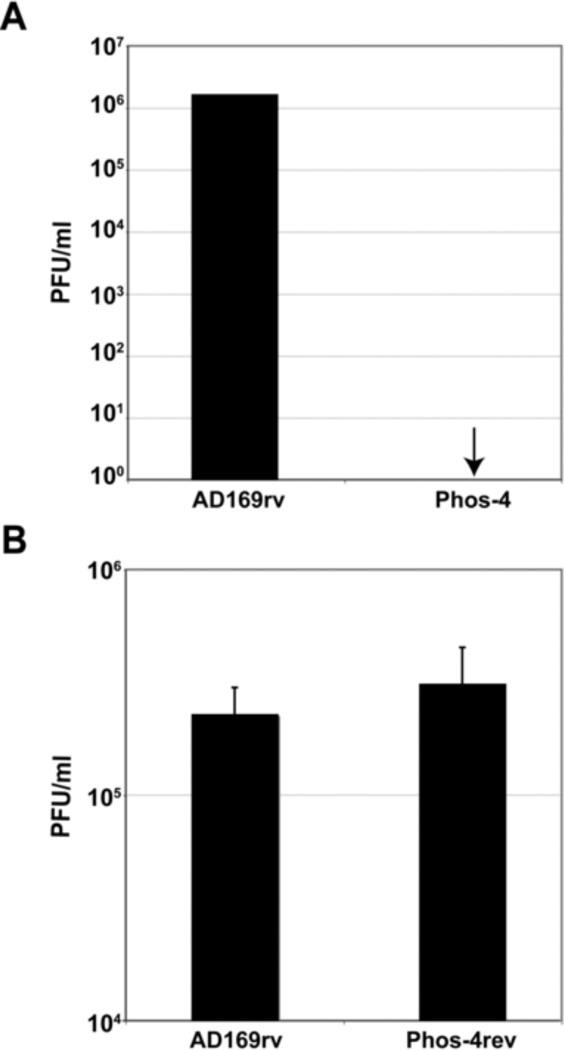
In our MS analysis, the segment of UL44 containing residues S413, S415 and S418 was the most highly phosphorylated in infected cells (Supplementary Table 3). In addition, these serines have both been predicted and found to be substrates for protein kinases CK1 and CK2 [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript]. To investigate the requirement for these serines in viral replication, the three codons for these residues were mutated to codons encoding alanine in the AD169rv BAC to create Phos-4 (Table 2). This BAC or the AD169rv BAC was electroporated into HFF cells. Complete cytopathic effect was observed in cell cultures electroporated with the parental BAC and infectious virus was readily detected (Fig. 7A). However, at no time could cytopathic effect be observed in cells electroporated with Phos-4, and no infectious virus was detected (Fig. 7A). To ensure that no other mutation that affects virus replication was present in Phos-4, the mutated segment of the BAC was replaced with wt sequence so that the alanine residues at positions 413, 415 and 418 were reverted to serine residues in Phos-4 to create Phos-4rev. Complete cytopathic effect could be observed in HFF cells transfected with this BAC. Peak titers of the reconstituted virus were comparable to those of AD169rv (Fig. 7B). These data indicate that one or more of these three serines located upstream of the NLS of UL44 is critical for viral replication.
Figure 7.
Mutation of serines at positions 413, 415 and 418 in UL44 yields a replication-defective BAC. (A) AD169rv and Phos 4 BACs were electroporated into HFF cells. The amount of virus in each transfected cell culture supernatant was determined by titration of cell culture supernatant on HFF cells. Data shown are representative of two independent experiments. (B) The mutations in the Phos 4 BAC were rescued to restore wt sequence to create BAC-Phos4rev. Virus reconstituted from electroporation of Phos4rev or AD169rv BACs were used to infect HFF cells at a MOI of 1 PFU/cell. The amount of virus produced from each infection 5 dpi was determined by titration of virus on HFF cells. Data shown represent the mean value and standard deviation of results obtained from three independent experiments.
Subcellular Localization of UL44 Expressed from Phos-4
In transfection studies, serines 413 and 418 were found to be important for enhanced nuclear import of UL44 and binding to importin α/β [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript]. To investigate the subcellular localization of the triply substituted UL44 in infected cells, the AD169rv and Phos-4 BACs were electroporated into HFF cells and immunofluorescence assays were performed using a monoclonal antibody recognizing UL44 (Fig. 8). Cells were also stained with Hoechst reagent to visualize the nucleus. UL44 could be visualized in a distinct structure within the nucleus in cells electroporated with parental BAC (Fig. 8, panels a-c). This structure is most likely a developing replication compartment (Ahn et al., 1999; Penfold and Mocarski, 1997). In cells electroporated with Phos-4, very little UL44 could be found in the nucleus. Rather, diffuse staining was observed throughout the cytoplasm (Fig. 8, panels d–f). Thus, consistent with and extending the work of Alvisi et al. in transfected cells [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript], the triple substitution makes nuclear entry of UL44 highly inefficient in infected cells, which suggests that the inability of Phos-4 to produce infectious virus is due to inefficient nuclear accumulation of UL44.
DISCUSSION
UL44 has long been described as a phosphoprotein (Gibson et al., 1981). Indeed, the accessory subunits of herpesvirus DNA polymerases are typically phosphorylated during infection (Chan and Chandran, 2000; Chang and Balachandran, 1991; Gibson et al., 1981; Marsden et al., 1987; Roeckel and Mueller-Lantzsch, 1985), but for many of these proteins, including UL44, the sites of phosphorylation and the significance of these modifications during infection had not been determined. For UL44, we were particularly interested in addressing the hypotheses that i) UL97 phosphorylates UL44 in infected cells (Krosky et al., 2003), and ii) UL97-mediated phosphorylation of UL44 is important for viral DNA synthesis and is thus the mechanism by which pharmacological or genetic inhibition of UL97 kinase activity affects viral DNA replication (Jacob et al., 2011; Krosky et al., 2003; Marschall et al., 2003; Mercorelli et al., 2008; Michel and Mertens, 2004; Wolf et al., 2001). In this study, we identified the sites on UL44 that are phosphorylated by UL97 and also by cdk1/cyclin B in vitro, as well as the sites of UL44 phosphorylation during infection. Additionally, we tested the effects of transient inhibition of UL97 and cdks on UL44 phosphorylation in infected cells. The coincidence of sites phosphorylated by UL97 in vitro and from infected cells, and our finding that maribavir inhibits phosphorylation of specific peptides of UL44 in infected cells adds to the evidence that UL44 is a substrate of UL97 during infection. However, the UL97 inhibitor, maribavir, did not reduce the overall incorporation of phosphate into UL44, and affected only two weakly labeled phosphopeptides. Thus, UL97 does not appear to be a major contributor to UL44 phosphorylation during HCMV infection, at least at 72 hpi. Additionally, substitutions of most of the sites phosphorylated by UL97 in vitro that are also phosphorylated in infected cells had only a modest effect on viral replication and little or no effect on DNA synthesis. In contrast, substitution of Ser 413, 415, and 418, which our analysis identified as the most extensively phosphorylated residues of UL44 (and thus highly unlikely to be mainly phosphorylated by UL97), resulted in failure of a mutant BAC to replicate. We discuss these results in terms of phosphorylation of UL44 by UL97, by cellular kinases, and the role of UL44 phosphorylation in viral replication.
UL44 Phosphorylation by UL97
As noted above, collectively our data add to previous evidence (Krosky et al., 2003) that UL44 is a substrate of UL97 during infection. Our analyses did not permit us to definitively identify the sites that are phosphorylated by UL97 during infection. Nevertheless, S402 is a likely candidate for such a site, as this residue was phosphorylated by UL97, but not cdk1, in vitro, and is phosphorylated during infection. However, S413, S415 or S418 are not likely candidates, since peptides containing these three sites were found to be phosphorylated 100% of the time, and the phosphopeptides affected by maribavir treatment were only modestly labeled by 32P. Additionally, MS analysis of cells infected with a UL97 null mutant virus readily detected phosphorylation at these sites (data not shown). Results from MS analyses comparing UL44 phosphorylation in cells infected with WT and the UL97 null mutant were too inconsistent to be able to interpret a failure to detect phosphorylation at a given site (data not shown).
Intriguingly, 32P labeling of UL44 was slightly elevated in infected cells transiently treated with maribavir. This observation may relate to a previous observation: When maribavir was present throughout viral infection, only the most acidic species, and therefore likely the most phosphorylated forms, of UL44 were detected (Krosky et al., 2003). On the other hand, when we transiently treated cells with maribavir and roscovitine, 32P incorporation into UL44 was lower than when roscovitine was used alone. Therefore, the kinase activity of UL97 may be more important for UL44 phosphorylation when the activities of cdks are reduced or absent.
UL44 Phosphorylation by Cellular Kinases
Our previous results (Krosky et al., 2003) and our phospholabeling inhibition and phosphotryptic peptide analyses of UL44 indicate that UL44 is largely phosphorylated by kinases other than UL97 during infection. We think it likely that these kinases include cdks. Several candidate sites that were phosphorylated by cdk1 in vitro were also phosphorylated in infected cells. Additionally, transient treatment of infected cells with roscovitine, an inhibitor of multiple cdks, including cdk1 (Meijer et al., 1997; Meijer and Raymond, 2003), inhibited phosphate incorporation into UL44 and decreased labeling of most phosphopeptides. It also seems likely that cellular kinses CK2 and possibly CK1 phosphorylate UL44 during infection (see below), and other kinases may also be involved.
Role of UL44 Phosphorylation in Viral Replication
It has been widely proposed that inhibition of UL97-mediated phosphorylation of UL44 is the mechanism by which maribavir affects viral DNA replication and viral yield (Jacob et al., 2011; Krosky et al., 2003; Marschall et al., 2003; Mercorelli et al., 2008; Michel and Mertens, 2004; Wolf et al., 2001). However, substitutions of sites within UL44 that were found to be phosphorylated in infected cells, including ones phosphorylated by UL97 in vitro other than those adjacent to the NLS (which are highly unlikely to be phosphorylated by UL97 in infected cells), had little effect on viral DNA synthesis, and only modestly affected virus production. Thus, these results suggest UL97-mediated phosphorylation of UL44 is not the principal mechanism by which UL97 -- and thus maribavir --influences viral DNA synthesis and virus production. Recent work from our lab suggests that UL97 likely modulates viral DNA synthesis through a mechanism involving phosphorylation of retinoblastoma protein family members in quiescent cells (Kamil et al., 2009). Nevertheless, the modest defects in viral replication kinetics of the viable Phos mutants raise the possibility that UL97 phosphorylation may impact UL44 function during infection. If so, these phosphorylation sites might affect a function of UL44 other than DNA synthesis -- perhaps, transcription of late genes (Isomura et al., 2008; Isomura et al., 2007).
UL44 contains sites for phosphorylation by CK2 and CK1 upstream of the C terminal NLS [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript]. Substitution of S413 or S418 with alanine results in decreased UL44 NLS function in transient transfection assays [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript]. Our data expand upon these findings, as substitution of serines 413, 415 and 418 in the context of the viral genome (the Phos-4 mutant), resulted in a loss of nuclear localization of UL44, and a virus that was unable to replicate. Although it has not yet been demonstrated that CK2 and CK1 phosphorylate these sites within UL44 during HCMV infection, a simple interpretation of our data together with those of Alvisi et al. [(Alvisi et al., 2005) and Alvisi, G., et al., accompanying manuscript] is that phosphorylation of the three serines upstream of the NLS by these protein kinases is required for UL44 nuclear localization in infected cells, and thus HCMV replication.
Notably, almost all of the phosphorylation sites of UL44 we identified during infection were located in the C-terminus of the protein. We and others have recently established that during HCMV infection the C-terminal segment of UL44 is required for viral replication (Kim and Ahn, 2010; Silva et al., 2010). Specifically, we found that this segment is crucial both for nuclear localization and at least one other function that is important for the proper formation of viral DNA replication compartments (Silva et al., 2010). In the present study, our results with the Phos4 mutant reinforce the importance of this segment for nuclear localization, and stand in contrast to a report to the contrary (Kim and Ahn, 2010; Silva et al., 2010). In addition, UL44 has been reported to interact with a number of viral and cellular proteins (Fulcher et al., 2009; Gao et al., 2008; Strang et al., 2010a; Strang et al., 2010b; Strang et al., 2009). It is possible that phosphorylation of UL44 can modulate binding to these, or other undiscovered, binding partners. Indeed phosphorylation at T427 can modulate binding to the cellular protein BRAP2 (Fulcher et al., 2009).
We caution that we limited our studies of UL44 phosphorylation in infected human foreskin fibroblasts at 72 hpi. UL44 appears to be phosphorylated throughout infection (Gibson, 1984). It is possible that the sites of phosphorylation on UL44, their prevalence, and the viral and cellular kinases that phosphorylate these sites, change as infection proceeds, as has been observed with the Epstein-Barr virus (EBV) homolog BMRF1 (Ohashi et al., 2007; Wang et al., 2005), or with different cell culture conditions (Chou et al., 2006). Thus, UL44 phosphorylation appears to be complex and there is clearly more to learn about the role of this important modification of UL44 during HCMV infection.
MATERIALS AND METHODS
Plasmids, BACs, and primers
The AD169rv bacterial artificial chromosome (BAC) clone of HCMV wild-type (wt) strain AD169varATCC (Borst et al., 1999; Hobom et al., 2000) was generously provided by Ulrich H. Koszinowski (Ludwig-Maximilians University). The pp71 expression plasmid, pCGN71 (Baldick et al., 1997), was kindly provided by Thomas Shenk (Princeton University). The Cre-expression plasmid, pBRep-Cre (Hobom et al., 2000), was kindly provided by Wolfram Brune (Robert Koch Institute). pEPkan-S was kindly provided by Nikolaus Osterrieder (Cornell University). The E. coli strain GS1783 used for mutagenesis of the wt AD169 BAC was a generous gift from Greg Smith (Northwestern University). pAB1-bactinPCRscript was kindly provided by Robert Kalejta (University of Wisconsin-Madison). The sequences of primers used to construct mutants are provided in Supplemental Table 4.
Expression and Purification of Proteins
Full-length UL44, cleaved from recombinant GST-UL44, was purified from Escherichia coli harboring the pD15-UL44 plasmid as previously described (Loregian et al., 2004a), with the addition of passing a high salt wash containing 1 M NaCl over the glutathione-Sepharose 4FastFlow column to remove a contaminating kinase. The preparation contained >95% full-length UL44 and small amounts of uncleaved fusion protein and degradation products of UL44 (Supplementary Fig. 1A). UL44ΔC290 was kindly provided by Jennifer Baltz and purified as previously described (Appleton et al., 2004). GST-UL97 was purified as previously described (He et al., 1997). His-Lamin A was purified as previously described (Hamirally et al., 2009). Quantitative amino acid analysis (performed by the Molecular Biology Core Facility, Dana-Farber Cancer Institute) was used to determine the concentration of proteins purified by our lab. Cdk1/cyclin B was purchased from Cell Signaling Technology.
In Vitro Kinase Reactions
Phosphorylation of UL44 by GST-UL97 in vitro kinase reactions was carried out using optimized kinase buffer containing 10 mM HEPES [pH 8.0], 25 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 200 µM unlabeled ATP (kinase buffer). To measure phosphorylation, 0.125 µL 32P-γATP (6000 Ci/mmol) was included in a 20 µL reaction. Reactions were conducted with the concentrations of protein indicated in the figure legend. In reactions where kinase inhibitors were included, 1 µM maribavir (a generous gift from John Drach and Karen Biron) or 500 nM CGP74514A (Calbiochem) was added. The reaction mixtures were incubated at 37°C for 60 min and the reactions were terminated by the addition of 5× SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The samples were heated at 100°C for 5 min and separated by 8% SDS-PAGE. Gels were dried onto blotting paper under vacuum and incorporated 32P label was assessed with a PhosphorImager (Molecular-Imager FX System; Bio-Rad).
To prepare UL97 phosphorylated UL44 for MS analysis, ~2.5 µg of purified UL44 was incubated with ~125 ng of GST-UL97 in kinase buffer lacking NaCl, making the effective salt concentration 86 mM. A negative control reaction included 1 µM maribavir to inhibit GST-UL97 kinase activity. For a large-scale preparation of cdk1 phosphorylated UL44, ~2.5 µg of UL44 was incubated with ~125 nM GST-cdk1/GST-cyclinB in kinase buffer lacking NaCl, making the effective salt concentration 123 mM. Reactions were incubated for 1.5 hours at 37°C and then resolved by 8% SDS-PAGE. Proteins were visualized with Coomassie blue. UL44 was excised from the gels and submitted for MS analysis.
MS Analysis
Phosphorylated UL44 samples were submitted to the Taplin MS Facility, Harvard Medical School. In some experiments, samples were also submitted to the MS Core Facility at Beth Israel Deaconess Hospital. At both facilities UL44 was analyzed for the identification of phosphorylation sites by LC/tandem MS. The relative amounts of phosphorylated and unphosphorylated peptides were estimated in one experiment by the Taplin Facility by measuring the peak intensities of the relevant peptides.
Cells and Viruses
Primary human foreskin fibroblast (HFF) cells, isolate Hs27 (American Type Tissue Culture Collection), were used to support HCMV replication and were cultured in Dubelcco’s modified Eagle’s medium (DMEM) (CellGro) supplemented with 10% fetal bovine serum (FBS, Sigma), 200 U penicillin and 200 U streptomycin per mL (Gibco; Carlsbad, CA) (DMEM-10%) at 37°C in 5% CO2. Viruses used were HCMV AD169rv, and mutants derived from it, which are described below. HCMV stocks were titrated using plaque assays.
Western Blot Analysis
Western blot analyses of UL97 and β-actin were performed as previously described (Hamirally et al., 2009). Western blot analysis of UL44 was performed using anti-UL44 primary antibody (αICP36, Virusys, Sykesville, MD) diluted 1:8,000.
Immunoprecipitations from Infected Cells
HFF cells (4.5 × 106) were either mock-infected or infected with AD169rv, at an MOI of ~3 PFU/cell and cell lysates were harvested at 72 hpi in EBC2 buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 2 mM EDTA, 0.5% NP-40) supplemented with protease and phosphatase inhibitors. Immunoprecipitations of UL44 were carried out as previously described (Strang et al., 2009) and visualized by colloidal blue staining. UL44 was excised from the gels and submitted for MS analysis as described above.
Orthophosphate Labeling
HFF cells (1.75 × 105) were seeded into 12-well plates in DMEM supplemented with 2% FBS. Twenty-four h post seeding, the 2% medium was removed, the monolayers were washed and replenished with DMEM containing 0.1% FBS. Infections, drug treatment (68 hpi), radiolabeling (70 hpi) were carried out in DMEM containing 0.1% FBS as previously described (Hamirally et al., 2009). At 72 hpi, cells lysates were lysed in 500 µL of EBC2 buffer supplemented with protease and phosphatase inhibitors. UL44 was immunoprecipitated as previously described (Strang et al., 2009), resolved by 8% SDS-PAGE, transferred to PVDF membrane and exposed to a Phosphorimager. The PVDF membrane was then analyzed by Western blotting for UL44 as described above. Experiments were initially performed using a series of three-fold dilutions of protein to ensure that quantification of UL44 protein levels was accurate. In multiple experiments, using multiple methods of detection, the quantified levels of UL44 protein accurately reflected the three-fold dilution series. For the experiments shown, we loaded the entire sample into one well of the SDS-PAGE gel. Quantifications of the phosphorimage and Western blot were performed using Quantity One software. We then calculated the ratio of the relative level of 32P label in each sample by dividing the amount of phosphate incorporation into UL44 by the UL44 protein level for each sample.
One-Dimensional Analysis of Tryptic Peptides
Immunoprecipitated UL44 from 32P-radiolabeled, infected cells, either untreated or treated with kinase inhibitors as described above, as well as UL44 radiolabeled using GST-UL97 in vitro (as described above) and unlabeled UL44 immunoprecipitated from infected cells, were excised from the PVDF membrane and digested in-membrane as described (Meisenhelder, 1999) in 50 µl digestion buffer (50 mM Tris-HCl [pH 8.0], 1% TX-100, 10% acetonitrile) containing 2.5 µg sequencing grade modified trypsin (Promega). Phospholabeled peptides of UL44 were resuspended in 25 µl of sample buffer (125 mM Tris-HCl and 6 M urea) and separated on an alkaline 40% acrylamide gel as previously described (West et al., 1984). The gel was dried, exposed to a PhosphorImager screen for ~5 weeks and was visualized by PhosphorImager analysis.
BAC Mutagenesis and Reconstitution of Virus
To construct a shuttle plasmid for mutagenesis of the UL44 gene, UL44 and flanking sequences were amplified by PCR from the AD169rv BAC DNA using primers upstream and downstream of the UL44 ORF. The resulting PCR product was subcloned into pCR2.1-Topo vector (Invitrogen), resulting in plasmid TOPO-UL44flanks. The KpnI-PstI restriction fragment of TOPO-UL44flanks was then inserted into pUC19, resulting in plasmid pUC19-44. The I-SceI-AphAI cassette containing a kanamycin resistance gene with a 5’ I-Sce I site was amplified by PCR from plasmid pEPkan-S (Tischer et al., 2006) using primers containing overlapping UL44 sequences and a unique BglII site. BglII-digested PCR product was ligated into pUC19-44, resulting in plasmid pUC19-44kan. Site directed mutagenesis using the QuikChange method (Stratagene; La Jolla, CA) was performed on pUC19-44kan to introduce S8A, S286A, S348A, S354A/S355A, S367A, S387A and S402A substitutions in different combinations and yielded plasmids pUC19-Phos-1, -2, -3 and -1/2. Restriction enzyme analysis and DNA sequencing confirmed the presence of each mutation and the absence of spurious mutations.
Mutagenesis of the HCMV BAC AD169rv was performed according to the protocol outlined in Tischer et al. (Tischer et al., 2006) as previously described (Strang et al., 2010a). Briefly, we constructed a bacmid, ΔUL44-Zeo, containing the majority of the UL44 ORF replaced by the Zeocin resistance cassette, which was PCR amplified from pZeo (Invitrogen) by using partially overlapping primers containing UL44 sequences, utilizing the two-step Red recombination method (Tischer et al., 2006). PCR products containing mutant UL44 ORFs were used for two-step Red recombination of ΔUL44-Zeo as described (Tischer et al., 2006) to generate BAC-Phos-1, -2, -3, and -1/2. For generation of BAC-Phos4 containing alanine substitutions in UL44 at residues 413, 415 and 418, mutagenic PCR primers were used to amplify a DNA sequence from plasmid pEP-KanS (Tischer et al., 2006). Red recombination was used to introduce the PCR product into the AD169rv BAC as described (Strang et al., 2010a; Tischer et al., 2006) to generate BAC-Phos-4. The resulting mutant BACs were sequenced to confirm the presence of mutations in the UL44 coding sequence. To generate the rescued version of BAC-Phos4, bacteria harboring the mutant BAC were transformed with a PCR product amplified using primers containing wt UL44 sequences. The PCR product was introduced into the mutant BAC as before using Red recombination (Tischer et al., 2006). Supplementary Table 4 lists all the primers used in this study.
To generate viruses, AD169rv BAC and the various mutant BACS were electroporated into HFF cells as described previously (Tischer et al., 2006), with plasmids pCGN71 (Baldick et al., 1997), expressing the viral transcriptional transactivator pp71, and pBRep-Cre (Hobom et al., 2000). Cells were maintained until ~90% cytopathic effect (CPE) was observed, at which time the supernatants of these cultures were collected as viral stocks, stored at −80°C, and titrated using plaque assays.
Viral Replication Kinetics
HFF cells (7.5 × 104 per well) were seeded in 24-well plates. Twenty-four h later, cells were infected with AD169rv or mutant viruses at an MOI of 1 PFU/cell (verified by back titration). Supernatants were harvested at 24 h intervals until 6 days post infection and stored at −80°C until titration. Viral titers were determined by plaque assays. Data are presented as the average viral titer ± standard deviation obtained from three independent experiments. Statistical analysis was performed with GraphPad Prism software using two-way ANOVA to determine the statistical significance of differences between the average viral titers of the different viruses in dividing cells followed by Bonferroni’s Multiple Comparison Post-Test to determine statistical significance of differences between the average viral titers on each day as compared to wt titers. Statistical significance was accepted at P < 0.05.
Viral DNA Accumulation Analysis by Quantitative PCR
HFF cells (7.5 × 104) were seeded in 24-well plates and 24 h later were infected with AD169rv or mutant viruses at a target MOI of 1 PFU/cell (verified by back titration). Total DNA was harvested in lysis buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 0.5% SDS, 100 mM NaCl) at various times after infection and stored at −80°C. Total DNA was then purified using the DNeasy Blood and Tissue kit (Qiagen) per the manufacturer’s instruction. Real-time quantitative PCR was used to determine the number of viral copies in the total DNA samples from infected cells as previously described (Saffert and Kalejta, 2007; Strang et al., 2010a), using primer and probe sets for UL83 (Gault et al., 2001) and β-actin (Hanfler et al., 2003). PCR reactions contained 4 µl of 150 µl of extracted DNA, 900 nM of each primer, 250 nM probe, 6 µl TaqMan Universal PCR Master Mix without AmpErase UNG (Applied Biosystems Inc.) and nuclease-free water to 12 µl. Real-time quantitative PCR was run on an ABI 7900HT and data were analyzed using the SDS 2.3 program (Applied Biosystems Inc.). Data are presented as the average ratio of UL83 copies to β-actin copies and when applicable, ± the standard deviation obtained from three independent experiments.
Immunofluorescence
HFF cells were electroporated with either BAC-AD169rv or BAC-Phos-4 as previously described (Tischer et al., 2006). Seventy-two h later, 5 × 104 HFF cells were plated onto glass coverslips. Cells were fixed and stained 24 h later using anti-UL44 antibody (Virusys) and Hoechst reagent (5mg/ml) as previously described (Strang et al., 2010a). Cells were imaged on an Axioplan 2 microscope (Zeiss) with a 63× objective and Hamamatsu CCD camera (model C4742-95). Images were deconvolved using the inverse filter algorithm in the Axiovision (Rel.4.5) software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health to D.M.C. (RO1 AI019838 and RO1 AI26077) and J.P.K (F32 AI075766), from the William Randolph Hearst Foundation to B.L.S., and from the Harvard Institute to Global Health to E.W.L., for which we are grateful. We thank all members of the Coen and Hogle labs for helpful discussions and technical advice, with special thanks to Jennifer Baltz, Martha Kramer, and My Sam. We thank Ross Tomaino (Taplin MS Facility) and John Asara (BIDMC MS Facility) for their efforts and helpful advice. We also thank Gualtiero Alvisi for helpful comments on a draft of this manuscript, and for providing a draft of his accompanying manuscript, and Jennifer Baltz, John Drach, Karen Biron, Ulrich Koszinowski, Thomas Shenk, Wolfram Brune, Nikolaus Osterrieder, Greg Smith, and Robert Kalejta for reagents. We gratefully acknowledge David Knipe for the use of microscopy facilities and Lynne Chang for assistance with microscopy and image processing. We gratefully acknowledge that immunofluorescence data for this study were analyzed in the Nikon Imaging Center at Harvard Medical School.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Advani SJ, Weichselbaum RR, Roizman B. cdc2 cyclin-dependent kinase binds and phosphorylates herpes simplex virus 1 U(L)42 DNA synthesis processivity factor. J. Virol. 2001;75:10326–10333. doi: 10.1128/JVI.75.21.10326-10333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani SJ, Weichselbaum RR, Roizman B. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase II alpha for post-DNA synthesis expression of late genes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4825–4830. doi: 10.1073/pnas.0730735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Jang WJ, Hayward GS. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J. Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvisi G, Jans DA, Guo J, Pinna LA, Ripalti A. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic (Copenhagen, Denmark) 2005;6:1002–1013. doi: 10.1111/j.1600-0854.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- Alvisi G, Roth DM, Camozzi D, Pari GS, Loregian A, Ripalti A, Jans DA. The flexible loop of the human cytomegalovirus DNA polymerase processivity factor ppUL44 is required for efficient DNA binding and replication in cells. J. Virol. 2009;83:9567–9576. doi: 10.1128/JVI.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton BA, Brooks J, Loregian A, Filman DJ, Coen DM, Hogle JM. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 2006;281:5224–5232. doi: 10.1074/jbc.M506900200. [DOI] [PubMed] [Google Scholar]
- Appleton BA, Loregian A, Filman DJ, Coen DM, Hogle JM. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell. 2004;15:233–244. doi: 10.1016/j.molcel.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Azzeh M, Honigman A, Taraboulos A, Rouvinski A, Wolf DG. Structural changes in human cytomegalovirus cytoplasmic assembly sites in the absence of UL97 kinase activity. Virology. 2006;354:69–79. doi: 10.1016/j.virol.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J. Biol. Chem. 2002;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, Pearson A, Coen DM. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology. 2004;324:184–193. doi: 10.1016/j.virol.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Baldick CJ, Jr, Marchini A, Patterson CE, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron KK. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 2006;71:154–163. doi: 10.1016/j.antiviral.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith AA, 3rd, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob. Agents Chemother. 2002;46:2365–2372. doi: 10.1128/AAC.46.8.2365-2372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Borst EM, Hahn G, Koszinowski UH, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan WA, Boldogh I, Chi P, Thompson EA, Albrecht T. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology. 1997a;231:239–247. doi: 10.1006/viro.1997.8489. [DOI] [PubMed] [Google Scholar]
- Bresnahan WA, Thompson EA, Albrecht T. Human cytomegalovirus infection results in altered Cdk2 subcellular localization. J. Gen. Virol. 1997b;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- Chan SR, Chandran B. Characterization of human herpesvirus 8 ORF59 protein (PF-8) and mapping of the processivity and viral DNA polymerase-interacting domains. J. Virol. 2000;74:10920–10929. doi: 10.1128/jvi.74.23.10920-10929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J. Virol. 1991;65:2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Van Wechel LC, Marousek GI. Effect of cell culture conditions on the anticytomegalovirus activity of maribavir. Antimicrob. Agents Chemother. 2006;50:2557–2559. doi: 10.1128/AAC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter LJ, Moss HW, Lang J, McGeoch DJ. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- de Wind N, Domen J, Berns A. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J. Virol. 1992;66:5200–5209. doi: 10.1128/jvi.66.9.5200-5209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl PF, Powell KL. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 1992;66:4126–4133. doi: 10.1128/jvi.66.7.4126-4133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger S, Stamminger T, Muller R, Graf L, Klebl B, Eickhoff J, Marschall M. Recruitment of cyclin-dependent kinase 9 to nuclear compartments during cytomegalovirus late replication: importance of an interaction between viral pUL69 and cyclin T1. J. Gen. Virol. 2011 doi: 10.1099/vir.0.030494-0. In press. [DOI] [PubMed] [Google Scholar]
- Fulcher AJ, Roth DM, Fatima S, Alvisi G, Jans DA. The BRCA-1 binding protein BRAP2 is a novel, negative regulator of nuclear import of viral proteins, dependent on phosphorylation flanking the nuclear localization signal. FASEB J. 2009;24:1454–1466. doi: 10.1096/fj.09-136564. [DOI] [PubMed] [Google Scholar]
- Gao Y, Colletti K, Pari GS. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 2008;82:96–104. doi: 10.1128/JVI.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault E, Michel Y, Dehee A, Belabani C, Nicolas JC, Garbarg-Chenon A. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 2001;39:772–775. doi: 10.1128/JCM.39.2.772-775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg E, Pagano JS. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta. 2008;1784:203–212. doi: 10.1016/j.bbapap.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershburg E, Raffa S, Torrisi MR, Pagano JS. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 2007;81:5407–5412. doi: 10.1128/JVI.02398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. Protein counterparts of human and simian cytomegaloviruses. Virology. 1983;128:391–406. doi: 10.1016/0042-6822(83)90265-9. [DOI] [PubMed] [Google Scholar]
- Gibson W. Synthesis, Structure and Function of Cytomegalovirus Major Nonvirion Nuclear Protein. In: Rapp F, editor. Herpesvirus: proceedings of a Burroughs Wellcome-UCLA symposium, held at Steamboat Springs; April 8–13, 1984; Colorado. New York City, NY: Alan R. Liss, Inc.; 1984. pp. 424–440. [Google Scholar]
- Gibson W, Murphy TL, Roby C. Cytomegalovirus-infected cells contain a DNA-binding protein. Virology. 1981;111:251–262. doi: 10.1016/0042-6822(81)90669-3. [DOI] [PubMed] [Google Scholar]
- Goldberg MD, Honigman A, Weinstein J, Chou S, Taraboulos A, Rouvinski A, Shinder V, Wolf DG. Human cytomegalovirus UL97 kinase and nonkinase functions mediate viral cytoplasmic secondary envelopment. J. Virol. 2011;85:3375–3384. doi: 10.1128/JVI.01952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfler J, Kreuzer KA, Laurisch K, Rayes N, Neuhaus P, Schmidt CA, Oettle H. Quantitation of cytomegalovirus (hCMV) DNA and beta-actin DNA by duplex real-time fluorescence PCR in solid organ (liver) transplant recipients. Med Microbiol Immunol. 2003;192:197–204. doi: 10.1007/s00430-002-0166-6. [DOI] [PubMed] [Google Scholar]
- He Z, He YS, Kim Y, Chu L, Ohmstede C, Biron KK, Coen DM. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J. Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Heineman TC, Cohen JI. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J. Virol. 1995;69:7367–7370. doi: 10.1128/jvi.69.11.7367-7370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel L, Chou S, Mocarski ES. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 2007;3:e6. doi: 10.1371/journal.ppat.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 2000;74:7720–7729. doi: 10.1128/jvi.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- Isomura H, Stinski MF, Kudoh A, Murata T, Nakayama S, Sato Y, Iwahori S, Tsurumi T. Noncanonical TATA sequence in the UL44 late promoter of human cytomegalovirus is required for the accumulation of late viral transcripts. J. Virol. 2008;82:1638–1646. doi: 10.1128/JVI.01917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura H, Stinski MF, Kudoh A, Nakayama S, Iwahori S, Sato Y, Tsurumi T. The late promoter of the human cytomegalovirus viral DNA polymerase processivity factor has an impact on delayed early and late viral gene products but not on viral DNA synthesis. J. Virol. 2007;81:6197–6206. doi: 10.1128/JVI.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob T, Van den Broeke C, Favoreel HW. Viral serine/threonine protein kinases. J. Virol. 2011;85:1158–1173. doi: 10.1128/JVI.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamil JP, Coen DM. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 2007;81:10659–10668. doi: 10.1128/JVI.00497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamil JP, Hume AJ, Jurak I, Munger K, Kalejta RF, Coen DM. Human papillomavirus 16 E7 inactivator of retinoblastoma family proteins complements human cytomegalovirus lacking UL97 protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16823–16828. doi: 10.1073/pnas.0901521106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi AJ, Spector DH. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 2008;82:394–407. doi: 10.1128/JVI.01681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev Med Virol. 2003;13:331–340. doi: 10.1002/rmv.402. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K, Tanaka M, Kanamori M, Nishiyama Y, Yamanashi Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 2003;77:2359–2368. doi: 10.1128/JVI.77.4.2359-2368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Matsumura T, Roizman B, Hirai K. Cellular elongation factor 1delta is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J. Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Ahn JH. Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus. J. Virol. 2010;84:8409–8421. doi: 10.1128/JVI.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosky PM, Baek MC, Jahng WJ, Barrera I, Harvey RJ, Biron KK, Coen DM, Sethna PB. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J. Virol. 2003;77:7720–7727. doi: 10.1128/JVI.77.14.7720-7727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuny CV, Chinchilla K, Culbertson MR, Kalejta RF. Cyclin-dependent kinase-like function is shared by the beta- and gamma- subset of the conserved herpesvirus protein kinases. PLoS Pathog. 2010;6:e1001092. doi: 10.1371/journal.ppat.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler E, Stuart AD, Chee MS. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- Loregian A, Appleton BA, Hogle JM, Coen DM. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 2004a;78:158–167. doi: 10.1128/JVI.78.1.158-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian A, Appleton BA, Hogle JM, Coen DM. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 2004b;78:9084–9092. doi: 10.1128/JVI.78.17.9084-9092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Freitag M, Suchy P, Romaker D, Kupfer R, Hanke M, Stamminger T. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology. 2003;311:60–71. doi: 10.1016/s0042-6822(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Marsden HS, Campbell ME, Haarr L, Frame MC, Parris DS, Murphy M, Hope RG, Muller MT, Preston CM. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J. Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- Meijer L, Raymond E. Roscovitine and other purines as kinase inhibitors. From starfish oocytes to clinical trials. Acc Chem Res. 2003;36:417–425. doi: 10.1021/ar0201198. [DOI] [PubMed] [Google Scholar]
- Meisenhelder J. Phosphopeptide Mapping and Identification of Phosphorylation Sites. Current protocols in protein science. 1999:13.19.19–13.19.10. doi: 10.1002/0471140864.ps1309s18. [DOI] [PubMed] [Google Scholar]
- Mercorelli B, Sinigalia E, Loregian A, Palu G. Human cytomegalovirus DNA replication: antiviral targets and drugs. Rev Med Virol. 2008;18:177–210. doi: 10.1002/rmv.558. [DOI] [PubMed] [Google Scholar]
- Michel D, Mertens T. The UL97 protein kinase of human cytomegalovirus and homologues in other herpesviruses: impact on virus and host. Biochim. Biophys. Acta. 2004;1697:169–180. doi: 10.1016/j.bbapap.2003.11.022. [DOI] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- Moffat JF, Zerboni L, Sommer MH, Heineman TC, Cohen JI, Kaneshima H, Arvin AM. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M, Horie K, Hoshikawa Y, Nagata K, Osaki M, Ito H, Sairenji T. Accumulation of Epstein-Barr virus (EBV) BMRF1 protein EA-D during latent EBV activation of Burkitt's lymphoma cell line Raji. Microbes Infect. 2007;9:150–159. doi: 10.1016/j.micinf.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Penfold ME, Mocarski ES. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology. 1997;239:46–61. doi: 10.1006/viro.1997.8848. [DOI] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. Human cytomegalovirus UL97 Kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 2005;79:15494–15502. doi: 10.1128/JVI.79.24.15494-15502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Gao N, Jairath S, Mulamba G, Krosky P, Coen DM, Parker BO, Pari GS. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 2008;82:5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechter S, Scott GM, Eickhoff J, Zielke K, Auerochs S, Muller R, Stamminger T, Rawlinson WD, Marschall M. Cyclin-dependent Kinases Phosphorylate the Cytomegalovirus RNA Export Protein pUL69 and Modulate Its Nuclear Localization and Activity. J. Biol. Chem. 2009;284:8605–8613. doi: 10.1074/jbc.M805693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeckel D, Mueller-Lantzsch N. Biochemical characterization of two Epstein-Barr virus early antigen-associated phosphopolypeptides. Virology. 1985;147:253–263. doi: 10.1016/0042-6822(85)90128-x. [DOI] [PubMed] [Google Scholar]
- Romaker D, Schregel V, Maurer K, Auerochs S, Marzi A, Sticht H, Marschall M. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J. Med. Chem. 2006;49:7044–7053. doi: 10.1021/jm060696s. [DOI] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 2007;81:9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Mahr JA, Orazio NI, Spector DH. Nuclear export of the human cytomegalovirus tegument protein pp65 requires cyclin-dependent kinase activity and the Crm1 exporter. J. Virol. 2007;81:11730–11736. doi: 10.1128/JVI.02760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, McElroy AK, Spector DH. Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J. Virol. 2003;77:13214–13224. doi: 10.1128/JVI.77.24.13214-13224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, McElroy AK, Yen J, Tamrakar S, Clark CL, Schwartz RA, Spector DH. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J. Virol. 2004;78:11219–11232. doi: 10.1128/JVI.78.20.11219-11232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Spector DH. Cyclin-dependent kinase activity is required for efficient expression and posttranslational modification of human cytomegalovirus proteins and for production of extracellular particles. J. Virol. 2006;80:5886–5896. doi: 10.1128/JVI.02656-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LA, Loregian A, Pari GS, Strang BL, Coen DM. The Carboxy-Terminal Segment of the Human Cytomegalovirus DNA Polymerase Accessory Subunit UL44 Is Crucial for Viral Replication. J. Virol. 2010;84:11563–11568. doi: 10.1128/JVI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Boulant S, Coen DM. Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. J. Virol. 2010a;84:1771–1784. doi: 10.1128/JVI.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Geballe AP, Coen DM. Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44. J. Gen. Virol. 2010b;91:2167–2175. doi: 10.1099/vir.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Sinigalia E, Silva LA, Coen DM, Loregian A. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J. Virol. 2009;83:7581–7589. doi: 10.1128/JVI.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan V, Talarico CL, Stanat SC, Davis M, Coen DM, Biron KK. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- Talarico CL, Burnette TC, Miller WH, Smith SL, Davis MG, Stanat SC, Ng TI, He Z, Coen DM, Roizman B, Biron KK. Acyclovir is phosphorylated by the human cytomegalovirus UL97 protein. Antimicrob. Agents Chemother. 1999;43:1941–1946. doi: 10.1128/aac.43.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamrakar S, Kapasi AJ, Spector DH. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 2005;79:15477–15493. doi: 10.1128/JVI.79.24.15477-15493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nishiyama Y, Sata T, Kawaguchi Y. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology. 2005;341:301–312. doi: 10.1016/j.virol.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- Trofe J, Pote L, Wade E, Blumberg E, Bloom RD. Maribavir: a novel antiviral agent with activity against cytomegalovirus. Ann. Pharmacother. 2008;42:1447–1457. doi: 10.1345/aph.1L065. [DOI] [PubMed] [Google Scholar]
- Wang JT, Yang PW, Lee CP, Han CH, Tsai CH, Chen MR. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 2005;86:3215–3225. doi: 10.1099/vir.0.81313-0. [DOI] [PubMed] [Google Scholar]
- Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–577. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- Weiland KL, Oien NL, Homa F, Wathen MW. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 1994;34:191–206. doi: 10.1016/0168-1702(94)90124-4. [DOI] [PubMed] [Google Scholar]
- West MHP, Wu RS, Bonner WM. Polyacrylamide gel electrophoresis of small peptides. Electrophoresis. 1984;5:133–138. [Google Scholar]
- Wolf DG, Courcelle CT, Prichard MN, Mocarski ES. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1895–1900. doi: 10.1073/pnas.98.4.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zydek M, Hagemeier C, Wiebusch L. Cyclin-dependent kinase activity controls the onset of the HCMV lytic cycle. PLoS Pathog. 2010;6:e1001096. doi: 10.1371/journal.ppat.1001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.