Summary
Phosphorylation or SUMOylation of the kainate receptor (KAR) subunit GluK2 have both individually been shown to regulate KAR surface expression. However, it is unknown if phosphorylation and SUMOylation of GluK2 are important for activity-dependent KAR synaptic plasticity. Here, we show that PKC-mediated phosphorylation of GluK2 at serine 868 promotes GluK2 SUMOylation at lysine 886 and that both these events are necessary for the internalization of GluK2 containing KARs that occurs during long-term depression of KAR-mediated synaptic transmission at rat hippocampal mossy fiber synapses. Conversely, phosphorylation of GluK2 at serine 868 in the absence of SUMOylation leads to an increase in KAR surface expression by facilitating receptor recycling between endosomal compartments and the plasma membrane. Thus, we describe a role for the dynamic control of synaptic SUMOylation in the regulation of KAR synaptic transmission and plasticity.
Keywords: SUMO, Kainate receptor, Phosphorylation, Synaptic plasticity, Long-term depression, Mossy fiber
Introduction
Synaptic plasticity is a cellular correlate of learning and memory and occurs through the processes of long-term potentiation (LTP) or long-term depression (LTD) 1. Changes in synaptic strength can be achieved by increases or decreases in the surface expression of neurotransmitter receptor proteins which has been shown to depend on interactions with specific intracellular binding partners. These interactions are often regulated by post-translational receptor modifications such as phosphorylation 2-4.
KARs are ionotropic glutamate receptors made up of tetrameric arrangements of five subunits (GluK15, previously known as GluR57 and KA12) and are found pre- and post--synaptically at many synapses throughout the brain 5-6. KAR surface expression at the plasma membrane depends on protein interactions at the intracellular c-termini of the receptor subunits 7-9, for example, the PDZ domain-containing proteins PICK1 and GRIP 10-11. In GluK2, amino acids 868-880 in the c-terminus are important for surface expression in heterologous systems or cultured neurons 9, 12.
Phosphorylation of KARs by PKC can enhance or depress KAR-mediated synaptic transmission 10, 13-16 and is involved in LTD in DRG neurons 16, the perirhinal cortex 13, 15, and the hippocampus 14. More recently, it has been shown that GluK2 can be SUMOylated in response to agonist binding 17, which leads to internalisation of GluK2 containing KARs. SUMOylation is a post-translational modification of lysine residues similar to ubiquitination, whereby a member of the Small Ubiquitin-like Modifier (SUMO) family is conjugated to target proteins, altering substrate function. SUMOylation is emerging as a major regulator of nuclear and extranuclear protein function in neurons 18-20. While the mechanisms that modulate SUMOylation remain largely unknown, protein phosphorylation has been reported to either enhance or inhibit SUMOylation, depending on the substrate protein 21-23.
The SUMOylation site at lysine 886 (K886) in GluK2 is located near to the PKC phosphorylation sites at serine 846 (S846) and 868 (S868). It has been demonstrated that PKC phosphorylation of S868 but not S846 promotes SUMOylation of K886 24. We therefore hypothesised that PKC phosphorylation of GluK2 may control SUMOylation and subsequent removal of GluK2 containing KARs during activity dependent LTD. We show that phosphorylation and SUMOylation are necessary for the PKC-dependent LTD of KARs at mossy fiber-CA3 synapses in the hippocampus. Furthermore, phosphorylation of S868 in the absence of SUMOylation results in an increase in KAR recycling between endosomal compartments and the plasma membrane.
Results
Phosphorylation promotes SUMOylation and loss of KARs
We have previously demonstrated SUMOylation of the KAR subunit GluK2 at hippocampal mossy fiber synapses 17. The functional consequences of SUMOylation were a removal of KARs from the postsynaptic membrane leading to a decrease in KAR-mediated synaptic transmission. Here we reproduce this finding by recording pharmacologically isolated KAR-mediated, synaptically evoked excitatory postsynaptic currents (KAR EPSCs) from CA3 neurons in hippocampal slices and included either SUMO-1 (active SUMO, 4.2 μM) or the conjugation-deficient mutant SUMO-1-ΔGG (inactive SUMO, 4.2 μM) in the recording pipette. We confirmed that inclusion of SUMO-1 caused a rapid decrease in the amplitude of the KAR EPSC to 46.6 ± 5.0% (Fig. 1a; n = 13; P < 0.001) of the initial amplitude obtained within 1 minute of rupturing the membrane inside the patch electrode whereas inclusion of SUMO-1-ΔGG had no effect on KAR EPSC amplitude (Fig. 1a; 103.4 ± 11.2%; n = 9; P > 0.05).
FIGURE 1. Phosphorylation promotes the SUMO-dependent removal of synaptic KARs.
a. Inclusion of 4.2 μM SUMO-1 in the patch solution caused a decrease in KAR EPSC amplitude compared to interleaved control experiments. Responses are normalised to the first minute. Example traces are taken as the average of the first minute (black) and 10-15 minutes (grey). Scale bars are 20 pA and 100 ms. b. Incubation of slices in PMA (1 μM) for 15 minutes prior to recording increased the depression in KAR EPSC amplitude mediated by SUMO whilst incubation with chelerythrine (5 μM) attenuated the SUMO-dependent depression in KAR EPSC amplitude. Data are plotted as mean ± s.e.m.
Phosphorylation of proteins can either facilitate or inhibit SUMOylation 21-23 and PKC-mediated phosphorylation of KARs regulates their subcellular localisation 13-14, 25. Since PKC-mediated phosphorylation of GluK2 promotes GluK2 SUMOylation 24, we reasoned that activation of PKC should facilitate, and inhibition reduce, the effects of SUMO on KAR EPSCs. To test this, we recorded KAR EPSCs from CA3 neurons following pre-incubation of the slices in either PMA (1 μM) or chelerythrine (5 μM) for a minimum of 15 minutes. In the presence of PMA (1 μM), inclusion of active SUMO in the recording pipette decreased the amplitude of KAR EPSCs to 22.9 ± 4.7%, a greater effect than seen in control conditions (Fig. 1b; n = 8; P < 0.05). In addition, in the presence of chelerythrine (5 μM), active SUMO no longer had any effect (Fig. 1b; 98.2 ± 6.0%, n=8; P>0.05). These results support the hypothesis that phosphorylation of GluK2 promotes SUMOylation and further suggests that phosphorylation is required for SUMO-dependent internalization of GluK2.
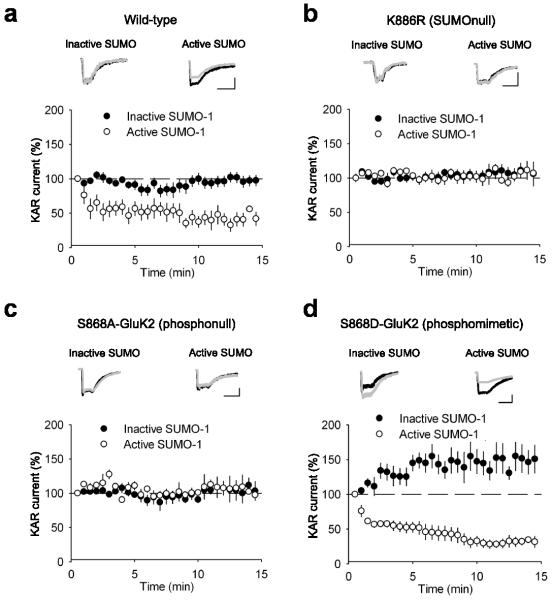
To determine the sites of phosphorylation and SUMOylation of GluK2 that regulate the surface expression of KARs, we expressed recombinant wild-type or mutated GluK2 where the putative PKC phosphorylation site S868 24-25 was altered to either a phosphomimetic aspartate or a non-phosphorylatable (phosphonull) alanine or the SUMOylation site K886 was altered to a non-SUMOylatable (SUMOnull) arginine 24. We first characterised kainate responses in HEK cells by expressing wild-type (WT) GluK2. This produced stable KAR-mediated response amplitudes to 300 ms applications of 100 μM kainate. Inclusion of inactive SUMO (SUMO-1-ΔGG) in the patch pipette had no effect on response amplitude (Fig. 2a; 106.3 ± 5.1%; n = 7; P > 0.05) but inclusion of active SUMO in the recording pipette induced a rapid depression of response amplitude (Fig. 2a; 52.5 ± 3.6%; n = 6; P < 0.0001). The speed of depression was faster than that seen in neurons but the magnitude was similar. The depression of KAR-mediated responses was directly due to SUMOylation of GluK2 as neither active nor inactive SUMO had any effect on KAR-mediated responses in HEK cells expressing the non-SUMOylatable (SUMOnull) GluK2 mutant K886R 17 (Fig. 2b; 106.6 ± 8.9% and 100.5 ± 12.6% inactive and active SUMO respectively; n = 6 for each; P > 0.05).
FIGURE 2. Phosphorylation of S868 on GluK2 promotes SUMOylation at K886 and subsequent removal of surface KARs.
a. Inclusion of 4.2 μM SUMO-1 in the patch solution caused a decrease in the KAR current amplitude in HEK cells expressing WT GluK2 whereas inclusion of inactive SUMO-1-ΔGG did not. KAR currents are responses to 300 ms applications of 100 μM kainate. Responses are normalised to the first minute. Example traces are taken as the average of the first minute (black) and 10-15 minutes (grey). Scale bars are 100 pA and 500 ms. b. SUMO-1 no longer caused a decrease in the KAR current amplitude evoked from HEK cells expressing K886R GluK2. c. SUMO-1 failed to decrease the amplitude of KAR currents evoked from HEK cells expressing S868A GluK2. d. Inclusion of SUMO-1 in the patch solution caused a decrease in the KAR current amplitude in HEK cells expressing S868D GluK2 which was comparable to the decrease seen in WT. However, inclusion of inactive SUMO-1 revealed an increase in KAR current amplitude. Data are plotted as mean ± s.e.m.
We next used the phosphomimetic and non-phosphorylatable mutations of serine 868 to test the role of phosphorylation in SUMO-mediated removal of surface KARs. In HEK cells expressing the S868A (phosphonull) GluK2 mutant, infusion of active SUMO via the recording pipette had no significant effect on the KAR mediated responses when compared to the inactive control (Fig. 2c; 98.2 ± 9.4% vs. 105.0 ± 8.3% inactive and active SUMO respectively; n = 6 for each; P > 0.05). However, in HEK cells expressing the S868D (phosphomimetic) GluK2 mutant, infusion of active SUMO caused a depression in KAR-mediated responses to 27.8 ± 3.5% (n = 6)compared to inactive SUMO (Fig. 2d; vs. 142.5 ± 11.2%; n = 6; P < 0.001), but not different from infusion of active SUMO with wild-type GluK2 (Fig. 2a). These data suggest that phosphorylation of GluK2 at S868 is required for SUMO-mediated removal of KARs from the plasma membrane.
A previous study from our labs reported that phosphorylation of S868 can enhance SUMOylation of GluK2 in Cos-7 cells 24. To confirm this finding we quantified the amount of SUMOylated GluK2 in HEK cells expressing wild-type GluK2 or the S868A, S868D or K886R mutants. Similar to the situation in neurons, some SUMOylation of wild-type GluK2 was detectable under basal conditions. However, SUMOylation of the S868D phosphomimetic mutant was increased compared to the wild-type (Supplementary Fig. 1), suggesting that phosphorylation of S868 enhances SUMOylation of GluK2.
Phosphorylation of GluK2 increases KAR EPSC amplitude
Surprisingly, infusion of inactive SUMO into HEK cells expressing the phosphomimetic S868D mutant of GluK2 led to an increase in the amplitude of the KAR-mediated current when compared to wild-type (Fig. 2d; 142.5 ± 11.2% vs. 106.3 ± 5.1%; P < 0.05). These data suggest that phosphorylation of S868 combined with receptor activation may increase surface expression of GluK2, which would directly oppose the increased removal of GluK2 by SUMOylation. Consistent with this interpretation, PMA (1 μM) caused an increase in the amplitude of the KAR EPSC recorded from CA3 neurons to 139.3 ± 12.2% (Fig. 3a; n = 7; P < 0.05). Furthermore, the PKC inhibitor chelerythrine (5 μM) caused a decrease in KAR EPSC to 68.5 ± 8.0% (Fig. 3b; n = 8; P < 0.01). PKC inhibition by infusion of the PKC inhibitory peptide PKC19-36 also caused a decrease in KAR EPSC confirming the role of PKC inhibition (Supplementary Fig. 2a; 57.4 ± 12.4%; n = 5; P < 0.05).
FIGURE 3. Phosphorylation of S868 on GluK2 increases surface expression of KARs.
a. PMA (1 μM) caused an increase in the KAR EPSC amplitude in hippocampal slices. Responses for each cell are normalised to the first 10 minutes. Example traces are taken as the average between 5-10 minutes (black) and 20-25 minutes (grey). Scale bars are 50 pA and 50 ms. b. Chelerythrine (5 μM) caused a decrease in the KAR EPSC amplitude. c. PMA caused an increase in the KAR current evoked from HEK cells expressing WT GluK2. Scale bars are 100 pA and 500 ms. d. PMA caused an increase in the KAR current evoked from HEK cells expressing K886R GluK2. e. Comparison of the timecourse of PMA-induced increase in KAR current between WT and K886R expressing HEK cells. f. PMA application did not increase the KAR current evoked from HEK cells containing S868A GluK2. Data are plotted as mean ± s.e.m.
To investigate this further we recorded from HEK cells expressing GluK2 and applied PMA. Perfusion of PMA (1 μM) onto HEK cells expressing wild-type GluK2 led to a significant increase in the amplitude of the KAR current to 170.3 ± 23.6% (Fig. 3c; n = 6; P < 0.05). Perfusion of PMA (1 μM) onto HEK cells expressing the non-SUMOylatable K886R GluK2 mutant also led to an increase in KAR-mediated response amplitude to 178.3 ± 38.3% (Fig. 3d; n = 6; P < 0.05). Interestingly, whereas the increase was not significant after 10 minutes of PMA application in HEKs expressing wild-type GluK2 (Fig. 3e; 146.0 ± 20.3%; n = 6; P > 0.05), it was significant after 10 minutes of PMA application in HEKs expressing K886R (Fig. 3e; 184.0 ± 40.6%; n = 6; P < 0.05) suggesting that the increase is faster in HEKs expressing the non-SUMOylatable K886R GluK2 mutant. We also tested the effects of PMA (1 μM) perfusion on HEK cells expressing the S868A non-phosphorylatable mutant. In this case, PMA no longer led to an increase in amplitude of the KAR current but to a small decrease (Fig. 3f; 77.5 ± 8.4%; n = 5; P > 0.05). These data indicate that, in the absence of SUMOylation, PKC-mediated phosphorylation of GluK2 at S868 leads to an increase in GluK2 surface expression.
GluK2 phosphorylation regulates kainate receptor recycling
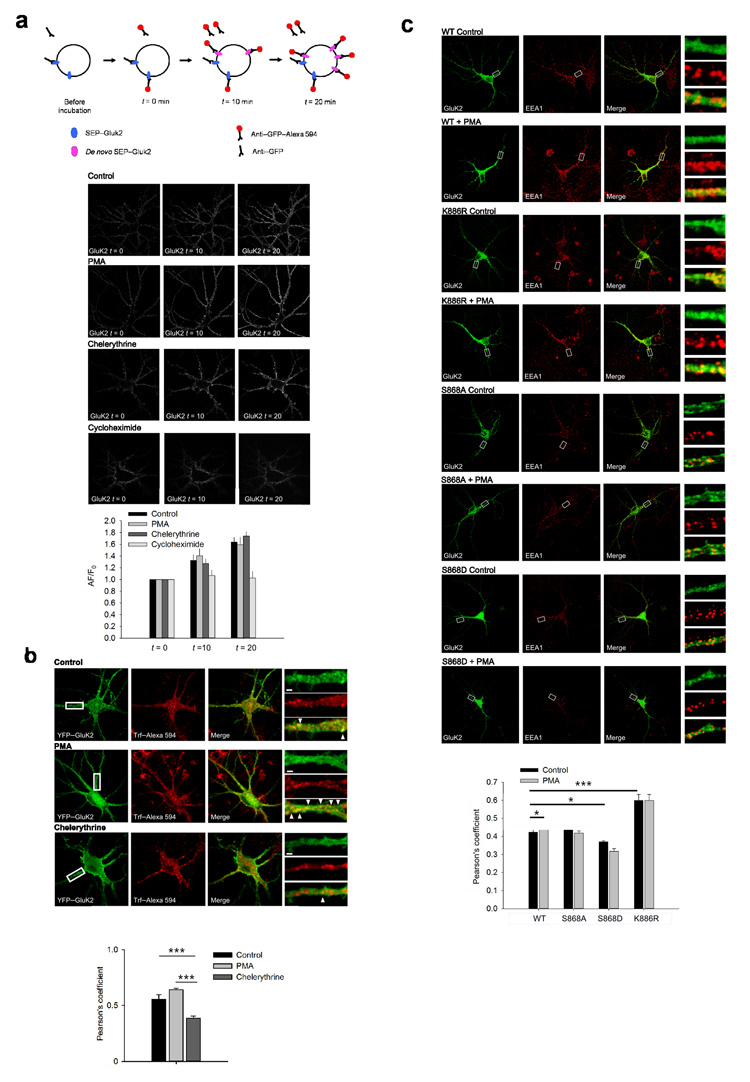
Our results implicate PKC in both increased and decreased GluK2 surface expression. We therefore reasoned that PKC phosphorylation might be involved in both exo- and endocytosis components of GluK2 trafficking. We first addressed whether the PKC-dependent increase in the amplitude of the KAR EPSC is due to changes in the rate of delivery and surface expression of de-novo synthesised GluK2 in neurons. We used Sindbis virus to express SEP-GluK2 26 in neurons and monitored the dynamics of SEP-GluK2 membrane insertion using anti-GFP antibody labelling. In these experiments live neurons were first incubated with unlabelled anti-GFP antibody for 5 minutes occupy and block the GFP epitope on all existing surface expressed SEP-GluK2. Following washing, to remove all unbound unlabelled anti-GFP antibody, the amount of newly inserted SEP-GluK2 was assessed at 0, 10 or 20 min using anti-GFP antibody directly coupled to Alexa594. We found that the amount of SEP-GluK2 inserted into the plasma membrane was unaffected after 10 or 20 min treatment with PMA or chelerythrine compared to control (Fig. 4a). As expected, inhibition of protein synthesis with cycloheximide prevented any increase in GluK2 inserted into the plasma membrane. These results suggest that PKC activation does not affect the rate of insertion of de-novo GluK2 in neurons.
FIGURE 4. PKC activation promotes KAR localisation in recycling pathways.
a. PKC activation does not alter the amount of exocytosis of KARs in hippocampal neurons when receptors already at the surface are excluded. Scheme illustrates the experimental procedure to exclude all initially surface expressed KARs from analysis. Representative images show GFP antibody binding to surface expressed SEP-GluK2 at intervals of 0, 10 and 20 minutes. Application of PMA (1 μM) or chelerythrine (5 μM) did not change the amount of KAR insertion into the plasma membrane in comparison to control. Inhibition of protein synthesis with cycloheximide prevented KAR insertion. b. Colocalization of GluK2 with TfR is increased by application of PMA but not chelerythrine. Representative images show increased GluK2 and TfR colocalization along the dendrites of hippocampal neurons following treatment with PMA. Scale bars are 2 μm. Arrowheads indicate puncta with high colocalization. c. Colocalization of wild-type GluK2 with EEA1 is increased by application of PMA (1 μM) but this is prevented by expression of the S868A, S868D or K886R GluK2 mutants. Representative images show increased GluK2 and EEA1 colocalization along the dendrites of hippocampal neurons following treatment with PMA. Data are plotted as mean ± s.e.m.
Since inserting newly synthesised receptors does not account for the increased surface expression of GluK2 we investigated whether there was increased recycling to the plasma membrane from early endosomes 27 by functional colocalization analysis with the transferrin receptor (TfR). In the presence of transferrin, the TfR is internalized into early endosomes and then constitutively sorted to recycling endosomes 28. As expected, after 5 min of pulse and 15 min of chase in the presence of either chelerythrine or vehicle, Alexa594-conjugated transferrin (Alexa-Trf) was localised predominantly in intracellular endosomes with a moderate colocalization with GluK2 (Fig. 4b). In the presence of the PKC inhibitor chelerythrine, GluK2 showed limited colocalization with TfR (Pearson’s coefficient 0.38 ± 0.01, n = 28) and this level of colocalization was increased in cells treated with PMA (Pearson’s coefficient 0.64 ± 0.01, n = 31). This suggests PKC activation increases the proportion of GluK2 sorted into recycling pathways. Intermediate levels of colocalization between GluK2 and TfR were observed under control conditions (Pearson’s coefficient 0.55 ± 0.04, n = 25), however this value was highly variable, likely due to differences in basal PKC activity.
Receptor recycling consists of both activity-dependent and independent components 29. In the presence of tetrodotoxin (TTX), to block the activity-dependent component of GluK2 recycling, the colocalization of GluK2 with TfR was significantly decreased (Pearson’s coefficient 0.44 ± 0.03, n = 19). Interestingly, this level of colocalization in the presence of TTX was unaffected by the addition of chelerythrine (Pearson’s coefficient 0.38 ± 0.09, n = 31, P > 0.05), suggesting that chelerythrine acts on the activity-dependent component of GluK2 recycling, leaving basal recycling unaffected. Thus, these results suggest that PKC phosphorylation regulates activity-dependent recycling of GluK2.
An increase in receptor recycling leads to greater numbers of receptors in early endosomal compartments. Therefore, PKC phosphorylation of GluK2 is expected to increase the amount of GluK2 associated with early endosomes. Consistent with our hypothesis, addition of PMA to cultured neurons transduced with wild-type GluK2 led to an increase in GluK2 colocalization with the early endosomal marker EEA1 (Fig. 4c; Pearson’s coefficient 0.42 ± 0.02 in control to 0.53 ± 0.04 (n = 12-15, P = 0.004). In contrast, PMA had no effect on the colocalization of EEA1 with S868A, S868D or K886R GluK2 mutants. Interestingly, in the absence of PMA the K886R GluK2 mutant showed an increased colocalization in comparison to wild-type (p < 0.001) whereas the phosphomimetic S868D GluK2 mutant showed a small decrease (p<0.05) and the non-phosphorylatable S868A GluK2 mutant showed no change (p>0.05). Importantly, the pattern of GluK2 expression was not dramatically altered by GluK2 mutations when colocalized with PSD95 staining (WT – 0.57 ± 0.02, n = 10; S868A – 0.59 ± 0.03 n = 10; S868D – 0.55 ± 0.08, n = 10; K886RR – 0.56 ± 0.02, n = 15) and the pattern of EEA1 expression was not affected by application of PMA (Fig. 4c), although chronic chelerythrine treatment did change EEA1 staining to a less punctate distribution (Supplementary Fig 3) preventing analysis of mutant GluK2 colocalization with EEA1 under these conditions. These data support the conclusion that PKC phosphorylation of S868 increases recycling of GluK2 between the plasma membrane and early endosomal compartments.
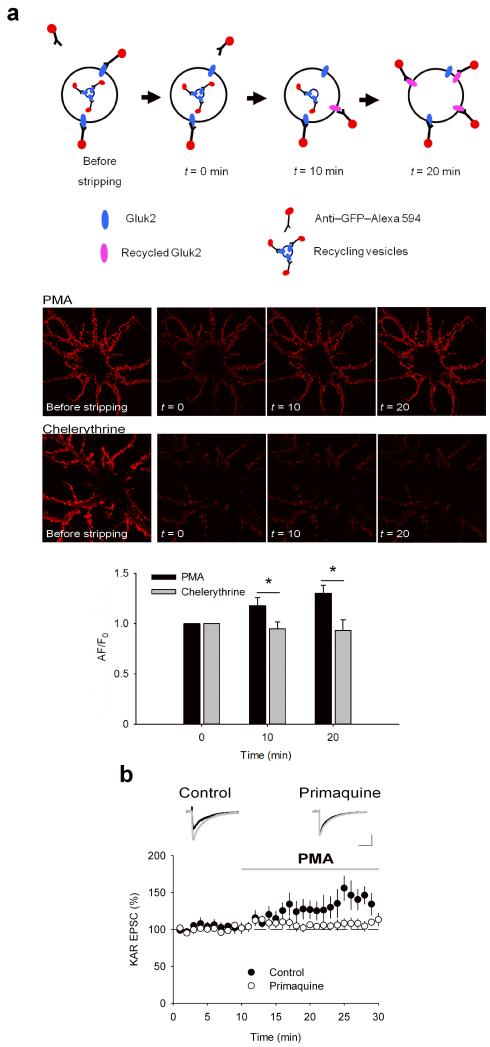
To address the influence of PKC activation on GluK2 recycling we compared recycling in neurons incubated in PMA or chelerythrine. Using live-cell antibody feeding we quantified the recycling of SEP-GluK2, again using SEP as an extracellular epitope for anti-GFP antibody. Neurons were incubated with Alexa594 coupled to anti-GFP antibody together with PMA or chelerythrine for 10 mins to allow internalisation of surface labelled receptors. The Alexa594-coupled anti-GFP antibody was then stripped from any remaining surface receptors and the reappearance (recycling) of Alexa594-tagged SEP-GluK2 at the plasma membrane after 10 or 20 min (Fig. 5a). The intracellular signal from internalized Alexa594-tagged SEP-GluK2 was diffuse and largely below detection threshold and was subtracted for analysis (see methods). The amount of Alexa594-tagged SEP-GluK2 that was previously surface expressed then internalised and returned back to the surface was increased at 10 and 20 min in cells treated with PMA. In contrast, GluK2 recycling was undetectable in cells treated with chelerythrine (Fig. 5a). These data indicate that PKC activation increases the rate of GluK2 recycling in neurons.
FIGURE 5. PKC activation enhances KAR recycling in neurons.
a. PMA but not chelerythrine increased the amount of KAR recycling. Scheme illustrates the experimental procedure to selectively analyse surface reinsertion of KARs. Representative images show an increase in surface GluK2 in hippocampal neurones after antibody stripping in the presence of PMA but not chelerythrine. b. Incubation of hippocampal slices in primaquine (1 μM) to inhibit exocytosis from early endosomes prevented the increase in KAR EPSC caused by application of PMA (1 μM). Responses for each cell are normalised to the first 10 minutes. Example traces are taken as the average between 5-10 minutes (black) and 20-25 minutes (grey). Scale bars are 20 pA and 50 ms. Data are plotted as mean ± s.e.m.
We further confirmed the effect of PKC phosphorylation of GluK2 on recycling using primaquine, an inhibitor of exocytosis from early endosomes 30. Application of primaquine to slices produced a dose-dependent decrease in KAR EPSC amplitude. 100 μM primaquine decreased KAR EPSC amplitude to 6.0 ± 1.1% after 10 minutes whereas 1μM primaquine decreased KAR EPSC amplitude to 57.0 ± 8.5% that stabilised after 60 minutes (Supplementary Fig. 4). Application of PMA following 1 hour pre-incubation with 1 μM primaquine failed to elicit an increase in KAR EPSC (Fig. 5b; 106.8 ± 6.5; n = 7; P > 0.05) in contrast to control conditions (data from Figure 3a is reproduced for comparison) indicating that exocytosis of GluK2 during receptor recycling is necessary for increased surface expression of KARs after activation of PKC.
To establish that PMA exerts its effects directly via phosphorylation of GluK2, we used live-cell antibody feeding experiments to quantify the PKC-dependent recycling in HEK cells expressing WT or the non-phosphorylatable S868A GluK2 mutant. We initially confirmed that GluK2 is recycled in HEK cells by comparing the colocalization of GluK2 and TfR (Supplementary Fig. 5) 17. In HEK cells expressing WT GluK2, application of PMAincreased the amount of recycled (reinserted) GluK2 at 10 and 20 min. In contrast, GluK2 recycling was undetectable in control cells or cells treated with chelerythrine (Fig. 6a). However, in HEK cells expressing the non-phosphorylatable S868A GluK2 mutant, there was no detectable reinsertion in the presence of either PMA or chelerythrine (Fig. 6b) demonstrating that phosphorylation of this specific residue on GluK2 is required for the enhancement of GluK2 recycling by PKC.
FIGURE 6. Phosphorylation of S868 on GluK2 enhances KAR recycling in HEK cells.
a. PMA but not chelerythrine increased the amount of KAR recycling compared to control in HEK cells expressing wild-type GluK2. Representative images show an increase in surface GluK2 on multiple HEK cells after antibody stripping in the presence of PMA but not chelerythrine. b. Neither PMA nor chelerythrine changed the amount of KAR recycling compared to control in HEK cells expressing S868A GluK2. Representative images show no change in surface GluK2 after antibody stripping in the presence of PMA or chelerythrine. Data are plotted as mean ± s.e.m.
Phosphorylation and SUMOylation are required for KAR LTD
Phosphorylation of KARs at mossy fiber-CA3 synapses is involved in KAR LTD 14 but the possible contribution of SUMOylation has not been investigated. Indeed, it is unknown if SUMOylation is involved in any form of activity-dependent synaptic plasticity. We induced KAR LTD by stimulating mossy fiber synapses for 5 minutes at a frequency of 1 Hz whilst maintaining the membrane potential at −70 mV. This produced a reduction in KAR EPSC amplitude to 56.1 ± 1.0% 30 minutes after induction of LTD (Fig. 7a; n = 9; P < 0.001). In this, and all other LTD experiments, there were no changes in paired pulse ratio after induction of LTD (Fig. 7a; 1.26 ± 0.15 vs 1.34 ± 0.19, baseline vs LTD respectively; n = 9; P > 0.05). Similar to previous findings 14, KAR LTD was not dependent on changes in postsynaptic [Ca2+] since it was insensitive to inclusion of BAPTA (10 mM) in the patch solution (Fig. 7b; 63.9 ± 3.4%, n = 7; P < 0.05), but was sensitive to blockade of mGluR5 receptors by bath perfusion with 30 μM MPEP (Fig. 7c; 94.3 ± 12.5%, n = 8; P > 0.05). We also confirmed that PKC phosphorylation is required for KAR LTD since inclusion of the PKC inhibitor peptide PKC19-36 (4.8 μM)in the recording pipette, but not the inactive (Glu27)PKC19-36 peptide 31 (4.8 μM), blocked KAR LTD (Fig. 7d; 100.1 ± 7.1%; n = 7; vs. 44.9 ± 4.9%; n = 5; P < 0.05). This was confirmed using chelerythrine (5 μM), which also completely blocked KAR LTD (Supplementary Fig. 2b).
FIGURE 7. KAR LTD at mossy fiber synapses is dependent on activation of mGluR5 and PKC.

a. 1 Hz stimulation for 5 minutes (300 pulses) induced LTD of KAR EPSCs at mossy fiber-CA3 synapses. Responses are normalised to the first 10 minutes. Example traces are taken as the average of the first 10 minutes (black) and 30-35 minutes after LTD (grey). Scale bars are 50 pA and 50 ms. Bar represents period of LTD induction. b. Inclusion of BAPTA (10 mM) in the patch solution had no effect on KAR LTD. c. Incubation in MPEP (30 μM) to block mGluR5 abolished KAR LTD. d. Inclusion of the PKC inhibitory peptide PKC19-36 but not the inactive (Glu27)PKC19-36 in the patch solution completely abolished KAR LTD. Data are plotted as mean ± s.e.m.
Having established and validated conditions for robust KAR LTD we tested the affect of protein SUMOylation on LTD expression. Inclusion of inactive SUMO-1-ΔGG had no effect on LTD compared to control conditions (Fig. 8a; 44.5 ± 6.8% after 30 minutes; n = 7; P < 0.001). However, infusion of the active SUMO peptide caused a decrease in KAR EPSC to 58.1 ± 8.6% (n = 5; P < 0.01) after which LTD was completely occluded (Fig. 8a; 100.0 ± 6.3% after 30 minutes; n = 8; P < 0.01 compared with inactive). In addition, we tested whether inhibiting SUMOylation blocked the expression of LTD. Inclusion of the active catalytic domain of the SUMO-specific isopeptidase SENP-1 caused an increase in KAR EPSC to 155.4 ± 10.3% (n = 6; P < 0.01) after which KAR LTD was completely blocked (Fig. 8b; 93.8 ± 8.1% after 30 minutes; n = 8; P > 0.05). However, infusion of an inactive point mutant of SENP-1 (SENP C603S)17 in the patch solution had no effect on LTD (Fig. 8b; 59.6 ± 9.0 % after 30 minutes; n = 6; P < 0.05 compared with active SENP-1).
FIGURE 8. SUMOylation is required for KAR LTD.
a. Inclusion of active but not inactive SUMO-1 in the patch pipette occluded KAR LTD at mossy fibre-CA3 synapses. Active SUMO-1 caused an initial decrease in KAR EPSC amplitude that stabilised before the induction of LTD. Responses are normalised to a 10 minute baseline after EPSC amplitude had stabilised. Example traces are taken as the average of the 10 minute baseline (black) and 30-35 minutes following LTD (grey). Scale bars are 50 pA and 50 ms. Bar represents period of LTD induction. b. Inclusion of active but not inactive SENP-1 in the patch pipette blocked KAR LTD at mossy fibre-CA3 synapses. Active SENP-1 caused an initial increase in KAR EPSC amplitude that stabilised before the induction of LTD. c. Inclusion of active SUMO-1 in the patch pipette caused a decrease in KAR EPSC amplitude in neurons in cultured hippocampal slices transduced with wild-type GluK2 and non-transduced neurons. Responses are normalised to the first minute. Example traces in panels c, e & g are taken as the average of the first minute (black) and 10-15 minutes (grey). d. Wild-type GluK2 transduced neurons exhibited normal KAR LTD in comparison to non-transduced neurons. Example traces in panels d, f & h are taken as the average of the last 5 minutes of baseline (black) and 30-35 minutes following LTD (grey). Scale bars are 20 pA and 50 ms for panels d-h. e & g. Reduction in the KAR EPSC by inclusion of active SUMO-1 in the patch pipette was prevented in neurons transduced with K886R GluK2 (e) or S868A GluK2 (g) in comparison to non-transduced neurons. f & h. Neurons transduced with K886R GluK2 (f) or S868A GluK2 (h) demonstrated no KAR LTD. Data are plotted as mean ± s.e.m.
To define whether GluK2 SUMOylation and phosphorylation is necessary for KAR LTD we virally expressed YFP-GluK2 constructs in CA3 neurons of acute hippocampal slices cultured for 24 hours to allow expression of wild-type, K886R or S868A GluK2 in CA3 neurons. Viral expression of GluK2 did not change KAR EPSC amplitude when comparing transduced and non-transduced neurons (non-transduced, 62.5 ± 11.7pA, n = 15; wild-type, 55.1 ± 10.0pA, n = 5; K886R, 45.5 ± 17.1pA, n = 5; S868A, 55.7 ± 9.4pA, n = 5; P > 0.05 for all comparisons with ANOVA). Infusion of the active SUMO peptide into CA3 neurons expressing wild-type GluK2 depressed KAR EPSCs in a similar fashion to non-transduced CA3 neurons (Fig. 8c; 49.2 ± 4.4% vs. 49.4 ± 7.1%; n = 5). In addition, KAR LTD was indistinguishable between wild-type GluK2 transduced and non-transduced neurons (Fig. 8d; 42.2 ± 12.6% vs. 32.1 ± 7.6%; n = 5). In contrast, infusion of the active SUMO peptide into neurons transduced with either non-SUMOylatable K886R GluK2 or non-phosphylatable S868A GluK2 did not depress KAR EPSCs (Fig. 8e,g; K886R – 16.3 ± 7.3%; S868A – 7.7 ± 12.7%; n = 5) and KAR LTD was completely blocked (Fig. 8f,h; K886R – 11.5 ± 10.9%; S868A – increased 3.7 ± 21.8). Taken together, these results indicate that SUMOylation of GluK2 at K886 and phosphorylation at S868 are required for KAR LTD.
Discussion
Our results demonstrate that PKC phosphorylation of GluK2 is necessary for SUMOylation-dependent internalisation of KARs from the synaptic membrane. Thus, we provide evidence for the dynamic interaction of two forms of post-translational protein modification controlling the subcellular localisation of KARs. Further, we show that this is the mechanism for the removal of KARs from the postsynaptic membrane during activity-dependent LTD.
The observations that the phophomimetic GluK2 mutant shows an increase in KAR EPSC under baseline stimulation conditions (Fig. 2d) and that PMA causes an increase in KAR responses in HEK cells expressing wild-type or the non-SUMOylatable GluK2 mutant (Fig. 3c,d), but not the non-phosphorylatable GluK2 mutant (Fig. 3f), suggest that phosphorylation of S868 is also involved in non-SUMOylation dependent trafficking of KARs. This is further supported by the increase in KAR recycling between the plasma membrane and intracellular compartments after phosphorylation of GluK2 at S868 (Figs. 4, 5 and 6). We propose that increased recycling leads to an increase in surface expressed KARs. Interestingly, either preincubation in PMA (Fig. 1c) or expression of the phosphomimetic S868D GluK2 mutant (Fig. 2d) produced a greater decrease in KAR response amplitude when SUMO was applied compared to control conditions. This suggests that PKC phosphorylation in the absence of SUMOylation promotes recycling and increased surface insertion of KARs but can also promote enhanced endocytosis through SUMOylation of GluK2 17. Thus, we propose that SUMOylation is the molecular switch that determines either enhanced or decreased KAR surface expression.
We show that phosphorylation of GluK2 at S868 by PKC has dual, competing effects on KAR localisation. Phosphorylation of GluK2 increases recycling and the proportion of KARs on the plasma membrane, but also promotes SUMOylation that leads to a loss of surface KARs. This is highlighted by the observation that PMA increases KAR EPSC amplitude faster in the SUMOnull K886R GluK2 mutant than in wild-type GluK2 (Fig. 3c,d). In addition, PMA causes an increase in KAR EPSC amplitude in CA3 neurons while chelerythrine leads to a decrease (Fig. 3a,b) strongly suggesting that the increase in receptor insertion outweighs the increase in SUMOylation-dependent internalisation under these conditions. Intriguingly, the equilibrium between these two events may help to explain why, in different recording conditions, different studies have shown opposing effects of PKC inhibitors at mossy fibre CA3 synapses 10, 14. Phosphorylation of S846 by PKC has been shown to increase KAR endocytosis 25 even though it does not promote SUMOylation of K886 24. Our data suggest that the phosphorylation of S868 and subsequent SUMOylation of K886 override the role of S846 phosphorylation. Furthermore, phosphorylation of the S868 residue on GluK2 has also been shown to increase KAR retention in the ER 25 providing evidence for an additional role of S868 phosphorylation in controlling KAR localization.
Crucially, we show that SUMOylation is critical for KAR LTD at mossy fiber-CA3 synapses. The degree of depression seen after LTD (Figs. 7 and 8) is comparable to the proportion of KARs internalised after infusion of SUMO into the cell (Fig. 1a). This suggests that the inclusion of SUMO in the patch solution occludes LTD via prior removal of SUMO sensitive KARs (Fig. 8a). Conversely, infusion of SENP blocks LTD by removing SUMO from GluK2 before KARs can be internalised (Fig. 8b). The dominant negative effect of transduced GluK2 (Fig. 8) implies that expressed mutant GluK2 supplants endogenous GluK2 at mossy fiber synapses. Interestingly, KAR LTD at mossy Fiber-CA3 synapses has also been shown to require the destabilisation of surface KARs containing the GluK5 subunit by SNAP-25 14. Our results do not directly address the role of GluK5 but taken together these results suggest that KARs removed from the synaptic membrane during LTD contain both GluK2 and GluK5 and that endocytosis during LTD requires dual synergistic processes initiated by PKC phosphorylation that leads to both the binding of PICK1 and SNAP-25 to GluK5 and the SUMOylation of GluK2.
In conclusion, our results demonstrate a role for SUMOylation in synaptic plasticity of KARs at mossy fiber synapses in the hippocampus opening the possibility that this form of post-translational protein modification may play a role in other forms of synaptic plasticity. SUMO proteins and the enzymes required for SUMO conjugation are found throughout dendrites and are concentrated at spines 17 and accumulating evidence suggests that many synaptic proteins other than KARs are also SUMOylated 32. Thus, post-translational protein modifications such as SUMOylation, phosphorylation and ubiquitination may dynamically interact to coordinate neurotransmitter receptor localisation and function.
Supplementary Material
Acknowledgements
We thank A. Randall for electrophysiological facilities to perform experiments on cultured preparations and S. Martin and J. Hanley for comments on previous versions of the manuscript. This work was funded by BBSRC (SELC, JRM, JMH), European Research Council (JMH), MRC (FAK, SK, JMH) and Wellcome Trust (JRM, JMH). IMG-G is an EMBO Fellow.
Appendix
Methods
Electrophysiological recordings in hippocampal slices
Transverse hippocampal slices (400-500 μm thickness) were prepared from p14 Wistar rats. Slices were cut in ice cold artificial cerebrospinal fluid (aCSF) containing (in mM): 119 NaCl, 10 Glucose, 26 NaHCO3, 2.5 KCl, 1 NaH2PO4, 0.5 CaCl2, 5 MgSO4 saturated with 95% O2 and 5% CO2. Slices were transferred to recording aCSF containing (in mM): 119 NaCl, 10 Glucose, 26 NaHCO3, 2.5 KCl, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgSO4 saturated with 95% O2 and 5% CO2 and heated to 35°C for 30 minutes. Slices were thereafter stored at room temperature. Cultured slices were transferred directly after preparation to tissue culture inserts (Millicell-CM, 0.4 μm pore size) and bathed in sterile MEM (Gibco) containing (in mM): 5 NaHCO3, 30 HEPES, 1 Glutamine, 1 CaCl2, 2 MgSO4, 13 Glucose and 20% Horse Serum adjusted to pH 7.28 with NaOH and 320mOsM. Slices were placed in an incubator at 35°C and 5% CO2 for one hour before being pressure injected at multiple sites within CA3 with Sindbis virus which resulted in 1020 successfully transduced neurons in each slice. Slices were then returned to the incubator for 2026 hours before recording. For electrophysiological recording, cultured and acute slices were immersed in recording aCSF at 35°C containing L689560 (5 μM), GYKI 53655 (40 μM) and picrotoxin (50 μM) to antagonise NMDAR, AMPAR and GABAAR respectively. Whole-cell patch clamp recordings were made from visually identified CA3 pyramidal neurons using borosilicate glass pipettes of resistance 26 MΩ filled with a solution containing (in mM): 117 CsMeSO4, 8 NaCl, 10 HEPES, 5 QX-314Cl, 4 MgATP, 0.3 NaGTP, 0.2 EGTA, 0.1 bestatin and 0.1 leupeptin at pH 7.4 and 280 mOsm. In cultured slices, transduced neurons were identified by combined differential interference contrast and fluorescent microscopy. Synaptic responses were recorded within 30 s of entering the whole-cell configuration and elicited using a bipolar tungsten electrode placed in the granule cell layer stimulated every 20 s with three pulses applied at 167 Hz or, for LTD experiments, with paired pulse stimulation (separation 40 ms). Series and input resistances were continuously monitored during experiments and recordings discarded if series resistance changed by >20%.
All data were sampled at 10 kHz, filtered at 5 kHz and recorded onto computer using Signal software (CED). Analyses were performed with scripts written in this software environment. Data are presented as mean ± s.e.m. Values used for statistical analyses were taken from the same timepoints as example traces. Statistical significance was determined using paired or unpaired two-tailed Student’s t-tests as appropriate.
Electrophysiological recordings in HEK cells
HEK cells were cultured in DMEM (Gibco) containing 10% fetal calf serum, 2 mM glutamine and 2% penicillin/streptomycin. Cells were split 24 hours prior to transfection onto 25 mm coverslips coated with 5 μg ml−1 poly-L-lysine in 35 mm dishes. Cells were transfected in plain DMEM with 2 μg plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturers’ instructions and 3 hours post-transfection, cells were returned to complete media. Electroyphysiology was performed 48 hours after transfection. Recordings were made using an Axopatch 200B amplifier and pClamp 10.2 software (Axon Instruments). Borosilicate patch electrodes had series resistances of 2-6 MΩ. Extracellular solution contained (in mM): 135 NaCl, 5 KCl, 30 Glucose, 2 CaCl2, 1 MgCl2, and 10 HEPES at pH 7.3 and 305 mOsm. Intracellular solutions contained (in mM): 117 CsMeSO4, 8 NaCl, 10 HEPES, 5 QX-314Cl, 4 MgATP, 0.3 NaGTP, 0.2 EGTA, 0.1 bestatin and 0.1 leupeptin at pH 7.4 and 280 mOsm. Before whole-cell configurations were obtained, cells were lifted from the coverslip into a laminar stream of extracellular solution containing Conconavalin A (0.3 mg ml−1) to block receptor desensitisation. Kainate (100 μM) was applied by multi-barrel fast perfusion system controlled by Clampex software (Axon Instruments). Analysis was performed off-line using Clampfit software (Axon Instruments). Experiments were performed at room temperature.
L689560, PMA, chelerythrine, (Glu27)PKC19-36, primaquine and kainic acid were purchased from Tocris. Picrotoxin, PKC19-36 and conconavalin A were purchased from Sigma. GYKI 53655 was purchased from Ascent Scientific.
Receptor Exocytosis Experiments
Dissociated hippocampal neuronal cultures were pretreated with PMA, chelerythrine or vehicle for 15 min at 37°C prior to live-cell imaging experiments in the continued presence of the drugs. At the beginning of the experiment, GFP reactive sites on the cell surface of neurons expressing SEP-GluK2 were blocked by pretreatment with an excess of unlabeled anti-GFP rabbit polyclonal antibody (rAb; 1:200, Invitrogen) at room temperature for 5 min (rather than at 37°C, in order to limit receptor endocytosis). Neurons were immediately placed on a microscope stage heated to 37 °C and incubated with diluted anti-GFP rAb directly coupled to Alexa594 (anti-GFP-Alexa594, 1:500) for 2 min to visualise the plasma membrane. Neurons were quickly washed twice and the fluorescence at time zero acquired. Cells were incubated either a further 10 or 20 min in the presence of PMA, chelerythrine or vehicle and then reprobed with anti-GFP-Alexa594 (1:200), rapidly washed twice and imaged to determine the SEP-GluK2 inserted during the 10 or 20 min incubation interval.
Receptor Recycling Experiments
SEP-GluK2 recycling was quantified by live-cell antibody feeding in the presence of PMA or chelerythrine. Cells were incubated in PMA or chelerythrine for 10 min with anti-GFP-Alexa594 (1:200) at 37 °C to allow the endocytosis of the labelled surface receptors. Cells were washed twice in aCSF and the total surface SEP-GluK2 determined by confocal microscopy. Cells were then washed with stripping buffer (recording buffer pH 3.2 plus MesNa 50 mM) for 30 seconds. Because MesNa is membrane impermeant it removes anti-GFP antibody from surface expressed SEP-GluK2 but not from SEP-GluK2 that has been internalised. Immediately after stripping, the cells were imaged (t = 0) and then incubated at 37 °C for 10 min and 20 min (t = 10 and t = 20) to allow recycling to occur in the presence of either PMA or chelerythrine.
The red fluorescence corresponding to anti-GFP labelled SEP-GluK2 returning to the plasma membrane was recorded at time 0, 10 and 20 min from the same cell as a series of Z stacks (0.25 μm spacing between single confocal slices) using a Zeiss LSM 510 confocal laser-scanning station with an oil-immersion 63× 1.4 NA objective (Zeiss). Alexa594 was visualized using a 543 nm HeNe laser line and a 585 nm long-pass filter, whereas SEP was excited with the 488 nm laser line from an argon–krypton laser and the emitted light was detected using a 505–550 nm band pass filter.
The efficiency of the stripping was normalized in each cell as the ratio of fluorescence pre-stripping/post-stripping. If the efficiency of stripping was not greater than 40%, then the analysis did not continue. Differences in expression were normalized to the mean of the fluorescence at time 0 and the rate of receptor reappearance at the cell surface within individual cells was then determined. To ensure that the red fluorescence from the area of interest came only from surface-expressed KARs all SEP-GluK2 experiments included a brief (10 s) pH 6 wash at the end of the experiment to quench surface SEP-GluK2 fluorescence. Any areas of interest that retained SEP fluorescence during the pH 6 wash were excluded from the analysis of the red Alexa594 fluorescence (Supplementary Fig. 5). In HEK cells, the same control was performed by incubating the cells with anti-rabbit Cy5 at the end of the experiment for 2 mins at room temperature (Supplementary Fig. 6). Statistical analysis of differences between experimental groups was performed using one way ANOVA followed by post-hoc Tukey test calculated using SigmaStat software. Data are presented as mean ± SD.
GluK2 colocalization analysis
48 hrs after transfection, HEK cells grown on coverslips were washed and pre-incubated with serum free DMEM for 2 hrs. For transferrin assays in neurons, the neurons were pre-incubated with B27 free Neurobasal during 1h. HEK293 or neurons were then incubated with 10 μg ml−1 of Alexa594 conjugated transferrin (Molecular Probes) in serum/B27-free medium for 5 min at room temperature. Cells were then washed with PBS twice and shifted to 37°C for 20 min to label recycling endosomes in the presence of PMA, chelerythrine or vehicle. Following incubation the cells were washed with PBS twice and fixed in 4.0% paraformaldehyde. For the EEA1 GluK2 colocalization experiment, 24 h after infection, the neurons were incubated with PMA, chelerythrine or vehicle for 30 min. For EEA1 or PSD95 GluK2 colocalization neurons were then washed with PBS, fixed in 4.0% paraformaldehyde and stained with specific primary antibodies against EEA1(BD Biosciences) or PSD95 (Millipore).
Three-dimensional volumes of z stacks (0.25 μm spacing between single confocal slices) were analysed using Image processing and analysis in Java (ImageJ). The degree of colocalization was assessed in whole cell volumes and sub-volumes by calculating the Pearson’s correlation coefficient in the region of interest using a semiautomated algorithm embedded in the JaCoP plugin 33 of the ImageJ software. The colocalization plugin also performed a two-step analysis to calculate the Pearson’s correlation coefficient for the original data and for a large set (approximately 500) of images randomized with a grain size determined by the point spread function of the microscope objective. If the Pearson’s correlation coefficient of the original image was not greater than 95% of the randomized images, then the samples were not used. In addition, cells displaying saturated or low, near-threshold signals were discarded, and user bias in setting analysis parameters was avoided by using an automated thresholding procedure 33. Histograms presenting the mean correlation coefficient (derived from 19 to 31 cells assessed per treatment condition) are shown with standard deviation bars in all figures. Statistical significance for differences between paired combinations of images was calculated using the two-tailed Student’s t-test.
GluK2 SUMOylation in HEK cells
HEK cells in 35mm dishes were transfected with 1μg plasmid DNA encoding YFP-myc-GluK2 WT, S868D, S868A or K886R along with FLAG-SUMO-1 and FLAG-Ubc9. 36 hours post-transfection, cells were washed twice with PBS and lysed in 500 μl lysis buffer (10 mM tris-HCl, 150 mM NaCl, 1% triton X-100, 0.1% SDS, 20 mM NEM and protease inhibitors (Roche)). Lysates were then sonicated briefly and GluK2 solubilised by rotation for 1 hour at 4°C. Lysates were then cleared by centrifugation at 16000g for 15 minutes. 200 μl cleared lysate was then taken and 600 μl lysis buffer lacking NEM added. GluK2 was immunoprecipitated by addition of 1 μg sheep anti-myc (made in-house) and left rotating at 4°C overnight. To each IP, 20μl (bed volume) of protein G sepharose (Sigma) pre-washed in lysis buffer (lacking NEM) was added, and IPs incubated for a further 2 hours at 4°C. After extensive washing in lysis buffer lacking NEM, beads were boiled in 50 μl 2× Laemmli buffer. IPs and corresponding inputs were then subjected to SDS-PAGE and Western blotting for FLAG (M2 monoclonal antibody, Sigma) or myc (9E10, Santa Cruz), respectively. Blots were analysed using ImageJ by normalizing the band representing SUMOylated GluK2 to total GluK2. Statistical significance was determined using two-tailed Student’s t-test.
Statistical significance is denoted by * < 0.05, ** < 0.01, ***< 0.001 throughout.
Footnotes
Author Contributions
SELC performed and analysed electrophysiology experiments; IMG-G performed and analysed imaging experiments; KAW and FAK performed and analysed biochemical experiments; SK prepared SUMO and SENP proteins for electrophysiological experiments; JMH and JRM supervised the project; SELC, IMG-G, JMH and JRM wrote the manuscript.
References
- 1.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 2.Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol Rev. 2005;57:253–277. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cognet L, Groc L, Lounis B, Choquet D. Multiple routes for glutamate receptor trafficking: surface diffusion and membrane traffic cooperate to bring receptors to synapses. Sci STKE. 2006;2006:pe13. doi: 10.1126/stke.3272006pe13. [DOI] [PubMed] [Google Scholar]
- 4.Santos SD, Carvalho AL, Caldeira MV, Duarte CB. Regulation of AMPA receptors and synaptic plasticity. Neuroscience. 2009;158:105–125. doi: 10.1016/j.neuroscience.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Isaac JT, Mellor J, Hurtado D, Roche KW. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacology and Therapeutics. 2004;104:163–172. doi: 10.1016/j.pharmthera.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Huettner JE. Kainate receptors and synaptic transmission. Prog Neurobiol. 2003;70:387–407. doi: 10.1016/s0301-0082(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 7.Ren Z, et al. Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J Neurosci. 2003;23:6608–6616. doi: 10.1523/JNEUROSCI.23-16-06608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaskolski F, Coussen F, Mulle C. Subcellular localization and trafficking of kainate receptors. Trends in Pharmacological Sciences. 2005;26:20–26. doi: 10.1016/j.tips.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Jaskolski F, et al. Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. J Neurosci. 2004;24:2506–2515. doi: 10.1523/JNEUROSCI.5116-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirbec H, et al. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron. 2003;37:625–638. doi: 10.1016/s0896-6273(02)01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coussen F, et al. Co-assembly of two GluR6 kainate receptor splice variants within a functional protein complex. Neuron. 2005;47:555–566. doi: 10.1016/j.neuron.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 12.Yan S, et al. A C-terminal determinant of GluR6 kainate receptor trafficking. J Neurosci. 2004;24:679–691. doi: 10.1523/JNEUROSCI.4985-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park Y, Jo J, Isaac JT, Cho K. Long-term depression of kainate receptor-mediated synaptic transmission. Neuron. 2006;49:95–106. doi: 10.1016/j.neuron.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 14.Selak S, et al. A role for SNAP25 in internalization of kainate receptors and synaptic plasticity. Neuron. 2009;63:357–371. doi: 10.1016/j.neuron.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Cho K, et al. Regulation of kainate receptors by protein kinase C and metabotropic glutamate receptors. J Physiol. 2003;548:723–730. doi: 10.1113/jphysiol.2003.040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivera R, Rozas JL, Lerma J. PKC-dependent autoregulation of membrane kainate receptors. EMBO J. 2007;26:4359–4367. doi: 10.1038/sj.emboj.7601865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8:948–959. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 21.Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle. 2003;2:528–530. doi: 10.4161/cc.2.6.597. [DOI] [PubMed] [Google Scholar]
- 22.Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]
- 23.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konopacki FA, et al. Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc Natl Acad Sci U S A. 2011;108:19772–19777. doi: 10.1073/pnas.1111575108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasu-Nishimura Y, Jaffe H, Isaac JT, Roche KW. Differential regulation of kainate receptor trafficking by phosphorylation of distinct sites on GluR6. J Biol Chem. 2010;285:2847–2856. doi: 10.1074/jbc.M109.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin S, Bouschet T, Jenkins EL, Nishimune A, Henley JM. Bidirectional regulation of kainate receptor surface expression in hippocampal neurons. J Biol Chem. 2008;283:36435–36440. doi: 10.1074/jbc.M806447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S, Henley JM. Activity-dependent endocytic sorting of kainate receptors to recycling or degradation pathways. EMBO J. 2004;23:4749–4759. doi: 10.1038/sj.emboj.7600483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron. 2002;36:435–449. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 29.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 30.van Weert AW, Geuze HJ, Groothuis B, Stoorvogel W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur J Cell Biol. 2000;79:394–399. doi: 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- 31.House C, Kemp BE. Protein kinase C contains a pseudosubstrate prototope in its regulatory domain. Science. 1987;238:1726–1728. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson KA, Nakamura Y, Henley JM. Targets and consequences of protein SUMOylation in neurons. Brain Res Rev. 2010;64:195–212. doi: 10.1016/j.brainresrev.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.