Abstract
Several lines of evidence suggest that defects in telomere maintenance play a significant role in the initiation of genomic instability during carcinogenesis. Although the general concept of defective telomere maintenance initiating genomic instability has been acknowledged, there remains a critical gap in the direct evidence of telomere dysfunction in human solid tumors. To address this topic, we devised a multiplex PCR-based assay, termed TAR (telomere-associated repeat) fusion PCR, to detect and analyze chromosome end-to-end associations (telomere fusions) within human breast tumor tissue. Using TAR fusion PCR, we found that human breast lesions, but not normal breast tissues from healthy volunteers, contained telomere fusions. Telomere fusions were detected at similar frequencies during early ductal carcinoma in situ and in the later invasive ductal carcinoma stage. Our results provide direct evidence that telomere fusions are present in human breast tumor tissue and suggest that telomere dysfunction may be an important component of the genomic instability observed in this cancer. Development of this robust method that allows identification of these genetic aberrations (telomere fusions) is anticipated to be a valuable tool for dissecting mechanisms of telomere dysfunction.
Keywords: breast cancer, retrotransposon
Defects in telomere maintenance have been suggested to play significant roles in the initiation of genomic instability via breakage–fusion–bridge cycles and aneuploidy, which are associated with the development of human cancers, including breast cancer (1, 2). A critical function of the telomere is to disguise the chromosome end from being recognized as a double-strand break, to prevent aberrant chromosomal end joining and recombination events. Cells disguise telomeric DNA by encapsulating or “capping” the chromosome ends with several telomere-associated proteins and unique telomere-specific structures (3). In healthy cells, telomere length is highly regulated in a tissue- and cell type-specific manner and is dependent on mitotic turnover rate, telomerase activity, and telomerase-associated factors (4, 5).
Several lines of evidence from mouse and human systems suggest that defects in telomere maintenance play an important role in the development of cancer (1, 2). Induction of telomere dysfunction by deficiency in the telomerase RNA component (mTER) in a p53 mutant mouse background results in significant levels of breast adenocarcinomas and colon carcinomas (6–8). Telomere dysfunction also has been reported in a human mammary epithelial cell culture model (9). In this model, late-passage human mammary epithelial cell escape a stress-associated senescence-like barrier and acquire genomic alterations, including telomere fusions (9). In addition, clinical studies have shown that a significant proportion of normal breast luminal cells and ductal carcinoma in situ (DCIS) tissues have shortened telomeric DNA lengths when assayed by telomere FISH (10). Several studies have reported that anaphase bridges, possibly formed as a consequence of telomere dysfunction, are present in early-stage tumors, including DCIS (7, 10, 11). But although the presence of anaphase bridges and shortened telomeres may delineate a significant population of cells at risk for telomere dysfunction, it does not provide conclusive evidence of telomere dysfunction in human cancers. In contrast, direct evidence of telomere dysfunction in the form of high levels of telomere fusions was recently reported in the bloodborne cancer chronic lymphocytic leukemia (12).
To directly test whether telomere fusions are present in human early- and later-stage breast tumors, we have developed a highly specific and sensitive PCR-based technique that we term TAR (telomere-associated repeat) fusion PCR. This technique allows us to detect and analyze telomere dysfunction within human solid tumor tissue. We discovered that telomere fusions are present at similar levels in DCIS and at the later invasive ductal carcinoma(IDC) stage. The presence of telomere end-to-end fusions is a fundamental indication of and a marker for loss of telomere function. This approach has allowed us to begin to elucidate mechanisms responsible for the origin of genomic instability involving defects in telomere maintenance from human tumor tissue.We propose that TAR fusion PCR could have significant applications in both clinical early detection strategies and investigations of basic mechanisms of telomere-driven genomic instability in human cancers.
Results
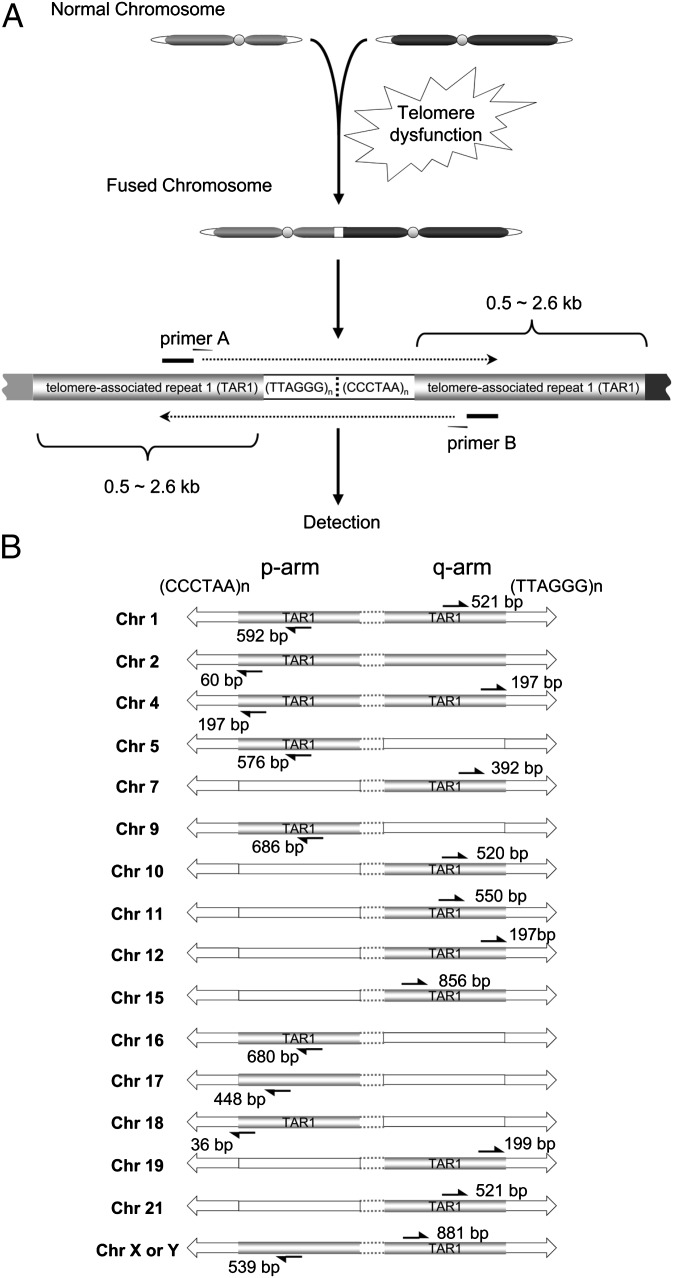
We have devised a multiplex PCR-based assay to detect and analyze chromosome end-to-end fusions from solid tumors, whose existence we report in human breast tumor tissue (Fig. 1A). To amplify individual telomere fusion junctions, we used a mixture of primer sets that anneal within telomere-associated repeat 1 (TAR1) distal chromosomal regions (Fig. 1B and Table S1). TAR1 regions are present within ∼2 kb of telomere variant repeat regions at chromosome ends and contain portions of sequence homology with other TAR1 regions (13, 14). We used two multiplex primer sets to increase the coverage of potential chromosome fusion combinations and reduce the total number of PCR reactions required. In addition, to increase fusion coverage, we used two primers in this assay that anneal within distal regions of Xp and 17p, which are not within known TAR1 regions (12, 15) (Fig. 1B). Assuming that TAR1 regions are essentially identical on homologous chromosome ends, we calculated that the multiplex PCR primer sets used in this study (Materials and Methods) cover ∼14% of the theoretically possible chromosome end-to-end fusion combinations.
Fig. 1.
Strategy for the detection of telomere fusions using TAR fusion PCR. (A) Fusion junctions were amplified by multiplex primer sets (e.g., primer A, primer B; Table S1) annealing within TAR1 regions adjacent to telomeric DNA sequences. The fusion junctions were detected by Southern blot analysis using a telomeric DNA [TTAGGG]4 probe. (B) Chromosome locations of multiplex PCR primers used for TAR fusion PCR. Black arrows represent primers and the distances (bp) from telomeric DNA starting positions, open arrows denote telomeric DNA, and open rectangles denote genomic regions currently not available in current human genome sequence databases.
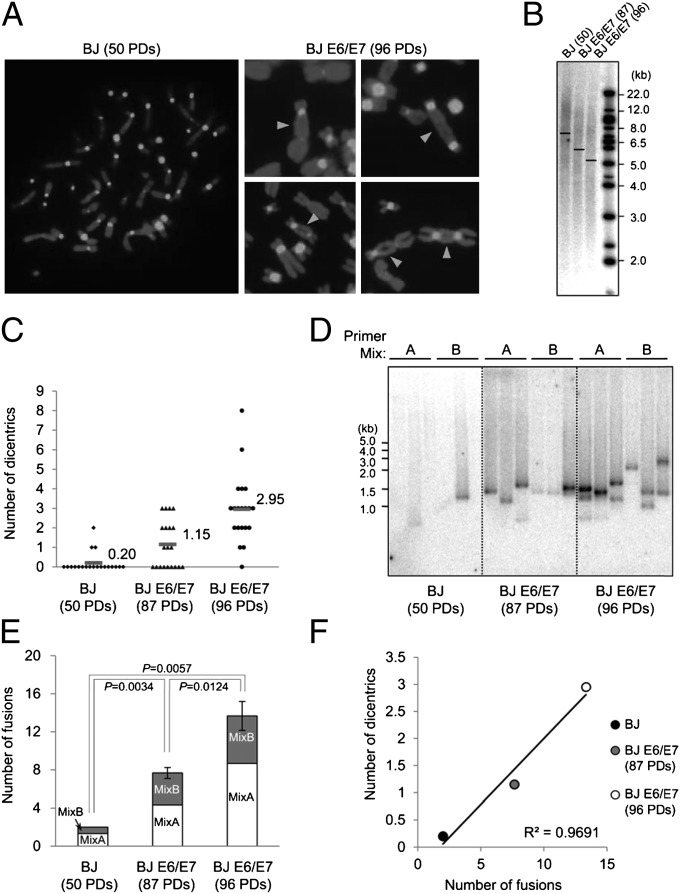
We developed the TAR fusion PCR assay using the well-characterized BJ foreskin fibroblast-expressing HPV16 E6/E7 (BJ E6/E7) telomere crisis model (16), which accumulates telomere fusions with increasing population doublings (PDs) (Fig. 2 A and C). We determined empirically that specific primer sequences, primer set combinations in mixes A and B, and PCR conditions were critical for specificity and sensitivity parameters to eliminate background signals from PCR artifacts and the vast majority of normal human genomic DNA. As a negative control for telomere fusions, we used primary BJ foreskin fibroblast cells, which have comparatively few cells with low numbers of telomere fusions (Fig. 2 A and C). As described previously (16), we found a decrease in mean telomere lengths with these cell populations on increasing cellular passage: BJ (50 PD; 7.6 kb), BJ E6/E7 (87 PD; 5.9 kb), and BJ E6/E7 (96 PD; 5.2 kb) (Fig. 2B). We performed TAR fusion PCR followed by Southern blot analysis using a 32P-labeled [TTAGGG]4 probe and found an increasing number of TAR fusion PCR products with increasing BJ E6/E7 PDs (Fig. 2 D and E). We found that TAR fusion PCR products appear as single bands because they likely represent individual and relatively rare fusion events between specific chromosome pairs within a single nucleus (Fig. 2D).
Fig. 2.
Telomere fusion analysis in cultured human male foreskin fibroblasts during telomere crisis. (A) Chromosome spreads were hybridized with a pancentromere probe to permit detection of dicentric chromosomes after telomere fusion. Chromosomes were stained with DAPI. (Left) A diploid BJ fibroblast metaphase spread at 50 PDs. (Right) Examples of several telomere fusions from BJE6/E7 (96 PDs) metaphase spreads. Arrowheads indicate fusion junctions between two chromosomes. (B) Southern blot analysis of mean telomere lengths. Mean telomere lengths are shown by a horizontal line in each lane: BJ (50 PD; 7.6 kb), BJ E6/E7 (87 PD; 5.9 kb), and BJ E6/E7 (96 PD; 5.2 kb). (C) Average number of dicentric chromosomes per metaphase. BJ cells at 50 PDs contained 0.20 dicentric chromosomes per metaphase, BJE6E7 cells at 87 PDs contained 1.15 decentric chromosomes per metaphase, and BJE6E7 cells at 96 PDs contained 2.95 dicentric chromosomes per metaphase. Twenty metaphase spreads were analyzed for each cell type at the referenced PD. (D) Southern blot analysis of TAR fusion PCR products using a 32P-labeled [TTAGGG]4 probe. Triplicate reactions are shown from multiplex primer mixes A and B. The variation in band sizes between separate PCR reactions using identical primer mixes, template DNA, and PCR conditions was likely due to an early PCR cycle competition between telomere fusions for PCR reagents. By excising and sequencing telomere fusion junctions, we determined that the variation in band sizes among triplicate reactions represented different telomere fusions owing to different chromosome combinations and size variations within the length of fusion junction sequences (Fig. 3). (E) Number of telomere fusions. The sum of the number of bands per six reactions is shown for each cell type from three independent TAR fusion PCR experiments using the same DNA sample. (F) Correlation between the metaphase telomere fusion analysis and TAR fusion PCR. The y-axis represents the average number of dicentric chromosomes per metaphase as determined in C; the x-axis represents the average number of TAR fusion PCR bands per six reactions as determined in E. White circles represent BJ (50 PDs); gray circles, BJ E6/E7 (87 PDs); black circles, BJ E6/E7 (96 PDs). Note that the frequency of telomere fusion junctions using TAR fusion PCR is highly correlated with the frequency of dicentric chromosomes shown in B with R2 = 0.96914 (P < 0.01).
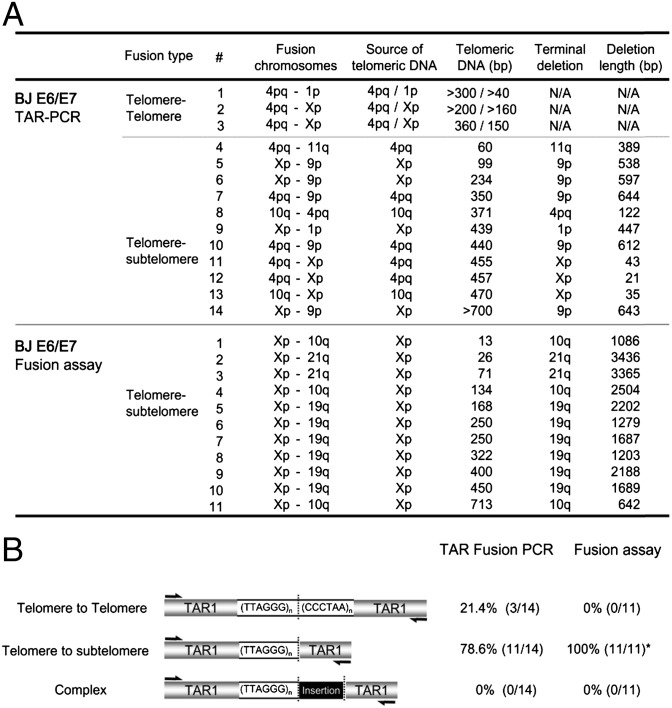
We then cloned TAR fusion PCR products to isolate specific fusion junctions and sequenced them to determine the composition of these products. We found that most TAR fusion PCR products contained shortened telomeric DNA, ∼60–700 bp long, adjacent to subtelomeric DNA at the fusion junctions (78.6%) (Fig. 3 A and B; see Fig. 6 A and B). Another class of less common telomere fusion junctions contained shortened telomeric DNA at the fusion junction (∼40–360 bp), presumably from both fused chromosome ends (Fig. 3 A and B). TAR fusion PCR-amplified telomere fusion junctions appeared as defined bands under these conditions (Fig. 2D), as opposed to smeared heterogeneous telomeric DNA, because they contained defined tracts of telomeric DNA and/or other distinct sequences (Fig. 3). We compared TAR fusion PCR with a previously reported method, fusion PCR, for chromosome end-to-end fusion junction analysis, and found similar results within fusions in this crisis cell model (12, 15). The only major qualitative difference between these two PCR-based methods is that with TAR fusion PCR, we found evidence of telomere-to-telomere fusion junctions (Fig. 3 A and B). Thus, we conclude that TAR fusion PCR amplifies actual telomere fusions. This conclusion is supported by the correlation between increasing accumulation of TAR fusion PCR products with increasing frequency of telomere fusions, as measured by the appearance of dicentric chromsomes on FISH analysis (Fig. 2F), and confirmation of chromosome fusion junctions by sequence analysis.
Fig. 3.
Telomere fusion junction sequence analysis from BJ E6/E7 cells. (A) Summary of telomere fusion junction sequence analysis using TAR fusion PCR and the fusion assay performed as previously described by Baird and coworkers (12, 15). Fusion chromosomes, chromosomes involved in fusion junction. Source of telomeric DNA, chromosome presumed to contribute telomeric DNA within the fusion junction. Telomeric DNA (bp), size of telomeric DNA at fusion junctions in bp. Terminal deletion, chromosome presumed to contain deletions within subtelomeric regions at fusion junctions. Deletion length (bp), chromosomal deletion size of subtelomeric regions. NA, not applicable. (B) Classification of telomere fusion types in BJ E6/E7 cells. Telomere–telomere fusion, fusion junctions containing telomeric DNA presumably from both chromosomal ends. Telomere–subtelomere fusion, junctions containing telomeric DNA presumably from one chromosome fused with another chromosome end with subtelomeric deletions. Complex fusion, fusion junctions containing nontelomeric DNA from interstitial chromosomal regions.
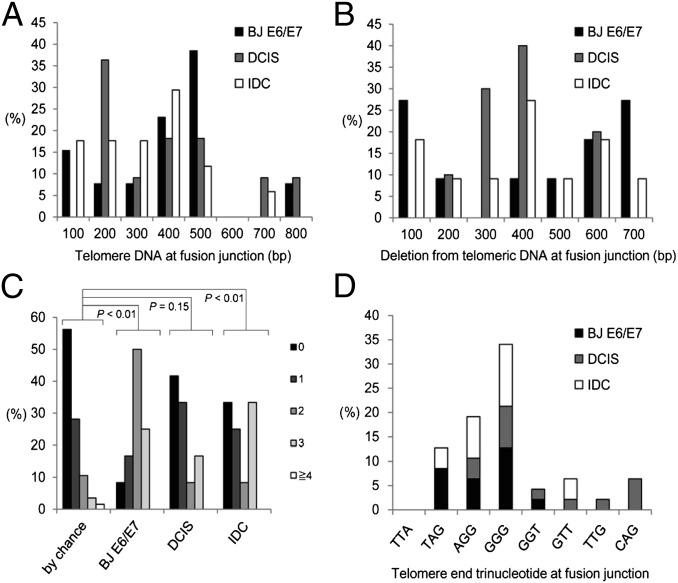
Fig. 6.
Characterization of telomere fusion junctions from BJ E6/E7 cells, DCIS, and invasive tumor tissue. (A) Size of telomeric DNA at fusion junctions. (B) Size of subtelomeric DNA deletion at fusion junctions. (C) Distribution of microhomology by chance and observed microhomology at telomere fusions junctions from BJ E6/E7 (96 PDs), DCIS, and IDC samples. The probability that a junction will have X nucleotides of microhomology by chance assuming an unbiased base composition is indicated. Black box, no shared nucleotides at junction; dark-gray box, one shared nucleotide at junction; medium-gray box, two shared nucleotides at junction; light-gray box, three shared nucleotides at junction; open box, greater than or equal to four shared nucleotides at junction. (D) Trinucleotide sequences of telomeric DNA termini at fusion junctions.
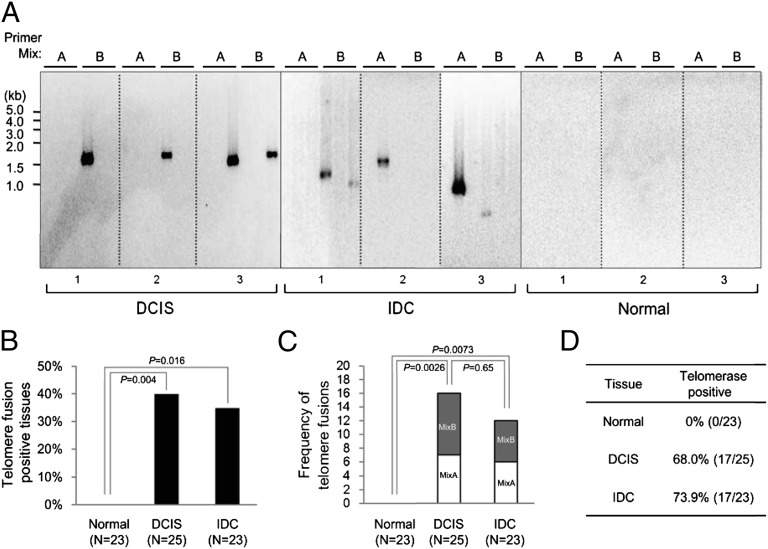
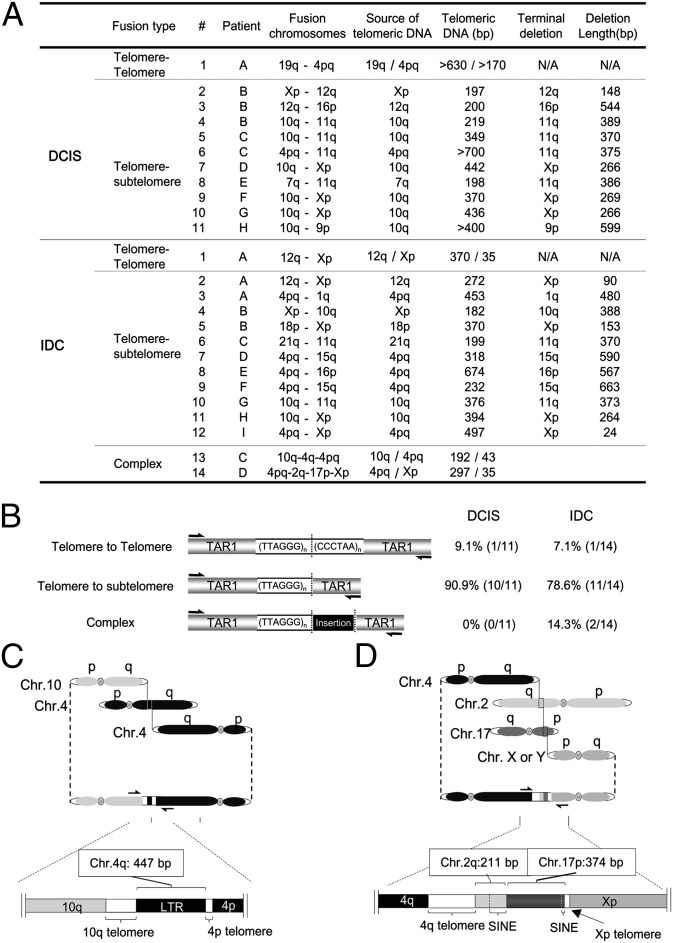
We next used TAR fusion PCR to determine whether human normal and tumor breast tissue contained telomere fusions. We found telomere fusions in ∼40% of both DCIS (n = 25) and IDC (n = 23) specimens, but no telomere fusions in normal controls (n = 23) (Fig. 4 A and B). The DCIS tissue that tested positive for telomere fusions was of grade 3, most likely owing to the high percentage of DCIS grade 3 tissue examined in this study (Table S2). We cannot rule out the possibility that other grades of DCIS are also positive for telomere fusions, given the relatively low percentage of other DCIS graded tissue examined in this study and the current limitations of primer coverage in TAR fusion PCR. We also found similar frequency of fusions within individual positive tumor tissues regardless of tumor stage (Fig. 4C). Therefore, the fraction of breast tumors positive for telomere fusion and the frequency of fusion accumulation within individual tumor tissues remained constant from the DCIS (grade 3) to IDC stage of breast carcinoma. Fewer TAR fusion PCR-amplified bands were detected in breast tumor tissue DNA (Fig. 4A) compared with later-passage BJ E6/E7 DNA (Fig. 2D). This lower detection rate likely reflects a reduced frequency of telomere fusions in breast tumor tissue per unit of DNA, especially considering the heterogeneous composition of tumor tissue that comprises both tumorigenic and nontumorigenic cells. Sequence analysis revealed that the majority of telomere fusion junctions from breast tumor tissue contained shortened telomeric DNA tracts (197 to ∼700 bp long) from one chromosome fused to subtelomeric DNA regions from another distal chromosome end, 90.9% for DCIS (grade 3) and 78.6% for IDC tumor tissue (Figs. 5 A and B and 6 A and B). We also found evidence of telomeric DNA–telomeric DNA fusion junctions containing telomeric DNA likely from both chromosomes involved in the fusion (Fig. 5 A and B). Interestingly, IDC also contained (at a lower frequency) complex fusion junctions with short regions (211–447 bp) of nontelomeric interstitial genomic DNA regions (Fig. 5 C and D). These short regions found within fusion junctions were composed of LTR and non-LTR short interspersed element retrotransposon elements (Fig. 5 C and D). Analysis of microhomology at telomere fusion junctions revealed that fusions from both the BJ E6/E7 telomere crises model and invasive breast tumor tissue contained significant levels of short regions of sequence microhomology (P < 0.01); however, fusion junctions within DCIS (grade 3) breast tumor tissue did not contain significant levels of microhomology (P = 0.15) (Fig. 6C). In addition, junction sequence analysis revealed that the majority of telomeric DNA found at fusion junctions ended in two or three G residues (Fig. 6D). These results indicate that telomere fusions are present in human breast tumor tissue, and fusion junction sequence analysis provides important clues into potential mechanisms of chromosome end-to-end fusion events during human breast carcinogenesis.
Fig. 4.
Telomere fusions in human breast tumor tissue. (A) Southern blot analysis of TAR fusion PCR products using a 32P-labeled [TTAGGG]4 probe. Six reactions from two primer sets, A and B, are shown for each patient sample. (B) Percentage of human breast tumor tissues containing telomere fusions. n, total number of tissue samples analyzed. (C) Frequency of fusions within individual telomere fusion positive tumor tissue. The sum of the number of bands per six reactions is shown for each tissue positive for telomere fusions from three independent experiments. (D) Percentage of human breast tissue positive for telomerase activity as assayed by TRAP.
Fig. 5.
Telomere fusion junction sequence analysis from breast tumor tissue. (A) Summary of telomere fusion junction sequence analysis using TAR fusion PCR. Fusion chromosomes, chromosomes involved in fusion junction. Source of telomeric DNA, chromosome presumed to contribute telomeric DNA within the fusion junction. Telomeric DNA (bp), size of telomeric DNA at fusion junctions in base pairs. Terminal deletion, chromosome presumed to contain deletions within subtelomeric regions at fusion junctions. Deletion length (bp), chromosomal deletion size of subtelomeric regions. NA, not applicable. (B) Classification of telomere fusion types in DCIS and invasive tumors. Telomere–telomere fusion, fusion junctions containing telomeric DNA presumably from both chromosomal ends. Telomere–subtelomere fusion, junctions containing telomeric DNA presumably from one chromosome fused with another chromosome end with subtelomeric deletions. Complex fusion, fusion junctions containing nontelomeric DNA from interstitial chromosomal regions. (C) Detailed molecular structure of complex telomere fusion #14 from invasive tissue described in A. (D) Detailed molecular structure of complex telomere fusion #13 from invasive tissue described in A. SINE, short interspersed elements. LTR, long terminal repeat. Base pairs (bp), size of insertion.
Discussion
We examined whether telomere dysfunction occurs in human breast cancer using our TAR fusion PCR technique. Our findings demonstrate that telomere fusions accumulate in a relatively early stage of breast carcinoma (DCIS grade 3) and remain at similar levels in the more advanced stage of IDC, as detected by TAR fusion PCR and confirmed by fusion junction sequence analysis. Telomere dysfunction may be limited during later stages of carcinogenesis by telomerase reactivation and other factors to prevent total genomic catastrophe, which would likely result in the loss of essential cellular functions required to maintain tumor cell viability. Our finding that telomere fusions appear in DCIS correlates with previous reports of telomere shortening, telomerase activation, and other genomic aberrations detected in DCIS (10, 11, 17, 18). These results provide evidence that telomere dysfunction is a prevalent event during breast carcinogenesis, and that telomere end-to-end fusions may be causal in driving genomic instability via breakage–fusion–bridge cycles.
Our TAR fusion PCR method uses only partial primer coverage of chromosome ends, owing to the absence of sequence information for the majority of chromosome termini in genomic sequence databases (Fig. 1B, open rectangles; http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/) (19). We estimated that our current method detects ∼14% of the theoretically possible telomere fusion combinations. However, sequence information at chromosome ends is likely to become available with future improvements in sequencing methodologies within repetitive regions. Despite this limitation, ∼40% of the breast tumor tissue (both DCIS and IDC) that we tested contained telomere fusion. Thus, even though our current method covers only ∼14% of the possible chromosomal end fusion combinations, telomere dysfunction appears to be a prevalent genetic aberration event in breast tumorigenesis. Although it is possible that certain chromosome ends are more frequently involved in end-to-end fusions during breast carcinogenesis, and that our current primer sets anneal at TAR1 regions within chromosome ends not currently available in the genomic sequence databases, we found no evidence for this in our fusion junction sequence analysis. In the future, it will be interesting to determine whether telomere fusion frequencies are related to genomic instability levels, tumor grade, and receptor status. However, this type of analysis will likely be complicated by increased TAR fusion PCR primer coverage, which may well result in a substantial increase in breast tumor tissue testing positive for telomere fusions. Based on our present findings, we cannot rule out the possibility that telomere fusions may be present within stromal cells as well, given that total tumor tissue was assayed with TAR fusion PCR. Future studies may involve the isolation of highly enriched tumor subpopulation for telomere fusion analysis to determine the specific cellular subpopulation containing telomere fusions within breast tumor tissue.
Our sequence analysis at fusion junctions has provided some insight into potential mechanisms responsible for chromosomal end-to-end fusions. The majority of telomere fusions contained relatively short regions of telomeric DNA (∼100–800 bp) from one or both chromosomal ends involved in the fusion event. The shortened tracts of telomeric DNA may indicate that telomeric DNA is relatively shortened before fusion. Moreover, the majority of telomeric DNA found at fusion junctions ended in two or three G residues. However, chromosome ends during or before end-to-end fusion may be processed by an active mechanism. Thus, the static composition of fusion junctions does not necessarily provide information regarding the structure of the chromosome end before fusion events, but may provide information regarding mechanistic pathways that connect dysfunctional chromosome ends. Interestingly, we found significant levels of microhomology at fusion junctions in both the telomere crisis model cells and invasive breast tissue, but not in DCIS grade 3 tissue (Fig. 6C). A portion of fusion junctions present in the crisis model cells and invasive tissue may be the result of mechanisms distinct from those generally found in DCIS, potentially through pathways involving DNA-PKcs–independent nonhomologous DNA end joining/microhomology-mediated end joining (20, 21).
In addition to the foregoing, we found small regions of LTR and non-LTR retrotransposon elements from interstitial chromosomal regions within fusion junctions in human breast invasive tissue (Fig. 5 C and D). These more complex fusion junctions may be the product of extensive breakage–fusion–bridge cycles that may occur during more advanced stages of breast carcinoma. Interestingly, similar retrotransposon elements are present at Drosophila chromosome ends and have been reported to integrate at dysfunctional mammalian telomeres in a Chinese hamster ovary cell line (22, 23). The presence of retrotransposon elements at spontaneous fusion junctions within human invasive breast tissue may indicate an evolutionary conserved pathway for potential retrotransposon integration events at human dysfunctional telomeres. The presence of nontelomeric DNA retrotransposon elements within fusion junctions also may provide important additional leads for genetic biomarkers associated with tumorigenesis and reveal potential mechanisms for genomic instability during breast tumorigenesis involving telomere dysfunction and other genomic interstitial regions vulnerable to breakage or recombination events.
In summary, our results suggest that the occurrence of telomere dysfunction may be an early and potentially highly frequent genetic aberration event in the development of breast cancer. Therefore, it will be important to investigate both the basic mechanistic studies regarding the molecular and cellular processes responsible for the generation of these fusion junctions, as well as the translational potential for clinical applications.
Materials and Methods
Normal and Tumor Breast Tissues.
Normal human breast tissues (20–65 y; mean, 29.5 ± 13 y) were obtained from the Susan G. Komen Tissue Bank at the Indiana University Simon Cancer Center, and human breast tumor tissues (DCIS, 30–80 y, mean, 55 ± 12 y; IDC, 31–86 y, mean, 47.6 ± 13.3 y) were obtained from the Cancer Center Tissue Bank at the Indiana University Simon Cancer Center using an Indiana University School of Medicine Institutional Review Board-approved protocol. Written informed consent was obtained from all participants involved in this study. Immediately after surgical removal, the tissue specimen was placed in an iced sterile container and transferred to Pathology for diagnosis and processing. Each aliquot of deidentified tumor tissue was assigned a unique barcode for identification. Each tumor tissue aliquot contained ∼100–250 mg of breast tumor tissue. All tumor tissues specimens were obtained by excisional biopsy and were reviewed histologically to confirm the absence or grade of breast carcinoma by a group of pathologists at the Indiana University Simon Cancer Center. The tumor cell content of tumor tissues used in this study ranged from 50% to 90%. Additional information on normal and tumor breast tissue is provided in Tables S2 and S3.
Cell Culture.
Foreskin fibroblast BJ cells were obtained from American Type Culture Collection (http://www.atcc.org/). Cells were maintained in advanced DMEM (Life Technologies) supplemented with l-glutamine, penicillin-streptomycin, and 5% FBS at 37 °C in 5% (vol/vol) CO2. BJ cells (60 PD) were infected with a pLXSN retrovirus expressing HPV16 E6 and E7 proteins (16) and selected on G418 (400 μg/mL) after 3 wk of continuous drug treatment.
TAR Fusion PCR.
Genomic DNA from cultured cells was isolated by a salt precipitation method as described previously (24). Genomic DNA was extracted from the majority of normal and tumor breast tissue obtained (∼80%) using the Agencourt DNAdvance Kit (Beckman Coulter), with ∼1.5 μg of genomic DNA obtained from each ∼20 mg of breast tissue sample as determined by a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Fusion PCR was performed as described by Baird and coworkers (12, 15). The TAR fusion PCR conditions outlined below were the same for both cultured cell and breast tissue DNA. Two-step touchdown PCR was performed in a 20-μL reaction mixture using 50 ng of DNA, multiple primers, 10% 7-Deaza-dGTP (Roche Diagnostics), and Advantage GC Genomic LA Polymerase Mix (Clontech). The cycling conditions were initially denatured at 94 °C for 3 min, followed by 10 cycles of 94 °C for 30 s and 72 °C (−0.4 °C each cycle to a “touchdown” at 68 °C) for 5 min, and then 20 cycles of 94 °C for 30 s and 68 °C for 5 min. Primer mix A contained eight primers that anneal within TAR1 regions (with the exception of Xp and 17p): 1p, 2p, 5p, 7q, 9p, 11q, 12q, 15q, 16p, 17p, 18p, Xp, and Xq (Fig. 1B). Primer mix B contained eight primers that anneal within TAR1 regions (with the exception of Xp and 17p): 1q, 2p, 4p, 4q, 5p, 7q, 10q, 12q, 17p, 18p, 19q, 21q, and Xp (Fig. 1B). Table S1 lists primer sequences. TAR fusion PCR products were then resolved on a 0.8% agarose gel, and Southern blot analysis was performed using a 32P-labeled [TTAGGG]4 probe (25).
To calculate the total number of theoretical telomere fusion combinations and the percentage of telomere fusions detectable by TAR Fusion PCR, we used the general equation C(n, 2) + n, where n is the number of unique chromosomal ends. This equation can be simplified to n (n + 1)/2. Therefore, in human somatic cells with 46 unique chromosomal ends, the total number of theoretical telomere fusion combinations is 46 × (46 + 1)/2 = 1,081. Each of the current TAR-fusion PCR primer mixes A and B covers 13 chromosomal arms (n = 13 for each primer mix); thus, the number of detectable fusion combinations is 13 × 14/2 = 91 for each mix. The two primer mixes share seven primers, and hence 7 × 8/2 = 28 combinations are detected by both mixes. As a result, the total number of detectable fusion combinations is 91 + 91 – 28 = 154, and the coverage rate is 154/1,081 × 100% = 14.2%.
Telomere Length and Telomerase Activity.
Genomic DNA was isolated and telomere length measured by in-gel hybridization as described previously (25). Mean telomere lengths were calculated using the Excel spreadsheet program TELORUN as described previously (25). Telomerase activity was measured by the telomeric amplification protocol (TRAP) using the TRAPeze Telomerase Detection Kit (Millipore). Cells were lysed with ice-cold 1× CHAPS lysis buffer [10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM EGTA, 0.1 mM benzamidine, 5 mM β-mercaptoethanol, 0.5% CHAPS, and 10% glycerol] containing RNase inhibitor at a final concentration of 100–200 U/mL. A reaction mixture of 50 μL containing 0.5 μg of protein extract, 10 μL of 5× TRAP reaction mix (Tris buffer, primers, dNTPs, and oligomer mix for amplification of 36-bp internal control band), and 2 U of Taq DNA polymerase was incubated for 30 min at 30 °C and then subjected to denaturation at 95 °C for 5 min. PCR involved 32 cycles of 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s. For direct visualization of the TRAP ladder, all of the PCR products were electrophoresed on a 12.5% nondenaturing polyacrylamide gel and stained with SYBR Green.
FISH.
Cultured cells were treated with 0.1 μM colcemid (Roche) for 4 h and fixed in 3:1 (vol/vol) methanol-acetic acid. Metaphase chromosomes were prepared using standard methods (26). A pancentromere probe was prepared by purification of a PCR product amplified with human genomic DNA (Roche) as a template, using the degenerate alpha satellite primer pairs 5′-ACA GAA GCA TTC TCA GAA-3′ and 5′-TTC TGA GAA TGC TTC TGT-3′, as reported previously (27). The pancentromere probe was labeled with spectrum-red dUTP fluorophore using nick translation (Abbott Laboratories) and hybridized to metaphase spreads. After hybridization, slides were washed in (50% formamide/2× SSC) at 42 °C, then washed in 2× SSC at 37 °C. Slides were mounted with DAPI (Vectashield; Vector Laboratories). Fluorescence images were captured using a SPOT RT camera mounted on a Leica CTR 5000 fluorescence microscope and processed using Adobe Photoshop.
Cloning and Sequencing of Telomere Fusion Junction.
DNA samples were purified from either PCR reactions or gel-purified DNA fragments using the GENECLEAN Turbo Kit (MP Biomedicals). Cloning and transformation were carried out with a TOPO XL PCR Cloning Kit (Life Technologies). Clones containing the TTAGGG repeat sequences were identified by colony hybridization with radiolabeled [TTAGGG]4 probe. Positive clones were isolated, and fusion junctions were sequenced. Sequence alignments were performed using the University of California Santa Clara Genome Bioinformatics database (http://genome.ucsc.edu/).
Supplementary Material
Acknowledgments
We thank Sumita Bhaduri-McIntosh, Brittney-Shea Herbert, Anna Malkova, Marc Mendonca, and Amber Mosley for their valuable comments during the preparation of this manuscript and Aaron Ernstberger for his valuable technical assistance. We also thank the Susan G. Komen for the Cure Tissue Bank and the Indiana University Cancer Center Tissue Bank at the Indiana University Simon Cancer for the normal and tumor breast tissue used in this study. This work was supported by the Indiana University Cancer Center, the American Cancer Society, the Showalter Foundation, the Susan G. Komen Foundation, the Avon Foundation, the Flight Attendant Medical Research Institute, and the Indiana Genomics Initiative (supported in part by Lilly Endowment, Inc.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120062109/-/DCSupplemental.
References
- 1.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Donate LE, Blasco MA. Telomeres in cancer and ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cifuentes-Rojas C, Shippen D. Telomerase regulation. Mutat Res. 2011;730:20–27. doi: 10.1016/j.mrfmmm.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 7.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 8.Artandi SE, et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci USA. 2002;99:8191–8196. doi: 10.1073/pnas.112515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanov SR, et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 10.Meeker AK, et al. Telomere shortening occurs in subsets of normal breast epithelium as well as in situ and invasive carcinoma. Am J Pathol. 2004;164:925–935. doi: 10.1016/S0002-9440(10)63180-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin K, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 12.Lin TT, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: Evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 13.Brown WR, et al. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 14.Riethman H. Human telomere structure and biology. Annu Rev Genomics Hum Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- 15.Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic Acids Res. 2010;38:1841–1852. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou Y, Misri S, Shay JW, Pandita TK, Wright WE. Altered states of telomere deprotection and the two-stage mechanism of replicative aging. Mol Cell Biol. 2009;29:2390–2397. doi: 10.1128/MCB.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbert BS, Wright WE, Shay JW. Telomerase and breast cancer. Breast Cancer Res. 2001;3:146–149. doi: 10.1186/bcr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heaphy CM, et al. Genomic instability demonstrates similarity between DCIS and invasive carcinomas. Breast Cancer Res Treat. 2009;117:17–24. doi: 10.1007/s10549-008-0165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler EE, Clark RA, She X. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat Rev Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 20.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 22.Capkova Frydrychova R, Biessmann H, Mason JM. Regulation of telomere length in Drosophila. Cytogenet Genome Res. 2008;122:356–364. doi: 10.1159/000167823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 24.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215–1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda N, Tanaka H, Herbert BS, Reed T, Gilley D. Shared environmental factors associated with telomere length maintenance in elderly male twins. Aging Cell. 2007;6:709–713. doi: 10.1111/j.1474-9726.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 26.Gardner L, Malik R, Shimizu Y, Mullins N, ElShamy WM. Geminin overexpression prevents the completion of topoisomerase IIα chromosome decatenation, leading to aneuploidy in human mammary epithelial cells. Breast Cancer Res. 2011;13:R53. doi: 10.1186/bcr2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 28.Greene FL, et al., editors. AJCC Cancer Staging Manual. 6th Ed. New York: Springer; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.