Abstract
Genome-wide association studies can identify common differences that contribute to human phenotypic diversity and disease. When genome-wide association studies are combined with approaches that test how variants alter physiology, biological insights can emerge. Here, we used such an approach to reveal regulation of cell death by the methionine salvage pathway. A common SNP associated with reduced expression of a putative methionine salvage pathway dehydratase, apoptotic protease activating factor 1 (APAF1)-interacting protein (APIP), was associated with increased caspase-1–mediated cell death in response to Salmonella. The role of APIP in methionine salvage was confirmed by growth assays with methionine-deficient media and quantitation of the methionine salvage substrate, 5′-methylthioadenosine. Reducing expression of APIP or exogenous addition of 5′-methylthioadenosine increased Salmonellae-induced cell death. Consistent with APIP originally being identified as an inhibitor of caspase-9–dependent apoptosis, the same allele was also associated with increased sensitivity to the chemotherapeutic agent carboplatin. Our results show that common human variation affecting expression of a single gene can alter susceptibility to two distinct cell death programs. Furthermore, the same allele that promotes cell death is associated with improved survival of individuals with systemic inflammatory response syndrome, suggesting a possible evolutionary pressure that may explain the geographic pattern observed for the frequency of this SNP. Our study shows that in vitro association screens of disease-related traits can not only reveal human genetic differences that contribute to disease but also provide unexpected insights into cell biology.
Keywords: HapMap, lymphoblastoid, pyroptosis, quantitative trait, polygenic adaptation
Human genetic variation has long been recognized as playing an important role in susceptibility to infectious disease (1). Recent technological progress has facilitated the development of systematic approaches to identify resistance and susceptibility alleles through genome-wide association studies (GWASs) of infectious diseases, such as leprosy (2–4), HIV (5, 6), tuberculosis (7), and meningococcal disease (8). These studies have identified common loci important for susceptibility and in some cases, highlighted the relevance of specific pathways in controlling infection. However, it is difficult to determine what aspects of pathogenesis are being affected by the variants, especially when susceptibility loci fall near genes of unknown function or gene-poor regions.
Examining variation in cellular phenotypes may provide a means of obtaining greater understanding of human variation to infection. Pathogen recognition and activation of appropriate inflammatory pathways are crucial for clearance of infection and can accurately be measured at the cellular level. One of the central mediators of inflammation is the cysteine protease caspase-1. Activation of caspase-1 in response to pathogens leads to a proinflammatory form of cell death termed pyroptosis (9) as well as cleavage of the proinflammatory cytokines IL-1β and IL-18 (10, 11). Alterations of the caspase-1 inflammatory response have been linked to multiple inflammatory diseases in humans, including inflammatory bowel disease (12), gout (13), and autoinflammatory cryopyrinopathies (14). Additionally, caspase-1–deficient mice have a reduced ability to control Salmonella typhimurium (15) and other bacterial infections (16) because of impaired pyroptosis (17). Thus, the cell death response to S. typhimurium is a relevant cellular phenotype that may inform understanding of both infectious and inflammatory diseases. Furthermore, the human and mouse data imply that there is a tradeoff: too little caspase-1 activation impairs the ability to fight infection, whereas too much activation predisposes to inflammatory disease.
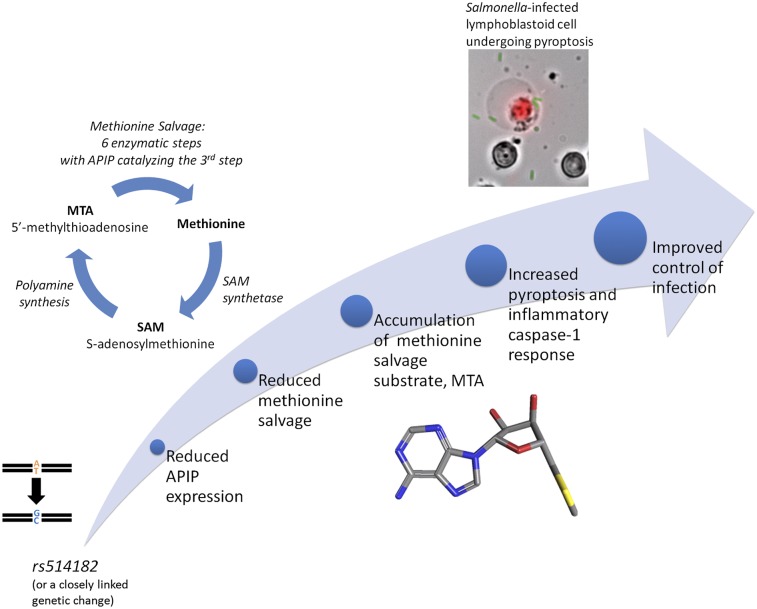
We developed an in vitro association screen of human host response to microorganisms named high-throughput human in vitro susceptibility testing (Hi-HOST) to identify genetic variants that affect cellular phenotypes that may modulate infectious and inflammatory disease (18). Similar approaches have also been applied to other cellular phenotypes, including sensitivity to chemotherapeutic drugs (19, 20), HIV infection (21), and sensitivity to a bacterial toxin (22). Lymphoblastoid cell lines (LCLs), collected and genotyped as part of the HapMap Project (23), were assayed for pyroptosis and other quantitative phenotypes of host response. Previously, we used Hi-HOST to show that a known nonsense mutation in the CARD8 gene, a reported caspase-1 inhibitor (24) associated with severity of rheumatoid arthritis (25), modulates cell death in response to S. typhimurium (18). In the report presented here, Hi-HOST was used as a true discovery tool to uncover variation altering expression of apoptotic protease activating factor 1 (APAF1)-interacting protein (APIP; National Center for Biotechnology Information gene 51074), a gene previously not known to regulate pyroptosis or be associated with phenotypic variation. Follow-up studies examining how APIP regulates pyroptosis unexpectedly revealed a role for the methionine salvage pathway in regulation of cell death. In patients with systemic inflammatory response syndrome, the same variant was associated with significantly altered risk of death.
Results and Discussion
Prioritizing Candidates from Hi-HOST Using Functional SNP Information.
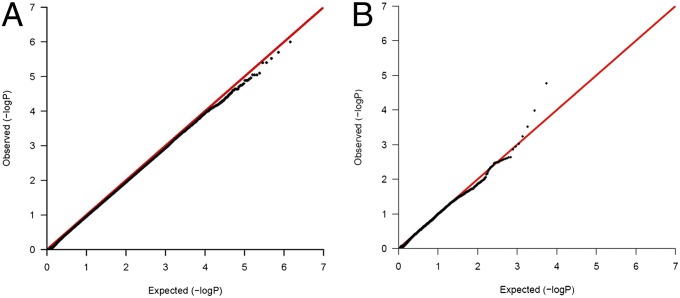
We measured variation in the amount of Salmonella-induced pyroptosis among genotyped LCLs from nearly 120 parent–offspring trios from Utah residents with ancestry from northern and western Europe (CEU) and Yoruba in Ibadan, Nigeria (YRI) (Table S1). We carried out genome-wide family-based association using pyroptosis as a quantitative trait (Materials and Methods and Dataset S1). A quantile–quantile (Q-Q) plot of P values for all HapMap phase 3 SNPs showed little deviation of observed vs. expected (null) distribution (1.4 million SNPs) (Fig. 1A). Thus, although there was no evidence of systematic bias, there were no SNPs with P values lower than neutral expectation. However, previous work has shown the value in examining functional subsets of SNPs, specifically SNPs associated with expression levels of nearby genes [cis-expression quantitative trait loci (cis-eQTL)], to enhance discovery from GWAS (26–28). Doing so also makes biological sense, because SNPs that alter nearby gene expression are more likely to affect cellular function as well. A Q-Q plot of just cis-eQTLs (identified in ref. 29; 5,474 SNPs) showed an upward deviation from neutral expectation for P values < 0.001, suggesting that true-positive results may be within this tail (Fig. 1B). Thus, filtering for cis-eQTLs identified deviation from the neutral model that was not otherwise detectable. The results encouraged additional follow-up studies to determine whether these SNPs played a role in regulating pyroptosis.
Fig. 1.
Q-Q plots show deviation from neutral expectations for cis-eQTLs associated with pyroptosis. (A) A Q-Q plot of P values for all SNPs for the pyroptosis association screen shows that most SNPs follow the expected distribution and are not associated with the pyroptosis phenotype. (B) A Q-Q plot of P values for only cis-eQTLs reveals P values lower than expected by chance for SNPs with P < 0.001.
Hi-HOST Identifies an SNP Near APIP Associated with Cell Death.
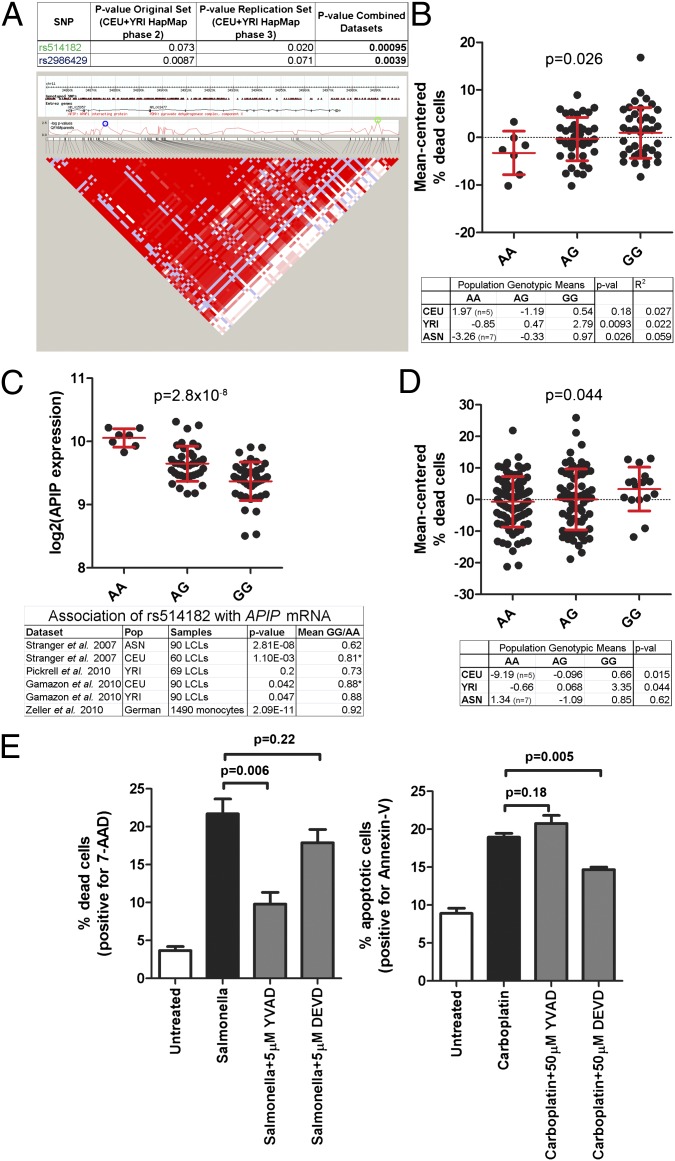
There were five cis-eQTLs with P < 0.001 for association with the pyroptosis phenotype (Table S2). These SNPs are undergoing additional study to determine if the implicated genes affect pyroptosis and determine mechanisms of action. The remainder of this manuscript focuses on the SNP from this group that has undergone the greatest breadth and depth of experimental analysis, rs514182 (Fig. 2A).
Fig. 2.
An SNP near the APIP gene (rs514182) is associated with Salmonellae-induced cell death, APIP expression, and carboplatin-induced cell death. (A) rs514182 is associated with pyroptosis. P values for family-based association analysis are given for the original screen (18), the replication screen, and the combined datasets. The genomic region around rs514182 shows extensive linkage disequilibrium (LD) encompassing the APIP and PDHX genes (ASN LCLs shown; similar LD is seen for CEU and YRI). The SNP with the lowest P value in the region is rs514182 (green circle), but several other SNPs also show an association, including rs2986429 (blue circle) within the first intron of APIP. (B) The derived (G) allele of rs514182 is associated with higher levels of Salmonellae-induced cell death assessed by 7-AAD. Mean-centered percentage dead cells are plotted for ASN LCLs, with mean and SD in red. (C) The derived allele of rs514182 is associated with lower APIP RNA levels in multiple datasets. APIP expression from the ASN LCLs is plotted. An association in the same direction is verified in the other studies listed (30, 79, 80). The mean expression level ratio of GG to AA is given for each study, except for CEU. *The ratio of GG to AG is given, because CEU only has a single AA individual. (D) The derived allele of rs514182 is associated with higher levels of carboplatin-induced cell death as assessed by alamarBlue cell growth inhibition. Mean-centered percentage dead cells are plotted for YRI LCLs. Note that for both S. typhimurium pyroptosis and carboplatin apoptosis in each population, the genotypic means follow the pattern of GG > AG > AA for all genotypes with at least 10 individuals. (E) S. typhimurium-infected and carboplatin-treated LCLs display differential sensitivity to peptide caspase inhibitors. LCL 12155 was untreated, infected with S. typhimurium at MOI 30, or treated with 20 μM carboplatin. Inhibitors of caspase-1 (Ac-YVAD-fmk; N-acetyl-Tyr-Val-Ala-Asp-fluoromethylketone) or caspase-3/7 (the executioner caspases of apoptosis activated by both caspase-9 and -8; Ac-DEVD-fmk; N-acetyl-Asp-Glu-Val-Asp-fluoromethylketone) were added at the indicated concentrations for the duration of the experiment. S. typhimurium cell death was measured at 3 h postinfection using 7-AAD and shows reduced cell death with the caspase-1 inhibitor. Carboplatin apoptosis was measured using annexin staining at 42 h and shows reduced apoptosis with the caspase-3 inhibitor. Graphs show the mean ± SEM from three (S. typhimurium) or four (carboplatin) independent experiments, and P values are from paired t tests.
The derived allele of rs514182 (G) was associated with more cell death (P = 9.5 × 10−4). We found that this association with pyroptosis was present not only in the screened CEU and YRI LCLs that we used to identify the SNP, but also the HapMap Han Chinese in Beijing, China and Japanese in Tokyo, Japan populations (Asian; ASN; P = 0.026) (Fig. 2B). Although the SNP is intergenic (and may be hitchhiking with a true causal variant), the derived allele is associated with lower mRNA levels of the nearby APIP gene in LCLs (29) (P = 2.8 × 10−8 in ASN LCLs) and primary monocytes (30) (P = 2.09 × 10−11) (Fig. 2C). Although the RNA differences seem modest, they translate into protein level differences (Fig. S1). Reduced APIP expression accompanied by increased Salmonellae-induced cell death suggested that APIP inhibits pyroptosis.
The association of rs514182 with pyroptosis was surprising, because APIP was previously identified as an inhibitor of a different cell death pathway, the intrinsic (caspase-9–dependent) apoptotic pathway (31). Apoptosis can be induced in LCLs by pharmacologic agents, such as the chemotherapeutic drug carboplatin (20, 32). Apoptosis induced by carboplatin is at least partially caspase-9–dependent (33), although caspase-8 may also be involved (34). We observed an association between rs514182 and sensitivity to carboplatin in the same LCLs (Fig. 2D). The association with both pyroptosis and apoptosis is remarkable in that the two processes differ markedly in morphology (osmotic lysis vs. fragmentation into apoptotic bodies), kinetics (<3 h vs. 2–3 d), and sensitivity to peptide inhibitors with varying caspase specificity (Fig. 2E). Thus, the association data are consistent with APIP inhibition of two distinct cell death programs and led us to perform extensive experimental studies to examine the significance of these associations.
APIP Inhibits Pyroptosis and Apoptosis.
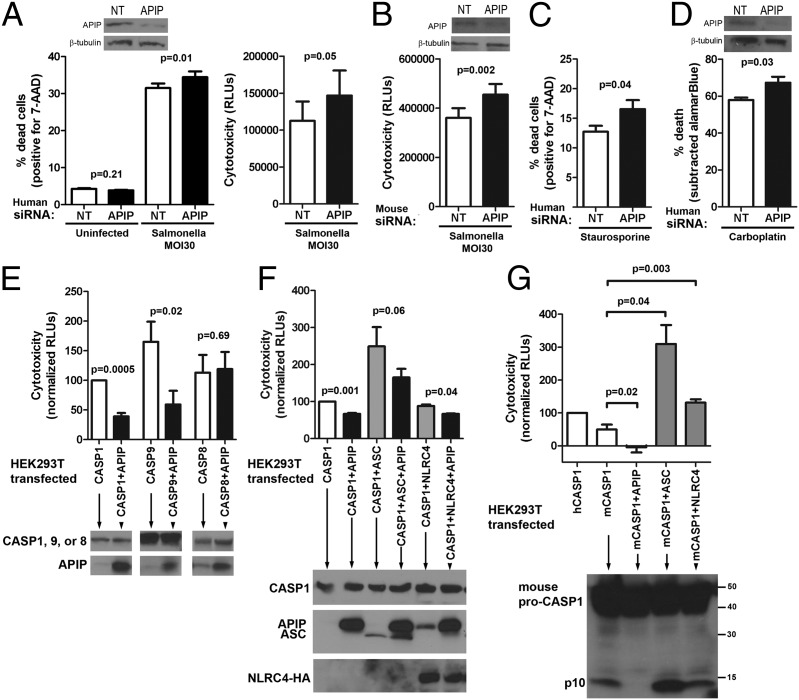
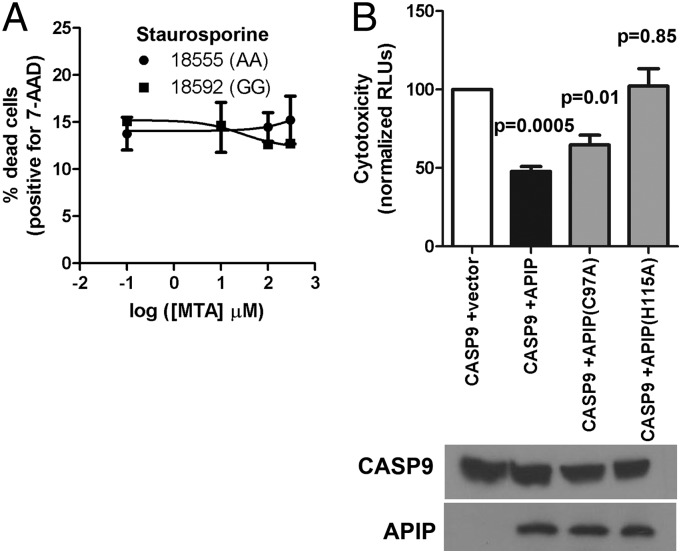
Experiments altering APIP expression levels showed that APIP inhibits both pyroptosis and apoptosis. Reducing APIP expression by RNAi caused higher levels of Salmonellae-induced pyroptosis in both human LCLs and mouse primary bone marrow-derived macrophages (BMMs) (Fig. 3 A and B). Despite efficient knockdown of expression, the size of the effect on pyroptosis is statistically significant but modest, suggesting that, although APIP inhibits pyroptosis, the amount of inhibition under these conditions is only a partial dampening of response. APIP knockdown had previously been shown to increase cisplatin-induced apoptosis (31). We found that APIP RNAi in LCLs increased cell death in response to staurosporine (Fig. 3C), a rapid activator of apoptosis (35), and also increased the effects of carboplatin in HEK-293T cells (Fig. 3D). The data confirmed previous reports (31, 36) of APIP inhibiting apoptosis and revealed a role in inhibition of pyroptosis.
Fig. 3.
APIP inhibits caspase-1– and -9–mediated cell death. (A) Human APIP RNAi in LCL 12155 results in more cell death in response to S. typhimurium (MOI 30) assessed at 3 h by 7-AAD staining or cytotoxicity measured by Cytotox-Glo. (B) Mouse APIP RNAi in BMMs results in more cell death in response to S. typhimurium (MOI 30) assessed at 1 h by cytotoxicity. (C) Human APIP RNAi in LCL 12155 results in more cell death in response to staurosporine (0.5 μM) assessed at 6 h by 7-AAD staining. (D) Human APIP RNAi in HEK-293T results in decreased viability in response to carboplatin (200 μM) assessed at 72 h. Viability was measured by alamarBlue and subtracted from untreated cells to obtain percent death. In A–D, knockdown of APIP protein was assessed by Western blot using β-tubulin as a loading control. (E) Overexpression of APIP in HEK-293T cells inhibits cell death induced by caspase-1 and -9 but not -8. Cells were transfected with the indicated human caspase constructs and either APIP or empty vector. At 23 h posttransfection, supernatants were assayed for cytotoxicity, and phase microscopy images (Fig. S2) were taken. Western blots showed equal expression of caspases with or without APIP. (F) Coexpression of ASC or NLRC4 does not affect APIP inhibition of caspase-1–mediated cell death assayed as described in E. (G) Autocleavage of mouse caspase-1 is inhibited by APIP. HEK293T cells were transfected with the indicated constructs and assayed as in E. APIP coexpression inhibited cell death and autocleavage of mouse caspase-1, whereas ASC and NLRC4 result in an increase in both cell death and autocleavage. In A–G, mean, SEM, and P values from paired t tests are from three to five independent experiments.
Overexpression experiments confirmed the inhibitory activity of APIP. HEK-293T cells have been used as an inflammasome reconstitution system, because they are easily transfected but do not express any endogenous caspase-1 or other known components of inflammasomes (37) (complexes that promote caspase-1 activation). Transfection of caspase-1 triggers cell death in 293T cells, and this transfection was partially inhibited by coexpression of APIP (Fig. 3E). When other known inflammasome components [apoptosis-associated speck-like protein containing a CARD (ASC) and NLR family CARD domain-containing protein 4 (NLRC4)] were coexpressed, the degree of APIP inhibition of cell death was similar (Fig. 3F). Thus, APIP inhibited caspase-1–mediated cell death, even in the absence of other known inflammasome components, whereas their coexpression did not enhance or diminish this activity. Furthermore, overexpression of APIP caused a major reduction in the autocatalytic cleavage of mouse caspase-1 as assessed by the p10 cleavage product (Fig. 3G). Mouse caspase-1 was used for these experiments, because its cleavage is more readily detected than human caspase-1, both for overexpression and endogenous protein in stimulated macrophages. In contrast, ASC and NLRC4 induced an increase in the p10 product. Inhibition of cell death by APIP was also observed for caspase-9 overexpression but not for caspase-8 overexpression (Fig. 3E). Therefore, overexpression studies showed that APIP inhibited caspase-1 and -9 cell death, consistent with the RNAi findings.
Previous studies of the mechanism of APIP inhibition of the intrinsic apoptotic pathway suggested that APIP could inhibit cell death through binding to the caspase recruitment domain (CARD) domain of APAF-1 (31). We speculated that a similar mechanism could be at work in inhibition of pyroptosis, because NLRC4, ASC, and caspase-1 all contain CARD domains. Coimmunoprecipitation studies did not reveal an interaction between APIP and NLRC4 or ASC, but a weak interaction between caspase-1 and APIP was detected in overexpressing cells (Fig. S3A). Consistent with these findings, caspase-1–dependent enrichment of endogenous APIP protein was observed by fluorescence microscopy in inflammasome foci (38, 39) in response to S. typhimurium infection (Fig. S3B). However, despite extensive efforts, we could detect no inhibition of caspase-1 by APIP using recombinant or cell-free systems (Fig. S3C). In the course of these experiments, a caspase-1–dependent APIP cleavage product was observed (Fig. S3D). The lack of in vitro inhibition and the appearance of the cleavage product suggest that the weak caspase-1–APIP binding may be an enzyme–substrate interaction and is likely not the mechanism of APIP inhibition of pyroptosis. Therefore, we explored possible indirect mechanisms by which APIP might inhibit pyroptosis.
APIP Regulates Pyroptosis Through Its Role in Methionine Salvage.
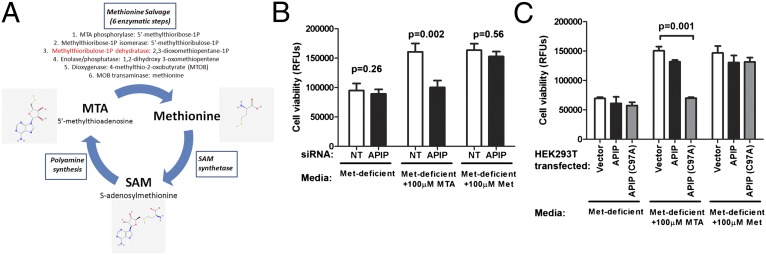
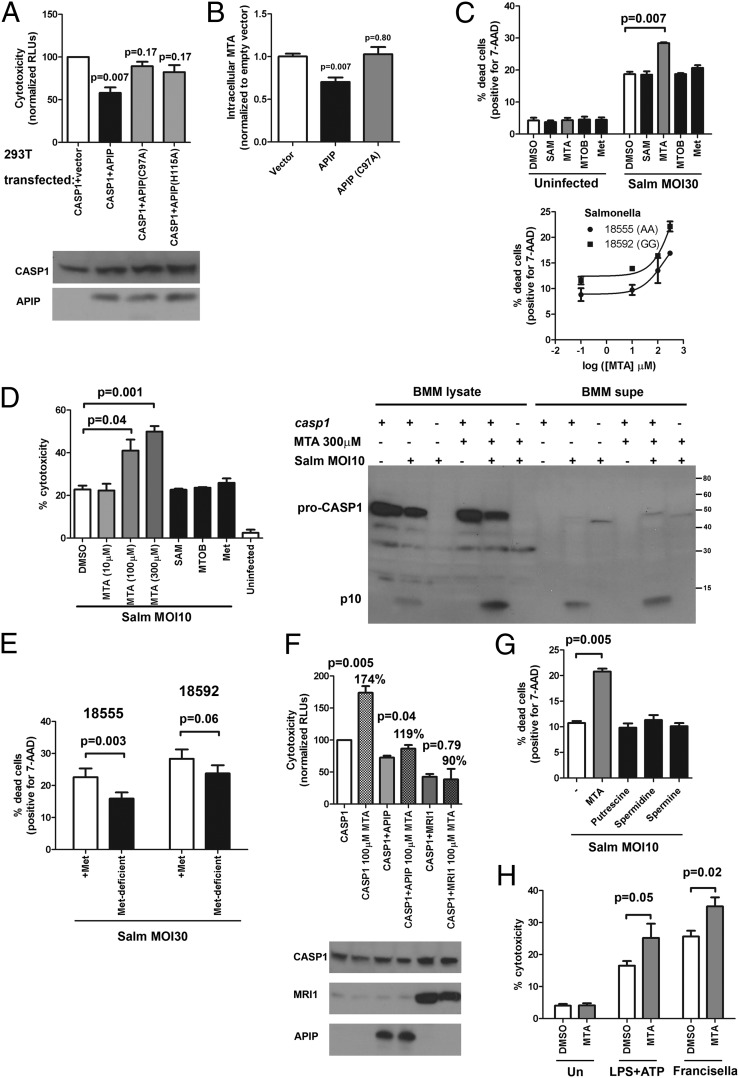
APIP is predicted to be a methylthioribulose-1-phosphate dehydratase, an enzyme in the methionine salvage pathway. The methionine salvage pathway is a highly conserved pathway for converting 5′-methylthioadenosine (MTA; Pubchem 439176), a byproduct of polyamine synthesis derived from S-adenosylmethionine (SAM; Pubchem 34755), back to methionine (Pubchem 6137) (Fig. 4A) (40). To determine if APIP was required for methionine salvage, viability was measured in cells provided either methionine or MTA as the source of methionine. Reduction of APIP expression caused no growth defect with methionine supplementation but decreased viability of cells supplemented with MTA (Fig. 4B). Furthermore, overexpression of a putative catalytic mutant of APIP (C97A) resulted in a dominant negative phenotype that rendered cells unable to use MTA as their source of methionine (Fig. 4C). This cysteine is conserved from yeast to humans, and the position in other class II aldolase family members (although often occupied by an acidic residue) is involved in a catalytic protonation/deprotonation step (41). Because class II aldolases function as dimers or tetramers (41), overexpression of the C97A mutant likely interferes with the endogenous APIP enzyme, resulting in the observed dominant negative phenotype. One well-studied member of this superfamily (class II fructose-1,6-bisphosphate aldolase) shows strong negative cooperativity among the subunits (42, 43). If APIP behaves similarly, substrate binding to a catalytically inactive subunit would decrease binding to the others, even if they were the WT polypeptide.
Fig. 4.
APIP is required for methionine salvage. (A) Metabolic cycle for methionine, SAM, and MTA. The major processes in the cycle are boxed in blue. The methionine salvage pathway converts MTA to methionine using the six indicated enzymatic steps, with APIP predicted to be the third step (highlighted in red). The product of each step of methionine salvage is listed after the enzyme. (B) Reducing APIP expression renders cells unable to use MTA as their source of methionine for growth. HEK-293T cells were treated with nontargeting (NT) or human APIP siRNA for 3 d and then grown in methionine-deficient media with the supplementation indicated. Cell viability was assessed with alamarBlue after 3 d. (C) Overexpressing a putative APIP catalytic mutant (C97A) renders cells unable to use MTA as their source of methionine for growth. HEK-293T cells were transfected, counted, and split the next day, and then, they were plated in methionine-deficient media with the supplementation indicated. Cell viability was assessed with alamarBlue after 48 h. For both B and C, mean, SEM, and P values for paired t tests are from four independent experiments.
Could altered methionine salvage be responsible for the effect of APIP on pyroptosis? The C97A mutant is unable to inhibit cell death induced by caspase-1 overexpression, although it still binds caspase-1 (Fig. 5A and Fig. S3E), indicating that overexpression of a caspase-1 binding partner or substrate alone is not responsible for APIP inhibition. This finding suggested that the enzymatic activity of APIP might be essential to its effect on cell death. Cells with varying amounts of APIP could have altered levels of intermediates in the methionine salvage pathway that might mediate the effect of APIP on pyroptosis. For example, yeast cells deleted for enzymes in the pathway (including the APIP ortholog) have been reported to accumulate MTA (44). We observed that overexpression of APIP decreased cellular MTA levels by 30% (P = 0.007), whereas the C97A mutant had no effect on MTA levels (Fig. 5B). Thus, varying the amount of APIP can affect the level of MTA and possibly, other cellular small molecules metabolized as a result of reactions in the methionine salvage pathway.
Fig. 5.
Methionine salvage regulates pyroptosis. (A) APIP mutants C97A and H115A are unable to inhibit cell death induced by caspase-1. HEK-293T cells were transfected and assessed for cytotoxicity after 23 h. (B) APIP overexpression reduces intracellular MTA. HEK-293T cells were transfected and extracted after 2 d, and relative MTA levels were quantified by liquid chromatography-coupled MS. (C) MTA increases cell death induced by Salmonella. LCL 12155 was treated with 300 μM SAM, MTA, MTOB, or methionine for 30 min before S. typhimurium infection. For MTA dose–response curves, LCL 18555 and 18592 were treated with the indicated MTA concentration beginning 30 min before infection. Cell death was measured at 2 h with 7-AAD. Mean, SEM, and P values are from two independent experiments. Data are representative of six different LCLs of varying rs514182 genotype. (D) MTA increases cell death and caspase-1 cleavage induced by S. typhimurium in BMMs. BMMs were treated with the indicated concentration of MTA or 300 μM SAM, MTOB, or methionine for 30 min before infection, and cytotoxicity was measured after 1 h. Immunoblotting of lysates and supernatants from infected BMMs revealed that MTA increased the intensity of the p10 caspase-1 autocleavage product. The p10 band was not observed in BMMs from caspase-1 −/− mice. (E) Methionine-deprivation attenuates pyroptosis in LCLs. LCL 18555 and 18592 were grown for 20 h in either methionine-deficient media or methionine-deficient media supplemented with 100 μM methionine before S. typhimurium infection. Cell death was measured at 2 h with 7-AAD. (F) MTA increases cell death induced by caspase-1 overexpression, but coexpression of APIP or MRI1 suppresses the effect. HEK-293T cells were transfected and assessed for cytotoxicity after 23 h. P values and percentages are for paired t tests comparing the same transfected plasmids ± MTA. (G) Polyamines have no effect on pyroptosis. LCL 18555 was treated with 300 μM MTA, putrescine, spermidine, or spermine beginning 30 min before S. typhimurium infection. Cell death was measured at 2 h with 7-AAD and baseline cell death subtracted. Mean, SEM, and P value are from two independent experiments. (H) MTA increases pyroptosis regardless of the inflammasome stimuli used. BMMs were stimulated overnight with either 1 μg/mL LPS (for LPS + ATP) or 20 ng/mL LPS (for Francisella or uninfected samples). Cells were treated with 300 μM MTA or DMSO for 30 min before infection with F. novicida U112 (MOI 30) or addition of 5 mM ATP. Cytotoxicity was measured after 5 h. For all panels, mean, SEM, and P values from paired t tests are from three to five independent experiments, except where indicated.
If MTA were mediating the effect of APIP on pyroptosis, then adding MTA to cells should have the same effect as decreasing APIP expression. Exogenous addition of MTA resulted in a dose-dependent increase in cell death in response to S. typhimurium (but not baseline viability) in both LCLs and mouse BMMs, with the BMMs also showing a clear increase in caspase-1 cleavage (Fig. 5 C and D). In contrast, no effect was seen with three other small molecules involved with methionine salvage: SAM, 4-methylthio-2-oxobutyrate (MTOB; the final precursor before conversion to methionine; Pubchem 473), and methionine itself. Although addition of MTA increased pyroptosis, decreasing MTA through growth of cells in methionine-deficient media had the predicted opposite effect in attenuating pyroptosis (Fig. 5E). Although growth of cells in methionine-deficient media reduced MTA levels to near the mass spectrometric detection limit, this treatment certainly could have other metabolic effects that could alter inflammatory cell death. Although exogenous addition of MTA results in more than a doubling in pyroptosis in BMMs, the effect of MTA depletion is more modest (and in line with the effect of APIP RNAi on pyroptosis and the effect of APIP overexpression on human caspase-1–mediated cell death). The moderate effects support the idea that MTA is not a central component of caspase-1 signaling but likely, an accessory regulator that takes part in the complex interplay between metabolism and immunity. Exogenous MTA also increased cell death because of caspase-1 overexpression, and interestingly, this effect was partially suppressed by coexpression of APIP or MRI1 (methylthioribose-1-phosphate isomerase; the enzyme that precedes APIP in the methionine salvage pathway) (Fig. 5F), consistent with these enzymes increasing flux through the pathway and thereby, reducing intracellular MTA levels. Our data support a model where APIP functions in methionine salvage, and alterations in metabolites, such as MTA, then regulate host response.
How might MTA regulate pyroptosis? One hypothesis is that it may be an indirect effect mediated by polyamines, because MTA is a known regulator of their synthesis (45). However, no effect on pyroptosis was noted with addition of polyamines (Fig. 5G). The fact that both MTA and APIP overexpression have an effect in the 293 overexpression assay (where no inflammasome components are present) suggests that MTA does not act on inflammasome activation or assembly. Consistent with this finding, MTA increases cell death in BMMs, regardless of the stimuli used for caspase-1 activation. Cell death in response to LPS + ATP [which activates caspase-1 through NLRP3 (NLR family, pyrin domain containing 3)] (13) and Francisella infection [which activates through AIM2 (absent in melanoma 2)] (46, 47) was increased by MTA (Fig. 5H). We hypothesize that MTA may directly or indirectly enhance caspase-1 activity. One possible mechanism under investigation is that MTA might regulate the redox state of the caspase-1 reactive site cysteine. This cysteine must be in a reduced state for proteolytic activity (48), and we speculate that MTA (which differs from adenosine only by the presence of sulfur and a methyl group) might aid in keeping the cysteine reduced. Although our data clearly support a model where APIP affects methionine salvage and levels of MTA, which subsequently regulate pyroptosis, other possible molecular mechanisms could explain the effect of MTA in modulating caspase-1 activity.
Although the inhibition of pyroptosis by APIP seems to be mediated by its role in methionine salvage, the mechanism involved in apoptosis is more complex. MTA did not alter apoptosis in response to staurosporine in LCLs (Fig. 6A), and the APIP C97A mutant was still able to inhibit cell death induced by caspase-9 overexpression (Fig. 6B). In contrast, mutating one of the putative zinc binding residues (H115A) of APIP resulted in a protein that is unable to inhibit both caspase-1 and -9 cell death (Figs. 5A and 6B). Zinc binding is required for catalysis of other class II aldolase family members; however, it also seems to play a structural role, because crystallization of proteins in this family requires the presence of zinc (49). Thus, although both mutants likely affect the function of APIP in methionine salvage, we suspect that H115A is also affecting protein conformation or stability in a way that renders APIP unable to inhibit cysteine-aspartic protease 9 (CASP9)-mediated cell death. Others have reported that MTA can both inhibit and increase apoptosis depending on the cell type (50, 51). Published studies showing APIP inhibition through binding to APAF-1 (31) and regulation of protein phosphorylation (36) provide alternative mechanisms where APIP could regulate apoptosis independent of its role in methionine salvage. For the regulation of apoptosis by APIP, multiple mechanisms are likely involved with their relative importance determined by cell type, metabolic state, and specific stimuli.
Fig. 6.
APIP inhibition of apoptosis is mechanistically distinct from pyroptosis. (A) MTA does not alter cell death in response to staurosporine in LCLs. LCL 18555 and 18592 were treated with the indicated MTA concentration beginning 30 min before staurosporine treatment (0.5 μM). Cell death was measured at 7 h with 7-AAD. Mean, SEM, and P values are from two independent experiments. (B) APIP mutant H115A is unable to inhibit cell death induced by caspase-9, but C97A retains partial function. HEK-293T cells were transfected and assessed for cytotoxicity after 23 h. Mean, SEM, and P values for paired t tests comparing caspase-1 + empty vector vs. each APIP plasmid are from three independent experiments.
APIP Variation and Human Health.
Some of the measured in vitro effects of APIP and MTA seem modest, consistent with methionine salvage playing an accessory role in regulation of pyroptosis. However, small differences can still have significant consequences on human health, particularly when combined with multiple other genetic variants that contribute to a quantitative trait.
There are two broad areas of human health that might be affected by variation in APIP inhibition of apoptosis and pyroptosis. (i) APIP variation may modulate apoptosis-induced response and toxicity from treatment with chemotherapeutic agents such as carboplatin. Genetic variants that reduce APIP expression might predispose to toxicity, whereas somatic mutations in tumors could modify chemotherapeutic response. (ii) Proinflammatory conditions, such as systemic inflammatory response syndrome (SIRS), that are likely to involve caspase-1 may also be regulated by genetic variation in APIP. Indeed, critically ill individuals meeting SIRS criteria carrying the procell death APIP allele (the derived G allele of rs514182 associated with lower APIP expression and greater pyroptosis) had one-half the odds of exhibiting a positive blood culture (odds ratio = 0.49, P = 0.01) and one-half the odds of dying during hospitalization (odds ratio = 0.44, P = 0.005) (Table 1). These findings are consistent with a stronger caspase-1 response being beneficial for controlling infection, although this association will need to be replicated in future studies. Recent studies in mice have shown that pyroptosis is a means of exposing intracellular bacteria to oxidative killing by neutrophils (17), and it may be that individuals with the high pyroptosis APIP allele are more efficient at bacterial clearance because of a similar mechanism. In contrast, MTA has been shown in mice to protect against endotoxic shock from LPS injection by dampening the inflammatory response, although caspase-1 was not examined in the study (52). Differences between LPS and live pathogens may help explain the apparent discrepancy. Furthermore, because it has recently been shown that resistance to LPS-induced shock in CASP1 KO mice is actually caused by a disruption of the neighboring CASP11 gene (53), the effect of MTA in LPS-induced shock vs. bacterial infection may be complex and depend on which inflammatory caspases are activated. Additional experiments exploring the effects of MTA and APIP in mice exposed to LPS vs. Salmonellae will be required to clarify the complex roles of APIP and methionine salvage during infection.
Table 1.
Association of rs514182 with SIRS-associated outcomes
| AA | AG | GG (low APIP and high cell death) | OR (95% CI)* | P value† | |
| In-hospital mortality | |||||
| Died (n = 128) | 18 (14.1%) | 48 (37.5%) | 62 (48.4%) | 0.44 (0.25–0.78) | 0.005 |
| Survived (n = 776) | 51 (6.6%) | 289 (37.2%) | 436 (56.2%) | ||
| Blood cultures | |||||
| Positive (n = 149) | 19 (12.8%) | 45 (30.2%) | 85 (57.1%) | 0.49 (0.28–0.85) | 0.01 |
| Negative (n = 755) | 50 (6.6%) | 292 (38.7%) | 413 (54.7%) |
Genotypes are relative to the + strand of chromosome 11. APIP coding sequence is on the − strand.
*Odds ratios (ORs) are for a G allele-dominant model [odds (GG + GA)/odds (AA)], with 95% confidence intervals (CIs) in parentheses. The G allele is associated with reduced odds of death and blood culture positivity.
†P values are from multiple logistic regression (adjusted for age and sex) with a G allele-dominant model.
One possible explanation for why methionine salvage would regulate pyroptosis is that MTA might serve as a signal for the nutritional status of the host for sulfur-containing amino acids. During infection and other proinflammatory states, there is increased methionine use to support leukocyte proliferation and synthesis of acute phase proteins and polyamines (54). We would predict that this increase would lead to higher levels of MTA and flux through the methionine salvage pathway, which could support a more robust pyroptotic response. Later in an infection (or throughout in a malnourished patient), depletion of methionine would decrease cell proliferation, and levels of MTA would drop, reducing pyroptosis. Such a metabolic switch for caspase-1 dampening could, perhaps, be a means of reducing tissue damage later in an infection or during starvation. Furthermore, severe sepsis may be a setting in which the inhibition by APIP of two central cell death pathways could play a role in survival and organ dysfunction. An infection could lead to pathogen activation of the caspase-1 pathway along with later activation of the intrinsic apoptotic pathway because of hypoxia in tissues during sepsis. In fact, APIP expression is known to be induced in hypoxic tissue (31), and APIP-induced expression could serve as a defense against tissue damage, although this defense might come at a price of less control of bacteremia. Hypoxia and inflammation also both play a role in cystic fibrosis, and a recent study found SNPs near APIP associated with lung disease severity (Fig. S4) (55).
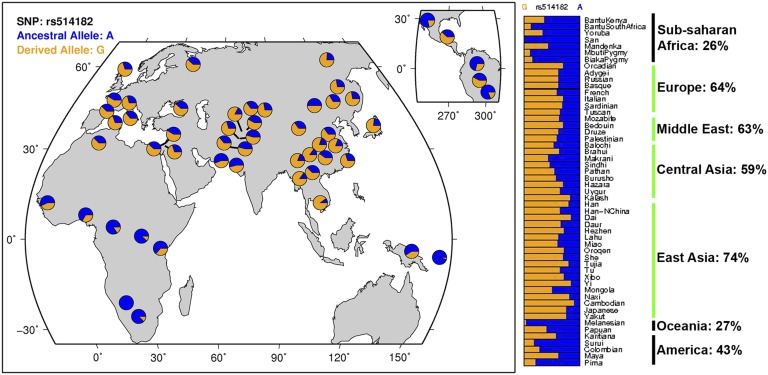
The association of rs514812 with mortality in SIRS patients suggests that there may be a survival advantage conferred by the procell death APIP allele. Therefore, variation in pyroptosis may have served as a substrate for adaptive human evolution. The fitness advantage of a more robust caspase-1 response would have become most apparent within the past 11,000 y, when development of agriculture and animal domestication allowed for increased population densities necessary for the rise of crowd epidemic diseases (56). These diseases include typhoid fever (a systemic illness caused by S. typhi) but also, numerous other activators of caspase-1 (57), including influenza, pertussis, and bubonic plague, that represent some of the greatest causes of death in human history. Strong natural selection for a more robust caspase-1 response to combat these infections would have led to an increase in frequency of the procell death APIP allele. However, this selection would not have been uniform across the globe. The development of agriculture occurred earlier in Europe and Asia (as early as 11,000 y ago) compared with sub-Saharan Africa and the Americas (∼4,000 y ago) (58). Areas of the world that acquired agriculture earlier and with more animal domestication should have higher frequencies for alleles that confer a stronger inflammatory response. Remarkably, this pattern is observed for the APIP SNP (Fig. 7) as well as the CARD8 variant that we previously characterized (rs2043211) (Fig. S5). Quantification of the correlation between SNP and geography (59) yields a Bayes factor of 125 for rs514182, which would place it in the top 0.7% of all Human Genome Diversity Project SNPs for this specific geographic pattern. Additionally, a truncation allele of a third caspase-1 inhibitor, CASP12, confers a threefold survival advantage in sepsis (60) and has reached near fixation in most non-African populations (61). Thus, alleles for reduced function or expression of three different direct and indirect caspase-1 inhibitors seem to have undergone stronger natural selection on continents that underwent earlier and more extensive agricultural acquisition. This idea of natural selection working through loss-of-function mutations is an extension of Maynard Olson’s “less-is-more hypothesis” (62). Not only can increased fitness occur through gene loss as described in the hypothesis, but a more potent response can be the outcome if the loss-of-function mutations are in inhibitors. Thus, the caspase-1 cell death response seems to be a trait that has undergone polygenic selection, with loss of function of inhibitors resulting in a stronger response beneficial against infection.
Fig. 7.
Population differentiation of rs514182 suggests adaptive evolution of caspase-1 response. The map and bar graphs display the geographic distribution of imputed APIP rs514182 allele frequencies from the HGDP Selection Browser (74, 75). Allele frequencies for the derived procell death allele in each continent are summarized to the right of bar graphs, with earlier adopters of agriculture shown in green. The observed pattern of higher derived allele frequencies in Europe and Asia vs. sub-Saharan Africa, the Americas, and Oceania may be a signature of selection acting on standing variation in Europe and Asia after settlement of the Americas.
In conclusion, our work shows that biological discoveries and clinically relevant associations can be found from modestly sized cell-based association studies when coupled with extensive experimental follow-up studies. Future screens at the cellular level are likely to be productive for understanding basic biology, human variation, and evolution.
Materials and Methods
Cells.
HapMap LCLs were purchased from the Coriell Institute. Cells were maintained at 37 °C in a 5% CO2 atmosphere in RPMI 1640 media (Invitrogen) supplemented with 10% FBS or 15% for the carboplatin screen, 2 mM glutamine, 100 U/mL penicillin-G, and 100 mg/mL streptomycin. BMMs were prepared from the femurs of C57BL/6 mice and casp1−/− (10) mice by culture with 85% supplemented RPMI + 15% L929-cell supernatant. HEK-293T cells were grown in DMEM supplemented with 10% FBS, 1 mM glutamine, 100 U/mL penicillin-G, and 100 mg/mL streptomycin.
Salmonella Infection Screen and Association Analysis.
Assaying LCLs for Salmonellae-induced cell death was conducted as previously described using S. typhimurium 14028s tagged with an inducible GFP plasmid (18). Briefly, overnight bacterial cultures were subcultured 1:33 and grown for 2 h and 40 min at 37 °C. Bacterial invasion was conducted for 1 h at a multiplicity of infection (MOI) of 30 followed by gentamicin (50 μg/mL) treatment. Cell death was assessed at 3.5 h after initial infection by staining with 7-aminoactinomycin D (7-AAD; Biomol) and measuring on a Guava Easycyte Plus flow cytometer (Millipore). Each LCL was assayed on three sequential passages.
In addition to the previously screened 173 CEU and YRI LCLs, we assayed 89 CEU and 90 YRI LCLs from HapMap phase 3 and the 90 ASN LCLs from HapMap phase 2. Genome-wide association analysis was conducted with PLINK (63) using HapMap phases 2 (2.9 million SNPs) and 3 (1.4 million SNPs) genotypes. For CEU and YRI trios, family-based association was conducted with QFAM parents (family-based association tests for quantitative traits), whereas analysis of unrelated ASN LCLs was carried out with basic association. In either case, adaptive permutation was used with default settings. No SNPs reached genome-wide significance after correcting for multiple hypothesis testing; however, P values were not used to strictly define statistically significant associations but as a tool for narrowing down candidate SNPs for additional experimental analysis. R2 values for the contribution of rs514182 to pyroptosis variation are from ANOVA of unrelated individuals. Q-Q plots were generated using an R script from ref. 64. Phenotypic and association data are available using GWAS Analyzer (http://go.nwrce.org/cgi-bin/variation/home.cgi) (65). Descriptive statistics were performed with GraphPad Prism 5. Linkage disequilibrium was examined with HaploView (66).
Carboplatin Screen and Association Analysis.
Cell growth inhibition was assessed using alamarBlue (BioSource International). Briefly, 1 × 104 cells in 100 μL were plated in 96-well round-bottom plates. After 24 h, treatment was started by adding 100 μL 20 μM carboplatin and continued for 72 h. Experiments were conducted at least two times in triplicate; 20 μL alamarBlue were added to each well 24 h before colorimetric detection to determine the percent cell growth inhibition relative to control according to the manufacturer’s protocol. Absorbance at 570 and 600 nM was recorded using a Synergy HT plate reader (BioTek).
SNP genotypes were from the International HapMap database (release 24). More than 2 million SNPs with minor allele frequency >5% and no Mendelian inheritance transmission errors were used. Percent cell survival was log2-transformed to achieve normality. P values for genotype–phenotype associations were obtained using the Quantitative Trait Disequilibrium Test (67) with covariates for sex (20) and HapMap panel (67, 68).
Cell Death Assays.
Three different assays of cell death/viability were used: 7-AAD, Cytotox-Glo (Pierce), and alamarBlue (Invitrogen). For LCLs, cells were stained with the membrane-impermeable nuclear dye 7-AAD (1.3 μg/mL), and the fractions living and dead were determined by flow cytometry. To confirm the effect of APIP RNAi in LCLs and also, measure cell death in adherent cells (BMMs and HEK-293Ts), Cytotox-Glo was used: 25 μL supernatant was added to 25 μL PBS and 25 μL AAF-Glo (alanyl-alanylphenylalanyl-aminoluciferin) substrate, mixed, and incubated for 15 min before measurement of luminescence with a Victor3 V Plate Reader (Perkin-Elmer). Cell death with this assay for BMMs was reported as either relative light units after subtraction of background reading from untreated cells or percent cytotoxicity in cases where cells were subsequently lysed by the addition of digitonin to obtain a 100% reading. For HEK-293T overexpression experiments, cytotoxicity measurements were background-subtracted (cells transfected with empty vector) and expressed as a percentage relative to caspase-1 + empty vector set to 100 to obtain normalized relative light units. Finally, carboplatin treatment of HEK-293Ts results in visible membrane blebbing/cell death but minimal release of intracellular proteins, and therefore, cell viability was measured by alamarBlue assay; 10× alamarBlue was added to cells, and fluorescence was measured after 3 h. Background (media only) was subtracted, and cell death was calculated relative to untreated cells. alamarBlue was also used for cell viability measurement for HEK-293Ts grown in methionine-deficient media and reported as background-subtracted relative fluorescence units.
Inhibitor and Methionine Experiments.
Caspase inhibitors (Ac-YVAD-FMK and Ac-DEVD-FMK; Calbiochem) and methionine pathway compounds (MTA, SAM, MTOB, methionine; Sigma) were added to cells at the indicated concentration for 30 min before infection or drug treatment. For methionine-deprivation of LCLs, cells were grown for 20 h in methionine-deficient RPMI (Invitrogen) (+10% FBS) before infection assay. LCLs cell death was assessed either by 7-AAD staining for S. typhimurium at 3 h postinfection or Guava Nexin Staining (Millipore) at 42 h for carboplatin treatment.
RNAi Experiments.
LCLs (2 × 105 cells) or BMMs (1 × 105 cells) were treated for 3 d in 500 μL Accell media (Dharmacon) with either nontargeting Accell siRNA #1 or a SmartPool directed against either human or mouse APIP (1 μM total siRNA; Dharmacon). Before infection, LCLs were plated at 1 × 105 in 100 μL RPMI complete media (without antibiotics) in 96-well plates, whereas BMMs were assayed in the same 24-well plates used for RNAi treatment in 500 μL RPMI complete media (without antibiotics). Salmonella infections were conducted as described above. Staurosporine (Biomol) was added at indicated concentrations. Cell death was assessed after 3 h for S. typhimurium and 7 h for staurosporine with 7-AAD.
HEK293Ts (1 × 105 cells) were treated for 3 d in 500 μL Accell media (Dharmacon) with either nontargeting Accell siRNA #1 or a SmartPool directed against human APIP (1 μM total siRNA; Dharmacon). Cells were trypsinized, counted, and replated in DMEM complete with indicated concentrations of drugs or RPMI complete without methionine (with 100 μM MTA or methionine as indicated). Viability was assessed with alamarBlue.
HEK-293T Overexpression Experiments.
HEK-293T cells were plated at 4 × 105 cells in six-well plates and transfected using Fugene6 according to the manufacturer’s instructions (Roche). Plasmids used included human APIP (Open Biosystems), human caspase-1-flag (variant-α; Origene), human caspase-9 (variant-α; Origene), human caspase-8 (variant C; Origene), mouse caspase-1 (Open Biosystems), human ASC (Open Biosystems), human NLRC4 (Open Biosystems), and mouse MRI1 (Open Biosystems). For assessing inhibition of caspase-induced cytotoxicity, 1 μg total plasmid was used for each transfection, with 400 ng caspase plasmid and 300 ng APIP plasmid; the remainder was made up with empty vector. If MTA was included in the experiment, 100 μM was added just before transfection; 23 h posttransfection, cytotoxicity was measured using Cytotox-Glo. Phase microscopy images were taken at 20× using a Nikon TS100. Expression levels were assessed by Western blot for human caspase-1 (1:200, SC-515; Santa Cruz Biotechnology), caspase-9 (1:1,000, C9; Cell Signaling Technology), caspase-8 (1:1,000, D35G2; Cell Signaling Technology), APIP (1:200, d-20; Santa Cruz Biotechnology), mouse caspase-1 (1:200, SC-514; Santa Cruz Biotechnology), ASC (1:1,000; Millipore), HA (hemagglutinin; 1:200, Y-11; Santa Cruz Biotechnology), and MRI1 (1:200; Santa Cruz Biotechnology).
Stimulation of BMMs with LPS + ATP and Francisella.
BMMs (50,000) in 96-well plates were prestimulated overnight with either 1 μg/mL LPS (for LPS + ATP) or 20 ng/mL LPS (for Francisella). Cells were treated with 300 μM MTA or DMSO for 30 min before the addition of 5 mM ATP or infection with F. novicida U112 (MOI30). F. novicida was spun onto cells for 10 min at 730 × g, and infection was allowed to proceed for 2 h before the addition of gentamicin (50 μg/mL). Cytotoxicity was measured by Cytotox-Glo 5 h after the addition of LPS or Francisella.
Immunoprecipitation.
HEK293T cells were plated at 4 × 106 in 10-cm dishes and transfected using Fugene6; 22 h posttransfection, lysates were collected in 50 mM Tris⋅HCl, pH 7.4, 1% Triton-X100, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and minicomplete protease inhibitor tablet (Roche). Immunoprecipitation was conducted using antiflag magnetic beads (Sigma) 1 h at 4 °C. Beads were washed three times and eluted with 3xFLAG peptide (Sigma).
Localization of APIP in BMMs.
BMMs were stained with fluorescein amidite (FAM)-YVAD-fmk (caspase-1 FLICA; Immunochemistry Technologies) beginning 5 min before infection with S. typhimurium at an MOI of 30. Cells were fixed after 1 h with 4% paraformaldehyde for 20 min. Blocking and permeabilization were carried out for 30 min with 0.2% saponin and 5% normal donkey serum in PBS. Primary antibody staining was carried out against APIP (1:200 d-20 or G14; Santa Cruz Biotechnology) and ASC (1:500; Millipore).The epitope for the d-20 antibody is within amino acids 50–100, whereas G14 is between 1 and 50. Both antibodies displayed similar enrichment in inflammasome foci with S. typhimurium infection. Several other antibodies displayed no enrichment in inflammasome foci, including: anti-lysosomal-associated membrane protein 2 (LAMP2), anti-protein disulfide isomerase (PDI), anti–pericentriolar material 1 (PCM-1), anti-perixosomal membrane protein 70 (PMP70), and anti-GAPDH. Secondary antibodies conjugated to Alexa Fluor 555 and Alexa Fluor 647 (Molecular Probes) were used for staining. Imaging was done with a Nikon TE2000 inverted fluorescence microscope with a 60× oil objective and NIS-Elements Software.
APIP Protein Purification and in Vitro Assays.
His6-tagged APIP was overexpressed by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to cultures of BL21(λDE3) in late log phase containing a pET-based expression vector for 10–12 h at 20°. Cells were harvested, resuspended in PBS, and lysed in a French pressure cell. The lysate was clarified by centrifugation, and the supernatant (containing soluble APIP) was loaded on a 5 mL HiTrap Chelating HP column (GE Healthcare). The column was washed with PBS containing 25 and 50 mM and developed with 300 mM imidazole. APIP fractions were dialyzed against PBS.
Measurement of caspase-1 activity was carried out using purified caspase-1 and YVAD-AFC (7-amido-4-trifluoromethylcoumarin) substrate (BioVision) in 100 mM Hepes, pH 7.5, 10% sucrose, 0.1% CHAPS, and 10 mM DTT as described (69). Cleavage of YVAD-AFC was monitored using excitation at 370 nM and emission at 480 nM on an EnVision Multilabel Plate Reader (Perkin-Elmer). Varying conditions, including caspase-1, substrate, EDTA, DTT, NaCl, phosphate, and glycerol concentrations, did not affect the inability of APIP to inhibit caspase-1. We did note that the addition of the Zinc2+ chelator TPEN (Sigma) resulted in some inhibition of caspase-1 activity. However, a size exclusion column (Microcon YM-10; Amicon) showed that the inhibitory activity was all in the small molecular weight fraction. We suspect that it is because of inhibition of caspase-1 by Zn2+ previously bound by APIP, because divalent cations are well-characterized caspase inhibitors (70). We also carried out assays of caspase-1 autocleavage using cells extracts from THP-1s (71), but we found no effect of recombinant APIP or HEK-293T APIP overexpression lysates.
MTA Quantification.
HEK-293T cells were transfected as described above in six-well plates. After 48 h, cells were rinsed with PBS, trypsinized, washed again with PBS, and counted by flow cytometry. Cell pellets were frozen in liquid nitrogen.
Extraction was carried out in methanol as described (72). Frozen cell pellets were extracted by the addition of 600 μL HPLC grade methanol (Acros) and allowed to thaw on ice. Cells were dispersed by pipetting, vortexed, and flash frozen in liquid nitrogen three times. Pellets were collected by centrifugation for 5 min at 9,000 × g at 4°. The supernatant was transferred to a glass vial for subsequent MS analysis. Pellets were washed with 200 μL MeOH and centrifuged as above, and the supernatant was combined and placed into glass inserts for autosampler vials.
Samples (5 μL) were injected by a Waters Acquity UPLC SDS system into a Thermo Hypersil Gold PFP column (2.1 × 100, 1.9 u). Retention of MTA was achieved at a flow rate of 0.3 mL/min using mobile phase A, consisting of 0.1% acetic acid in water, and mobile phase B, consisting of 0.1% acetic acid in methanol. A three-step gradient used started at 0% B for 1 min, continued at 0–100% B over the next 2 min, was held at 100% B for 1 min, returned to starting conditions in 0.1 min, and was allowed to reequilibrate for an additional 2 min. The column was interfaced with a standard electrospray ionization source on a Waters Premier-XE tandem mass spectrometer. Positive electrospray ionization mode was used. Data were obtained using multiple reaction monitoring using the ion transitions 298.1 > 136.1. Instrumental settings are as follows: cone voltage = 25V, collision energy = 25, capillary voltage = 3.5 kV, extractor = 5, desolvation temperature = 350 °C, and source temperature = 120 °C. Issues with carryover from injection to injection were alleviated by using 500 μL 100% isopropanol as a weak needle washing solution and 500 μL 50:50 acetonitrile:methanol as a strong needle wash solution. MTA peak area was divided by the number of cells in each sample and normalized to cells transfected with empty vector in each experiment.
Clinical Study of SIRS.
The SIRS cohort was enrolled from the intensive care units of Harborview Medical Center (Seattle, WA). Inclusion criteria included admission to an intensive care unit for more than 24 h and presence of three of four criteria for SIRS (73). Exclusions included admission for major trauma, chronic treatment with antiinflammatory medications, history of cancer, massive transfusion, presence of an advanced directive against resuscitation, and an age of <18 or >90 y. All subjects were followed for the presence of SIRS criteria and the development of a microbiologically documented or clinically suspected infection. These studies were approved by the Division of Human Subjects Research, University of Washington (Seattle, WA). Genotyping for rs514182 was conducted with a Taqman-based assay (Applied Biosystems). We limited our analyses to subjects of European descent to minimize confounding because of population-specific allele frequencies. Genotype frequencies indicated no significant deviation from Hardy–Weinberg equilibrium (P = 0.26). To identify associations between APIP genotypes and clinical outcomes in SIRS patients, we used multiple logistic regression adjusted for age (quartiles) and sex using Stata SE 11 (StataCorp).
Evolutionary Analysis.
Maps, bar graphs, and genotype frequencies for rs514182 and rs2043211 were downloaded from the Human Genome Diversity Project (HGDP) Selection Browser (74–76). Quantification of the correlation between allele frequency and geographic distribution (Europe and Asia vs. sub-Saharan Africa, the Americas, and Oceania) was conducted using bayenv (59). The covariance matrix used was based on 2,331 autosomal HGDP SNPs from ref. 77. For each SNP (rs514182, rs2043211, and the 640,704 HGDP SNPs), 30,000 runs and a seed value of −69,213 were used (similar results were obtained regardless of the seed value). Although the allele frequencies for rs514182 were imputed, they are in general agreement with the directly genotyped HapMap phase 3 panel (78).
Supplementary Material
Acknowledgments
We thank Joe Pickrell for supplying the imputed HGDP genotype frequencies for all HapMap SNPs. This project was funded by National Institute of Allergy and Infectious Diseases Grant U54 AI057141 (to the Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research). D.C.K. was funded as a Wyeth Fellow of the Life Sciences Research Foundation. E.R.G., A.L.S., and M.E.D. were supported by National Institute of General Medical Sciences Grant UO1GM61393.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13886.
See Author Summary on page 13900 (volume 109, number 35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206701109/-/DCSupplemental.
References
- 1.Vannberg FO, Chapman SJ, Hill AV. Human genetic susceptibility to intracellular pathogens. Immunol Rev. 2011;240:105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 2.Wong SH, et al. Leprosy and the adaptation of human toll-like receptor 1. PLoS Pathog. 2010;6:e1000979. doi: 10.1371/journal.ppat.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SH, Hill AV, Vannberg FO. India-Africa-United Kingdom Leprosy Genetics Consortium Genomewide association study of leprosy. N Engl J Med. 2010;362:1446–1447. doi: 10.1056/NEJMc1001451. [DOI] [PubMed] [Google Scholar]
- 4.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 5.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shrestha S, et al. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS. 2009;23:673–677. doi: 10.1097/QAD.0b013e328325d414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thye T, et al. African TB Genetics Consortium Wellcome Trust Case Control Consortium Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davila S, et al. International Meningococcal Genetics Consortium Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet. 2010;42:772–776. doi: 10.1038/ng.640. [DOI] [PubMed] [Google Scholar]
- 9.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 11.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 12.Villani AC, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 14.Savic S, Dickie LJ, Battellino M, McDermott MF. Familial Mediterranean fever and related periodic fever syndromes/autoinflammatory diseases. Curr Opin Rheumatol. 2012;24:103–112. doi: 10.1097/BOR.0b013e32834dd2d5. [DOI] [PubMed] [Google Scholar]
- 15.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko DC, et al. A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease. Am J Hum Genet. 2009;85:214–227. doi: 10.1016/j.ajhg.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang RS, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeuillet C, et al. In vitro whole-genome analysis identifies a susceptibility locus for HIV-1. PLoS Biol. 2008;6:e32. doi: 10.1371/journal.pbio.0060032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martchenko M, Candille SI, Tang H, Cohen SN. Human genetic variation altering anthrax toxin sensitivity. Proc Natl Acad Sci USA. 2012;109:2972–2977. doi: 10.1073/pnas.1121006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razmara M, et al. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 25.Fontalba A, et al. Deficiency of the NF-kappaB inhibitor caspase activating and recruitment domain 8 in patients with rheumatoid arthritis is associated with disease severity. J Immunol. 2007;179:4867–4873. doi: 10.4049/jimmunol.179.7.4867. [DOI] [PubMed] [Google Scholar]
- 26.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeller T, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho DH, et al. Induced inhibition of ischemic/hypoxic injury by APIP, a novel Apaf-1-interacting protein. J Biol Chem. 2004;279:39942–39950. doi: 10.1074/jbc.M405747200. [DOI] [PubMed] [Google Scholar]
- 32.Huang RS, et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin Cancer Res. 2011;17:5490–5500. doi: 10.1158/1078-0432.CCR-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh S, Chhipa RR, Vijayakumar MV, Bhat MK. DNA damaging drugs-induced down-regulation of Bcl-2 is essential for induction of apoptosis in high-risk HPV-positive HEp-2 and KB cells. Cancer Lett. 2006;236:213–221. doi: 10.1016/j.canlet.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 34.Itoh M, Chiba H, Noutomi T, Takada E, Mizuguchi J. Cleavage of Bax-alpha and Bcl-x(L) during carboplatin-mediated apoptosis in squamous cell carcinoma cell line. Oral Oncol. 2000;36:277–285. doi: 10.1016/s1368-8375(00)00010-5. [DOI] [PubMed] [Google Scholar]
- 35.Voth DE, Howe D, Heinzen RA. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun. 2007;75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho DH, et al. Suppression of hypoxic cell death by APIP-induced sustained activation of AKT and ERK1/2. Oncogene. 2007;26:2809–2814. doi: 10.1038/sj.onc.1210080. [DOI] [PubMed] [Google Scholar]
- 37.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 38.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broz P, et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life. 2009;61:1132–1142. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- 41.Ashida H, Saito Y, Kojima C, Yokota A. Enzymatic characterization of 5-methylthioribulose-1-phosphate dehydratase of the methionine salvage pathway in Bacillus subtilis. Biosci Biotechnol Biochem. 2008;72:959–967. doi: 10.1271/bbb.70651. [DOI] [PubMed] [Google Scholar]
- 42.De Montigny C, Sygusch J. Functional characterization of an extreme thermophilic class II fructose-1,6-bisphosphate aldolase. Eur J Biochem. 1996;241:243–248. doi: 10.1111/j.1432-1033.1996.0243t.x. [DOI] [PubMed] [Google Scholar]
- 43.Sauvé V, Sygusch J. Molecular cloning, expression, purification, and characterization of fructose-1,6-bisphosphate aldolase from Thermus aquaticus. Protein Expr Purif. 2001;21:293–302. doi: 10.1006/prep.2000.1380. [DOI] [PubMed] [Google Scholar]
- 44.Pirkov I, Norbeck J, Gustafsson L, Albers E. A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. FEBS J. 2008;275:4111–4120. doi: 10.1111/j.1742-4658.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- 45.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 49.Hall DR, et al. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J Mol Biol. 1999;287:383–394. doi: 10.1006/jmbi.1999.2609. [DOI] [PubMed] [Google Scholar]
- 50.Li TW, et al. S-Adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol Pharmacol. 2009;76:192–200. doi: 10.1124/mol.108.054411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ansorena E, et al. S-adenosylmethionine and methylthioadenosine are antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells. Hepatology. 2002;35:274–280. doi: 10.1053/jhep.2002.30419. [DOI] [PubMed] [Google Scholar]
- 52.Hevia H, et al. 5′-methylthioadenosine modulates the inflammatory response to endotoxin in mice and in rat hepatocytes. Hepatology. 2004;39:1088–1098. doi: 10.1002/hep.20154. [DOI] [PubMed] [Google Scholar]
- 53.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 54.Grimble RF, Grimble GK. Immunonutrition: Role of sulfur amino acids, related amino acids, and polyamines. Nutrition. 1998;14:605–610. doi: 10.1016/s0899-9007(98)80041-5. [DOI] [PubMed] [Google Scholar]
- 55.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamkanfi M, Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol. 2011;187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- 58.Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: Origins. New York: Garland; 2004. [Google Scholar]
- 59.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saleh M, et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 61.Xue Y, et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 65.Fong C, et al. GWAS analyzer: Integrating genotype, phenotype and public annotation data for genome-wide association study analysis. Bioinformatics. 2010;26:560–564. doi: 10.1093/bioinformatics/btp714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 67.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stark AL, et al. Population differences in the rate of proliferation of international HapMap cell lines. Am J Hum Genet. 2010;87:829–833. doi: 10.1016/j.ajhg.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Calvo M, et al. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 70.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 71.Kahlenberg JM, Dubyak GR. Differing caspase-1 activation states in monocyte versus macrophage models of IL-1beta processing and release. J Leukoc Biol. 2004;76:676–684. doi: 10.1189/jlb.0404221. [DOI] [PubMed] [Google Scholar]
- 72.Stevens AP, Dettmer K, Wallner S, Bosserhoff AK, Oefner PJ. Quantitative analysis of 5′-deoxy-5′-methylthioadenosine in melanoma cells by liquid chromatography-stable isotope ratio tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;876:123–128. doi: 10.1016/j.jchromb.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 73.Levy MM, et al. International Sepsis Definitions Conference 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med. 2003;29:530–538. doi: 10.1007/s00134-003-1662-x. [DOI] [PubMed] [Google Scholar]
- 74.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 78.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gamazon ER, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]