Abstract
The Wnt antagonist Sost has emerged as a key regulator of bone homeostasis through the modulation of Lrp4/5/6 Wnt coreceptors. In humans, lack of Sclerostin causes sclerosteosis and van Buchem (VB) disease, two generalized skeletal hyperostosis disorders that result from hyperactive Wnt signaling. Unlike sclerosteosis, VB patients lack SOST coding mutations but carry a homozygous 52 kb noncoding deletion that is essential for the transcriptional activation of SOST in bone. We recently identified a putative bone enhancer, ECR5, in the VB deletion region, and showed that the transcriptional activity of ECR5 is controlled by Mef2C transcription factor in vitro. Here we report that mice lacking ECR5 or Mef2C through Col1-Cre osteoblast/osteocyte-specific ablation result in high bone mass (HBM) due to elevated bone formation rates. We conclude that the absence of the Sost-specific long-range regulatory element ECR5 causes VB disease in rodents, and that Mef2C is the main transcription factor responsible for ECR5-dependent Sost transcriptional activation in the adult skeleton.
Keywords: osteocytes
Several rare genetic disorders that interfere with Wnt signaling have provided strong evidence that the “canonical” Wnt signaling pathway is critical in bone (1). The Wnt coreceptor LRP5 has been described as a modulator of bone mass where loss-of-function mutations cause osteoporosis-pseudoglioma syndrome (OPPG) (2), an autosomal recessive disorder characterized by low bone mass and skeletal fragility; conversely, gain-of-function Lrp5 alleles cause high bone mass (HBM) (3). Similar hyperactive osteoblast activity due to elevated Wnt signaling was observed when Sost, a secreted Wnt inhibitor, was mutated in knockout (KO) mice or in sclerosteosis patients who suffer from generalized hyperostosis (4–6). Lrp5 gene targeting or SOST overexpression in transgenic (TG) mice causes osteopenia (3, 7), whereas TG overexpression of G171V Lrp5 allelic variant causes HBM, similar to the Sost KO phenotypes (4, 8, 9). The recapitulation of the human phenotypes in mouse models supports the conclusion that canonical Wnt signaling plays a critical role in bone metabolism, and points to Sost and Lrp5 as key regulators of bone mass.
The skeletal phenotype describing sclerosteosis patients is similar to what has been documented for van Buchem (VB) disease. Although both VB and sclerosteosis map to the same locus on human chromosome 17 that includes the SOST transcript, the Sclerostin transcription unit was not affected in VB. All VB patients examined to date carry a 52-kb noncoding deletion, 35 kb downstream of SOST that results in the absence of postnatal SOST transcript and protein (10, 11). Although both sclerosteosis and VB are caused by sclerostin deficiency, the VB phenotype is a result of dysregulated SOST transcription. To identify the potential transcriptional regulatory elements responsible for Sost transcription in bone, we have characterized the expression of a human SOST transgene or an engineered allele corresponding to VB in mice. Only the wild-type (WT) SOST allele faithfully expressed SOST in the adult bone and thereby caused osteopenia, whereas the TG mice carrying the VB deletion allele were indistinguishable from WT (7). Cross-species sequence comparisons combined with in vitro and in vivo enhancer assays identified an evolutionarily conserved candidate enhancer element, termed ECR5, that drove reporter expression in UMR-106 cells and in the skeletal anlage of the murine embryo at embryonic day (E) 14.5 (7).
Despite wide interest in the manipulation of Sost as a skeletal anabolic therapy, only a modest amount of data has been generated describing the upstream regulatory pathways responsible for its transcription. We have shown in vitro that Sost transcription is controlled by both its proximal promoter and the distal enhancer, ECR5. Specifically, ECR5, which drives reporter gene expression in mature osteoblastic cells, mediates responsiveness to parathyroid hormone (PTH) (12) and TGF-β (13). PTH suppresses the activity of ECR5-luciferase constructs independently of the SOST promoter and repression occurs via a myocyte enhancer factor 2 (Mef2)-responsive element (12). Among the four Mef2 family members of transcription factors (Mef2A–D), only Mef2C and Mef2D were shown to be robustly expressed in mineralized bone (14), and we determined that Mef2C and Sost colocalize in UMR-106 cells and mouse osteocytes (12). The activity of ECR5 enhancer was increased by exogenous Mef2C and was inhibited by dominant-negative Mef2C cotransfection. Finally, siRNA-mediated knockdown of Mef2C–D significantly suppressed endogenous Sost expression (12). Together, these results identified Mef2C as a transcriptional regulatory protein in bone, and position it upstream of Sost as a critical transcriptional modulator of the ECR5 candidate enhancer to control bone-specific Sost expression.
In this study, we extend our in vitro results using TG and KO mice. We show that ECR5 is sufficient to drive TG reporter expression in osteocytes, in adult mice. We also show that targeted deletion of ECR5 reduces tissue-specific expression of Sost, increasing bone formation rates and causing HBM. As in VB patients, the hyperostosis in ECR5 KO mice (ECR5KO) is milder than observed in sclerosteosis patients and murine Sost KO models, suggesting that the 338-bp ECR5 noncoding deletion is sufficient to recapitulate the phenotypes observed in VB disease. Using a combination of Mef2C;Col1-Cre and Mef2C;Col1-Cre;ECR5-TG compound mice, we show that ECR5-mediated osteocyte expression of Sost is dependent on Mef2C. In addition, targeted ablation of Mef2C in osteoblasts and osteocytes also causes HBM, indicating that Mef2C is a critical regulatory protein in bone and controls Sost expression via ECR5, by functioning as a modulator of Wnt signaling and bone homeostasis. As such, the discovery and characterization of the Mef2C-Sclerostin transcriptional axis has important implications for the anabolic treatment of disorders in which bone loss is a significant component.
Results
ECR5 Drives Transgenic Reporter Expression in Osteoblasts and Osteocytes.
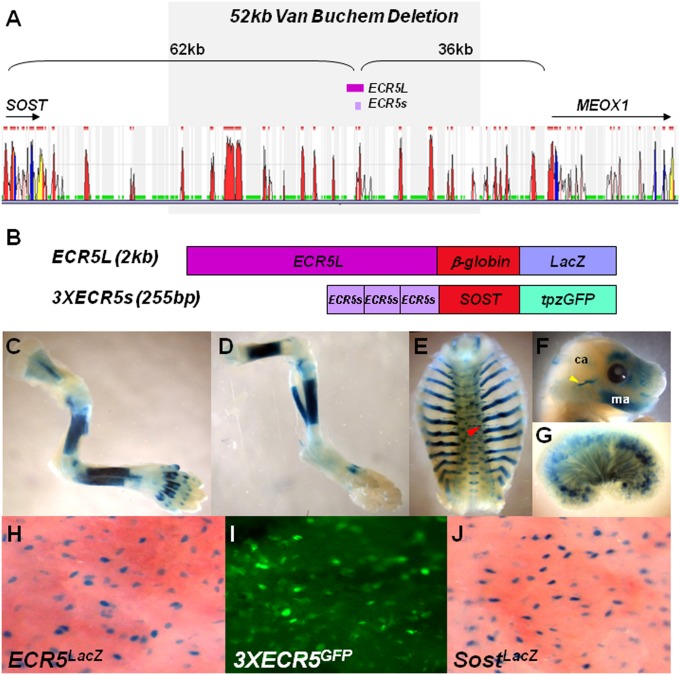
To determine whether ECR5 functions as a tissue-specific enhancer in vivo, we generated two ECR5 TG constructs (Fig. 1 A and B). ECR5L included a 2-kb ECR5 fragment upstream of the mouse β-globin minimal promoter driving β-galactosidase (LacZ) (Fig. 1B; ECR5LacZ). 3XECR5s included three tandem copies of a 255bp ECR5 fragment upstream of the 2kb SOST promoter driving topaz-green fluorescent protein (tpzGFP) (Fig. 1B; 3XECR5GFP). ECR5LacZ expression was compared to SostLacZ (LacZ replaced Sost in SostKO) and was found throughout the skeleton (Fig. 1 C–F). In a minority of ECR5LacZ lines, TG expression was also observed in the vasculature and the kidney (Fig. 1 E and G). We also observed kidney expression in 3XECR5GFP but no expression in the vasculature.
Fig. 1.
TG expression of ECR5. (A) Using multiple sequence alignment, a 255-bp element, ECR5, was identified 62 kb downstream of Sost transcriptional start site. (B) Two TG constructs were used to generate TG mice, ECR5L and 3XECR5. (C–F) ECR5L expressed LacZ in the entire mouse neonate skeleton. Here we show representative images of forelimbs (C), hindlimbs (D), ribs (E), and head (F). Lower expression was observed in the calvaria (ca) relative to the mandible (ma). Nonskeletal tissues expressing LacZ included the thoracic vasculature (red arrow) (E) and the kidney (G). At higher resolution, in the calvaria, ECR5L (H), 3XECR5 (I), and LacZ expressed from the endogenous mouse Sost locus (J), marked the osteocytes.
In neonatal calvaria, ECR5LacZ (Fig. 1H) and 3XECR5GFP (Fig. 1I) expression highlighted osteoblasts and osteocytes, and was indistinguishable from endogenous Sost expression (Fig. 1J). Both ECR5LacZ and 3XECR5GFP exhibited highly similar skeletal expression pattern in all lines examined (five ECR5LacZ and two 3XECR5GFP lines), with varying degrees of expression intensity. No dramatic differences were noted between ECR5LacZ and 3XECR5GFP expression in the neonatal skeleton, with the exception that 3XECR5GFP also expressed in hypertrophic chondrocytes. Previously, we also observed TG expression in the hypertrophic chondrocytes of SOST TG mice, suggesting that this expression may be specific to the human promoter. These findings allowed us to conclude that the 255-bp ECR5 element is sufficient to drive osteoblast/osteocyte-specific expression in neonatal mice, independently of the SOST promoter. In the adult mice, 3XECR5GFP expression was more robust in osteocytes than ECR5LacZ, suggesting that the SOST promoter is also required for high levels of osteocyte-specific expression of Sost.
Mef2C is Required for ECR5-mediated Sost Expression in Osteocytes.
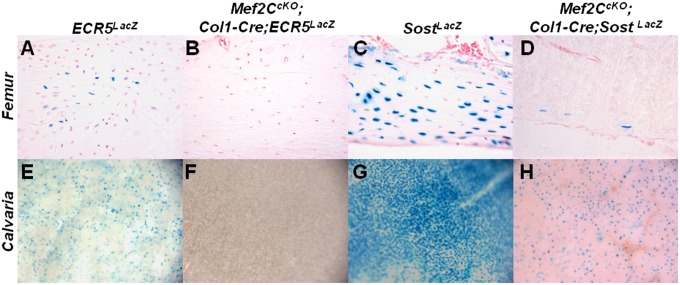
Comparative sequence and transcription factor binding site analysis predicted a Mef2 binding site within the ECR5 element, and this element was shown to be essential for ECR5 activity in vitro (12). To determine whether Mef2C is required for ECR5-mediated transcriptional activation of Sost in vivo, we compared LacZ expression from either the ECR5LacZ or the SostLacZ allele to that of animals where the Mef2C gene has also been removed in osteoblasts and osteocytes with a Col1-Cre TG (15). This Col1-Cre TG is expressed in both osseous and nonosseous tissues: embryonically in the head and limbs, and postnatally in the skeletal system, predominantly in osteoblasts and osteocytes (16). In femora and calvaria, the LacZ expression from ECR5LacZ was abolished in the absence of Mef2C (Fig. 2 A, B, E, and F). Similarly, in the femurs of SostLacZ mice, >95% of osteocytes positive for Sost did not express LacZ when Mef2C was deleted (Fig. 2 C and D). A significant reduction was also observed in calvaria; however, a larger number of Sost-positive cells were retained than in the femur (Fig. 2 G and H).
Fig. 2.
Mef2C is required for ECR5-dependent Sost expression, in vivo. ECR5LacZ transgenic mice that were also mutant for Mef2C did not express LacZ in the femur (B) or calvaria (F) relative to the ECR5LacZ controls (A and E). Similar results were obtained in Mef2CcKO; Col1-Cre; SostKO double knockout animals, where LacZ expression was dramatically reduced in the femur (D) and calvaria (H) relative to SostLacZ controls (C and G).
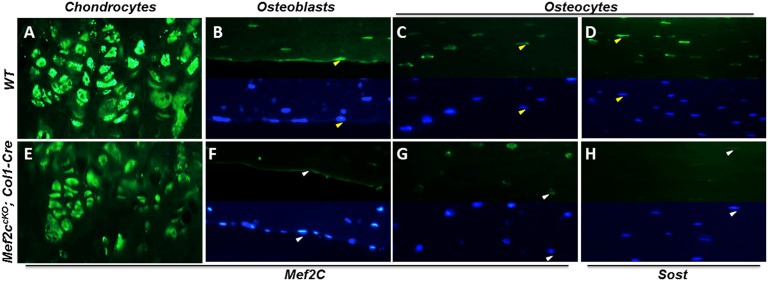
We also examined Mef2C and Sost expression in cortical bone of WT and Mef2CcKO; Col1-Cre. We found Mef2C mRNA levels to be reduced by ∼70% (Fig. S1A) and Sost by ∼50% (Fig. S1B) in Mef2CcKO;Col1-Cre relative to WT controls. Growth plate chondrocytes robustly expressed Mef2C protein in WT and the expression was unchanged in the Mef2CcKO;Col1-Cre (Fig. 3 A and E). Excision of Mef2C by Col1-Cre eliminated Mef2C protein expression in most osteoblasts and osteocytes relative to WT (Fig. 3 B, C, F, and G); particularly on the periosteal surface, all osteoblasts were negative for Mef2C in Mef2CcKO;Col1-Cre (Fig. 3F). The same bone regions that failed to express Mef2C down-regulated Sost expression, primarily in osteocytes (Fig. 3 D and H). These results support a model where ECR5 directs gene expression in osteocytes, and this ECR5-dependent expression requires Mef2C (Fig. 2 B and F). Similarly, Mef2C is required for the majority of mouse Sost expression in these cells (Fig. 2 D and H).
Fig. 3.
Mef2CcKO;Col1-Cre mice have reduced Mef2C and Sost bone expression. Mef2C protein expression is unchanged in chondrocytes (A and E). On the periosteal surface, most osteoblasts were positive for Mef2C in the WT (B; positive cells are marked by yellow arrows) and this expression was absent in Mef2CcKO;Col1-Cre (F; negative cells are marked by white arrows). Fewer cortical osteocytes expressed Mef2C in Mef2CcKO;Col1-Cre (G; white arrows point to Mef2C negative cells) relative to WT (C, yellow arrows point to Mef2C positive cells). A significant reduction in Sost-positive cortical osteocytes was also observed in Mef2CcKO;Col1-Cre (H; white arrows mark Sost negative cells) relative to WT control (D; yellow arrows mark Sost-positive cells). In B–D and F–H, Upper is the Mef2C protein signal in the green channel, and Lower is the same image showing the cell nuclei (blue channel/DAPI); arrows point to the same cells visualized in the green and blue channel.
ECR5KO Mice Have Increased Bone Mass.
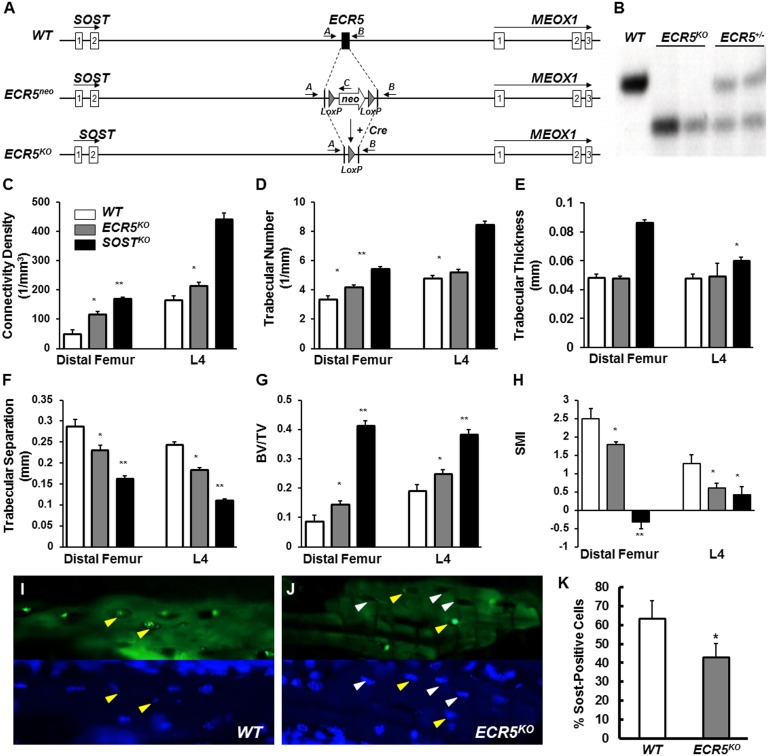
To determine whether deleting ECR5 causes VB, we made ECR5KO mice (Fig. 4 A–B). KO mice did not differ in size or weight from their same-sex control littermates, and no overt defects were noted at birth or throughout their lives. At 6 mo of age, microscale computed tomography (μCT) analysis of distal femur and lumbar area (L4) showed that trabecular bone volume fraction (BV/TV) in ECR5KO was 41% (P < 0.000001) higher than WT (Fig. 4G). ECR5KO had significantly higher connectivity density (Conn. D., Fig. 4C), trabecular number (Tb.N., Fig. 4D), and lower trabecular separation (Tb. Sp., Fig. 4F), and structure model index (SMI; Fig. 4H) compared with WT controls. In the trabecular bone compartment, a significant increase in osteoblast surface (Ob.S) and no significant change in osteoclast surface (Oc.S) was observed. We did observe a significant difference in osteoclast surface to bone surface ratio in both SostKO and ECR5KO mice.
Fig. 4.
ECR5KO mice have HBM due to reduced number of Sost-expressing osteocytes. ECR5 was replaced by a neo cassette, which was subsequently removed by Cre recombinase to generated ECR5KO mice (A). ECR5 targeting was confirmed by Southern blot (B). μCT analysis showed significant differences between ECR5KO mice and WT control littermates (C–H) consistent with ECR5KO mice having HBM. Fewer osteocytes expressed Sost in ECR5KO mice (L) compared with WT (I); (Upper) protein signal (green channel); (Lower) the same section visualizing nuclei with DAPI (blue channel). Quantitative assessment of the number of cortical osteocytes expressing Sost in ECR5KO relative to WT identified a ∼30% reduction in Sost-positive cells, in ECR5KO mice (K). Yellow arrows point to Sost-positive cells; white arrows point to Sost-negative osteocytes.
Bone formation (BFR/TV, Table 1) in ECR5KO mice was significantly increased for trabecular bone (more than twofold in ECR5KO relative to approximately threefold in SostKO) at the distal femur. Bone formation was also stimulated at the endocortical surface of the femur midshaft in SostKO, but not in ECR5KO mice, as demonstrated by femoral cortical bone morphology via μCT analysis: Bone area fraction (BA/TA 68.9%) was significantly increased in SostKO (P < 1 × 10−10), compared with 47.5% in ECR5KO and 46.2% in WT (Fig. 4E). Consistent results were obtained through histomorphometric characterization of the cancellous bone compartment at the distal femurs, supporting a HBM phenotype in ECR5KO mice due to elevated bone formation rates (Table 1). In humans, the hyperostosis phenotype in VB disease has been described to be significantly milder than those for sclerosteosis (10). Consistent with the human data, we found the ECR5KO mice to have a significant increase in bone mass (BV/TV, more than twofold, 104% increase, P < 0.001), which is less dramatic than in SostKO (more than threefold, 265% increase, P < 1 × 10−11); therefore, ECR5 contributes to the VB phenotypes found in the human disease.
Table 1.
Bone phenotying based on histomorphometric indices in the cancellous bone compartment of the distal femurs of 6-mo-old ECR5KO, SostKO, and Mef2CcKO;Col1-Cre male mice compared with WT controls
| Histomorphometric index | WT | ECR5KO | % change | SostKO | % change | WT | Mef2CcKO | % change |
| BV/TV | 7.79 ± 4.02 | 15.894 ± 5.833* | +104 | 28.45 ± 2.79* | +265 | 0.127 ± 0.02 | 0.201 ± 0.05* | +58 |
| BS/BV | 70.90 ± 20.54 | 48.86 ± 12.54* | −31 | 39.89 ± 4.82* | −44 | 56.72 ± 9.92 | 40.39 ± 7.68* | −29 |
| Tb.Dm (Plate) | 30.33 ± 8.39 | 43.82 ± 12.57* | +44 | 50.85 ± 6.77* | +68 | — | — | — |
| Tb.N (Plate) | 2.53 ± 1.16 | 4.26 ± 1.67* | +68 | 5.63 ± 0.94* | +122 | — | — | — |
| Tb.Sp (Plate) | 438.3 ± 211.7 | 222.6 ± 83.80* | −49 | 131.81 ± 29.82* | −70 | — | — | — |
| Tb.Dm (Rod) | — | — | — | — | — | 0.072 ± 0.010 | 0.097 ± 0.02* | +35 |
| Tb.N (Rod) | — | — | — | — | — | 5.652 ± 0.87 | 5.299 ± 0.81 | −6 |
| Tb.Sp (Rod) | — | — | — | — | — | 0.108 ± 0.02 | 0.086 ± 0.01* | −20 |
| Ob.S | 2.62 ± 1.11 | 5.956 ± 1.89* | +127 | 6.143 ± 1.68* | +134 | 1.402 ± 1.55 | 3.326 ± 4.40 | +137 |
| Ob.S/BS | 14.42 ± 3.93 | 17.730 ± 6.54* | +23 | 17.16 ± 2.62 | +19 | 0.130 ± 0.15 | 0.046 ± 0.02 | −65 |
| Oc.S | 1.64 ± 0.76 | 1.404 ± 0.364 | −14 | 1.623 ± 0.38 | −1 | 1.743 ± 1.25 | 1.219 ± 0.73 | −30 |
| Oc.S/BS | 8.92 ± 3.29 | 4.426 ± 1.87* | −50 | 4.608 ± 0.89* | −48 | 0.067 ± 0.04 | 0.049 ± 0.02 | −27 |
| MAR (μm/d) | 2.06 ± 0.36 | 2.634 ± 0.324* | +28 | 2.938 ± 0.165* | +42 | 0.981 ± 0.15 | 1.368 ± 0.33* | +39 |
| BFR/BS | 0.79 ± 0.19 | 1.188 ± 0.40* | +50 | 1.035 ± 0.28 | +31 | 0.073 ± 0.02 | 0.108 ± 0.13 | +48 |
| BFR/BV | 56.29 ± 23.36 | 59.89 ± 32.58 | +6 | 40.062 ± 7.96 | −29 | 3.027 ± 1.46 | 2.829 ± 1.66 | −7 |
| BFR/TV | 3.87 ± 1.41 | 8.371 ± 2.84* | +116 | 11.46 ± 2.57* | +196 | 0.306 ± 0.12 | 0.736 ± 0.22* | +141 |
Data represent mean ± SD for parameters measured. Group size n = 5–10. *P < 0.05. BFR, bone formation rate; BS, bone surface; BV, bone volume; MAR, mineral apposition rate; Ob.S, osteoblast surface; Oc.S, osteoclast surface; Tb.Dm, trabecular diameter; Tb.N, trabecular number; Tb.Sp, trabecular separation; TV, total volume; —, no measurements were conducted.
ECR5 Is Necessary for Robust Osteocyte Sost Expression in Mice.
Sost primarily marked periosteal/cortical and trabecular osteocytes, consistent with its known expression (Fig. S2). No Sost expression was detected in proliferating or hypertrophic chondrocytes. ECR5KO expressed Sost in a significantly lower percentage of osteocytes (Fig. 4J) relative to WT (Fig. 4I). To determine the number of osteocytes expressing Sclerostin ∼1000 matrix-embedded osteocytes were examined per genotype. Of the 1071 WT osteocytes, 693 (63.4%) expressed Sclerostin (Fig. 4 I and K). Of the 984 examined ECR5KO osteocytes, 425 (43.11%) were positive for Sclerostin (Fig. 4 K and J); 32% less positive (P < 0.02) ECR5KO osteocytes relative to WT. We found no changes in Sost expression in ECR5KO mice relative to WT in kidney, brain, or liver (Fig. S3). These data indicate that deleting ECR5 reduces the probability that an osteocyte will express high levels of Sost, which quantitatively reduces Sost expression by ∼50% (Fig. S1B). This reduction is sufficient to boost bone formation and cause HBM consistent with VB phenotypes.
Mef2C Inactivation in Bone Causes HBM Due to Elevated Bone Formation.
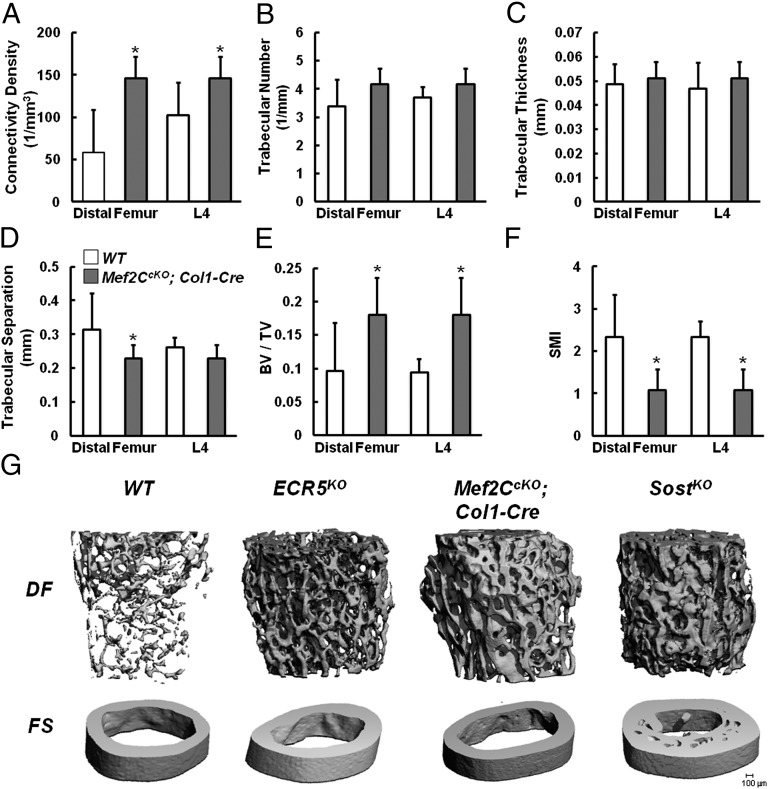
We next examined whether targeting Mef2C in osteoblasts/osteocytes phenocopies ECR5KO and causes HBM in mice. Mef2CcKO;Col1-Cre mice were born at normal Mendelian ratios and were indistinguishable from their control littermates at birth. After weaning, these mice were slightly smaller, and their tails displayed swelled protrusions due to increased ossification. Analysis of Mef2CcKO;Col1-Cre femora and lumbar vertebrae by μCT revealed that osteoblast inactivation of Mef2C results in HBM. In every parameter measured (Fig. 5 A–F), the absolute numbers were highly similar to those determined for ECR5KO. Mef2CcKO;Col1-Cre mice had significantly increased Connectivity Density (Fig. 5A), BV/TV (Fig. 5E), and decreased SMI (Fig. 5F) relative to WT controls, and similar to ECR5KO, did not display a significant difference in the trabecular thickness (Fig. 5C). BV/TV was increased ∼twofold in Mef2CcKO;Col1-Cre mice (Fig. 5E), highly similar to the ECR5KO (Fig. 4G).
Fig. 5.
Mef2CcKO;Col1-Cre mice have HBM. μCT analysis of Mef2CcKO;Col1-Cre mice showed significant differences between Mef2CcKO;Col1-Cre mice and WT control littermates (A–F) consistent with Mef2CcKO;Col1-Cre mice having HBM. Mef2CcKO;Col1-Cre mice had significantly increased connectivity density (A), bone volume/total volume ratio (BV/TV) (E), and significantly decreased trabecular separation in the distal femur (D) and SMI (F). No significant differences were observed in trabecular number (B) or thickness (C) or lumber trabecular separation (D). Quantitatively and qualitatively, the bones of Mef2CcKO;Col1-Cre mice had higher BMD than WT and less than SostKO and were indistinguishable from ECR5KO mice (G).
To determine the cellular mechanism by which the HBM phenotype is generated in Mef2CcKO;Col1-Cre mice, we examined the cancellous bone compartment of the distal femurs of 6-mo-old male mice using dynamic histomorphometry. Similar to ECR5KO and SostKO, Mef2CcKO;Col1-Cre mice also displayed a slight decrease in the osteoclast surface to bone surface ratio; however, this difference was not significant. We also observed a significant increase in the mineral apposition rate (MAR) and the bone formation rate (BFR/TV, Table 1), supporting that the removal of Mef2C in osteoblasts and osteocytes phenocopies VB (Fig. 5G). Recently, it has been reported that Mef2CcKO; Dmp1-Cre mice display HBM; however, this phenotype was shown to be driven primarily by a reduction in osteoclast activity (19). To further examine whether reduced osteoclast activity in Mef2CcKO;Col1-Cre contributes to the HBM phenotype, we measured CTX-1 and RANKL serum levels but found no significant differences between SostKO, ECR5KO, and Mef2CcKO;Col1-Cre mice and WT (Fig. S4). Furthermore, we examined the expression of activated β-catenin in the femurs of WT, SostKO, and Mef2CcKO;Col1-Cre mice and found significantly elevated levels of β-catenin in both SostKO and Mef2CcKO;Col1-Cre mice, consistent with an up-regulation of Wnt signaling as a shared mechanism for the HBM in these genetically distinct mice (Fig. S5).
Discussion
Until recently, members of the Mef2 family of transcriptional factors (Mef2A–D) have only been described as contributors to muscle and cardiovascular function (15, 20). In 2007, Arnold et al. reported that Mef2C functions as an early regulator of chondrocyte hypertrophy (14) and showed that Mef2C indirectly controls endochondral bone formation (14). They also reported that Mef2A, -C, and -D are expressed in the emerging endochondral bone, in the late embryonic spongiosa (14). Subsequently, we showed that Sost expression is modulated by PTH through a Mef2 responsive element, present in the distal ECR5 Sost enhancer (12). The results of the study presented here reveal the following insights into the molecular basis of transcriptional regulation of Sost in bone: (i) ECR5 is sufficient to drive tissue-specific expression in osteoblasts and osteocytes in vivo (Fig. 1J). (ii) SOST promoter is not required for neonatal expression of ECR5-dependent TGs, but is needed for high levels of TG expression in adult osteocytes (Fig. 2A vs. 2C and 2E vs. 2G). (iii) In the absence of ECR5 ∼30% less osteocytes express Sost (Fig. 5). (iv) ECR5KO mice have HBM due to elevated bone formation rates resembling VB phenotypes (Fig. 4 E–J). (v) Mef2C is important in osteocytes for the activity of ECR5-dependent Sost transcription (Fig. 2B, D, F, and H). (vi) Mef2C deletion from osteoblasts and osteocytes yields HBM and increased bone formation, and displays bone parameters that are similar to ECR5KO (Fig. 5 A–F), suggesting that Mef2C is the main transcriptional regulator of ECR5-dependent Sost expression.
Although several regulatory sequences have been characterized and shown to have tissue-specific activity in bone, such as Col1a1(3.6) (21, 22) and Dmp1 (23, 24) the regulatory sequences in these constructs were derived from promoter proximal regions only. ECR5 represents a distal enhancer that has osteoblast- and osteocyte-specific activity in vivo and is located ∼62 kb away from the Sost transcriptional start site. ECR5 is located in the VB region that is deleted from both alleles in VB, and the evidence we present indicates that the human phenotypes are, in part, dependent on the absence of ECR5 and its interaction with the SOST promoter; we also show that ECR5KO recapitulates VB in mice. Currently, there is only one other report of a distal transcriptional enhancer: the RankL distal regulatory enhancer (DCR) that affects bone mass. When deleted, DCR caused HBM due to reduced osteoclast activity, leading to a lower rate of bone remodeling similar to that observed in humans and mice with hypoparathyroidism (25, 26).
Because Sost is an inhibitor of an anabolic pathway, there is great interest in elucidating how Sost is regulated transcriptionally and posttranslationally, since modulating Sost levels has profound consequences downstream of Wnt signaling ranging from promoting bone formation (Sost deficiency) (4, 27) to accelerating bone loss (Sost overexpression) (7, 28), to enhancing bone repair (29). Here, we provide genetic evidence that Mef2C is important in bone for transcriptional activation of Sost in osteocytes, and hence functions as an upstream regulatory protein and modulator of Wnt signaling. In addition to Sfrp2/3 (19), Sost represents a Mef2C transcriptional target in bone; therefore, other Mef2C-dependent transcripts will likely be found in osteoblasts and osteocytes. It is also likely that, through its known corepressor partners, the type II HDAC proteins (14, 20), Mef2C may also serve as a key intermediary in the transmission of extracellular signals to the genome in bone. Further studies of Mef2C-Sost-Wnt signaling axis are likely to reveal basic mechanisms of bone formation and homeostasis, and it should be possible to modulate complex metabolic phenotypes through the manipulation of Sost and Mef2C activities in animal models.
Materials and Methods
Generation of ECR5KO, ECR5L, 3XECR5, and compound Mice.
A 338-bp ECR5 region (101782666-101782996; chr 11) was replaced with a floxed Neo cassette using Velocigene (30) and homologous recombination (31–33) in BAC RP23_252b10. The modified BAC was digested with SgrA1 and electroporated into ES line F1H4 (C57BL/6–129SvJ hybrid). Targeted ES cell clones were identified using Loss of Allele assay and verified by Southern blot. Two clones (VG1369B-C4/VG1369B-D2) were used to generate mice; both lines had the same HBM phenotype. ECR5L and 3XECR5 were PCR amplified from human genomic DNA using 5′-GCCAGTCTACTGCCATTGTCC-3′ 5′-GGGCAGAGATTTCTAGGGGTG-3′ and 5′-AATTCTAGCCACTCCCAGGCA-3′ 5′-AATTCGGCTCCCCCTCATGGCTGGT-3′ primer sets, respectively, and cloned into pCR2.1 vector. ECR5L was cloned into β-globin LacZ vector (gift from M. Nobrega, University of Chicago, Chicago, IL), and three copies of ECR5s were cloned in combination with the human SOST promoter we described (7) and topazGFP (gift from D. Rowe, University of Connecticut, Farmington, CT). Plasmid DNA was prepared and injected to generate TG mice as described (34). Pups were genotyped by PCR.
SostKO mice (Sosttm1(KOMP)Vlcg) were generated by a LacZ replacement of the entire Sost ORF. ECR5LacZ or SostKO mice were mated to the described Mef2CcKO (15) and Col1-Cre transgenic mice (16) to generate Mef2CcKO;Col1-Cre; ECR5LacZ and Mef2CcKO;Col1-Cre; SostKO mice. Genotyping was carried out by PCR. All animal experiments were carried out in accordance with guidelines set by the Institutional Animal Care and Use Committees at University of California, Berkeley, and Lawrence Livermore National Laboratory.
μCT and Histomorphometry.
Distal femurs, midfemoral cortical bone, and lumbar (L4) vertebrae were scanned using VivaCT-40 and μCT35 (Scanco Medical) with an isotropic voxel size at 10 or 6 μm. For the metaphysic trabecular bone in distal femur, transverse CT slices were evaluated in the region starting 0.1mm proximal to the growth plate and extending 2mm proximally. Trabecular bone was separated from cortical bone with manually drawn contour lines, and trabecular bone volume fraction (BV/TV, %), trabecular thickness (Tb.Th, mm), trabecular number (Tb.N, mm−1), trabecular separation (Tb.Sp, mm), connectivity density (ConnD, 1/mm3), and structure model index (SMI) were determined (SMI quantifies the characteristic form of a three-dimension structure as an index of plates or rods in the bone composition). For the femoral midshaft cortical bone, total cross-sectional area (bone plus bone marrow area) (TA, mm2), cortical bone area (BA, mm2), bone marrow area (MA, mm2), and bone area fraction (BA/TA, %) were calculated.
Bone Histomorphometry.
Bone histomorphometry was performed using semiautomatic image analysis (Bioquant Image Analysis) as described (36).
Sost-Positive Cell Scoring.
Two animals, 16 slide sections, 250 cells per scoring for up to ∼1,000 cells per genotype were examined. Slides were imaged, encompassing the entire mineralized area of the section. Cells were scored as positive if both DAPI and Sost immunostain were present, negative if DAPI was present without any evidence of immunostain. Percentages (% positive) were calculated for each replicate of 250 cells, and compared using Student’s paired t test. Data are expressed as an average % positive of all replicates + SD.
Supplementary Material
Acknowledgments
We thank Dr. Schwarz for providing the Mef2CcKO; and Deepa Murugesh, Cindy Thomas, and Salustra Urbin for technical assistance with animal breeding, tissue collection, and sample preparation. G.G.L. and N.M.C. were supported by NIH Grants HD47853 and DK075730; D.C.G. was supported by NIH Grants DK075730 and AR057547. This work performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207188109/-/DCSupplemental.
References
- 1.Yadav VK, Ducy P. Lrp5 and bone formation: A serotonin-dependent pathway. Ann N Y Acad Sci. 2010;1192:103–109. doi: 10.1111/j.1749-6632.2009.05312.x. [DOI] [PubMed] [Google Scholar]
- 2.Gong Y, et al. Osteoporosis-Pseudoglioma Syndrome Collaborative Group LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 3.Cui Y, et al. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, et al. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res. 2008;23:860–869. doi: 10.1359/jbmr.080216. [DOI] [PubMed] [Google Scholar]
- 5.Brunkow ME, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balemans W, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- 7.Loots GG, et al. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Wesenbeeck L, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babij P, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18:960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 10.Balemans W, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39:91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staehling-Hampton K, et al. A 52-kb deletion in the SOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am J Med Genet. 2002;110:144–152. doi: 10.1002/ajmg.10401. [DOI] [PubMed] [Google Scholar]
- 12.Leupin O, et al. Control of the SOST bone enhancer by PTH using MEF2 transcription factors. J Bone Miner Res. 2007;22:1957–1967. doi: 10.1359/jbmr.070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loots GG, et al. TGF-β regulates sclerostin expression via the ECR5 enhancer. Bone. 2012;50:663–669. doi: 10.1016/j.bone.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold MA, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 17.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner JC, et al. Bone mineral density in sclerosteosis; affected individuals and gene carriers. J Clin Endocrinol Metab. 2005;90:6392–6395. doi: 10.1210/jc.2005-1235. [DOI] [PubMed] [Google Scholar]
- 19.Kramer I, Baertschi S, Halleux C, Keller H, Kneissel M. Mef2c deletion in osteocytes results in increased bone mass. J Bone Miner Res. 2012;27:360–373. doi: 10.1002/jbmr.1492. [DOI] [PubMed] [Google Scholar]
- 20.Potthoff MJ, Olson EN. MEF2: A central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 21.Bogdanovic Z, et al. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J Bone Miner Res. 1994;9:285–292. doi: 10.1002/jbmr.5650090218. [DOI] [PubMed] [Google Scholar]
- 22.Zha L, et al. Collagen1alpha1 promoter drives the expression of Cre recombinase in osteoblasts of transgenic mice. J Genet Genomics. 2008;35:525–530. doi: 10.1016/S1673-8527(08)60072-7. [DOI] [PubMed] [Google Scholar]
- 23.Kalajzic I, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, et al. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galli C, et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer I, Loots GG, Studer A, Keller H, Kneissel M. Parathyroid hormone (PTH)-induced bone gain is blunted in SOST overexpressing and deficient mice. J Bone Miner Res. 2010;25:178–189. doi: 10.1359/jbmr.090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler DG, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ominsky MS, et al. Inhibition of sclerostin by monoclonal antibody enhances bone healing and improves bone density and strength of nonfractured bones. J Bone Miner Res. 2011;26:1012–1021. doi: 10.1002/jbmr.307. [DOI] [PubMed] [Google Scholar]
- 30.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 32.Narayanan K, Williamson R, Zhang Y, Stewart AF, Ioannou PA. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 1999;6:442–447. doi: 10.1038/sj.gt.3300901. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 34.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 35.Collette NM, Genetos DC, Murugesh D, Harland RM, Loots GG. Genetic evidence that SOST inhibits WNT signaling in the limb. Dev Biol. 2010;342:169–179. doi: 10.1016/j.ydbio.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ionova-Martin SS, et al. Changes in cortical bone response to high-fat diet from adolescence to adulthood in mice. Osteoporos Int. 2011;22:2283–2293. doi: 10.1007/s00198-010-1432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.