Abstract
In cells that exhibit apical growth, mechanisms that regulate cell polarity are crucial for determination of cellular shape and for the adaptation of growth to intrinsic and extrinsic cues. Broadly conserved pathways control cell polarity in eukaryotes, but less is known about polarly growing prokaryotes. An evolutionarily ancient form of apical growth is found in the filamentous bacteria Streptomyces, and is directed by a polarisome-like complex involving the essential protein DivIVA. We report here that this bacterial polarization machinery is regulated by a eukaryotic-type Ser/Thr protein kinase, AfsK, which localizes to hyphal tips and phosphorylates DivIVA. During normal growth, AfsK regulates hyphal branching by modulating branch-site selection and some aspect of the underlying polarisome-splitting mechanism that controls branching of Streptomyces hyphae. Further, AfsK is activated by signals generated by the arrest of cell wall synthesis and directly communicates this to the polarisome by hyperphosphorylating DivIVA. Induction of high levels of DivIVA phosphorylation by using a constitutively active mutant AfsK causes disassembly of apical polarisomes, followed by establishment of multiple hyphal branches elsewhere in the cell, revealing a profound impact of this kinase on growth polarity. The function of AfsK is reminiscent of the phoshorylation of polarity proteins and polarisome components by Ser/Thr protein kinases in eukaryotes.
Keywords: hyphal growth, protein phosphorylation, peptidoglycan, cytoskeleton, tip extension
How cells establish polarity is a fundamental question in developmental biology. It typically involves the initial deposition of a landmark protein at a cellular locus, followed by reinforcement of the polarization mark by assembly of larger multiprotein complexes. In eukaryotes, these complexes include broadly conserved proteins involved in the reorganization and polarization of the cytoskeleton and other cellular constituents (1, 2). Among the most pronounced cases of cell polarity are those in which growth or extension of the cell is targeted to a specific subcellular site, resulting in polar or apical growth. Important examples of polarized growth in eukaryotic cells include neuronal dendrites in animals, root hairs and pollen tubes in plants, bud emergence in Saccharomyces cerevisiae, the hyphal growth of filamentous fungi, and the elongation of fission yeast. However, in evolutionary terms, the most ancient forms of polarized growth are found in Bacteria (3, 4), most strikingly in the filamentous bacteria Streptomyces, which, in analogy to filamentous fungi, grow as branching hyphae and ramify into mycelial networks (5, 6).
The shape and integrity of Streptomyces hyphae are, like for most bacteria, maintained by the peptidoglycan cell wall, but the spatial control of cell wall assembly differs from other groups of bacteria. Conventional rod-shaped bacteria like Escherichia coli and Bacillus subtilis grow by intercalating new peptidoglycan along the lateral cell wall, and this cell elongation is orchestrated by a cytoskeleton formed by the actin-homologous MreB proteins, which interact directly with the cell wall biosynthetic machinery (reviewed in, e.g., refs. 7, 8). In sharp contrast, streptomycetes grow by tip extension and hyphal branching. This apical mode of growth is independent of MreB (9), and instead depends on the coiled-coil protein DivIVA, which is localized in large assemblies at growing hyphal tips (10). divIVA is essential for growth, and overexpression of divIVA is sufficient to trigger hyper-branching, showing that DivIVA is a key determinant of polarized growth in Streptomyces (10, 11). Together with the nonessential coiled-coil protein Scy (12), DivIVA forms an apical multiprotein complex, here termed the bacterial polarisome by analogy with the polarisome complex that directs cell polarity in yeasts and filamentous fungi (13).
It was previously demonstrated that small foci of DivIVA mark the sites of new branches before visible outgrowth occurs (11), although it remained unclear how such polarity marks are established (5). However, we have recently shown that the foci of DivIVA that trigger branching are primarily created by a unique polarisome splitting mechanism, in which the apical polarisome, visualized as a tip focus of DivIVA-EGFP, splits to deposit a small daughter polarisome, which is left behind on the lateral membrane as the tip grows away (14). Each daughter focus acts as a polarity mark, growing in size and ultimately initiating the outgrowth of a new hyphal tip. An obvious benefit of the polarisome splitting mechanism is that it appears to bypass kinetic barriers and other constraints that may be associated with de novo nucleation of new DivIVA clusters (5, 14–16). Intriguingly, a recent study showed that the polarisome scaffold protein SPA-2 in Neurospora crassa also exhibits tip-focus splitting behavior, in which small foci of SPA-2 detach from the main apical cluster, remain on the lateral wall, and mark the sites of new lateral branches (17). This observation suggests that branch site selection in filamentous fungi could be determined by a polarisome splitting mechanism similar to the one we have characterized in Streptomyces.
A polarisome splitting mechanism for branch-site selection may also facilitate regulation of cell polarity and hyphal branching. Hyphal morphology is dependent on growth conditions, and the ability to control tip extension and branching in response to internal and external cues should be of large adaptive value (18). Still, the regulation of polarized growth is very poorly understood in streptomycetes and fungi. Here, we show that cell polarity and branch-site selection in Streptomyces is regulated by a eukaryotic-type Ser/Thr protein kinase (STK) that is located at hyphal tips, directly targets the cell polarity determinant DivIVA, and affects the development of new daughter polarisomes and hyphal branches.
Despite the prominent roles of STKs in eukaryotic signal transduction, the importance of bacterial STKs was, for a long time, largely overlooked, and overshadowed by the histidine kinases that target response regulators in conventional bacterial two-component signal transduction systems (19). However, it is now clear, for example, from genomics and phosphoproteomics, that STKs are extensively used by bacteria in a variety of regulatory roles (reviewed recently in ref. 20). For example, in B. subtilis, the STK PrkC controls germination of spores in response to muropeptides released from bacterial cell walls, and, in Streptococcus pneumoniae, the STK StkP is involved in coordination of growth and cell division (21, 22). These two bacterial species have only two and one STKs, respectively, but the phylogenetic distribution of STKs among bacterial taxa is uneven; some groups encode only a few per genome whereas others have dozens—or, in some cases, even hundreds—of STK genes (23). The Actinobacteria are an ancient and deeply branching bacterial phylum in which STKs are particularly widespread and abundant. For example, Streptomyces coelicolor encodes at least 34 and Mycobacterium tuberculosis encodes 11 STKs (20, 24–26). In S. coelicolor, a recent phosphoproteomic survey detected at least 40 phosphoproteins (27), but the number of substrates is anticipated to be much larger, underlining the fundamental importance of actinobacterial STKs and the need for improved understanding of their substrates and biological functions. In this study, we show that the kinase AfsK, which has previously only been implicated in control of secondary metabolism (28), plays a vital role in regulating cell polarity, apical growth, and branch-site selection in Streptomyces, reminiscent of the involvement of Ser/Thr kinases in control of cell polarity in eukaryotes.
Results
DivIVA Phosphorylation Increases Dramatically When Cell Wall Synthesis Is Blocked.
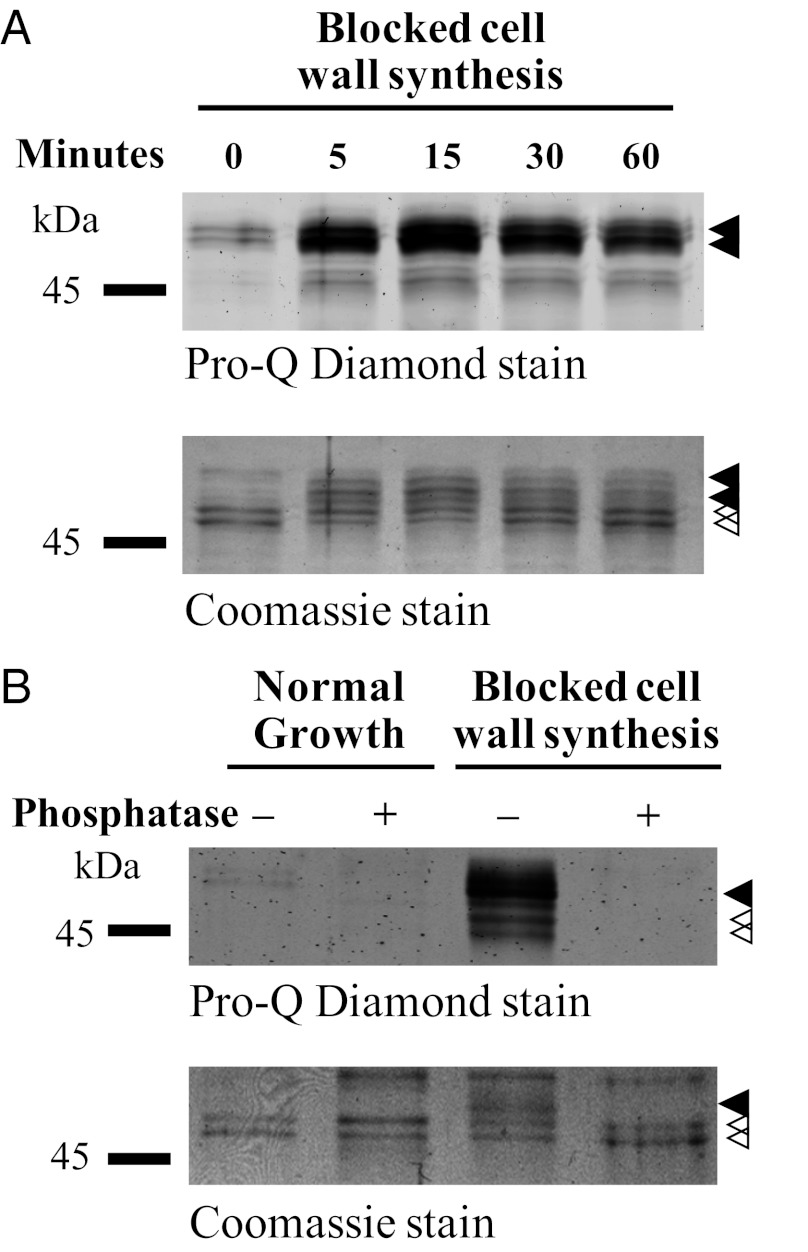
Our strategy to identify regulatory mechanisms that control apical growth in Streptomyces was to perturb the system by exposing growing S. coelicolor mycelium to different stress conditions and to monitor how the DivIVA protein responds. Western blot analysis revealed a clear mobility shift of DivIVA when cell wall synthesis was blocked by bacitracin, which arrests the export of the peptidoglycan precursor lipid II (29). One possible cause of this mobility shift could be a posttranslational modification of DivIVA, such as phosphorylation. To address this possibility, we introduced into the WT strain a thiostrepton-inducible divIVA allele encoding an N-terminally FLAG-tagged version of the protein, which is known to coimmunoprecipitate with the native DivIVA (30). This allowed analysis of immunoprecipitated FLAG-DivIVA/DivIVA mixtures from growing and bacitracin-treated mycelium by staining with the phosphorylation-specific stain Pro-Q Diamond. The presence of DivIVA with and without FLAG tag gives rise to a double band in the Coomassie-stained SDS/PAGE gel (Fig. 1A, open arrowheads). A weak signal from Pro-Q Diamond staining of more slowly migrating species suggested a low level of DivIVA phosphorylation during growth (Fig. 1A, closed arrowheads). Addition of bacitracin led, within 5 min, to phosphorylation of a large fraction of DivIVA, as shown by the relative amount of DivIVA that shifted mobility to the position coinciding with strong Pro-Q Diamond staining.
Fig. 1.
DivIVA is subject to phosphorylation. (A) Time course of DivIVA phosphorylation in response to the arrest of cell wall synthesis induced by bacitracin. Bacitracin (50 μg/mL) was added to growing cultures of WT S. coelicolor expressing FLAG-divIVA from a thiostrepton-inducible promoter. At the times indicated, cells were lysed, cell extracts were prepared, and FLAG-DivIVA/DivIVA was immunoprecipitated by using anti-FLAG affinity gel. (B) Phosphatase treatment of DivIVA. WT S. coelicolor expressing FLAG-divIVA was incubated with 50 μg/mL bacitracin for 60 min before harvest, preparation of cell extracts, and immunoprecipitation. The immunoprecipitated FLAG-DivIVA/DivIVA was analyzed before and after treatment with lambda protein phosphatase. Closed arrowheads indicate phosphorylated DivIVA and open arrowheads indicate nonphosphorylated DivIVA.
To confirm that the effect we observe was caused by phosphorylation, we treated immunoprecipitated FLAG-DivIVA/DivIVA from growing and bacitracin-treated mycelium with lambda protein phosphatase. This treatment abolished both the Pro-Q Diamond staining and the mobility shift (Fig. 1B). Next, we determined whether extensive phosphorylation is triggered only in response to bacitracin. We tested a number of different antibiotics inhibiting different steps in cell wall synthesis (vancomycin, phosphomycin, and penicillin G) and antibiotics inhibiting DNA and protein synthesis (novobiocin, kanamycin, and thiostrepton) and analyzed cell extracts by Western blotting. Our results showed that bacitracin and vancomycin induced a mobility shift of a large fraction of DivIVA, and that phosphomycin and penicillin G also caused some mobility shift (S. coelicolor is relatively insensitive to both phosphomycin and penicillin G; Fig. S1A). In contrast, the antibiotics that inhibit DNA and protein synthesis did not induce phosphorylation of DivIVA. These results demonstrate that DivIVA is indeed subject to phosphorylation, that there is a low but significant basal level of DivIVA phosphorylation (as detailed later) during undisturbed vegetative growth in liquid medium, and that DivIVA phosphorylation increases dramatically when cell wall synthesis is blocked.
Previous studies have shown that Streptomyces has a signal transduction system, the CseB/CseC-σE system, which is involved in sensing and responding to changes in the integrity of the cell envelope, and that inducers of this system include antibiotics that inhibit cell wall synthesis such as bacitracin and vancomycin (31). To test whether the CseB/CseC-σE system might be involved in mediating the increase in DivIVA phosphorylation observed when cell wall synthesis is blocked as part of this general cell envelope stress response, we analyzed immunoprecipitated DivIVA material from a sigE-null mutant. The results showed that bacitracin-induced DivIVA phosphorylation does not depend on the σE-mediated cell envelope stress response (Fig. S1B).
C-Terminal Region of DivIVA Is Target of Multiple Phosphorylations.
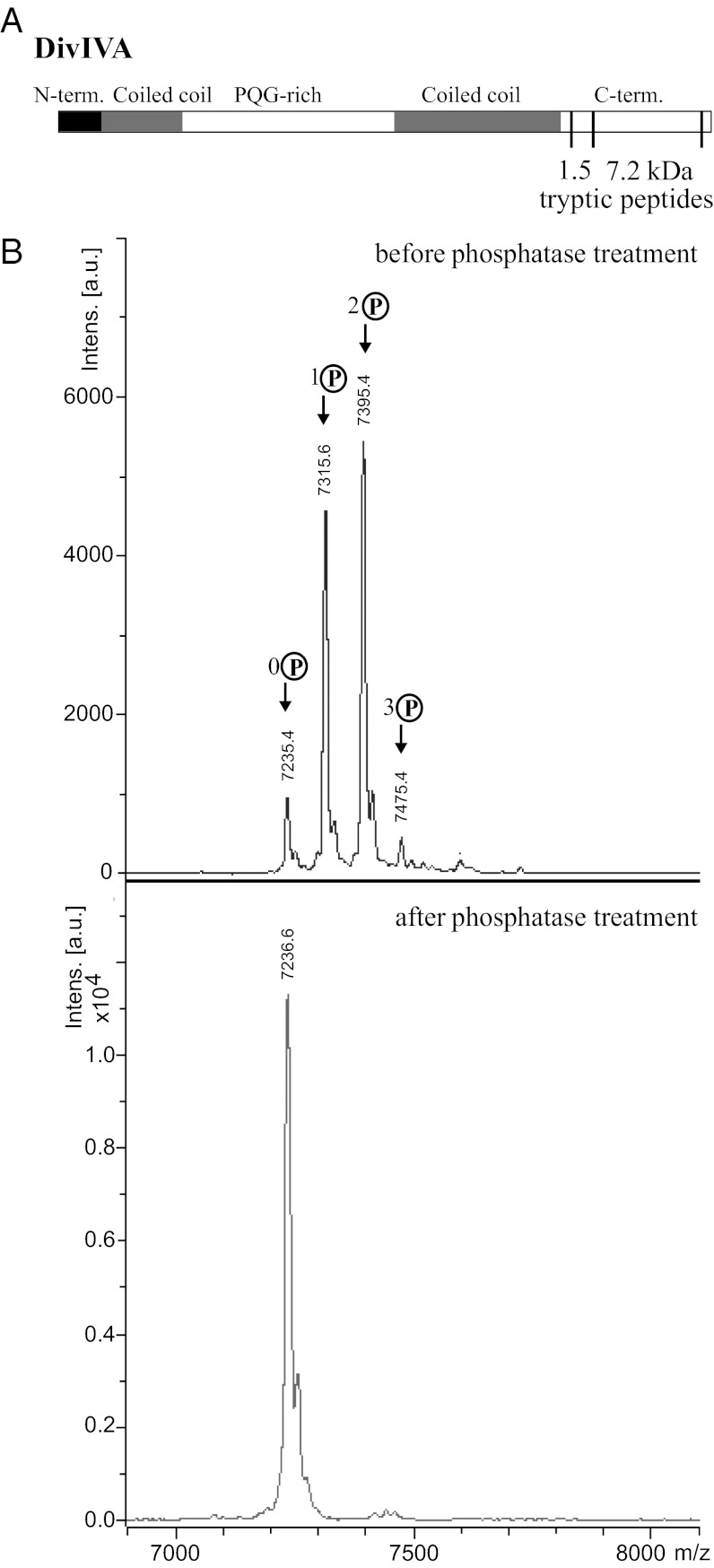
To confirm and extend our results, we used MS to further characterize the phosphorylation of DivIVA. DivIVA was immunoprecipitated from cultures that had been exposed to bacitracin to block cell wall synthesis, and the protein was digested with trypsin and analyzed by MALDI-TOF. A 7.2-kDa tryptic peptide that contains most of the C-terminal region of DivIVA was found to be singly, doubly, and triply phosphorylated, with the doubly phosphorylated species the most abundant (Fig. 2B). After treatment with calf intestinal alkaline protein phosphatase, the three peaks corresponding to the phosphorylated forms of DivIVA disappeared, leaving only the peak corresponding to the nonphosphorylated form (Fig. 2B). Further analysis showed that another DivIVA tryptic peptide was also multiply phosphorylated. This second peptide is 1.5 kDa in size and sits immediately N-terminal to the 7.2-kDa tryptic peptide in the primary amino acid sequence of DivIVA (Fig. 2A). Thus, the C-terminal region of DivIVA becomes highly phosphorylated in response to the inhibition of cell wall synthesis in S. coelicolor.
Fig. 2.
S. coelicolor DivIVA is multiply phosphorylated in the C-terminal region. (A) Schematic showing the positions within the DivIVA primary sequence of the 7.2-kDa phosphorylated peptide (residues 315–389) relative to the 1.5-kDa phosphorylated peptide (residues 301–314) described in the text. (B) Upper: MALDI mass spectrum of a 7.2-kDa tryptic fragment derived from the C-terminal region of DivIVA showing 0 to 3 phosphorylations (+80, +160, and +240 Da). Lower: Disappearance of the phosphorylated species upon treatment of the protein with calf intestinal alkaline phosphatase.
DivIVA Kinase Is AfsK.
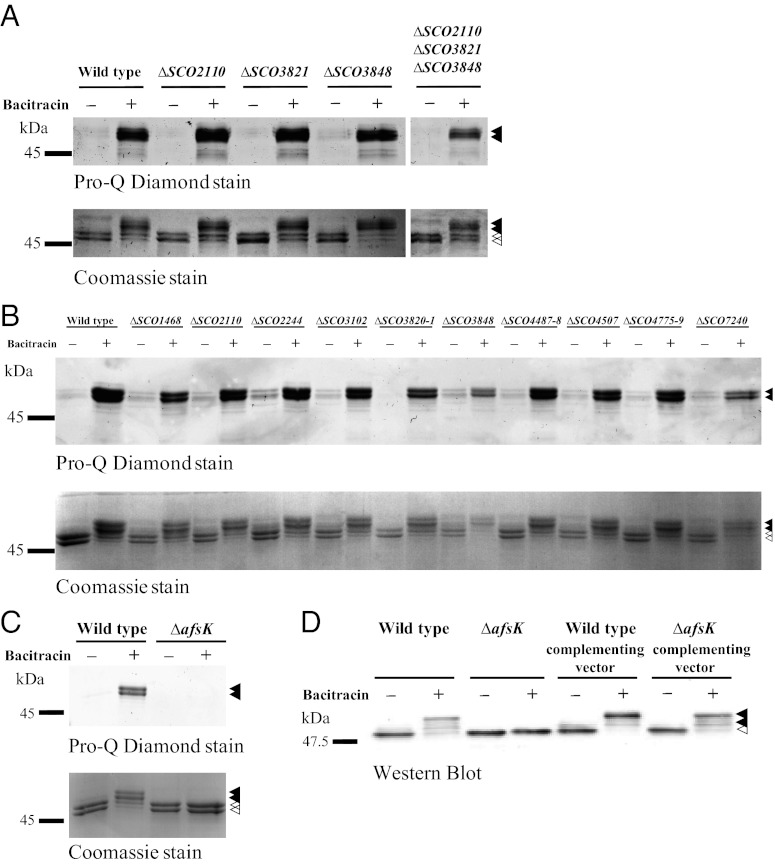
We next attempted to identify the kinase responsible for DivIVA phosphorylation. Multiple reports show that STKs carrying PASTA domains play important regulatory roles in Mycobacterium and Corynebacterium. PASTA domains are known to bind peptidoglycan components and β-lactam antibiotics (21, 32), and actinobacterial STKs carrying such domains (PknA and PknB) have been reported to phosphorylate several proteins involved in cell wall growth and cell division, including the mycobacterial DivIVA-orthologue Wag31 (e.g., refs. 24, 33–35). These reports prompted us to investigate the three PASTA domain-containing STKs in S. coelicolor (SCO2110, SCO3821, and SCO3848), of which SCO3848 shows microsynteny with mycobacterial pknB. Accordingly, we constructed SCO2110-, SCO3821-, and SCO3848-null mutants and found that both the basal level of DivIVA phosphorylation during growth and the strongly increased level seen after bacitracin treatment occurred normally in each of the three mutants (Fig. 3A). To rule out the possibility of redundancy, we constructed a triple mutant lacking all three of these kinases. Again, basal DivIVA phosphorylation during growth and the dramatic increase in phosphorylation caused by the inhibition of cell wall synthesis occurred normally, even in the absence of all three kinases (Fig. 3A). Thus, DivIVA phosphorylation in S. coelicolor is mediated by some route other than PknA/PknB-like PASTA domain-containing STKs.
Fig. 3.
The DivIVA kinase is AfsK. The phosphorylation state of DivIVA, before and after the inhibition of cell wall synthesis, was analyzed in (A) single, double, and triple mutants corresponding to three PASTA domain-containing STKs of S. coelicolor, SCO2110, SCO3821, and SCO3848, (B) 13 other constructed STK mutants, and (C) a constructed afsK mutant. Growing cultures of WT S. coelicolor and of the STK mutants, each expressing FLAG-divIVA, were incubated with 50 μg/mL bacitracin for 30 min before harvest, preparation of cell extracts, and immunoprecipitation of FLAG-DivIVA/DivIVA. (D) Complementation of the afsK-null mutant restores DivIVA phosphorylation. afsK was cloned into the integrative vector pMS82 to create pKF256, which was introduced into the afsK null mutant and into WT S. coelicolor. The phosphorylation state of DivIVA, before and after the inhibition of cell wall synthesis, was analyzed in each strain by Western blot analysis of crude cell extracts. Closed arrowheads indicate phosphorylated DivIVA and open arrowheads indicate nonphosphorylated DivIVA.
The S. coelicolor genome carries at least 34 genes predicted to encode STKs (25). Accordingly, we began systematically disrupting these genes, introducing the divIVA allele encoding the N-terminally FLAG-tagged version of the protein into the resulting mutants, and examining the pattern of DivIVA phosphorylation in FLAG-DivIVA/DivIVA mixtures immunoprecipitated from each strain. Including the three PASTA domain kinases described earlier, we examined 17 STKs for their involvement in DivIVA phosphorylation (Table S1 and Fig. 3). In mutants for 16 of these kinases, we observed the normal pattern of phosphorylation (Fig. 3 A and B). However, no DivIVA phosphorylation occurred in a constructed afsK mutant (SCO4423) during normal growth or after cell wall synthesis was arrested with bacitracin (Fig. 3C). Complementation of the afsK mutant restored DivIVA phosphorylation to the WT pattern (Fig. 3D), showing that the afsK-encoded kinase is required for the basal level of DivIVA phosphorylation and the high levels induced by arresting peptidoglycan synthesis.
DivIVA Is Phosphorylated by AfsK in Vitro.
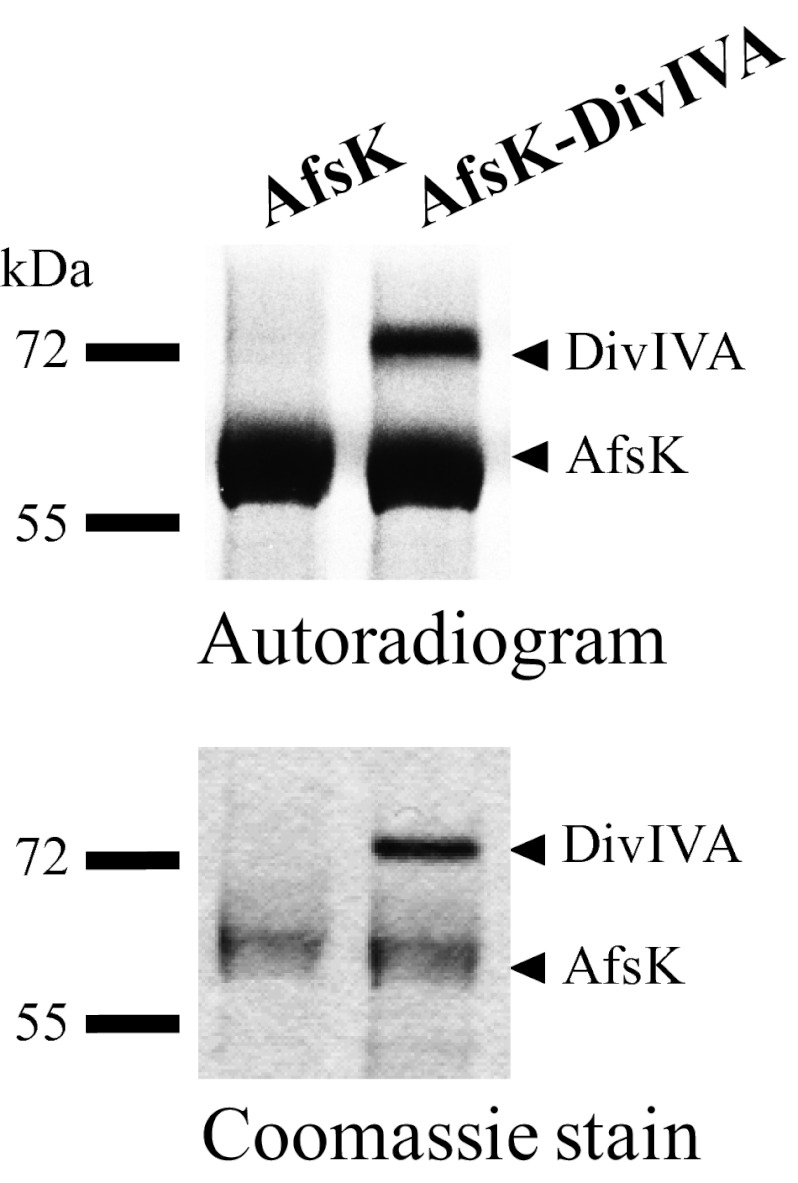
These results led us to investigate whether AfsK directly phosphorylates DivIVA. To address this question, we cloned, overexpressed, and purified the kinase domain of AfsK (amino acids 1–311) and DivIVA as GST-tagged fusion proteins and used them to establish an in vitro phosphorylation system. When the kinase domain of AfsK was incubated with γ-labeled ATP, it underwent autophosphorylation, as revealed by autoradiography, and, when this was mixed with purified DivIVA, the kinase was indeed able to phosphorylate DivIVA (Fig. 4). Thus, we conclude that the absence of DivIVA phosphorylation in the afsK mutant arises because DivIVA is a direct substrate for AfsK.
Fig. 4.
In vitro phosphorylation of DivIVA by AfsK. Recombinant AfsK and DivIVA were incubated with [γ-33P]ATP. Samples were separated by SDS/PAGE, visualized by autoradiography (Upper), and Coomassie-stained (Lower). Lower bands in the autoradiogram illustrate the autokinase activity of AfsK, whereas upper bands reflect DivIVA phosphorylation. In control experiments, DivIVA alone did not show any autophosphorylation activity.
AfsK Kinase Colocalizes with Its Substrate DivIVA at Tips of Growing Vegetative Hyphae.
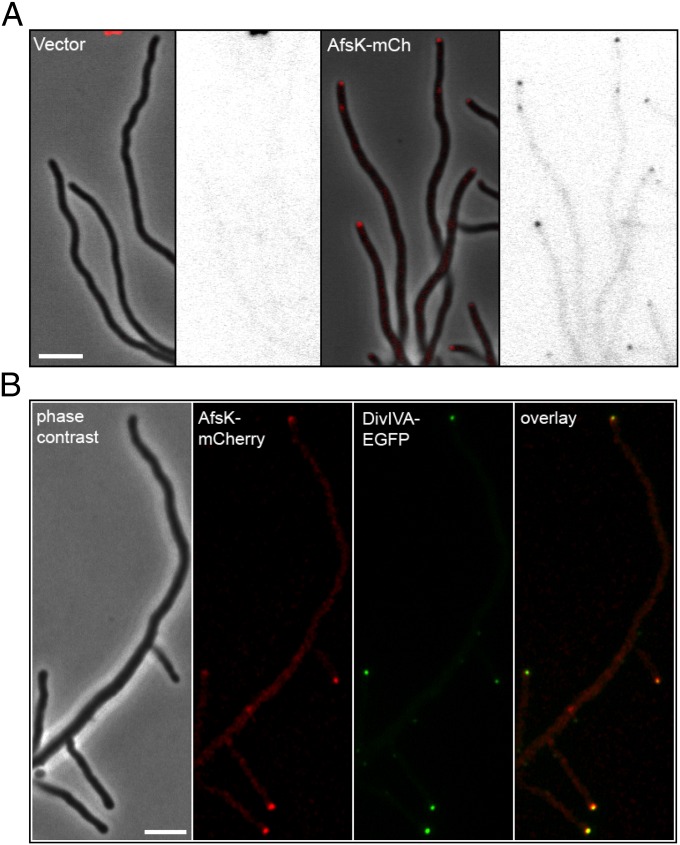
DivIVA shows a distinctive subcellular localization, with strong accumulation at the tips of growing hyphae (10). It was therefore of interest to determine whether AfsK would show a similar distribution and colocalize with its substrate. We investigated this question by creating a fusion between AfsK and the red fluorescent protein mCherry. The translational fusion was expressed from the afsK promoter and was integrated at the chromosomal attϕBT1 site in the WT strain and its congenic afsK-null mutant. The afsK-mCherry allele restored the ability to phosphorylate DivIVA to the afsK mutant—both the basal level seen during growth and the high level induced by bacitracin-treatment (Fig. S2)—showing that the fusion protein is functional. In both strain backgrounds, this hybrid protein showed clear accumulation as foci at the tips of vegetative hyphae, although we also observed weak fluorescence along the hyphae (Fig. 5A). The colocalization of AfsK with DivIVA at the hyphal tips was further confirmed by examining a strain expressing divIVA-egfp and afsK-mCherry (Fig. 5B). Thus, a substantial fraction of the AfsK kinase colocalizes with its substrate DivIVA at hyphal tips.
Fig. 5.
The DivIVA kinase AfsK localizes to hyphal tips. (A) S. coelicolor WT strain carrying empty vector pKF210 with promoterless mCherry (Left) or plasmid pKF255 expressing a translational afsK-mCherry fusion (Right). Representative images of growing hyphae are shown as phase-contrast image with overlaid fluorescence in red, and as the fluorescence image alone in inverted grayscale. (B) Colocalization of DivIVA and AfsK demonstrated by using an S. coelicolor strain producing both DivIVA-EGFP (green) and AfsK-mCherry (red). A series of images were collected for each channel, moving focus 0.2 μm between each image. The z-stacks were deconvolved by using Volocity software, and a central focal plane through the middle of the cells is shown as (from left to right) phase-contrast image, mCherry fluorescence, EGFP fluorescence image, and overlay of the fluorescence channels. (Scale bar, 4 μm.)
AfsK Regulates Branching of Growing Hyphae.
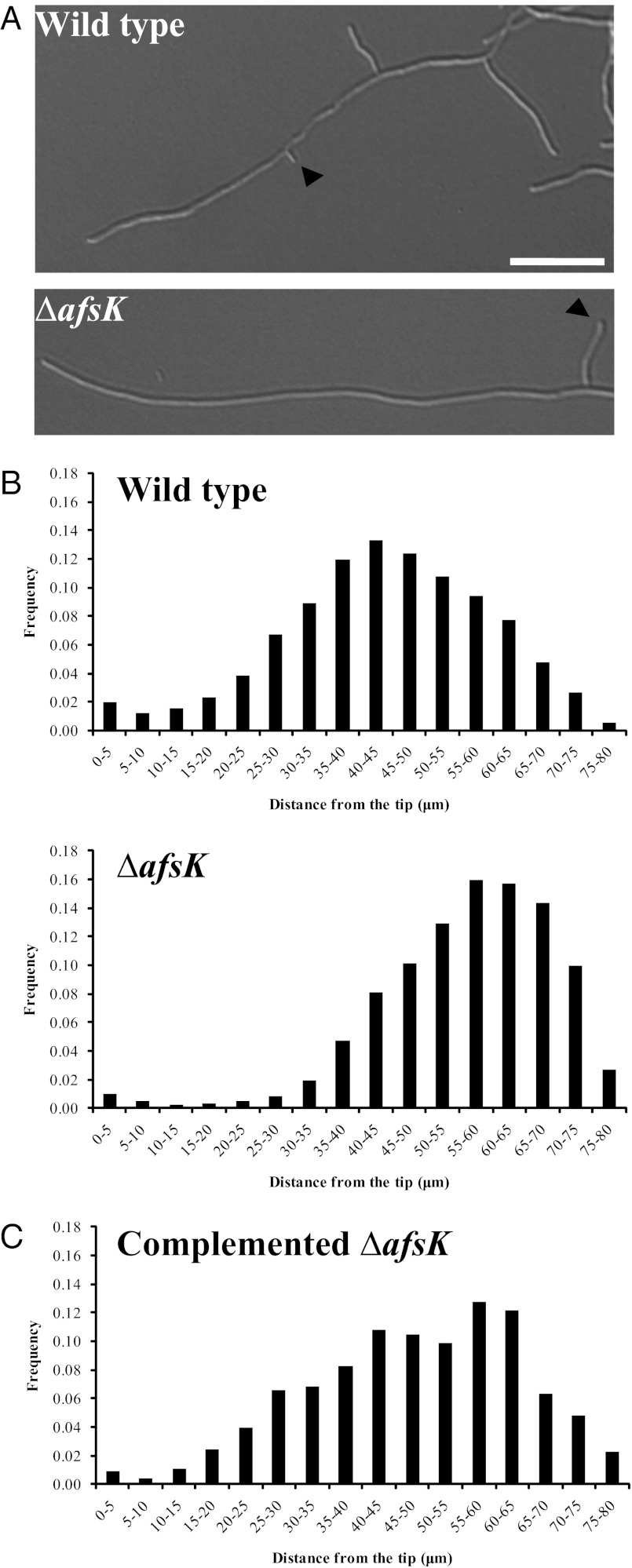
With the discovery that DivIVA is directly phosphorylated by AfsK, we wondered whether disruption of afsK would influence hyphal branching and the underlying polarisome splitting mechanism. Previously reported afsK mutant phenotypes in S. coelicolor have only concerned decreased synthesis of antibiotics (28). We analyzed liquid cultures of our S. coelicolor afsK deletion mutant microscopically in comparison with the WT and discovered that the afsK mutant strain does indeed have a previously unrecognized phenotype: it exhibits an altered tip-to-branch distribution, shifting the average to a longer distance than in the WT (Fig. 6). This effect is quantified in Fig. 6B. The effect is also clearly apparent when comparing time-lapse image sequences of growing hyphae of the afsK mutant and its congenic afsK+ parent (Movies S1 and S2). To confirm that the effect on hyphal branching was caused by the absence of afsK, we complemented the afsK mutant strain and found that reintroducing the afsK gene largely restored WT branching behavior (Fig. 6C). These results show that loss of the AfsK kinase affects the normal regulation of lateral branch formation, and because the vast majority of hyphal branches emerge from DivIVA foci deposited by polarisome splitting (14), this suggests that AfsK modulates this mechanism for the development of new daughter polarisomes.
Fig. 6.
The afsK mutant has a branching phenotype. (A) Representative DIC images of WT S. coelicolor and the congenic afsK mutant grown in YEME. Arrows indicate the first lateral branch behind the hyphal tip. (Scale bar, 10 μm.) (B) Histograms of distances between the tip and lateral branches at the moment of branch development in cultures of S. coelicolor WT and the congenic afsK mutant, and (C) the complemented afsK mutant grown for 15 to 18 h in YEME at 80 μm trim (Experimental Procedures). The number of tip-to-branch distances measured per strain were 1,097 (WT), 875 (afsK mutant), and 281 (complemented mutant).
Constitutively Active AfsK Profoundly Affects Apical Growth, DivIVA Localization, and Hyphal Branching.
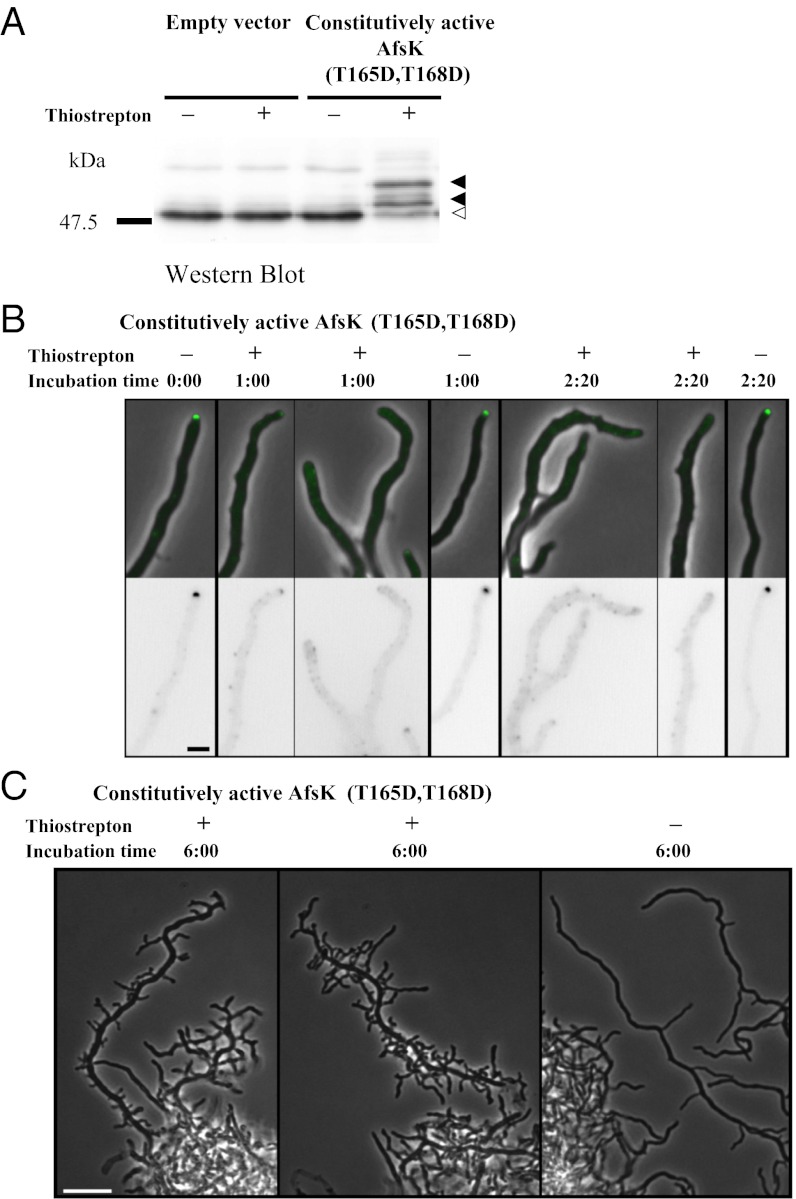
The data described earlier show that AfsK regulates branch-site selection and hyphal morphology during normal growth, even when its activity, as reflected in the basal level of DivIVA phosphorylation, is relatively low. However, when peptidoglycan synthesis is blocked, there is a pronounced up-regulation of AfsK-dependent DivIVA phosphorylation (as described earlier). This raised the possibility that such high levels of AfsK activity would strongly affect hyphal growth and branching, but these effects cannot be evaluated when growth is simultaneously blocked by bacitracin. We therefore engineered a strain in which AfsK kinase activity could be induced in normally growing hyphae. This was achieved by creating a constitutively active mutant version of AfsK in which two threonines in the activation loop of the kinase (T165 and T168) were changed to aspartates to mimic the autophosphorylation of AfsK that leads to its activation. As in other STKs, the conserved residues T165 and T168 of S. coelicolor and Streptomyces avermitilis AfsK are required for activation of the kinase, and T168 has been shown to undergo autophosphorylation in S. coelicolor (36, 37). Most importantly, in both species, T165D and T168D phosphomimic mutations result in a constitutively active kinase (36, 37). The mutant afsK(T165D,T168D) allele was placed under control of the thiostrepton-inducible tipAp promoter in the integrative vector pIJ6902 to create pKF275 (Table S1). When strains carrying pKF275 were grown in the absence of thiostrepton, they showed basal levels of DivIVA phosphorylation similar to those of control strains that carried only the empty vector pIJ6902 (Fig. 7A). However, addition of thiostrepton to cultures of pKF275-carrying strains led to a dramatic increase in the level of phosphorylated DivIVA, as detected by the mobility shift of a major part of the DivIVA protein population seen in Western blots (Fig. 7A).
Fig. 7.
Engineered expression of a constitutively active version of the AfsK kinase induces high levels of DivIVA phosphorylation and profoundly affects hyphal tip extension and branching. (A) Levels of DivIVA phosphorylation induced by expression of the afsK(T165D, T168D) allele from the thiostrepton-inducible tipAp promoter in plasmid pKF275. A strain carrying empty vector pIJ6902 was used as control. Growing cultures were split in two, and thiostrepton was added to one (+) whereas a mock addition of DMSO was made to the other (−). Extracts of cells harvested after 2.5 h were separated by SDS/PAGE, and DivIVA was detected by immunoblotting. Phosphorylated species of DivIVA (closed arrowheads) migrate more slowly than unphosphorylated DivIVA (open arrowhead). (B) The effects of overproduction of constitutively active AfsK(T165D, T168D) on DivIVA-EGFP localization are illustrated by typical examples of hyphae from strain K338. Images captured before addition of thiostrepton (10 μg/mL), the inducer of tipAp-afsK(T165D, T168D) expression, 1 h and 2 h 20 min after addition of thiostrepton or mock. EGFP fluorescence is shown in inverted grayscale (Lower) or shown in green overlaid on phase-contrast images (Upper). (Scale bar, 2 μm.) (C) Typical examples of hyperbranched hyphal morphology developing after overexpression of afsK(T165D, T168D) for 6 h (Left) compared with the uninduced control sample (Right). (Scale bar, 10 μm.)
The thiostrepton-induced hyperphosphorylation strongly affected DivIVA localization, as detected by using the DivIVA-EGFP fusion. Before thiostrepton addition, the majority of hyphae carried detectable DivIVA-EGFP foci at the tips, but when expression of the constitutively active AfsK was induced, the majority of these foci dissolved or were strongly reduced in intensity (Fig. 7B), leading to decreased average fluorescence intensity at the hyphal tip, and an increased fraction of hyphae without detectable apical foci. Cultures of pKF275-carrying strains that did not receive thiostrepton showed normal DivIVA localization (Fig. 7B), and, similarly, the control strain carrying the empty vector pIJ6902 was not affected by addition of thiostrepton and showed normal DivIVA localization to hyphal tips. These observations show that strong up-regulation of AfsK activity stimulates disassembly of polarisome structures and dissociation of DivIVA from hyphal tips.
The induction of the constitutively active AfsK further caused dramatic changes in hyphal growth and morphology. When cultures containing the tipAp-afsK(T165D,T168D) construct were incubated in the presence of the inducer thiostrepton, growth was impeded (Fig. S3). However, despite the arrest of growth at existing hyphal tips, multiple new hyphal branches started to emerge from the lateral walls distal to these tips, giving rise to conspicuous hyphal structures decorated by multiple branch initials (Fig. 7C). Such effects were not seen in control strains carrying the empty vector pIJ6902, which carried on growing without any detectable effect of thiostrepton (Fig. S3). Thus, induction of high AfsK activity causes disappearance of apical DivIVA foci, growth inhibition, and subsequent initiation of multiple new lateral branches. This gave the cultures a characteristic appearance, with unusually dense and compact hyphal pellets, from which emerge hyperbranched and irregularly shaped hyphal structures (representative images in Fig. 7C), strikingly different from the regular and loose hyphal pellets and long tip-to-branch distances seen in control cultures (representative image in Fig. 7C). In summary, AfsK kinase activity has strong effects on cell polarity, tip extension, subcellular localization of DivIVA, and initiation of new hyphal branches.
Discussion
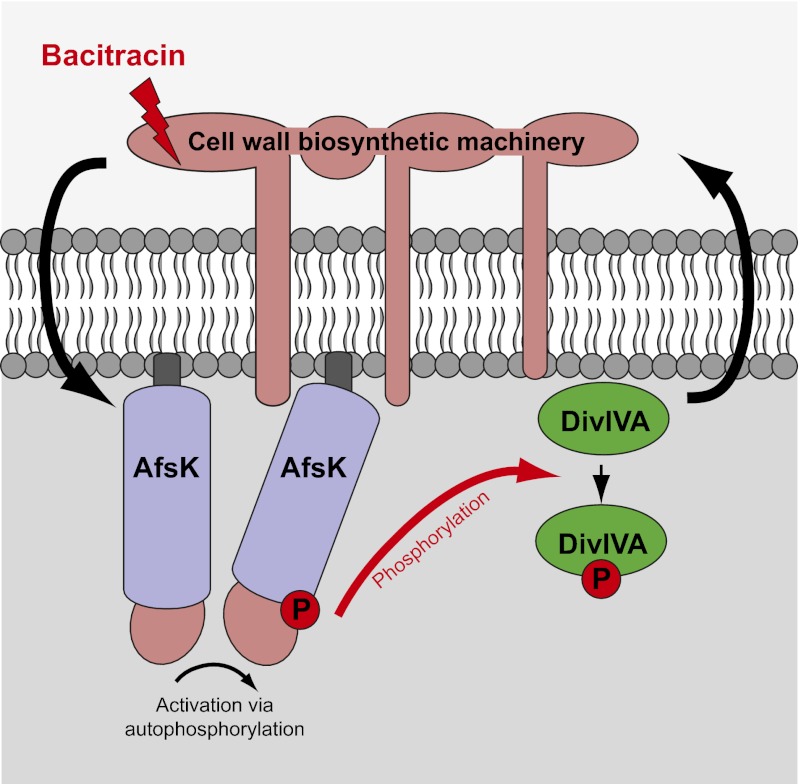
This study shows that the Ser/Thr kinase AfsK is part of the apparatus that controls polar growth in Streptomyces, and that it directly phosphorylates the cell polarity determinant DivIVA. Our data indicate dual roles for the AfsK kinase. First, during normal growth, it modulates the control of hyphal branching and the development of daughter polarisomes. Second, AfsK is involved in a stress response when cell wall synthesis is arrested; under such conditions, AfsK is strongly activated and causes profound reconfiguration of DivIVA localization, apical growth, and hyphal branching. This discovery represents an intriguing prokaryotic parallel to the widespread and broadly conserved roles of STKs in controlling cell polarity in eukaryotes (1, 2), and particularly to the control of polar growth by kinases targeting polarisome components in fungi (references in ref. 13).
Induction of a constitutively active form of AfsK causes the disappearance of the DivIVA foci that normally mark growing hyphal tips (Fig. 7). No concomitant degradation or decrease in cellular DivIVA content is observed, suggesting that high AfsK activity causes disassembly of the DivIVA-containing apical polarisome. DivIVA is a self-assembling coiled-coil protein that forms oligomers and higher-order complexes and is involved in polar targeting in a range of Gram-positive bacteria (15, 30, 38–40). The AfsK-mediated phosphorylation of DivIVA in Streptomyces occurs on two trypsin-generated fragments in the C-terminal domain (Fig. 2). Although this C-terminal domain is not conserved outside of Streptomyces orthologues (10), it lies just downstream of the conserved second coiled-coil segment, which is known to be important in the oligomerization of B. subtilis DivIVA (40). It is therefore possible that the AfsK-mediated phosphorylation influences oligomerization, acting as a means to control the assembly or disassembly of multimeric complexes or higher-order structures formed by DivIVA in the cell. Such a role for STKs in controlling assemblages of coiled-coil proteins is well known in eukaryotes, a classic example being the disassembly of the nuclear lamina mediated by cyclin-dependent kinases (41). In addition, it was recently reported that the assembly and subcellular localization of the coiled-coil protein RsmP in Corynebacterium glutamicum is affected by phosphorylation (42). However, it cannot be excluded that phosphorylation may also influence other aspects of DivIVA behavior, such as its interaction with other proteins or the membrane. The function of B. subtilis DivIVA depends on direct interactions with MinJ and the nucleoid-associated protein RacA (15, 43), and the function of S. coelicolor DivIVA is also likely to depend on the direct recruitment of other proteins to the cell poles (5, 6). Further, crystal structures of B. subtilis DivIVA show how the oligomers may interact with the membrane via an exposed phenylalanine residue in the highly conserved N-terminal part of the protein (40), and the polar and septal targeting of the B. subtilis DivIVA appears to be explained by a preference of the oligomers for negatively curved membrane surfaces (15, 44).
The decreased branching observed in afsK mutants could be explained by an effect of AfsK-mediated phosphorylation on the stability of the apical DivIVA clusters. Most branches in S. coelicolor are formed by a tip-focus splitting mechanism in which the DivIVA-containing apical polarisome splits to leave smaller foci behind along the lateral hyphal walls as the tip extends—foci that act as seeds for new branches (14). Although only a small fraction of the DivIVA molecules in the cell are detectably phosphorylated during normal growth (Fig. 1A), this low basal activity obviously has significant impact on hyphal branching. As AfsK colocalizes with the DivIVA foci at hyphal tips, it is possible that the low level of DivIVA phosphorylation seen during normal growth affects polarisome splitting (perhaps by controlling the initial size of new daughter polarisomes) and thereby modulates the pattern of hyphal branching.
Hyperactivity of AfsK inhibits growth at the original hyphal tips, but, paradoxically, it also induces the subsequent formation of multiple short lateral branches distal to these tips (Fig. 7). This observation, that growth can be initiated at new sites while being prevented at the original tip, could also be explained by the localization of AfsK to hyphal tips. A high level of DivIVA phosphorylation promotes the complete disassembly of the apical protein complexes. The released DivIVA molecules could then diffuse away and gradually be dephosphorylated, allowing them to form new foci that are capable of establishing new branches distal to the original tips. We suggest this provides S. coelicolor with a mechanism to dismantle the apical growth apparatus at hyphal tips that encounter problems with cell wall synthesis, for example through exposure to an antibiotic or by hitting a physical obstacle in the soil. Such conditions would strongly activate AfsK, leading to disassembly of the apical DivIVA complex and liberation of DivIVA molecules that can then direct emergence of new branches elsewhere, leading, for example, to growth around an obstacle. In the simplest scenario, DivIVA molecules that are liberated from the original tip could join small daughter foci that have previously been deposited along the lateral wall by polarisome splitting (14), accelerating their maturation into polarisomes competent to trigger branch outgrowth. Alternatively, the release of large quantities of soluble DivIVA from the disassembly of apical foci could trigger the spontaneous nucleation of new DivIVA foci.
Orthologues of AfsK are found among only streptomycetes and a few closely related mycelial actinomycetes, suggesting that its function is related to the filamentous or hyphal growth habit. Further, the AfsK-mediated phosphorylation of S. coelicolor DivIVA differs in several important ways from the previously observed phosphorylation of the mycobacterial DivIVA ortholog, Wag31 (34). First, the role of phosphorylation of mycobacterial Wag31 is poorly understood, but it seems to promote localization of Wag31 to cell poles and stimulate polar growth and cell wall synthesis (45, 46). In contrast, the activation of the DivIVA kinase in S. coelicolor has the opposite effect, promoting the disassembly of DivIVA foci and the inhibition of growth at existing hyphal tips. Second, different kinases are involved, which are likely to be activated by different stimuli. The essential PASTA-domain kinases PknA and PknB act on Wag31 in mycobacteria, and the reports so far describe activity only during undisturbed growth (34, 46). In contrast, AfsK, the DivIVA kinase in S. coelicolor, is weakly active during vegetative growth and strongly activated in response to the arrest of cell wall synthesis. Third, the site of phosphorylation is different, with a single threonine close to the first coiled-coil domain being targeted in M. tuberculosis, whereas it is the C-terminal domain of S. coelicolor DivIVA that is phosphorylated on multiple residues.
AfsK was one of the first bacterial STKs to be investigated, and an afsK disruption mutant of S. coelicolor was reported to grow and sporulate normally, while showing reduced production of blue-pigmented antibiotic actinorhodin (28). The effect on antibiotic production appears to be mediated by the transcription factor AfsR, which is directly phosphorylated by AfsK in vitro and activates transcription of afsS, encoding a small pleiotropic regulator of antibiotic synthesis in Streptomyces (47, 48). In this study, we report a completely different role for AfsK in the control of hyphal growth and branching, one that does not involve AfsR (as an afsR mutant shows normal hyphal branching and normal levels of DivIVA phosphorylation). Strikingly, the conditions we used to induce high AfsK activity (i.e., addition of bacitracin or the engineered expression of a constitutively active kinase) did not trigger overproduction of actinorhodin (Fig. S3). In summary, the function of AfsK in controlling polar growth and branching is not obviously related to the previously inferred role of AfsK in secondary metabolism.
Overall, our findings show that communication between the polarity determinant DivIVA and the cell wall biosynthetic machinery is bidirectional, with DivIVA directing cell wall synthesis (11), and the biosynthetic machinery communicating back to DivIVA via AfsK-mediated phosphorylation. All three components—the cell wall biosynthetic machinery, AfsK, and DivIVA—localize to growing hyphal tips. What, then, are the signals that lead to the activation of AfsK? AfsK has an N-terminal STK domain and a C-terminal putative sensory portion carrying PQQ domain repeats. These PQQ domain repeats are predicted to form a β-propeller structure similar to WD40 domains and may interact with a ligand, although the general function of PQQ domains is not known (25). Further, AfsK does not have a predicted transmembrane segment, and is reported to be cytoplasmic but loosely associated with the membrane (28). AfsK activity (at least as reflected in the level of DivIVA phosphorylation) is strongly stimulated by antibiotics like bacitracin and vancomycin, which block the lipid II cycle of peptidoglycan biosynthesis, raising the possibility that AfsK can sense the accumulation of intermediates in peptidoglycan biosynthesis. This would provide a mechanism to sense the capacity of the hyphal tip to sustain extension during normal growth and during stress conditions, and via AfsK-mediated phosphorylation transduce this information to the polarisome that directs apical growth and branching.
Experimental Procedures
Bacterial Strains, Plasmids, and General Methods.
Properties of bacterial strains and plasmids used in this study are described in Table S1. Details of plasmid construction are described in SI Experimental Procedures. Oligonucleotide primers are listed in Table S2. Media, culture conditions, and general genetic manipulations were as described previously for E. coli (49) and Streptomyces (50).
Analysis of DivIVA Phosphorylation by Immunoprecipitation, Pro-Q Diamond Staining, and MS.
The appropriate S. coelicolor strains were grown in yeast extract–malt extract (YEME) medium for 15 to 22 h. For expression of FLAG-divIVA from the thiostrepton-inducible promoter tipAp, strains were grown in the presence of 0.1 μg/mL of thiostrepton. The exact details of the following procedures are described in SI Experimental Procedures. Briefly, hyphae were harvested by centrifugation, washed twice, and resuspended in appropriate buffer. Cell extracts were prepared in a buffer designed to reduce phosphatase activity, and cell lysates were prepared by sonication or by bead beating. Cell lysates were cleared by centrifugation. The cleared cell lysates were used for immunoprecipitation by using anti-FLAG M2 affinity beads (Sigma-Aldrich), essentially as described by Wang et al. (30). Eluted proteins were separated by SDS-PAGE, and phosphorylated proteins were detected by using Pro-Q Diamond phosphoprotein gel stain (Molecular Probes). When appropriate, the material eluted from the affinity beads was dephosphorylated for 10 min at 30 °C by using lambda protein phosphatase (Sigma-Aldrich). Identification of phosphorylated trypsin fragments of DivIVA was done by using MALDI-TOF MS.
In Vitro Phosphorylation of DivIVA.
In vitro phosphorylation was carried out in 20-μL reactions containing the recombinant AfsK (1 μg) and/or DivIVA (4 μg) and 200 μCi/mL (65 nM) [γ-33P]ATP (3,000 Ci/mmol; PerkinElmer) in phosphorylation buffer (25 mM Tris-HCl, pH 6.8, 1 mM DTT, 5 mM MgCl2, 1 mM EDTA). The reaction was carried out for 30 min at 37 °C and stopped by addition of Laemmli sample loading buffer and incubated at 100 °C for 5 min before analysis by SDS-PAGE. After electrophoresis, gels were washed in 10% (wt/vol) trichloroacetic acid for 10 min at 90 °C and then stained with Coomassie stain, dried, and visualized by autoradiography overnight.
Western Blotting.
Cell lysates from S. coelicolor cultures grown in YEME (17% sucrose) were prepared by bead beating (six times, 6.0 m/s, 30 s; FastPrep-24; MP Biomedicals) in lysate buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) supplemented with complete EDTA-free protease inhibitor mixture (Roche). Proteins were separated by SDS/PAGE and transferred onto Immobilon-P PVDF membrane (Millipore) as described previously (10). The membrane was incubated overnight at 4 °C with anti-DivIVASC antiserum from rabbit, diluted 1:5,000 (30), then washed three times with 5% (wt/vol) nonfat dry milk in PBS solution before incubation for 1 h at room temperature with pig anti-rabbit IgG linked to horseradish peroxidase (1:1,000; DakoCytomation). The membrane was washed six times in PBS solution with 0.05% Tween, proteins were visualized by SuperSignal West Pico chemiluminescence substrate (Pierce), and results were captured by using a Digital Science Image Station 440CF (Kodak).
Microscopy.
Hyphae were prepared for microscopy as described previously (10). For differential interference contrast , phase-contrast, and fluorescence microscopy, liquid cultures of S. coelicolor were grown for 15 to 18 h in YEME from pregerminated spores. Samples of these cultures were spotted directly onto microscope slides coated with 1% (wt/vol) agarose in PBS solution and mounted with a coverslip. To obtain images for measuring the distance between the hyphal tip and lateral branches, samples were observed through a DIC 63× objective of a Nikon Eclipse 800 microscope equipped with a Pixera ProES600 camera and images were taken with Pixera software and processed with ImageJ (National Institutes of Health). The analysis of hyphal branching is further described in SI Experimental Procedures.
For fluorescence microscopy, equipment and imaging were as described previously (51). Deconvolution of fluorescence images used the iterative restoration algorithm in Volocity 3DM (Perkin-Elmer) and a calculated point spread function, and was carried out on z-stacks of more than 50 images with 0.2-μm spacing, captured with a 100× NA 1.4 lens. Live-cell time-lapse microscopy was performed as described previously (11).
Supplementary Material
Acknowledgments
The authors thank Erik Andreasson and Marit Lenman for invaluable assistance and advice on the analysis of phosphoproteins; Elisabeth Olsson, Jade Leiba, and Natalia Berges for excellent technical assistance; Paul Dyson and Katerina Petrickova for gifts of strains; and Keith Chater, Joe McCormick, and Nora Ausmees for critically reading the manuscript. This work was supported by postdoctoral stipends from the Carl Trygger Foundation (to S.C.) and Wenner-Gren Foundation (to S.-B.W.), John Innes Centre doctoral studentships (to A.M.H., J.L.P., and Y.-G.J.), Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BB/J004561/1 (to M.J.B.), the John Innes Foundation (M.J.B.), Swedish Research Council Grants 621-2007-4767 and 621-2010-4463 (to K.F.), the O. E. och Edla Johansson Foundation (K.F.), and the Crafoord Foundation (K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 13906 (volume 109, number 35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207409109/-/DCSupplemental.
References
- 1.Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCaffrey LM, Macara IG. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harb Perspect Biol. 2009;1:a001370. doi: 10.1101/cshperspect.a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown PJ, Kysela DT, Brun YV. Polarity and the diversity of growth mechanisms in bacteria. Semin Cell Dev Biol. 2011;22:790–798. doi: 10.1016/j.semcdb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown PJ, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flärdh K. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol. 2010;13:758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 7.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 9.Heichlinger A, et al. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J Bacteriol. 2011;193:1533–1542. doi: 10.1128/JB.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 11.Hempel AM, Wang SB, Letek M, Gil JA, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walshaw J, Gillespie MD, Kelemen GH. A novel coiled-coil repeat variant in a class of bacterial cytoskeletal proteins. J Struct Biol. 2010;170:202–215. doi: 10.1016/j.jsb.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Moseley JB, Goode BL. The yeast actin cytoskeleton: From cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards DM, Hempel AM, Flärdh K, Buttner MJ, Howard M. Mechanistic basis of branch-site selection in filamentous bacteria. PLOS Comput Biol. 2012;8:e1002423. doi: 10.1371/journal.pcbi.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araujo-Palomares CL, Riquelme M, Castro-Longoria E. The polarisome component SPA-2 localizes at the apex of Neurospora crassa and partially colocalizes with the Spitzenkörper. Fungal Genet Biol. 2009;46:551–563. doi: 10.1016/j.fgb.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Harris SD. Branching of fungal hyphae: Regulation, mechanisms and comparison with other branching systems. Mycologia. 2008;100:823–832. doi: 10.3852/08-177. [DOI] [PubMed] [Google Scholar]
- 19.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 20.Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beilharz K, et al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci USA. 2012;109:E905–E913. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galperin MY, Higdon R, Kolker E. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol Biosyst. 2010;6:721–728. doi: 10.1039/b908047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010;75:1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 25.Petrícková K, Petrícek M. Eukaryotic-type protein kinases in Streptomyces coelicolor: Variations on a common theme. Microbiology. 2003;149:1609–1621. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- 26.Prisic S, et al. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker JL, et al. Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3(2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics. 2010;10:2486–2497. doi: 10.1002/pmic.201000090. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto A, Hong SK, Ishizuka H, Horinouchi S, Beppu T. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 29.Stone KJ, Strominger JL. Mechanism of action of bacitracin: complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc Natl Acad Sci USA. 1971;68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SB, et al. Domains involved in the in vivo function and oligomerization of apical growth determinant DivIVA in Streptomyces coelicolor. FEMS Microbiol Lett. 2009;297:101–109. doi: 10.1111/j.1574-6968.2009.01678.x. [DOI] [PubMed] [Google Scholar]
- 31.Hong HJ, Paget MS, Buttner MJ. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol Microbiol. 2002;44:1199–1211. doi: 10.1046/j.1365-2958.2002.02960.x. [DOI] [PubMed] [Google Scholar]
- 32.Maestro B, et al. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585:357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiuza M, et al. The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J Biol Chem. 2008;283:36553–36563. doi: 10.1074/jbc.M807175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang CM, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz C, et al. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: Evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol Microbiol. 2009;74:724–741. doi: 10.1111/j.1365-2958.2009.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomono A, et al. Self-activation of serine/threonine kinase AfsK on autophosphorylation at threonine-168. J Antibiot (Tokyo) 2006;59:117–123. doi: 10.1038/ja.2006.18. [DOI] [PubMed] [Google Scholar]
- 37.Rajkarnikar A, Kwon HJ, Ryu YW, Suh JW. Two threonine residues required for role of AfsKav in controlling morphogenesis and avermectin production in Streptomyces avermitilis. J Microbiol Biotechnol. 2007;17:1563–1567. [PubMed] [Google Scholar]
- 38.Letek M, et al. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol. 2008;190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen L, et al. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol. 2007;189:7896–7910. doi: 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliva MA, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimi T, Butin-Israeli V, Adam SA, Goldman RD. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol. 2010;75:525–531. doi: 10.1101/sqb.2010.75.045. [DOI] [PubMed] [Google Scholar]
- 42.Fiuza M, et al. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. J Biol Chem. 2010;285:29387–29397. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bramkamp M, et al. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol. 2008;70:1556–1569. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- 44.Eswaramoorthy P, et al. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. mBio. 2011;2:e00257-11. doi: 10.1128/mBio.00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jani C, et al. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 2010;10:327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol. 2007;369:322–333. doi: 10.1016/j.jmb.2007.02.096. [DOI] [PubMed] [Google Scholar]
- 48.Lee PC, Umeyama T, Horinouchi S. afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2) Mol Microbiol. 2002;43:1413–1430. doi: 10.1046/j.1365-2958.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 50.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 51.Salerno P, et al. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J Bacteriol. 2009;191:6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]