Abstract
Sustained Toll-like receptor (TLR) stimulation continuously activates antimicrobial genes but paradoxically represses inflammatory genes. This phenomenon, termed TLR tolerance, is essential for preventing fatal inflammatory conditions such as sepsis, but its underlying mechanisms are unclear. We report here that NF-κB binding nucleic acids of gene promoters are tolerogenic motifs, which selectively recruit an NcoR–Hdac3–deacetylated-p50 repressosome to inflammatory genes. Genome-wide analyses of TLR4-induced genes revealed that NF-κB motifs were the only regulatory elements significantly enriched in tolerizable genes. Mutating the NF-κB motifs of tolerizable genes converted them into nontolerizable ones, whereas inserting NF-κB binding motifs into nontolerizable genes conferred the tolerance. Although NF-κB p50 was essential for assembling the repressosome, genetic disruption of the NcoR–Hdac3 interaction alone was sufficient to completely abolish TLR4 tolerance and to render mice vulnerable to sepsis. Thus, the specificity of TLR tolerance is dictated by evolutionally conserved nucleic acid motifs that bound by NF-κB and the NcoR repressosome.
Toll-like receptor activation induces the expression of over a thousand genes that encode inflammatory cytokines, antimicrobial proteins, and regeneration and metabolic regulators; these molecules in turn mediate inflammation, antimicrobial immunity, and tissue regeneration seen in patients with infectious diseases (1–3). However, uncontrolled or prolonged activation of TLRs can have devastating consequences, which include the development of septic shock and fatal inflammatory diseases (4, 5). Fortunately, TLR activation is tightly controlled by several signal-specific and gene-specific regulators (1, 5–10). These regulators ensure that prolonged or repeated exposure of TLRs to their ligands does not lead to sustained activation of the receptors; instead, it renders them insensitive or hyporesponsive to subsequent ligand stimulation. This phenomenon is referred to as TLR tolerance, or lipopolysaccharide (LPS) tolerance when LPS is the ligand involved (1, 11–15). Recent genomic profiling of LPS responses reveals that LPS tolerance is a gene-specific phenomenon, i.e., it selectively targets one class of genes (i.e., inflammatory genes) but not others (e.g., antimicrobial genes); in fact, the expression of antimicrobial genes is further up-regulated in LPS tolerized cells (1, 14). Thus, LPS target genes have been divided into two classes: (i) tolerizable (T) genes or class T genes that are sensitive to LPS tolerance, and (ii) nontolerizable (NT) genes or class NT genes that are not sensitive to LPS tolerance (1). Because it is the inflammatory genes, not the antimicrobial genes, that cause deleterious inflammatory responses, the selective inactivation of class T (inflammatory) but not class NT (antimicrobial) genes ensures that the host is able to continuously build up its antimicrobial immunity without causing fatal inflammatory diseases, even in the face of chronic or prolonged infections. However, the molecular mechanisms through which prolonged LPS exposure activates antimicrobial genes, but paradoxically suppresses inflammatory genes are unknown. We report here that NF-κB binding motifs of gene promoters dictate the specificity of TLR-induced gene repression by recruiting an NcoR-containing repressosome.
Results
NF-κB Motifs Specify TLR-Induced Gene Repression.
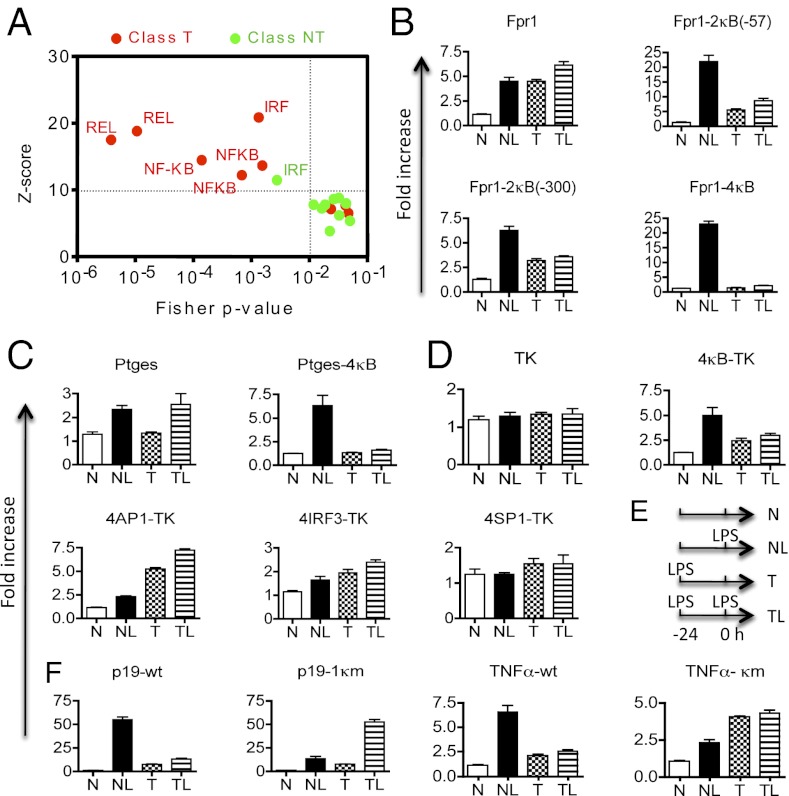
Our motif-based bioinformatic analyses of 508 reported LPS-induced genes (371 T and 137 NT genes) from the murine genome (1, 16) (Fig. 1A) using the oPOSSUM program (17) revealed that NF-κB binding motifs were the only ones that were significantly enriched among tolerizable genes, but not nontolerizable genes. The oPOSSUM program assesses which, if any, of the transcription factor binding motifs are statistically over- or underrepresented in a selected group of genes, based on a Z score and a Fisher probability (P) value it calculates (17). A Z score of 10 or greater and a Fisher P value of 0.01 or smaller are considered statistically significant (17). Interferon regulatory factors (IRF) are proteins regulating transcription of interferons. Although IRF binding motifs were also enriched in murine class T genes as reported (18), they were significantly enriched in the murine NT genes that we analyzed as well (Fig. 1A). The activator protein 1 (AP-1) is a heterodimeric transcription factor of either c-Fos, c-Jun, ATF, or JDP. Unexpectedly, AP1 sites were not significantly enriched in either T or NT genes, suggesting that it was not shared by a significant majority of LPS target genes. To confirm these findings, we selected the top 100 T and top 100 NT murine genes (based on their levels of expression in LPS-treated macrophages) and determined which one contained confirmed functional NF-κB binding sites based on published literature, and the VISTA and oPOSSUM transcription factor databases. We found that 71 (71%) of tolerizable genes possessed known NF-κB binding sites, whereas only 2 of the top 100 nontolerizable genes might contain an NF-κB site. The exact number of NF-κB target genes in the murine or human genome is unknown but is estimated to be in the range of 250–300 (19, 20). However, a recent ChIP sequencing analysis of the murine genome revealed thousands of NF-κB p65 binding sites in LPS-treated macrophages (21). Because ∼50% of these sites are located in the intergenic regions that may not regulate gene expression and because the ChIP sequencing analysis detects sites that NF-κB bind indirectly through other factors, the exact number of genes directly regulated by NF-κB in LPS-treated cells remains to be determined. Nonetheless, for the top LPS-induced T genes, ∼69% have NF-κB binding sites detectable by the ChIP sequencing analysis, whereas for the top LPS-induced NT genes, only ∼14% of them do.
Fig. 1.
NF-κB binding motifs of gene promoters specify LPS-induced tolerance. (A) Motif-based bioinformatic analysis of 508 LPS-induced genes (371 T and 137 NT genes) from the murine genome (1, 16) using the oPOSSUM program reveals specific enrichment of NF-κB binding motifs in tolerizable (red) but not nontolerizable (green) genes. Putative transcription factor-binding motifs located between 5000 bp upstream and 5000 bp downstream of the transcription start site of each gene were analyzed. Each data point represents a transcription factor-binding motif; only those that are significantly enriched in a class of genes are labeled (Upper Left quadrant). REL or NF-κB represents the binding site for the NF-κB/Rel family of transcription factors. IRF represents a binding site for the IRF family of transcription factors. (B–D) NF-κB motif insertion into the promoter constructs of nontolerizable genes converts them into tolerizable ones. The RAW246.7 macrophages were transiently transfected with the promoter reporter plasmids as indicated. They were either (i) left untreated for 36 h (naïve, N), or (ii) rested for 24 h and treated with 100 ng/mL LPS (L) for an additional 12 h (NL), or (iii) treated with 100 ng/mL LPS for 24 h and then rested in fresh media for an additional 12 h (tolerized, T), or (iv) treated with 100 ng/mL LPS for 24 h and restimulated with 100 ng/mL LPS for another 12 h (TL). The luciferase activities were quantified at the end of the culture as described in Materials and Methods. 2κB and 4κB, two and four identical NF-κB binding motifs, were inserted, respectively; 4AP1, 4IRF3, and 4SP1, four identical binding sites of AP1, IRF3, and SP1, respectively, were inserted. Numbers in parentheses of the gene construct names indicate the location where the NF-κB binding motifs were inserted. (E) Schematic illustration of the LPS stimulation time line. All luciferase assays were performed 12 h after the last LPS stimulation. i.e., at the +12 h. (F) Mutations of NF-κB binding motifs of tolerizable genes abolish tolerance. Mutations were introduced into an NF-κB binding motif (at −95) of the Il23p19 promoter region (−1,180/+110), and deletion of −218/−50 nucleotides of the Tnf promoter region (−512/+61) removed its NF-κB motifs. Wild-type (WT) and mutated (κm) promoter constructs were tested as in B above. Error bars represent SD of the mean. Experiments were repeated at least three times with similar results.
These unexpected results indicate that NF-κB motifs may specify the TLR-induced gene repression. To test this hypothesis, we performed mutagenesis analyses of NF-κB binding motifs in cloned promoters of both class T and class NT genes. A total of 10 genes were selected, which included eight NT genes, i.e., Bpil2 (bactericidal/permeability-increasing protein-like 2), Fpr1 (formyl peptide receptor 1), Ptges (prostaglandin E synthase), Camp (cathelicidin antimicrobial peptide), Orm1 (orosomucoid 1), TK (herpes simplex virus thymidine kinase), and Slfn1 (Schlafen 1), and three T genes, i.e., Il23p19 (interleukin 23 p19 or p19), Tnf (tumor necrosis factor α), and Hdc (histidine decarboxylase) (Fig. 1 and Fig. S1A). Each promoter construct contains up to 2 kb of the 5′ genomic sequence of the corresponding gene plus a firefly luciferase cDNA sequence. TK is not an LPS target gene, which serves as a control for the LPS-activated genes. Upon stimulation with LPS, all but the TK promoter constructs responded by driving luciferase expression (Fig. 1 B–F and Fig. S1A). However, only the promoters of the tolerizable genes (Tnf, p19, and Hdc) were sensitive to LPS tolerance, which was characterized by a marked reduction of the LPS-induced response in the TL (tolerized + LPS) group compared with the NL (naïve + LPS) group (Fig. 1F). Remarkably, adding NF-κB motifs to each of the nontolerizable promoters converted them into tolerizable ones. The degree of tolerance appeared to be a function of the number of the NF-κB sites added, with promoters that had four NF-κB sites the most tolerized (Fig. 1 B and C). Importantly, adding NF-κB sites to the non-LPS responsive TK promoter converted it to an LPS-responsive tolerizable one. By contrast, adding AP1, IRF3, or SP1 binding sites into the TK promoter did not confer tolerance although AP1 did make it LPS responsive (Fig. 1D). On the other hand, mutating the endogenous NF-κB sites in the tolerizable p19, Tnf, and Hdc gene promoters converted them into nontolerizable ones (Fig. 1F and Fig. S1A).
The NF-κB motif addition or mutation not only changed the tolerogenicity but also responsiveness of promoters to LPS. The vast majority of T genes are primary response genes, whereas most NT genes are secondary response genes (Fig. S1 B and C) (22, 23). Remarkably, NF-κB motif addition converted secondary response genes into primary ones, whereas NF-κB motif mutation had the opposite effect (Fig. S1 B and C). Thus, the NF-κB motif dictates both the tolerogenicity and responsiveness of TLR target genes.
If NF-κB motifs mediate LPS-induced tolerance, the tolerized cells should be defective in their responses to non-LPS ligands that activate NF-κB. Indeed, responses to both double-stranded DNA [poly(dA-dT)poly(dT-dA)] and double-stranded RNA (poly I:C) were significantly reduced in cells pretreated with LPS (Fig. S2 A and B). Mutation of the NF-κB motif of the Tnf promoter abolished the tolerance to RNA (Fig. S2B), indicating that NF-κB motifs are involved in mediating LPS-induced receptor cross-tolerance.
NF-κB p50 Is Required for the Repressive Epigenetic Modification of Tolerizable Genes.
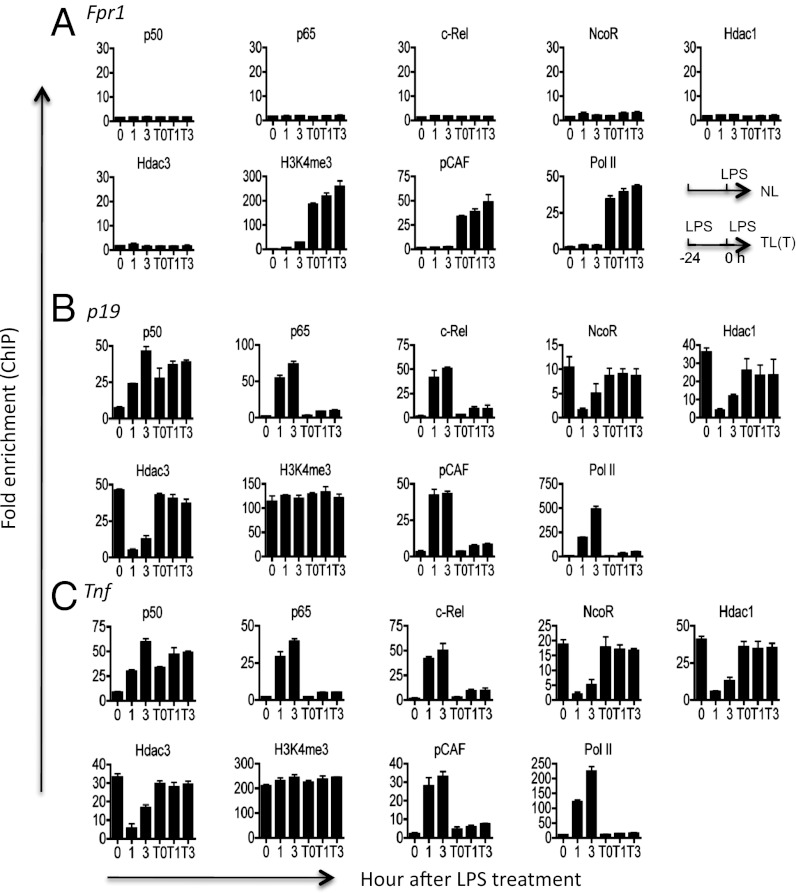
LPS tolerance is associated with epigenetic modifications of class T genes (1). By chromatin immunoprecipitation (ChIP), we detected striking differences between T and NT genes. Whereas tolerized T genes (p19 and Tnf) were occupied by several transcriptional repressors such as nuclear receptor corepressor (NcoR), histone deacetylase 1 (Hdac1), and Hdac3 in addition to p50, NT genes (Fpr1 and Bpil) were totally devoid of these factors (Fig. 2 and Figs. S2C and S3). The recruitment of transcriptional activators such as pCAF to NT genes was also significantly delayed, which is consistent with the secondary response nature of these genes. Although both p65 and c-Rel were also recruited to the T gene promoters during the initial LPS stimulation, p50 was the primary NF-κB protein remaining in tolerized cells (Fig. 2 and Fig. S2D) (14, 24). Importantly, the LPS-induced derepression of the inflammatory genes, characterized by a rapid removal of the NcoR–Hdac complex (25, 26), occurred only in naïve but not tolerized cells. Using Nfkb1 knockout cells that do not express p50, we confirmed a previous report that p50 is essential for LPS tolerance (Fig. S4) (14). However, p50 deficiency did not affect the expression of other proteins tested in our ChIP assays (Fig. S4).
Fig. 2.
Selective binding of NF-κB, NcoR, and histone deacetylases to tolerizable genes and the lack of LPS-induced gene derepression in tolerized cells. Naïve or tolerized (T) bone-marrow–derived macrophages were stimulated with 100 ng/mL LPS for 0, 1, or 3 h as indicated. Cells were then fixed and chromatin IP was performed for the indicated genes and factors as described in Materials and Methods. Tolerance was induced by pretreating cells with 100 ng/mL LPS for 24 h. T0, T1, and T3 indicate tolerized cells restimulated with LPS for 0, 1, and 3 h, respectively. Experiments were repeated three times with similar results.
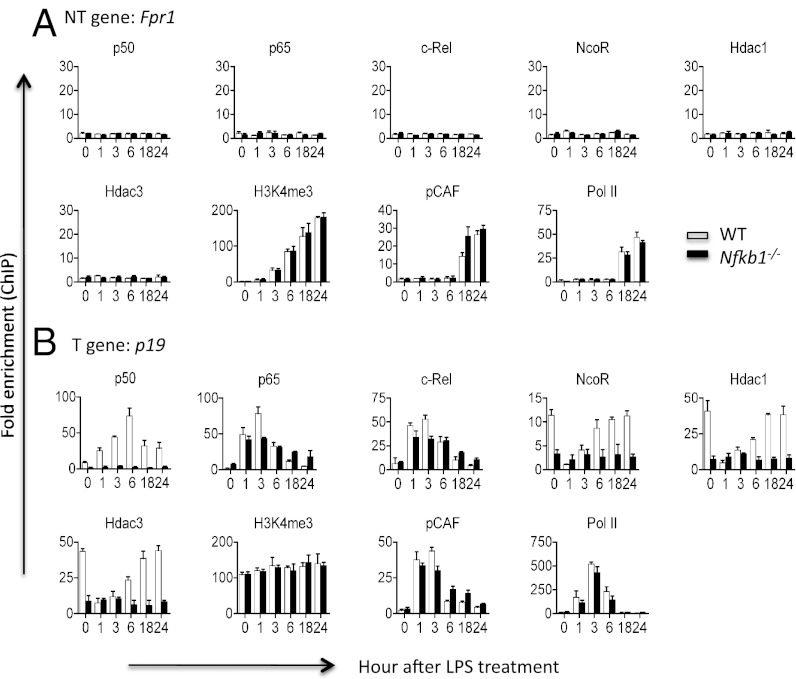
To test the hypothesis that NF-κB p50 is required for the epigenetic modification of the LPS-tolerized genes, we performed ChIP on both WT and Nfkb1 knockout cells that did not express p50 (27). We found that Nfkb1 gene mutation completely abolished the binding of transcriptional repressors (NcoR, Hdac1, and Hdac3) to T genes, but had no detectable effect on the epigenetic modification of NT genes (Fig. 3 and Fig. S5). These results indicate that p50 is involved in the repressive modification of the T genes.
Fig. 3.
Nfkb1 null mutation prevents NcoR and histone deacetylase binding to tolerizable genes, while having no effect on nontolerizable genes. Bone-marrow–derived macrophages from WT and Nfkb1−/− mice (n = 4) were either left untreated (0) or treated with 100 ng/mL LPS for up to 24 h as indicated. Cells were then fixed and chromatin IP was performed for the indicated genes and factors as described in Materials and Methods. Error bars depict SD of the mean. Experiments were repeated three times with similar results.
Because nuclear p65 and c-Rel levels were reduced after tolerance induction, we determined whether restoring their levels would abolish LPS tolerance using a retroviral gene transfer system. Thus, although the total levels of p65 and c-Rel were not altered by LPS tolerance (Fig. S6 A and B), the nuclear p65 and c-Rel levels were reduced in tolerized bone-marrow–derived macrophages (BMMs) and RAW264.7 macrophages (TL compared with NL condition) (Fig. S6D). To restore the nuclear levels of p65 and c-Rel in TL cells to pretolerance levels (as in NL cells), we first expressed p65 and c-Rel using the retroviral Migr1 vector. The nuclear p65 and c-Rel levels in TL cells treated with the respective NF-κB retroviruses were effectively restored compared with the NL groups (Fig. S6D). However, this restoration of nuclear p65 and c-Rel did not abolish LPS tolerance as measured by the TNFα gene expression (Fig. S6E).
LPS Induces the Formation of a Stable NcoR–Hdac3–Deacetylated-p50 Repressosome.
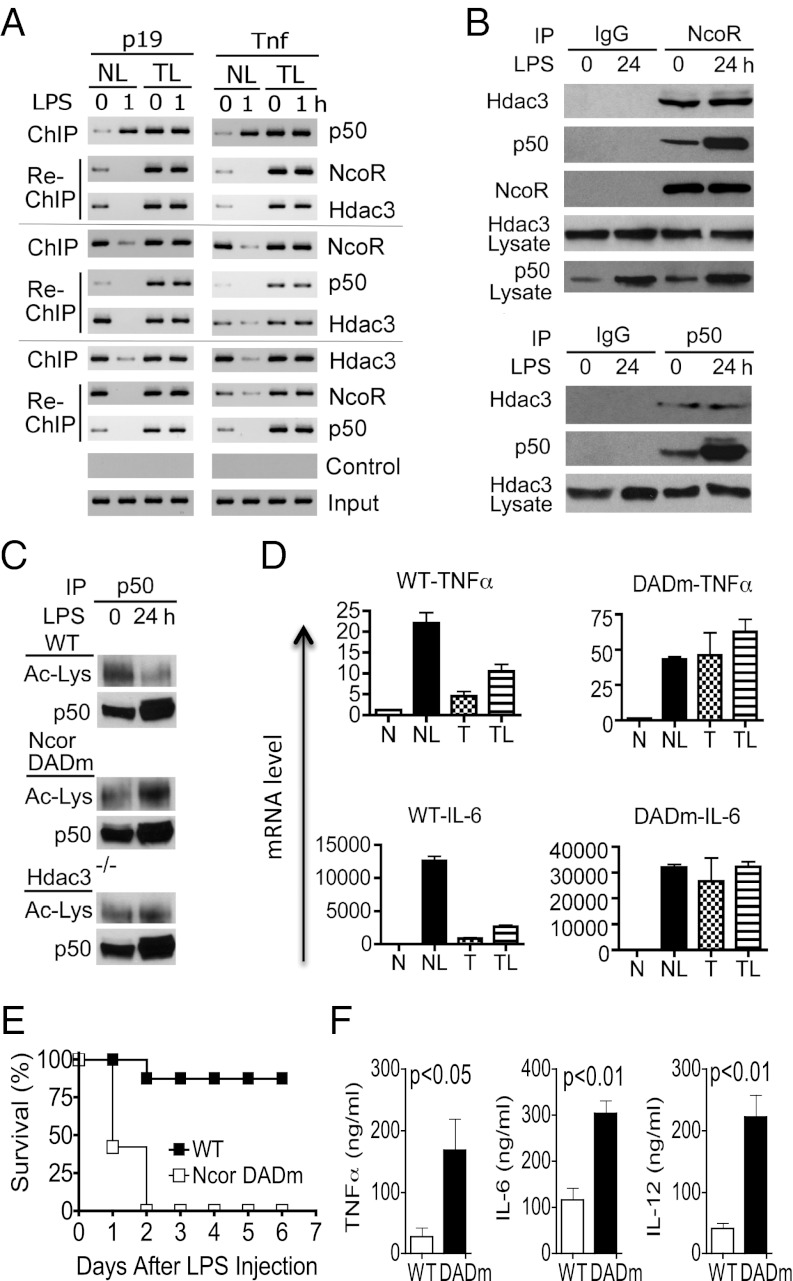
NF-κB p50 may repress gene expression through two distinct mechanisms: (i) by passively blocking binding of activating NF-κB dimers to DNA or (ii) by actively orchestrating the formation of repressosomes, which in turn inhibit gene transcription. To test the latter possibility in TLR tolerance, we searched for repressor complexes by ChIP, re-ChIP (also known as sequential ChIP), and coimmunoprecipitation (co-IP) in naïve and LPS-tolerized macrophages. In the re-ChIP, the precipitates from the first round of ChIP with anti-p50, anti-Hdac3, or anti-NcoR were subjected to a second round of ChIP with different antibodies so that multiple factors present in the same DNA complex could be identified (Fig. 4A). We found that only tolerized cells, but not naïve cells, contained a stable DNA-binding repressosome consisting of p50, NcoR, and Hdac3 (Fig. 4 A and B). The repressosome in LPS-tolerized cells remained bound to T gene promoters even after LPS challenge. By contrast, although a small amount of p50–NcoR–Hdac3 complex was detected in naïve cells, it was not stable because it completely dissociated from T gene promoters upon LPS stimulation (Fig. 4A). The stability of the repressosome may relate to both the quality and quantity of the p50 protein. The p50 in the tolerized cell nuclei were far more deacetylated and abundant than that in the naïve cell nuclei (Fig. 4C). The LPS-induced p50 deacetylation was likely mediated by Hdac3 because it was significantly reduced in Hdac3−/− macrophages and macrophages from deacetylase activation domain mutant (DADm) mice that carry a single amino acid substitution (Y478A) within the NcoR DAD. The mutant NcoR protein in the latter mice is stable but unable to associate with or activate Hdac3 (28).
Fig. 4.
A stable TLR4-induced repressosome mediates gene-specific tolerance and prevents sepsis. (A) Visualizing the TLR4-induced repressosome by re-ChIP. Naïve (NL) and tolerized (TL) bone-marrow–derived macrophages were either left untreated (0) or treated with 100 ng/mL LPS for 1 h as indicated. Tolerization was performed by pretreating cells with 100 ng/mL LPS for 24 h before the experiment. Cells were then fixed, and ChIP was performed using anti-p50, anti-NcoR, anti-Hdac3, or control IgG (control) as indicated. Re-ChIP was then carried out using the precipitates from the first round of ChIP for the indicated genes (p19 and Tnf) and factors (p50, NcoR, and Hdac3). (B) Detecting the TLR-induced repressor complex by coimmunoprecipitation. Bone-marrow–derived macrophages were either left untreated or treated with 100 ng/mL LPS for 24 h as indicated. Nuclear extracts were prepared and co-IP was performed using anti-NcoR, anti-p50, or control IgG. Western blots were performed using antibodies for the indicated proteins for both immunoprecipitates and total nuclear preparations (lysate). (C) Reduced p50 acetylation in tolerized cells. Bone-marrow–derived macrophages from WT, NcoR DADm, and Hdac3−/− mice were either left untreated or treated with 100 ng/mL LPS for 24 h to induce tolerance. Nuclear extracts were prepared and used for IP with anti-p50. The precipitated p50 was then subjected to electrophoresis and Western blot using an antiacetyllysine antibody (Ac-Lys) or anti-p50. (D) Lack of LPS tolerance in NcoR DADm macrophages. Bone-marrow–derived macrophages of the indicated genotypes were either (i) left untreated for 26 h (naïve, N), or (ii) rested for 24 h and treated with 100 ng/mL LPS for an additional 2 h (NL), or (iii) treated with 100 ng/mL LPS for 24 h and then rested in fresh media for an additional 2 h (tolerized, T), or (iv) treated with 100 ng/mL LPS for 24 h and restimulated with 100 ng/mL LPS for another 2 h (TL). TNFα and IL-6 mRNA levels were determined by real-time PCR. Error bars represent SD of the mean. (E) NcoR DAD mutation renders mice hypersensitive to septic shock. Sex- and age-matched WT (n = 9) and NcoR DADm mice (n = 8) were injected intraperitoneally with LPS (30 mg/kg) on day 0 and their survival rates recorded. The difference between the two groups was statistically significant (P < 0.0001). (F) Sera from the two groups of mice as treated in E were collected 36 h after the LPS injection. Concentrations of IL-6, TNFα, and IL-12p40 in the sera were determined by ELISA. Error bars depict SD of the mean. Experiments were repeated at least three times with similar results.
NcoR-Containing Repressosome Is Essential for LPS Tolerance.
Although we showed that both NcoR and Hdac3 were recruited to the repressosome in a p50-dependent manner, whether and how these proteins contribute to LPS tolerance is unknown. To address this issue, we first tested LPS tolerance in NcoR DADm macrophages in which NcoR–Hdac3 interaction is genetically disrupted (28). We found that NcoR DADm macrophages were not able to develop LPS tolerance and expressed high levels of TNFα and IL-6 even 24 h after the LPS tolerization (Fig. 4D). These results indicate that NcoR binding to Hdac3 is essential for LPS tolerance.
NcoR–Hdac3 Interaction Is Essential for Preventing Septic Shock.
Next, we tested the consequence of disrupting NcoR–Hdac3 interaction in LPS tolerance in mice. Following the injection of LPS, the vast majority of wild-type (WT) C57BL/6 mice survived and exhibited no detectable signs of illness 3 d later (Fig. 4E). By contrast, all NcoR DADm mice died of septic shock within 2 d of LPS challenge. Consistent with these clinical data, serum inflammatory cytokines that are known to mediate septic shock (TNFα, IL-6, and IL-12) were significantly higher in NcoR DADm mice than in WT mice (Fig. 4F). These results establish that NcoR–Hdac3 interaction is essential for generating LPS tolerance and for preventing septic shock.
NF-κB p50 Is Required for Hdac-Induced Gene Repression.
To determine whether p50 is required for Hdac-induced gene repression in LPS tolerized cells, we expressed Hdac1 and Hdac3 with or without p50 in LPS-treated macrophages. We found that Hdac1 and Hdac3 could suppress Tnf promoter only when p50 was coexpressed, indicating that the Hdac-induced repression is dependent on p50 (Fig. S7).
Discussion
Results reported here establish that the NF-κB binding motif of a gene promoter dictates its sensitivity to LPS tolerance. Thus, LPS activates two classes of genes: tolerizable genes that are controlled by NF-κB and nontolerizable genes that are not. This NF-κB motif-based division of TLR target genes allows the host to selectively tolerize one class while continuously activating another class of genes. Although p50:p50 homodimer can compete with stimulatory NF-κB dimers for DNA binding to repress gene expression, our data show that a stable repressosome induced by p50 is essential for generating LPS tolerance. These unique findings explain why TLR tolerance is gene specific and how selective use of NF-κB for inflammatory but not antimicrobial genes makes it possible to selectively tame down deleterious but not protective innate immune responses in the face of chronic or prolonged infections.
It should be pointed out that although ∼71% of tolerizable genes possess confirmed NF-κB binding motifs, other tolerizable genes may not contain them. On the other hand, although the vast majority of nontolerizable genes may not be regulated by NF-κB, ∼2% of them may. Therefore, in addition to consensus NF-κB binding motifs, other regulatory elements/mechanisms may be responsible for specifying TLR-induced gene repression. These may relate to the inducibility and the epigenetic state of the target genes, the use of other transcriptional regulators/elements, and the variation in the sequence and location of NF-κB binding motifs.
Chronic or repeated exposure of cells to LPS leads to a state of hyporesponsiveness, termed LPS or endotoxin tolerance, which is characterized primarily by a reduced expression of inflammatory genes (1, 9, 11–14); the expression of noninflammatory genes such as antimicrobial genes is not affected or further up-regulated in tolerized cells (1). The molecular mechanisms of LPS tolerance and suppression are not well understood, but two classes of negative regulators have been implicated: (i) gene-specific regulators that inhibit TLR4 target gene transcription and (ii) signal-specific regulators that block TLR-mediated signaling. Gene-specific regulators selectively target a group of genes but not others, which may explain why inflammatory genes are down-regulated, whereas antimicrobial genes are up-regulated in LPS-tolerized macrophages (1). By contrast, signal-specific regulators affect all genes downstream of a signaling pathway, leading to global inhibition of TLR target gene expression (1). We recently discovered that de novo synthesis of Bcl-3 in an NF-κB–dependent manner constitutes a negative feedback loop for NF-κB regulation (7). Newly synthesized Bcl-3 proteins enter the nucleus where they bind free p50 homodimers to terminate NF-κB transcriptional activity (7). A major finding from this work is that p50 homodimers exert their repressive effect through recruiting an NcoR-containing repressosome.
The signal-specific mechanisms of TLR suppression involve primarily competitive inhibition of TLR signaling molecules by their homologs or splice variants. These include the LPS-inducible IRAKM, a homolog of IRAK1, and IRAK2c/d, a splice variant of IRAK2, which interfere with the function of IRAK1 and IRAK4 (10). MyD88s is a splice variant of MyD88, which is unable to interact with IRAK4. Other negative regulators include TOLLIP, A20, and SIGIRR (single Ig IL-1R–related molecule). TOLLIP interacts with TLR2 and TLR4 as well as IRAK1 and interferes with the activation of NF-κB. A20 is an LPS-inducible inhibitor of NF-κB, which has recently been shown to play a critical role in the termination of TLR responses. The NF-κB regulatory functions of A20 are mediated through two ubiquitin-editing domains (29, 30). The N-terminal domain of A20 contains a deubiquitinating activity that removes K63-polyubiquitin chains from both RIP1 and TRAF6, thereby inhibiting the recruitment and activation of the TAK1/TAB2/TAB3 complex. In addition, a C-terminal ubiquitin ligase domain of A20 mediates the K48 polyubiquitination of RIP1, leading to its proteasomal degradation. SIGIRR directly binds to the TLR–IL-1R signaling complex to negatively regulate the proximal receptor signaling (31, 32). Additionally, TLR down-regulation and decoy receptor expression have also been reported, which may contribute to TLR suppression under certain circumstances (33). Because deficiency in any one of the negative regulators described above significantly diminishes the degree of TLR tolerance or suppression, it is likely that multiple nonredundant mechanisms are required to maintain TLR tolerance.
Materials and Methods
Mice.
Wild-type C57BL/6 mice were obtained from The Jackson Laboratory. The NcoR DADm mutant C57BL/6 mice (28) and the Hdac3–FLOX genetically modified C57BL/6 mice (34) were as previously described. When crossed with the mice expressing Cre recombinase from endogenous Lyz locus (The Jackson Laboratory), the myeloid-specific Hdac3 knockout mice were generated. The Nfkb1 knockout mice were obtained from The Jackson Laboratory. Age- and sex-matched wild-type littermates were used as controls. All procedures were preapproved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Cell Culture and Transfection.
The murine RAW264.7 macrophage cells (ATCC) were cultured in DMEM containing 10% (vol/vol) FBS, 2 mM glutamine, and 100 units/mL penicillin/100 microgram/ml streptomycin. Transfections were performed using Lipofectamine LTX according to the manufacturer’s instructions (Invitrogen).
To generate BMMs, bone marrow cells were harvested from mice and cultured for 7 d in 70% (vol/vol) D10 (DMEM containing 10% (vol/vol) FBS, 100 μg/mL streptomycin and 100 units/mL penicillin) and 30% (vol/vol) l-929 cell culture supernatant. Cells were washed twice with cold DPBS, collected with 5 mM EDTA in DPBS, and replated on tissue-culture plates. One day later, macrophages were either left untreated (naïve, N) or stimulated with 100 ng/mL LPS (L) for 24 h (tolerized, T); after additional washing with warm DPBS, cells were given either fresh media (N, T) or 100 ng/mL LPS (NL, TL).
Quantitative PCR.
Total RNA was isolated using RNeasy kits (Qiagen) primed with random hexamer oligonucleotides and reversely transcribed using Invitrogen reverse transcriptase II. Real-time quantitative PCR was performed using SYBR Green Master mix (Applied Biosystems). All data were normalized to 18s rRNA or Hprt mRNA.
Chromatin Immunoprecipitation (IP).
Chromatin immunoprecipitation was performed using the ChIP assay kit from Millipore according to the manufacturer’s instructions with minor modification. In brief, cells were first fixed with 1.5% (vol/vol) formaldehyde at room temperature for 15 min without stirring and lysed in lysis buffer, and DNA was broken up by sonication. After preclearance for 1 h at 4 °C with salmon sperm DNA-saturated protein A-agarose beads, chromatin solutions were immunoprecipitated overnight at 4 °C using 2 μg specific antibodies or control IgG. After washing with low salt buffer, high salt buffer, LiCl buffer, and 1× TE, the protein A-agarose beads were directly mixed with 200 μL 10% (wt/vol) Chelex-100 (Bio-Rad) slurry. After boiling for 10 min and a proteinase K treatment for 30 min, the beads were boiled again for 10 min to abolish proteinase K activity, which was followed by centrifugation at 12,000 × g for 1 min at 4 °C. The supernatant was collected and used for ChIP PCR. One percent of the sonicated chromatin preparations was used as a positive control to ensure equal input. All quantitative ChIP PCR data were normalized to those of the control IgG samples (which were set as 1) to calculate fold enrichments.
For re-ChIP, after washing twice with 1× TE, the precipitated protein A-agarose beads were suspended in 75 μL TE containing 10 mM DTT, and incubated at 37 °C for 30 min. After centrifugation at 800 × g for 2 min, the supernatant was diluted 20 times and used for the next round of ChIP. The PCR products were then analyzed on 2% agarose gels.
Promoter Constructs and Luciferase Assay.
Nontolerizable and tolerizable gene 5′ genomic fragments, which include Fpr1 (−1,817/+57), Ptges (−434/+47), Bpil2 (−425/+41), and p19 (−1,180/+110), were amplified by PCR from C57BL/6 murine genomic DNA, and cloned into the pGL3-basic vector (Promega). Transcription factor-absent sites were identified using the Transcriptional Element Search System program (www.cbil.upenn.edu/tess/). NF-κB binding site mutagenesis was performed using the QuikChange kit (Stratagene), according to the manufacturer’s instructions. Two (2×) consecutive consensus NF-κB binding motifs (gggaatttcc-gggaatttcc) were inserted into transcription factor-absent sites in the NT gene promoters by Klenow fragment (Promega). For the four (4×) NF-κB binding motif insertions, an additional pair of the consensus NF-κB binding motifs was inserted into the pGL3-basic plasmid at the KpnI/SacI site. The NT gene promoters were then inserted downstream. For the TK gene promoter, the promoter-containing NF-κB–Luc (Clontech) was used as the 4× NF-κB construct. Its Nhe I/BglII DNA fragment was replaced with either a random sequence, or four AP1, four IRF3, or four SP1 consensus sites. RAW264.7 cells were transfected with reporter constructs using Lipofectamin LTX (Invitrogen). The luciferase activities of whole cell lysates were analyzed using the dual-luciferase reporter assay system (Promega). Cotransfection with the Renilla-Luciferase expression vector pRL-TK (Promega) was performed for all reporter assays. For all samples, the data were normalized for transfection efficiency by dividing firefly luciferase activity by that of the Renilla luciferase.
ELISA.
Antibodies used in ELISA were purchased from BD Pharmingen and eBioscience including purified and biotinylated rat antimouse IL-6, TNFα, and IL-12. Quantitative ELISA was performed using paired mAbs specific for corresponding cytokines according to the manufacturer’s recommendations.
Western Blot.
Nuclear protein extract was prepared using a Nuclear Extract kit (Active Motif) according to the manufacturer’s instructions. Total cell lysate was prepared by suspending cells in the RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.4, 0.1% (wt/vol) SDS, 1% (vol/vol) Triton X-100, 1% (wt/vol) sodium deoxycholate, 100 μM Na3VO4, 5 mM EDTA, 1 mM PMSF) supplemented with 1× complete protease inhibitors mixture (Roche). Equal quantities of proteins were separated by SDS/PAGE, transferred to a nitrocellulose membrane, and blotted with specific antibodies. The membrane was developed using Pierce SuperSignal reagent.
Statistical Analyses.
The differences in cytokines, mRNA, and promoter activities were analyzed by two-tailed Student t test. The differences in survival rate were analyzed by Mann–Whitney u test.
Primers.
Primers used for quantitative real-time PCR:
| Gene | Forward primer | Reverse primer |
| Tnfα | ctgtagcccacgtcgtagc | ggttgtctttgagatccatgc |
| IL6 | caaagccagagtccttcaga | ttggtccttagccactcctt |
| Bpil2 | tccagctttggactcatcct | gtctgggaggaggcaatgta |
| p19 | aataatgtgccccgtatcca | aggctcccctttgaagatgt |
| Cxcl2 | agtttgccttgaccctgaag | ctttggttcttccgttgagg |
| IL1β | gagaaccaagcaacgacaaa | caaaccgtttttccatcttct |
| Nfkbia | cctggccagtgtagcagtct | agaggctaggtgcagacacg |
| Nfkbiz | gtggcaggtagagcaggaag | ccttgggcaacagcaatatg |
| Pim1 | tcaaggacacagtctacacgg | agcgatggtagcgaatcc |
| Hprt | cttcctcctcagaccgcttt | ataacctggttcatcatcgctaa |
| Peli1 | agccttaactgtgggcttga | cctgcacagcacatatggag |
Primers used for ChIP:
| Gene | Forward primer | Reverse primer |
| Tnfα | Aaccctctgcccccgcgatg | tcctcgctgagggagcttctgc |
| p19 | Tagccacaacaacctcacca | aagcggcttcctgatttctt |
| Bpil2 | Attgagtaattgtagtgaggcaattatg | cctcaagcccaggatgagtc |
| Fpr1 | Ggatatactctcagggtccttg | tgaactttccaacagctctgg |
Primers used for site-directed mutagenesis or NF-κB site insertion:
| Vector | Site | Type | Sequence |
| pGL3-TNFα | −660 | 2κB mutation-F | aggcttgtgaggtccgcgcactatcagggctgagttcattc |
| 2κB mutation-R | gaatgaactcagccctgatagtgcgcggacctcacaagcct | ||
| pNFκB-Luc | 27 | 4κB mutation-F | atcggctagcgcgcactatcgcgcactatcgcgcactatcgcgcactatcagatctatcg |
| 4κB mutation-R | cgatagatctgatagtgcgcgatagtgcgcgatagtgcgcgatagtgcgcgctagccgat | ||
| pGL3-Fpr1 | −57 | 2κB insertion-F | cctgttacccaatgcatttctgggaatttccgggaatttccgctgtttcccctcctcc |
| 2κB insertion-R | ggaggaggggaaacagcggaaattcccggaaattcccagaaatgcattgggtaacagg | ||
| pGL3-Fpr1 | −300 | 2κB insertion-F | cctcatagcttttccatatgatgggaatttccgggaatttcccagaaggttcttgggcac |
| 2κB insertion-R | gtgcccaagaaccttctgggaaattcccggaaattcccatcatatggaaaagctatgagg | ||
| pGL3-basic | 5 | 4κB insertion-F | atcgggtaccgggaatttccgggaatttccgggaatttccgggaatttccgaattcgagctcatcg |
| 4κB insertion-R | cgatgagctcgaattcggaaattcccggaaattcccggaaattcccggaaattcccggtacccgat | ||
| pGL3-basic | 27 | 4AP1 insertion-F | atcgggtaccggtgactcagtggtgactcagtggtgactcagtggtgactcagtgaattcatcg |
| 4AP1 insertion-R | cgatgaattcactgagtcaccactgagtcaccactgagtcaccactgagtcaccggtacccgat | ||
| pGL3-basic | 27 | 4IRF3 insertion-F | atcggctagcgaaacggaaattgaaacggaaattgaaacggaaattgaaacggaaattagatctatcg |
| 4IRF3 insertion-R | cgatagatctaatttccgtttcaatttccgtttcaatttccgtttcaatttccgtttcgctagccgat | ||
| pGL3-basic | 27 | 4SP1 insertion-F | atcggctagcgggggcggggccgggggcggggccgggggcggggccgggggcggggccagatctatcg |
| 4SP1 insertion-R | cgatagatctggccccgcccccggccccgcccccggccccgcccccggccccgcccccgctagccgat | ||
| pGL3-Hdc | −37 | κB mutation-F | gggcggggctaaaggaggcgtatagagaccgcattaaataag |
| κB mutation-R | cttatttaatgcggtctctatacgcctcctttagccccgccc | ||
| pGL3-Slfn1 | −55 | 2κB insertion-F | gaaaccggggttctccaaaagggaatttccgggaatttccctgaaatcgggacctagag |
| 2κB insertion-R | ctctaggtcccgatttcagggaaattcccggaaattcccttttggagaaccccggtttc |
Supplementary Material
Acknowledgments
The authors thank Dr. Sridhar Hannenhalli for his valuable assistance in the bioinformatic analysis of LPS target genes, Dr. Douglas Golenbook (University of Massachusetts) for providing the HEK293–TLR4 cells, and Dr. Andrew Bowie (Trinity College, Ireland) for the Ifnb1 luciferase reporter plasmid. This work was supported by National Institutes of Health Grants GM085112, DK070691, and AI50059 (to Y.H.C.). R.J.C. is supported by Science Foundation Ireland Grant 08/IN.1/B1843.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119842109/-/DCSupplemental.
References
- 1.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 4.Broad A, Jones DE, Kirby JA. Toll-like receptor (TLR) response tolerance: A key physiological “damage limitation” effect and an important potential opportunity for therapy. Curr Med Chem. 2006;13:2487–2502. doi: 10.2174/092986706778201675. [DOI] [PubMed] [Google Scholar]
- 5.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: New mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Brint EK, et al. ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat Immunol. 2004;5:373–379. doi: 10.1038/ni1050. [DOI] [PubMed] [Google Scholar]
- 7.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 8.Kinjyo I, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 9.Medvedev AE, Sabroe I, Hasday JD, Vogel SN. Tolerance to microbial TLR ligands: Molecular mechanisms and relevance to disease. J Endotoxin Res. 2006;12:133–150. doi: 10.1179/096805106X102255. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 11.Kastenbauer S, Ziegler-Heitbrock HW. NF-kappaB1 (p50) is upregulated in lipopolysaccharide tolerance and can block tumor necrosis factor gene expression. Infect Immun. 1999;67:1553–1559. doi: 10.1128/iai.67.4.1553-1559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum JT, Mandell RB. The effect of endotoxin and endotoxin tolerance on inflammation induced by mycobacterial adjuvant. Yale J Biol Med. 1983;56:293–301. [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler-Heitbrock HW. Molecular mechanism in tolerance to lipopolysaccharide. J Inflamm. 1995;45:13–26. [PubMed] [Google Scholar]
- 14.Bohuslav J, et al. Regulation of an essential innate immune response by the p50 subunit of NF-kappaB. J Clin Invest. 1998;102:1645–1652. doi: 10.1172/JCI3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myrianthefs P, Styliano W, Venetsanou K, Baltopoulos G. LPS tolerance in septic ICU patients: Laboratory and technical considerations. Surg Infect (Larchmt) 2004;5:63–64, author reply 64–65. doi: 10.1089/109629604773860327. [DOI] [PubMed] [Google Scholar]
- 16.Mages J, Dietrich H, Lang R. A genome-wide analysis of LPS tolerance in macrophages. Immunobiology. 2007;212:723–737. doi: 10.1016/j.imbio.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Ho Sui SJ, et al. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kollipara RK, Perumal NB. Motif prediction to distinguish LPS-stimulated pro-inflammatory vs. antibacterial macrophage genes. Immunome Res. 2010;6:5. doi: 10.1186/1745-7580-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 20.Martone R, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler-Heitbrock HW, et al. Tolerance to lipopolysaccharide involves mobilization of nuclear factor kappa B with predominance of p50 homodimers. J Biol Chem. 1994;269:17001–17004. [PubMed] [Google Scholar]
- 25.Ghisletti S, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell. 2009;35:48–57. doi: 10.1016/j.molcel.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 28.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 30.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 31.Thomassen E, Renshaw BR, Sims JE. Identification and characterization of SIGIRR, a molecule representing a novel subtype of the IL-1R superfamily. Cytokine. 1999;11:389–399. doi: 10.1006/cyto.1998.0452. [DOI] [PubMed] [Google Scholar]
- 32.Wald D, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 33.Nomura F, et al. Cutting edge: Endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J Immunol. 2000;164:3476–3479. doi: 10.4049/jimmunol.164.7.3476. [DOI] [PubMed] [Google Scholar]
- 34.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.