Abstract
The role of astrocytes in neurodegenerative processes is increasingly appreciated. Here we investigated the contribution of astrocytes to neurodegeneration in multiple sulfatase deficiency (MSD), a severe lysosomal storage disorder caused by mutations in the sulfatase modifying factor 1 (SUMF1) gene. Using Cre/Lox mouse models, we found that astrocyte-specific deletion of Sumf1 in vivo induced severe lysosomal storage and autophagy dysfunction with consequential cytoplasmic accumulation of autophagic substrates. Lysosomal storage in astrocytes was sufficient to induce degeneration of cortical neurons in vivo. Furthermore, in an ex vivo coculture assay, we observed that Sumf1−/− astrocytes failed to support the survival and function of wild-type cortical neurons, suggesting a non-cell autonomous mechanism for neurodegeneration. Compared with the astrocyte-specific deletion of Sumf1, the concomitant removal of Sumf1 in both neurons and glia in vivo induced a widespread neuronal loss and robust neuroinflammation. Finally, behavioral analysis of mice with astrocyte-specific deletion of Sumf1 compared with mice with Sumf1 deletion in both astrocytes and neurons allowed us to link a subset of neurological manifestations of MSD to astrocyte dysfunction. This study indicates that astrocytes are integral components of the neuropathology in MSD and that modulation of astrocyte function may impact disease course.
Keywords: lysosome, brain, glia
Multiple sulfatase deficiency (MSD) is a lysosomal storage disorder (LSD) caused by mutations in the sulfatase modifying factor 1 (SUMF1) gene that results in aberrant posttranslation modification of sulfatases (1). Sulfatases are a family of enzymes required for the turnover and degradation of sulfated compounds. Eight known metabolic disorders are caused by the deficiency of individual sulfatase activities. Six are LSDs (five mucopolysaccharidoses and metachromatic leukodystrophy); the remaining two disorders are caused by deficiency of nonlysosomal sulfatases. The impaired activity of all sulfatases in MSD is responsible for a very severe phenotype that combines all the clinical symptoms found in each individual sulfatase deficiency (2). As in many other LSDs, progressive and severe neurodegeneration is a prominent feature and is the most difficult challenge for therapy (3). Previous studies identified impaired autophagy in neurons as a crucial component in the pathogenic mechanisms leading to neurodegeneration in MSD as well as in other LSDs (4, 5). This lysosomal accumulation of undegraded substrates results in defective degradation of autophagosomes and causes a block of the autophagy pathway leading to an accumulation of ubiquitinated proteins, dysfunctional mitochondria, and neurodegeneration. To date, research has focused on neurons, but the contribution of nonneuronal cells to the neuropathology of MSD has remained largely unexplored.
Astrocytes play crucial roles in adult CNS homeostasis (6), including synaptic glutamate uptake (7), maintenance of extracellular potassium (8), and nutrient support for neurons (9). Here we demonstrate that, like neurons, astrocytes that lack Sumf1 suffer from severe lysosomal storage and autophagy impairment with consequential cytoplasmic accumulation of protein aggregates. By using in vivo and ex vivo approaches, we demonstrate that astrocyte dysfunction alone can lead to neuronal degeneration and in turn can impact the pathological manifestation of MSD significantly. Thus, this study proposes a mechanism of neurodegeneration in an LSD.
Results
Lysosomal Storage and Autophagy Dysfunction in Astrocytes of Sumf1−/− Mice.
We tested whether Sumf1−/− mice had astrocyte abnormalities. Transmission electron microscopy of Sumf1−/− brain revealed that 3-mo-old cortical astrocytes presented massive cytoplasmic vacuolization (Fig. 1A). The vacuoles were filled with amorphous or electron-dense material and were surrounded by a single or a double membrane (Fig. 1A and Fig. S1A). Subsequently, coimmunolabeling of astrocytes with GFAP and ubiquitin antibodies showed the accumulation of ubiquitin-positive aggregates in the cytoplasm of astrocytes of Sumf1−/− mice compared with controls (Fig. 1B). A similar phenotype was observed in neurons and microglia of Sumf1−/− mice (Fig. S1 B–D). To evaluate if this substrate accumulation was the consequence of impaired autophagy (10, 11), we isolated cortical astrocytes from control and Sumf1−/− mice that harbor a transgene that expresses a GFP-tagged LC3 protein (12), a well-established marker for autophagosomes (13). The cytoplasm of astrocytes from Sumf1−/− mice had abundant accumulation of large GFP+ autophagosome vesicles (Fig. 1C). Moreover, we observed a cytoplasmic increase of P62/SQSTM1, a known substrate of autophagy whose accumulation induces cellular toxicity (Fig. 1D) (14, 15). These data suggest that astrocytes from Sumf1−/− mice accumulate cytosolic substrates because of lysosomal/autophagic dysfunction.
Fig. 1.
Lysosomal storage and autophagy dysfunction in Sumf1−/− astrocytes. (A) Lysosomal storage in Sumf1−/− astrocytes. Electron micrograph shows large vacuoles (arrows) in the cytoplasm of astrocytes of 3-mo-old Sumf1−/− mice (Right). Astrocytes from control mice did not show signs of vacuolization (Left). (B) Astrocytes from Sumf1−/− mice accumulate ubiquitin-positive cytoplasmic aggregates. Frozen cortical tissue isolated from 3-mo-old Sumf1−/− (Lower) and control (Upper) mice immunostained with ubiquitin (green) and GFAP (red) antibodies. Insets show enlargements of the merge. (C) (Left) Primary astrocytes isolated from Sumf1−/−; GFP-LC3+/− and Sumf1+/+; GFP-LC3+/− were stained with GFP (green) and GFAP (red) antibodies. (Right) Histogram shows the number of GFP+ vesicles per cell. AV, autophagosomes. (D) (Left) Primary astrocytes isolated from Sumf1−/− and control mice were stained with GFAP (green) and P62/SQSTM1 (red) antibodies. (Right) Histogram shows the quantification of P62+ puncta per cell. Astrocytes were isolated from at least three mice per genotype, and at least 15 cells per mouse were counted. Error bars represent SD. *P ≤ 0.05, Student’s t test. (Scale bars: 2 μm in A; 10 μm in B; 5 μm in C and D.)
Lysosomal Storage in Astrocytes Causes Degeneration of Cortical Neurons in Vivo.
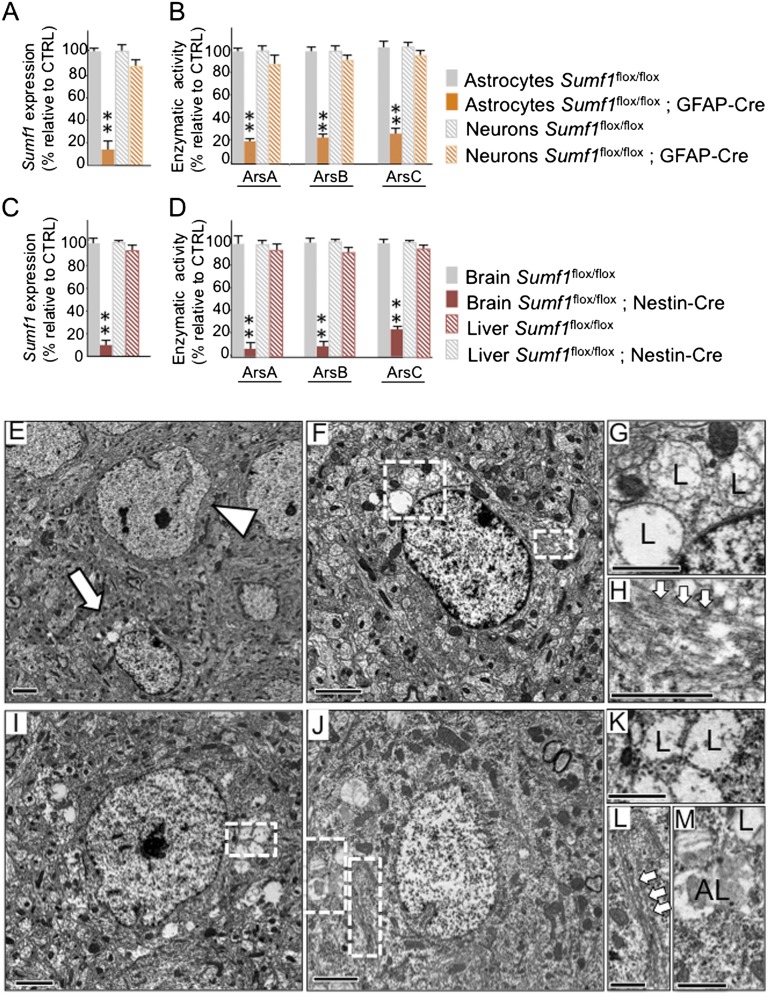
To examine the relationship between astrocyte dysfunction and neurodegeneration in MSD, we generated two mouse lines, one in which Sumf1 was deleted in both neurons and glia, obtained by crossing a transgenic mouse that expresses the Cre recombinase under the control of the nestin promoter with the Sumf1flox/flox mouse (Sumf1flox/flox; Nestin-Cre) (Fig. S2), and the other in which Sumf1 was deleted only in astrocytes, obtained using a transgenic mouse that expresses the Cre recombinase under the control of the GFAP promoter (Sumf1flox/flox; GFAP-Cre). We selected a GFAP-Cre line recently shown to be astrocyte specific (16). We first verified that this transgenic line is indeed specific for astrocytes by crossing it with a ROSA26-EYFP reporter mouse line, and we observed that the YFP signal colocalized with the astrocyte marker GFAP but not with the neuronal marker NeuN (Fig. S3). Furthermore, primary glial cultures isolated from cortex of postnatal day 1 (P1) Sumf1flox/flox; GFAP-Cre mice showed an almost complete lack of Sumf1 mRNA expression (Fig. 2A) and virtually no residual sulfatase activities (Fig. 2B), whereas primary cortical neurons isolated at embryonic day 16.5 (E16.5) had normal levels of Sumf1 mRNA and normal sulfatase activities (Fig. 2 A and B).
Fig. 2.
Lysosomal storage in Sumf1flox/flox; GFAP-Cre and Sumf1flox/flox; Nestin-Cre mouse lines. (A) Lack of Sumf1 expression in Sumf1flox/flox; GFAP-Cre astrocytes. Reverse transcription followed by quantitative PCR (RT-qPCR) of Sumf1 gene expression in astrocytes and neurons isolated from Sumf1flox/flox and Sumf1flox/flox; GFAP-Cre mice. (B) Lack of sulfatase activities in Sumf1flox/flox; GFAP-Cre astrocytes. The enzymatic activities of ArsA, ArsB, and ArsC were measured in astrocytes and neurons isolated from Sumf1flox/flox and Sumf1flox/flox; GFAP-Cre mice. (C) Lack of Sumf1 expression in Sumf1flox/flox; Nestin-Cre brain. RT-qPCR of Sumf1 was performed in whole-brain homogenates and, as control, in liver homogenates from Sumf1flox/flox and Sumf1flox/flox; Nestin-Cre mice. (D) Lack of sulfatase activities in Sumf1flox/flox; Nestin-Cre brain. Enzymatic activities of ArsA, ArsB, and ArsC were measured in whole-brain homogenates and liver homogenates from Sumf1flox/flox and Sumf1flox/flox; Nestin-Cre mice. All enzymatic activities are shown as percentage of activity ± SEM relative to the control genotype. Values represent mean of n = 3 mice per genotype. *P ≤ 0.05, **P ≤ 0.01, unpaired Student’s t test. (E–H) Lysosomal storage in Sumf1flox/flox; GFAP-Cre astrocytes. (E) Electron micrograph of cortical tissue isolated from a 6-mo-old Sumf1flox/flox; GFAP-Cre mouse showing a healthy neuron (arrowhead) in proximity to a vacuolized astrocyte (arrow). (F) Higher magnification of the astrocyte in E. (G) Enlargement of the large boxed area in F showing lysosomes (L) filled with floccular material. (H) Enlargement of the small boxed area in F showing GFAP fibrils (arrows) inside the cytoplasm of the astrocyte. (I–M) Lysosomal storage in Sumf1flox/flox; Nestin-Cre neurons and astrocytes. Electron micrograph of cortical tissue from a 6-mo-old Sumf1flox/flox; Nestin-Cre mouse showing a vacuolized neuron (I) and astrocyte (J). (K) Enlargement of the boxed area in I shows vacuolized lysosomes (L). (L and M) Enlargements of the boxed areas in J showing GFAP filaments (arrows) (L) and vacuolized lysosomes (L) and autolysosomes (AL) (M). (Scale bars: 2 μm in E, F, I, and J; 1 μm in G, H, and K–M.)
Conversely, both the expression levels of Sumf1 and the activities of sulfatases were almost undetectable in whole-brain lysates of Sumf1flox/flox; Nestin-Cre mice (Fig. 2 C and D), indicating that the recombination occurred in both neurons and glial cells.
Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre mice were born following Mendelian ratios and were indistinguishable in appearance from control littermates. Starting at age 3 mo, progressive weight loss occurred in Sumf1flox/flox; Nestin-Cre mice, whereas no difference was observed between Sumf1flox/flox; GFAP-Cre mice and controls (Fig. S4A). As early as age 6 mo, both genotypes showed signs of neurodegeneration that included abnormal limb-clasping reflexes (Fig. S4B), tremors, epileptic episodes, and behavioral abnormalities (see below) (Fig. S4C).
Ultrastructural analysis of brain slices isolated from 6-mo-old Sumf1flox/flox; GFAP-Cre mice showed neurons with normal cytoplasm in close proximity to astrocytes with enlarged vacuoles containing floccular material, which are typical signs of lysosomal storage (Fig. 2 E–H). As expected, in Sumf1flox/flox;Nestin-Cre mice the cytoplasm of both neurons and astrocytes presented extensive vacuolization (Fig. 2 I–M).
Regional neurodegeneration was evaluated qualitatively using Nissl staining in sagittal brain sections isolated from Sumf1flox/flox; Nestin-Cre, Sumf1flox/flox; GFAP-Cre, and control mice at age 6 mo. We observed decreased cellular density in cortical regions in both Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre mice as compared with controls (Fig. S5A). To perform a quantitative analysis, we undertook a grid-based neuronal count analysis using NeuN immunohistochemistry on cortical slices isolated from 6- and 12-mo-old mice. At both time points, we observed a significant decrease in the number of cortical neurons in all examined layers of the cortex (II–V) in Sumf1flox/flox; Nestin-Cre mice compared with control mice (Fig. 3 A and C and Fig. S5 B and C). Surprisingly, decreased neuronal number also was observed in the cortex of Sumf1flox/flox; GFAP-Cre mice compared with control mice (Fig. 3 A and C and Fig. S5 B and C). Interestingly, in Sumf1flox/flox; GFAP-Cre mice this decrease was evident in the more superficial layers (II– IV), but no significant changes were observed in deeper cortical layers. The phenotype was more pronounced in 12-mo-old mice, suggesting progressive neurodegeneration. These observations suggest that lysosomal storage in cortical astrocytes is sufficient to trigger degeneration of cortical neurons. We then analyzed Purkinje cells of the cerebellum, because their degeneration is documented in several LSDs (17–21). Calbindin staining showed a dramatic loss of Purkinje cells in Sumf1flox/flox; Nestin-Cre mice at age 12 mo, but no significant differences were found in Sumf1flox/flox; GFAP-Cre mice as compared with control mice at the same age (Fig. 3 B and D), even though there was expression of Cre in the Bergmann glia of these mice (Fig. S3C). These data indicated that Purkinje cell degeneration likely was cell autonomous. Interestingly, in younger (3-mo-old) Sumf1flox/flox; Nestin-Cre mice we observed massive lysosomal vacuolization in Purkinje cells, but cortical neurons appeared to be less affected (Fig. S6).
Fig. 3.
Lysosomal storage in astrocytes induced cortical neuronal degeneration. (A) NeuN immunostaining of frontal cortex of 12-mo-old mice of the indicated genotypes. Dashed lines mark the different cortical layers. (B) Calbindin immunostaining of Purkinje cells in cerebellar sections from 12-mo-old mice of the indicated genotypes. Boxed areas represent enlargements showing detail of Purkinje cell layer. (C and D) Quantification of neurons in the different cortical layers (C) and of Purkinje cells (D) in Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre mice. Values are expressed as percent ± SEM relative to control. *P ≤ 0.05; **P ≤ 0.01, Student’s t test. (Scale bars: 200 μm.)
Lysosomal Storage in Astrocytes Impairs Their Ability to Support Cortical Neuron Survival.
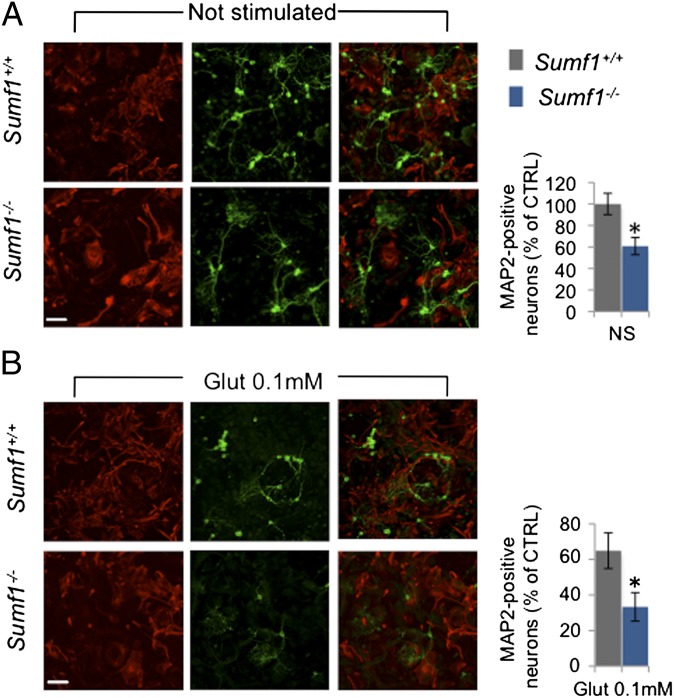
To confirm that lack of Sumf1 in astrocytes can trigger death of cortical neurons, we cocultured cortical neurons onto a layer of astrocytes. Wild-type cortical neurons isolated from embryos at E15.5 were plated at similar densities on a layer of cortical astrocytes isolated from newborn wild-type or Sumf1−/− mice. After 12 d of culture, the neurons plated on wild-type astrocytes formed extensive neuronal networks, whereas the neurons plated on Sumf1−/− astrocytes were significantly fewer and appeared less branched (Fig. 4A). Furthermore, challenging neurons with glutamate (0.1 mM for 24 h) further decreased the survival of the neurons cocultured with Sumf1−/− astrocytes as compared with wild-type astrocytes (33 ± 6% in Sumf1−/− vs. 65 ± 10% in wild type) (Fig. 4B). A similar difference was obtained after stimulation with a higher dose of glutamate (0.2 mM) (Fig. S7A). However, treatment with NMDA (0.1 mM), which causes neuronal toxicity independently of astrocytes, resulted in comparable cell death (Fig. S7B). Together these data indicate that lack of Sumf1 impairs the ability of astrocytes to provide support and protection to cortical neurons.
Fig. 4.
Lysosomal storage in astrocytes induces cortical neuron degeneration in an ex vivo coculture assay. (A and B) (Left) MAP2 (green) and GFAP (red) immunostaining of neurons and astrocytes, respectively, untreated (A) or treated with glutamate 0.1 mM for 24 h (B). (Right) Histograms represent quantification of MAP2+ neurons after 12 d in culture (A) and quantification of MAP2+ neurons after glutamate stimulation relative to the same sample without stimulation (B). Data represent mean ± SEM from three independent coculture experiments. *P ≤ 0.05, Student’s t test. (Scale bars: 20 μm.)
Lysosomal Storage in Neurons Elicits Neuroinflammation in MSD.
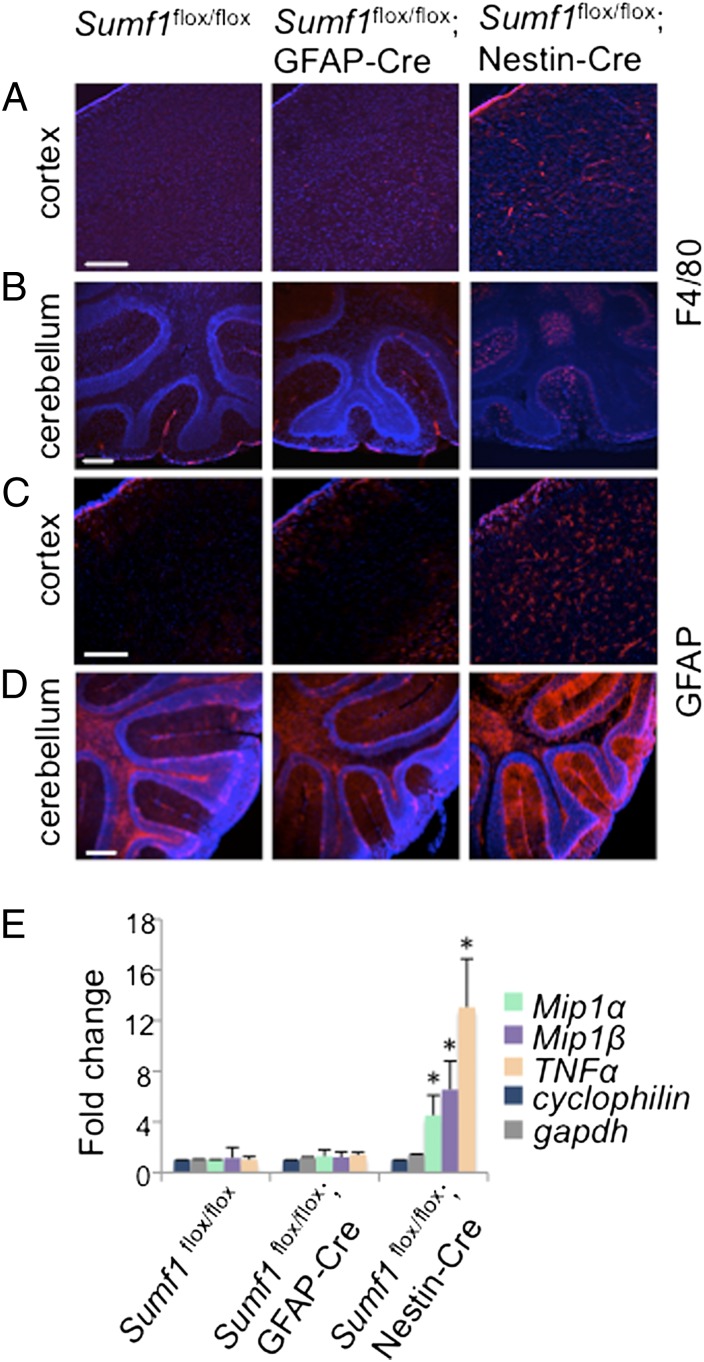
Neuroinflammation is implicated in the progressive nature of several neurodegenerative diseases and is described in almost all LSDs with neurological involvement (21–27). However, it is still unclear whether macrophage and astrocyte activation in LSDs is triggered by their intracellular storage or represents a response to neuronal damage. To address this question, we examined neuroinflammation in our mouse models. We observed strong microglial activation in different brain regions of Sumf1flox/flox; Nestin-Cre mice (Fig. 5 A and B), although in this model the microglia were not subjected to the Cre-mediated Sumf1 deletion. We did not observe any microglial activation in Sumf1flox/flox; GFAP-Cre brains, suggesting that direct neuronal dysfunction triggers microglia activation. Similarly, Sumf1flox/flox; Nestin-Cre brains presented widespread astrogliosis, and the Sumf1flox/flox; GFAP-Cre brains did not (Fig. 5 C and D), even though in both models astrocytes presented lysosomal vacuolization. Furthermore, the mRNA levels of the chemokines Mip1α and Mip1β and of the cytokine TNF-α were significantly up-regulated in Sumf1flox/flox; Nestin-Cre mice but not in Sumf1flox/flox; GFAP-Cre mice compared with control mice (Fig. 5E). These observations clearly indicate that lysosomal storage in neurons, rather than in astrocytes or microglia, triggers neuroinflammation in MSDs.
Fig. 5.
Neuroinflammation in Sumf1flox/flox; Nestin-Cre but not in Sumf1flox/flox; GFAP-Cre mouse brain. (A and B) F4/80 immunostaining in cortical (A) and cerebellar (B) sections isolated from 6-mo-old mice of the indicated genotype. (C and D) GFAP immunostaining of cortical (C) and cerebellar (D) sections from 6-mo-old mice of the indicated genotype. (E) RT-qPCR of Mip1α and Mip1β chemokines and TNF-α cytokine in Sumf1flox/flox, Sumf1flox/flox; GFAP-Cre, and Sumf1flox/flox; Nestin-Cre brain extracts from 6-mo-old mice. Values represent means ± SEM of three mice for each group. *P ≤ 0.05, Student’s t test. (Scale bars: 200 μm.)
Astrocyte Dysfunction Contributes to MSD Behavioral Abnormalities.
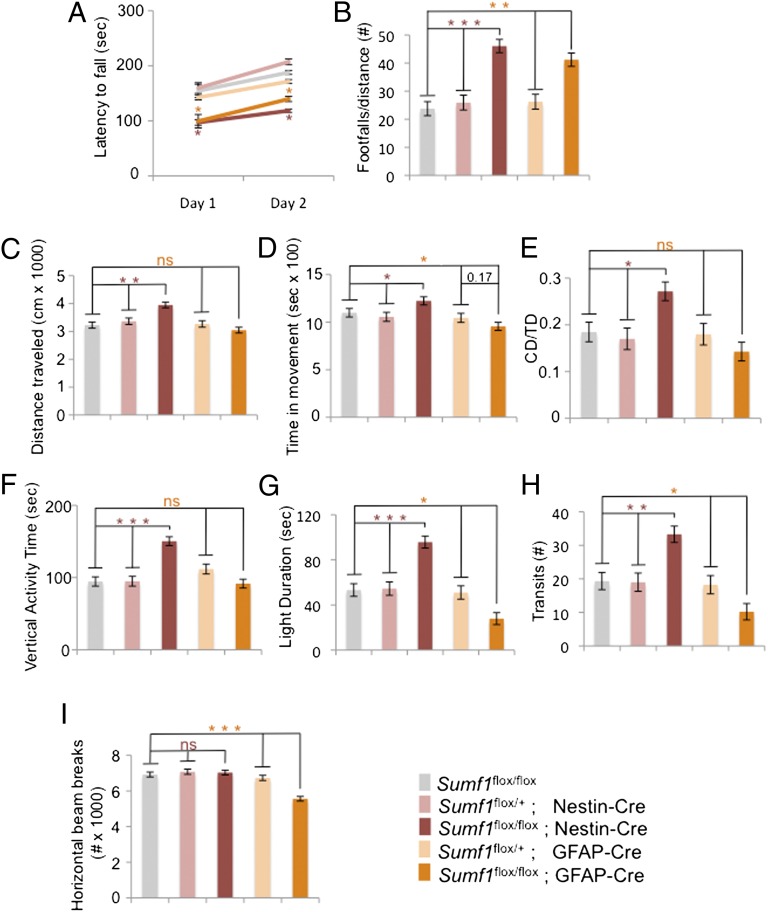
To study the neurological phenotype in MSD without the influence of peripheral organ dysfunction and to understand the contribution of astrocytes, we performed a panel of behavioral tests in Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre and their littermate controls. At age 7 mo, mice from both genotypes showed a statistically significant impairment in motor performance as assessed by decreased latency to fall in the Rotarod test and increased number of footfalls in the parallel-rod test (Fig. 6 A and B). However, Sumf1flox/flox; Nestin-Cre mice were unable to improve their performance on the Rotarod after several trials, but the Sumf1flox/flox; GFAP-Cre mice showed signs of improvement (Fig. 6A). The Sumf1flox/flox; Nestin-Cre mice also presented a specific hyperactive behavior demonstrated in both open-field and light/dark tests (Fig. 6 C–H), but the Sumf1flox/flox; GFAP-Cre mice were significantly hypoactive and more anxious than control mice because they spent less time in the light and had a reduced number of transits in the light/dark test and reduced number of horizontal beam breaks in the open-field test (Fig. 6 G–I). Therefore, although concomitant storage in neurons and glia causes a more severe phenotype, the lysosomal dysfunction in astrocytes alone is sufficient to cause neurological impairment.
Fig. 6.
Astrocyte dysfunction contributes to MSD neurological impairment. (A and B) Locomotor activity. (A) Rotarod test. Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox;GFAP-Cre mice showed decreased latency to fall compared with the respective controls. (B) Parallel rod test. Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre mice presented an increased number of footfalls compared with the respective controls. (C–H) Sumf1flox/flox; Nestin-Cre mice showed hyperactive behavior as demonstrated by an increase in total distance traveled (C); increased time spent in movement (D); increased distance traveled in center (CD) relative to total distance (TD) traveled (E); increased vertical activity in the open-field assay (F); and increased time spent in the light (G) and increased number of transits between the two sides of the light/dark box in the light/dark test (H). Sumf1flox/flox; GFAP-Cre mice show hypoactive behavior as demonstrated by decreased time spent in the light (G), decreased number of transits between the two sides of the light/dark box in the light/dark test (H), and decreased number of horizontal beam breaks in the open-field test (I). The number of mice of each genotype used was Sumf1flox/flox, n = 9; Sumf1flox/+; Nestin-Cre, n = 8; Sumf1flox/+; GFAP-Cre, n = 8; Sumf1flox/flox; Nestin-Cre, n = 10; Sumf1flox/flox; GFAP-Cre, n = 10. The control genotypes were Sumf1flox/flox and Sumf1flox/+; Nestin-Cre for Sumf1flox/flox; Nestin-Cre mice and Sumf1flox/flox and Sumf1flox/+; GFAP-Cre for Sumf1flox/flox; GFAP-Cre mice. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. ns, not significant. Values represent mean ± SEM.
Discussion
Astrocytes are the main neural cell type responsible for the maintenance of brain homeostasis. Their processes form extensive networks that modulate neuronal activity through the expression of various receptors for neurotransmitters, several transporters, cytokines, and growth factors. Indeed, they play critical roles in neurotransmitter trafficking and recycling, nutrient and ion metabolism, and protection against oxidative stress (28). Consistent with such a variety of fundamental functions exerted by astrocytes to support neurons, astrocyte impairment has been found to contribute to neuronal dysfunction in several neurodegenerative diseases, such as amyotrophic lateral sclerosis, Alzheimer’s disease, and Huntington disease (29–31). Neurodegeneration is one of the most common and prominent features in LSDs (32, 33), but very little is known about the role of astrocytes in their neuropathogenesis.
Here we studied a severe type of LSD, multiple sulfatase deficiency. To analyze the contribution of astrocytes to the neurological phenotype of MSD, we generated a mouse line with conditional knockout of the Sumf1 gene and crossed that mouse line with a transgenic mouse line expressing the Cre recombinase gene under the control of the GFAP promoter to delete Sumf1 specifically in astrocytes. We observed that Sumf1 deletion in astrocytes caused lysosomal/autophagic dysfunction and in turn led to the accumulation of cytosolic substrates in their cytoplasm. As a consequence, we found both in vivo and in vitro that astrocytes lacking the Sumf1 gene contributed directly to neurodegeneration because they lost their ability to support neuronal survival and function. This supportive function appeared to be particularly important for the survival of cortical neurons, although we cannot exclude the possibility that other neuronal populations also were affected in Sumf1flox/flox; GFAP-Cre mice.
Recent studies performed on a model of LSD, Niemann–Pick type C disease, showed that selective deletion of the Npc1 gene in astrocytes did not induce neurodegeneration, and indeed this disease is caused largely by cell-autonomous degeneration of Purkinje cells (34, 35). Consistent with this observation, we found that although lysosomal dysfunction in astrocytes may represent a significant determinant for cortical neuronal degeneration, a cell-autonomous pathway accounted for Purkinje cell death, because these neurons degenerated only in the Sumf1flox/flox; Nestin-Cre mice. In support of this model, in 3-mo-old Sumf1flox/flox; Nestin-Cre mice we observed a massive lysosomal vacuolization in Purkinje cells, but this vacuolization was barely detectable in cortical neurons (Fig. S6). This difference may be caused by higher glycosaminoglycan metabolism in Purkinje cells than in cortical neurons. Together these data suggest that neurodegeneration in LSDs involves both cell-autonomous and nonautonomous pathways.
Lysosomal enzymes either are targeted to the lysosome or are secreted in a mannose-6-phosphate receptor–dependent fashion (36). Secreted enzymes can be taken up by the surrounding cells or even by distant cells through the circulation. This mechanism of secretion and uptake most likely accounts for the milder cellular phenotype observed in Sumf1flox/flox; Nestin-Cre mice as compared with Sumf1−/− mice (compare vacuolization in Fig. 2 I and J with that observed in Fig. 1A and Fig. S1B). In Sumf1flox/flox; Nestin-Cre mice sulfatases may be secreted by nonrecombined cells such as microglia and blood vessel cells and taken up by neurons and astrocytes. However, our data indicate that uptake of enzymes from healthy cell types is not able to compensate for the genetic defect. Indeed, even if hematopoietic stem cell transplantation (HSCT) has demonstrated some efficacy for some LSDs, proving that donor-derived macrophages and microglia can progressively become a stable source of endogenous enzyme in the CNS, this approach has been unsatisfactory for many other LSDs (37). Currently, new promising therapies combine gene therapy and HSCT to obtain hematopoietic donor cells that express supraphysiological levels of the absent enzyme, resulting in increased levels of enzyme secretion (38).
Because most brain cell types are affected by the lysosomal storage in LSDs that present neurological involvement, the activation of the inflammatory system could be either the consequence of the intracellular storage of astrocytes and microglia or the secondary effect of neuronal dysfunction. Neuroinflammation has been proposed to induce oxidative stress, excitotoxicity, and metabolic failure in neurodegenerative diseases. In this scenario the reactive astrocytes and microglia would exacerbate neuronal deterioration at a later stage of disease progression (39–41). We observed microglial activation and astrogliosis only in Sumf1flox/flox; Nestin-Cre mice. This observation suggests at least two important conclusions: (i) microglial activation and astrogliosis are triggered by neuronal dysfunction, probably as an attempt to clear dying cells, and (ii) astrogliosis does not represent the main mechanism through which astrocytes contribute to neurodegeneration in LSDs, because no signs of astrogliosis were observed in Sumf1flox/flox; GFAP-Cre mice. Most likely, aberrant lysosomal storage in astrocytes hampers more fundamental and supportive functions needed for neuronal survival.
Comprehensive descriptions of the neurological phenotype of the MSD mouse model had not been possible in previous studies because of the early lethality caused by the complex systemic phenotype (21, 42). The generation of the conditional Sumf1 mouse line allowed us to overcome this limitation. We observed distinct behavioral phenotypes in Sumf1flox/flox; Nestin-Cre and Sumf1flox/flox; GFAP-Cre mice: Although they both showed significant impairment in motor performance, the deletion of the Sumf1 gene specifically in astrocytes caused hypoactivity and anxiety-like behavior, whereas its absence in both neurons and glia was responsible for hyperactivity and reduced motor-learning ability (Fig. S8). Interestingly, these neurological symptoms represent common features observed in different phases of disease manifestations in LSDs: Fear and anxiety generally are reported as early symptoms, whereas hyperactivity, learning difficulties, and progressive neurodegeneration occur at a later stage of the disease (43).
Thus, our data suggest that the progressive neurological deterioration found in LSDs could result from at least two independent insults to neurons, one non–cell-autonomous insult caused by astrocyte dysfunction and another, more severe, cell-autonomous insult caused by the lysosomal engulfment and dysfunction.
Materials and Methods
Mice.
To generate a conditional null mutant of the mouse Sumf1 gene (Sumf1flox mice), we used gene targeting to insert loxP sites on either side of exon 4 (Fig. S2A, green arrows). Cre-mediated excision of exon 4 is predicted to cause a frameshift mutation and the incorporation of multiple stop codons. The conditional targeting vector for Sumf1 gene was constructed using a BAC containing the C57BL/6J Sumf1 genomic clone. A fragment spanning from intron 3 to intron 6 of Sumf1 was subcloned into PL253 (a pBluescript-derived plasmid) via recombineering (44) using EL350 bacteria cells. Subsequently the loxP site upstream of the exon 4 was cloned into the plasmid by introducing a floxed neomycin-resistance (Neo) cassette (amplified from PL452 plasmid) via homologous recombination. Then the Neo cassette was removed via Cre recombinase, leaving a single loxP site at the targeted locus. The insertion of the second loxP site was accomplished using a Neo cassette flanked with two FRT sites (Fig. S2A, black arrows) and one loxP site following the second FRT (amplified from PL451 plasmid); the Sumf1 targeting vector was linearized with NotI and electroporated into Bruce4 mouse ES cells. The G418-resistant clones were screened by Southern Blot using the strategy shown in Fig. S2. Euploid clones that had undergone homologous recombination were injected into albino C57BL6/J blastocysts. Germline transmission of the floxed allele in offspring was confirmed by Southern blot (Fig. S2B). To remove the Neo cassette, the resulting mice were crossed with mice expressing FLP recombinase (strain B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/JRainJ; Jackson Laboratories). GFAP-Cre mice [strain name B6.Cg-Tg(GFAP-cre)8Gtm] were obtained from the National Cancer Institute (NCI) Mouse Repository. Nestin-Cre mice [strain name B6.Cg-Tg(Nes-cre)1Kln/J] were purchased from Jackson Laboratories. The R26-stop-EYFP mutant mice [strain name B6.129 × 1-Gt(ROSA)26Sortm(EYFP)Cos/J] (Jackson Laboratories) have a loxP-flanked stop sequence followed by EYFP inserted into the ROSA locus. The transgenic line expressing the GFP-tagged LC3 was a generous gift from N. Mizushima (Tokyo University, Tokyo, Japan). All animal procedures were approved by the Baylor College of Medicine on the Use and Care of Animals.
Genotyping.
Genotyping was performed on DNA isolated from tail biopsy at the time of weaning. PCR primers for Sumf1flox were forward, 5′-TGGAGTGGGCAGGTGGAGTCAT- 3′, and reverse, 5′-CACAGCACGCAGGAACTGTGAG- 3′. Predicted PCR products were 180 bp for the wild-type allele and 250 bp for the targeted allele. Genotyping of FLP recombinase mice, R26-stop-EYFP mutant mice, and Nestin-Cre mice was performed as described by Jackson Laboratories protocols. Genotyping of GFAP-Cre mice was performed as reported by NCI Mouse Repository. Genotyping of GFP-LC3 mice was described previously (12).
Neuronal Quantification.
For quantification of cortical neurons and Purkinje cells, five mouse brains for each genotype were divided sagittally into two parts and embedded in paraffin blocks. For cortical neurons count, six sections (6 μm thick) per brain were cut in a sagittal plane 500 μm lateral to the midline and were stained with NeuN antibody. The sections then were photographed, and a 200-μm2 grid was applied using Adobe Photoshop software. Subsequently four squares per section were selected from an area of the motor cortex below the bregma according to stereotaxic coordinates (45). The number of NeuN+ dots per square was counted using the cell-counter program (ImageJ software; National Institutes of Health) with a fixed threshold. The cortical layers were defined as follows: layers II and III, 150 μm from the surface to a depth of 350 μm; layer IV, 350–550 μm from the surface; layer V, 550–750 μm from the surface (46). To count Purkinje cells, at least three sagittal sections (6 μm thick) per brain were cut and stained with calbindin antibody. Cells were quantified within a defined region of the cerebellum (lobe VIII). The experiments were performed with the operator blinded to genotype.
Electron Microscopy and Morphological Criteria.
Ultrathin (∼70-nm) sections were obtained using an RMC MT6000-XL ultramicrotome (Eiko) and a Diatome Ultra45 diamond knife and were collected on 150-mesh hexagonal copper grids. The ultrathin sections then were stained with Reynold's lead citrate for 4 min. The air-dried samples were examined on an Hitachi H7500 transmission electron microscope, and images were captured using a Gatan US1000 digital camera and Digital Micrograph v1.82.366 software.
Following the morphological criteria in ref. 47, autophagosomes were identified as double-membrane vesicles with intact cytoplasmic material inside. Lysosomes were identified as single-membrane vacuoles containing amorphous and degraded material. Autolysosomes were single-membrane vacuoles containing partially degraded cytoplasmatic material.
The different cell populations of the brain were distinguished according morphological criteria (48). Briefly, glial cells were distinguished from neurons by their smaller, more irregular shape and by the prominent differences in the nucleus (absence of nucleolus and with a condensed envelope). Microglia cells are very similar to macrophages in morphology; they are the smallest cells within the CNS and have an irregular shape and a darker cytoplasm. Astrocytes are quite similar to oligodendrocytes in appearance, but their cytoplasm contains characteristic intermediated-sized filaments composed of GFAP instead of the microtubular filaments found in oligodendrocytes.
Astrocytes and Neuronal Cultures.
Astrocyte cultures were prepared from cortex isolated from Sumf1−/− or Sumf1+/+ P1 or P2 pups. They were plated at 100 μg/mL on poly-d-lysine–coated coverslips and cultured in DMEM/F12 with l-glutamine and 15 mM Hepes, supplemented with 10% (vol/vol) heat- inactivated horse serum (HS), 10% (vol/vol) FBS, and 1 mM sodium pyruvate. After 2 wk in culture, the astrocytes were examined by immunofluorescence. For glia/neuron coculture, cortical neurons were isolated from wild-type mouse embryos at E15.5 and were plated on top of Sumf1−/− or Sumf1+/+ astrocytes at 90% of confluence (∼2 wk in culture). To inhibit further proliferation of glial cells, cytosine arabinoside (10 μM) was added 2 d after neurons were plated. The coculture was cultured in MEM supplemented with 10% (wt/vol) glucose-bicarbonate buffer, 5% (vol/vol) FBS, 5% (vol/vol) heat-inactivated HS, and 1 mM sodium pyruvate.
Immunofluorescence and Immunohistochemistry.
For staining of mouse brain sections, mice were injected i.p. with 20 mg/mL Avertin (Winthrop Laboratories) and then were subjected to intracardial perfusion using 4% (wt/vol) paraformaldehyde (PFA) in PBS. Brains were dissected and postfixed with buffered 4% (wt/vol) PFA overnight at 4 °C, then were washed with PBS and cryoprotected in successive sucrose solutions diluted with PBS (5% for 30 min, 10% for several hours, and 20% overnight at 4 °C; all wt/vol), and finally were embedded in OCT (Sakura). Cryostat sections were cut at 30 or 50 μm (floating sections). For immunofluorescence floating sections were blocked and permeabilized in 3% (wt/vol) BSA, 5% goat serum in PBS + 0.3% Triton X-100 for 3 h and then were incubated with the primary antibody overnight. Sections were washed three times with 3% BSA in PBS + 0.3% Triton X-100 and then were incubated for 3 h with secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 633. For immunofluorescence of astrocyte culture or astrocyte/neuron coculture, cells were washed twice in PBS, fixed with cold-buffered 4% (wt/vol) PFA for 15 min at room temperature, permeabilized with PBS + 0.2% Triton X-100 for 30 min at room temperature, and blocked in 3% (wt/vol) BSA, 5% (vol/vol) HS in PBS + 0.1% Triton X-100. The primary antibody was added overnight; then cells were washed with PBS and incubated with secondary antibodies. Images were captured using a Leica SP5 confocal microscope, and quantitative analysis performed using Image J software.
For immunohistochemistry, after being postfixed with buffered 4% (wt/vol) PFA overnight at 4 °C, brains were dehydrated in a graded series of ethanol, cleared with xylene, and infiltrated with paraffin. Paraffin-embedded blocks were cut on a microtome in 6-μm sections. Immunohistochemistry was performed using the Vectastain ABC kit (Vector Labs) following the manufacturer’s instructions. Signal was developed using 0.05% 3,3-diaminobenzidine tetrahydrochloride in 0.02% H2O2. Bright-field images were taken using a Zeiss Axioplan 2 imaging microscope. Images were acquired with a high-resolution color digital camera (Axio cam) using the Zeiss AxioVision software.
Sulfatase Enzymatic Assays.
Specimens were disrupted by three freeze–thaw cycles. Protein concentration was measured in total homogenates. For all enzyme activity assays, 30 μg of protein was incubated at 37 °C for 3 h under the specific assay conditions. For the arylsulfatase A (ARSA) assay, homogenates were incubated with 0.05 M p-nitrocatechol sulfate in 0.25 M acetate buffer (pH 5) containing 0.85 M NaCl and 0.25 mM Na pyrophosphate in an incubation volume of 0.3 mL. The reaction was stopped with 0.7 mL 0.64 N NaOH, and absorbance was read at 515 nm on a Perkin-Elmer spectrophotometer.
For arylsulfatase B (ARSB) and arylsulfatase C (ARSC) enzymatic activity, 4-methylumbelliferyl (4-MU) sulfate was used as substrate. ARSB activity was determined by incubating cell homogenates with 6.25 mM 4-MU sulfate in 0.375 mM AgNO3, 0.1 M NaOAc buffer (pH 5) in 80 μL of incubation mixture. ARSC was measured by incubating cell homogenates with 0.2 mM 4-MU sulfate in 0.25% Triton X-100, 0.05 M phosphate buffer (pH 8) in an incubation volume of 0.2 mL. For ARSB and ARSC the reactions were stopped with 2 mL glycine-carbonate buffer (pH 10.7), and fluorescence was read at 365 nm (excitation) and 450 nm (emission) on a Turner fluorometer. Statistical analyses of the measurements were analyzed using Student’s t test. Data are shown as mean ± SEM.
Nissl Staining.
For the Nissl staining 6-μm paraffin-embedded sections were stained in 0.1% cresyl violet solution for 5 min, then were rinsed quickly in distilled water, and were differentiated in 95% ethyl alcohol for 15 min; finally they were cleared in xylene and mounted with permanent mounting medium.
Behavioral Assays.
All behavioral studies were carried out with the observer blinded to genotype. The mice were 7 mo old. The number of mice used per genotype is noted in the figure legends. For all studies, the mice were allowed to habituate to the testing room for 30 min before testing.
Accelerating Rotarod.
Mice were placed on an accelerating Rotarod apparatus (Ugo Basile) for eight trials (four trials/d on two consecutive days) with a 60-min rest interval between trials. Each trial lasted a maximum of 5 min, during which the rod accelerated linearly from 4 to 40 rpm. The amount of time for each mouse to fall from the rod was recorded for each trial. Data are shown as mean ± SE and analyzed with two-way ANOVA (genotype × trial) with repeated measures and Tukey’s post hoc analysis.
Parallel rod footslip test with ANY-maze.
The parallel rod (ANY-Maze) footslip test was used to assess lack of motor coordination in mice. The apparatus was used in conjunction with a special version of the ANY-maze to assess ataxia and locomotor activity simultaneously. Footslips were detected when a paw touched a metal plate below the parallel rod floor, thereby completing a circuit that was registered by the system. Locomotor activity was measured by the ANY-maze tracking software using an overhead camera. Test time was set to 10 min. Data are shown as mean ± SE and were analyzed by one-way ANOVA with Tukey’s post hoc analysis.
Open-field assay.
The open-field apparatus (Accuscan/Fusion) consisted of a clear, open Plexiglas box (40 × 40 × 30 cm) with photo beams that recorded horizontal and vertical movements of the mouse. Overhead lighting was set at 200-lux illumination and a white noise generator (Lafayette Instruments) maintained a 60-dB background noise. Mice were placed in the center of the box. Activity was quantified over a 30-min period by a computer-operated Digiscan optical animal activity system (Accuscan Electronics). Data are shown as mean ± SE and were analyzed by one-way ANOVA with Tukey’s post hoc analysis.
Light/dark box.
The light/dark box (Accuscan/Fusion) consisted of a clear Plexiglas chamber (36 × 20 × 26 cm) with an open top separated from a covered black chamber (15.5 × 20 × 26 cm) by a black partition with a small opening. Overhead lighting was maintained at 700-lux illumination, and white noise was maintained at 60 dB (Lafayette Instruments). Mice were placed in the illuminated side and allowed to explore freely for 10 min. A hand-held computer (Psion Workabout mx; Psion Teklogix) and the Observer program (Noldus Information Technologies) were used to score the number and latency of entries and the time spent in each compartment. An entry was scored with a mouse placed all four feet into either the light or the dark compartment. Data are shown as mean ± SE and were analyzed by one-way ANOVA with Tukey’s post hoc analysis.
Gene Expression Analysis.
Freshly dissected hemibrains cut along the midline from 6-mo-old mice (n = 3 mice per genotype) were placed in 2 mL TRIzol (Invitrogen) on ice and were homogenized immediately with a Polytron homogenizer. Total RNA was extracted following the manufacturer’s instructions. DNA contamination was removed with on-column DNase digestion using the Qiagen RNeasy Mini kit. Quantitec (Qiagen) was used to synthesize first-strand cDNA from 1 μg purified total RNA. Quantitative PCR was performed using Applied Biosystems 7300 Real-Time PCR System. Quantitative PCR reactions were conducted in triplicate, and the results were averaged for each sample and normalized to cyclophilin levels.
Antibodies.
The following antibodies were used: polyclonal rabbit anti-GFAP (1:500) (Dako); mouse anti-GFAP (1:500) (Sigma); mouse anti-NeuN (1:500) (clone A60; Millipore); rabbit anti-ubiquitin (1: 250) (Wako); chicken anti-GFP (1:500) (Abcam); mouse anti-SQSTM1 (1:1,000) (BD Bioscience); mouse anti-calbindin (1:200) (Sigma); mouse anti-MAP2 (1:200) (Millipore); mouse anti-F4/80 (1:100) (AbD Serotec).
Supplementary Material
Acknowledgments
We thank G. Diez-Roux and H. Y. Zoghbi for critical reading of the manuscript; A. M. Adesina for the help with the morphological analysis; and the behavioral, histology, and electron microscopy cores at Baylor College of Medicine. Support for this work was provided by the Italian Telethon Foundation (to C.D.M., C.S., and A.B.). Confocal microscopy was supported by Grant P30 HD024064 from the Intellectual and Developmental Disabilities Research Center at the Baylor College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 13898 (volume 109, number 35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209577109/-/DCSupplemental.
References
- 1.Cosma MP, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 2.Diez-Roux G, Ballabio A. Sulfatases and human disease. Annu Rev Genomics Hum Genet. 2005;6:355–379. doi: 10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 3.Platt FM, Walkley SU. Lysosomal Disorders of the Brain. Oxford, UK: Oxford Univ Press; 2004. pp. 50–75. [Google Scholar]
- 4.Ballabio A, Gieselmann V. Lysosomal disorders: From storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Settembre C, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 6.Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 2005;50:427–434. doi: 10.1002/glia.20207. [DOI] [PubMed] [Google Scholar]
- 7.Maragakis NJ, Rothstein JD. Glutamate transporters in neurologic disease. Arch Neurol. 2001;58:365–370. doi: 10.1001/archneur.58.3.365. [DOI] [PubMed] [Google Scholar]
- 8.Walz W. Role of astrocytes in the clearance of excess extracellular potassium. Neurochem Int. 2000;36:291–300. doi: 10.1016/s0197-0186(99)00137-0. [DOI] [PubMed] [Google Scholar]
- 9.Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 11.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 12.Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 2008;4:61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- 13.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zatloukal K, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weidemann A, et al. The glial cell response is an essential component of hypoxia-induced erythropoiesis in mice. J Clin Invest. 2009;119:3373–3383. doi: 10.1172/JCI39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko DC, et al. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 2005;1:81–95. doi: 10.1371/journal.pgen.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer A, et al. Sulfamidase deficiency in a family of Dachshunds: A canine model of mucopolysaccharidosis IIIA (Sanfilippo A) Pediatr Res. 1998;44:74–82. doi: 10.1203/00006450-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Ellinwood NM, et al. A model of mucopolysaccharidosis IIIB (Sanfilippo syndrome type IIIB): N-acetyl-alpha-d-glucosaminidase deficiency in Schipperke dogs. J Inherit Metab Dis. 2003;26:489–504. doi: 10.1023/a:1025177411938. [DOI] [PubMed] [Google Scholar]
- 20.Sleat DE, et al. A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J Neurosci. 2004;24:9117–9126. doi: 10.1523/JNEUROSCI.2729-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Settembre C, et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci USA. 2007;104:4506–4511. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farfel-Becker T, et al. Spatial and temporal correlation between neuron loss and neuroinflammation in a mouse model of neuronopathic Gaucher disease. Hum Mol Genet. 2011;20:1375–1386. doi: 10.1093/hmg/ddr019. [DOI] [PubMed] [Google Scholar]
- 23.Wada R, Tifft CJ, Proia RL. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc Natl Acad Sci USA. 2000;97:10954–10959. doi: 10.1073/pnas.97.20.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohmi K, et al. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc Natl Acad Sci USA. 2003;100:1902–1907. doi: 10.1073/pnas.252784899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu YP, Matsuda J, Kubota A, Suzuki K, Suzuki K. Infiltration of hematogenous lineage cells into the demyelinating central nervous system of twitcher mice. J Neuropathol Exp Neurol. 2000;59:628–639. doi: 10.1093/jnen/59.7.628. [DOI] [PubMed] [Google Scholar]
- 26.Jeyakumar M, et al. Central nervous system inflammation is a hallmark of pathogenesis in mouse models of GM1 and GM2 gangliosidosis. Brain. 2003;126:974–987. doi: 10.1093/brain/awg089. [DOI] [PubMed] [Google Scholar]
- 27.Macauley SL, Pekny M, Sands MS. The role of attenuated astrocyte activation in infantile neuronal ceroid lipofuscinosis. J Neurosci. 2011;31:15575–15585. doi: 10.1523/JNEUROSCI.3579-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bélanger M, Magistretti PJ. The role of astroglia in neuroprotection. Dialogues Clin Neurosci. 2009;11:281–295. doi: 10.31887/DCNS.2009.11.3/mbelanger. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka K, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagele RG, et al. Contribution of glial cells to the development of amyloid plaques in Alzheimer’s disease. Neurobiol Aging. 2004;25:663–674. doi: 10.1016/j.neurobiolaging.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Bradford J, et al. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci USA. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neufeld EF. Lysosomal storage diseases. Annu Rev Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 33.Scriver CR, Beaudet AL, Sly WS, Valle D. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver CR, et al., editors. New York: McGraw-Hill; 2001. pp. 3371–3896. [Google Scholar]
- 34.Yu T, Shakkottai VG, Chung C, Lieberman AP. Temporal and cell-specific deletion establishes that neuronal Npc1 deficiency is sufficient to mediate neurodegeneration. Hum Mol Genet. 2011;20:4440–4451. doi: 10.1093/hmg/ddr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elrick MJ, et al. Conditional Niemann-Pick C mice demonstrate cell autonomous Purkinje cell neurodegeneration. Hum Mol Genet. 2010;19:837–847. doi: 10.1093/hmg/ddp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni X, Canuel M, Morales CR. The sorting and trafficking of lysosomal proteins. Histol Histopathol. 2006;21:899–913. doi: 10.14670/HH-21.899. [DOI] [PubMed] [Google Scholar]
- 37.Orchard PJ, et al. Hematopoietic cell therapy for metabolic disease. J Pediatr. 2007;151:340–346. doi: 10.1016/j.jpeds.2007.04.054. [DOI] [PubMed] [Google Scholar]
- 38.Biffi A, et al. Correction of metachromatic leukodystrophy in the mouse model by transplantation of genetically modified hematopoietic stem cells. J Clin Invest. 2004;113:1118–1129. doi: 10.1172/JCI19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allan SM, Rothwell NJ. Inflammation in central nervous system injury. Philos Trans R Soc Lond B Biol Sci. 2003;358:1669–1677. doi: 10.1098/rstb.2003.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farina C, Aloisi F, Meinl E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 2007;28:138–145. doi: 10.1016/j.it.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Sofroniew MV. Reactive astrocytes in neural repair and protection. Neuroscientist. 2005;11:400–407. doi: 10.1177/1073858405278321. [DOI] [PubMed] [Google Scholar]
- 42.Settembre C, et al. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22:2645–2650. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staretz-Chacham O, Choi JH, Wakabayashi K, Lopez G, Sidransky E. Psychiatric and behavioral manifestations of lysosomal storage disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1253–1265. doi: 10.1002/ajmg.b.31097. [DOI] [PubMed] [Google Scholar]
- 44.Copeland NG, Jenkins NA, Court DL. Recombineering: A powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 45.Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Press, Waltham, MA: Academic; 1944. [Google Scholar]
- 46.Hampton DW, et al. Cell-mediated neuroprotection in a mouse model of human tauopathy. J Neurosci. 2010;30:9973–9983. doi: 10.1523/JNEUROSCI.0834-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross PC, Mercer KL. 1993. in Cell and Tissue Ultrastructure: A Functional Perspective (Freeman, New York, NY) pp 124–142.