Abstract
A persistent question in biology is how cis-acting sequence elements influence trans-acting factors and the local chromatin environment to modulate gene expression. We reported previously that the DNA transposon 1360 can enhance silencing of a reporter in a heterochromatic domain of Drosophila melanogaster. We have now generated a collection of variegating phiC31 landing-pad insertion lines containing 1360 and a heat-shock protein 70 (hsp70)-driven white reporter to explore the mechanism of 1360-sensitive silencing. Many 1360-sensitive sites were identified, some in apparently euchromatic domains, although all are close to heterochromatic masses. One such site (line 1198; insertion near the base of chromosome arm 2L) has been investigated in detail. ChIP analysis shows 1360-dependent Heterochromatin Protein 1a (HP1a) accumulation at this otherwise euchromatic site. The phiC31 landing pad system allows different 1360 constructs to be swapped with the full-length element at the same genomic site to identify the sequences that mediate 1360-sensitive silencing. Short deletions over sites with homology to PIWI-interacting RNAs (piRNAs) are sufficient to compromise 1360-sensitive silencing. Similar results were obtained on replacing 1360 with Invader4 (a retrotransposon), suggesting that this phenomenon likely applies to a broader set of transposable elements. Our results suggest a model in which piRNA sequence elements behave as cis-acting targets for heterochromatin assembly, likely in the early embryo, where piRNA pathway components are abundant, with the heterochromatic state subsequently propagated by chromatin modifiers present in somatic tissue.
Keywords: epigenetics, position-effect variegation
Transposable elements (TEs) are major structural features of nearly all eukaryotic genomes, and can play a role as cis-acting regulatory features with profound influence over the gene regulatory networks of their host (1). However, if left unchecked, the deleterious effects of TE mobilization can become insurmountable for the system, damaging genome integrity. Thus, silencing mechanisms that prevent TE mobilization are fundamental to the faithful propagation of genetic information. Position-effect variegation (PEV), a mosaic expression pattern resulting from silencing in some cells that normally express a gene, has frequently been used as an indicator of heterochromatic environments (2, 3). We reported previously that the DNA transposon 1360 can enhance reporter silencing in repeat-rich heterochromatic regions (4). To gain insight into the mechanisms involved, we sought to identify sequence elements present in 1360 that contribute to 1360-sensitive PEV, using an adjacent heat-shock protein 70 (hsp70)-driven white gene as the reporter.
We used the phiC31 landing pad system, where phiC31 integrase mediates recombination between a landing pad (inserted in the genome) containing phage attachment (attP) sites and a donor construct containing bacterial attachment (attB) sites (5). This system allows us to maintain a constant genomic context for multiple transgenic 1360 sequence constructs. A genomic test site identified at the base of chromosome arm 2L (site 1198) indeed exhibits 1360-dependent silencing, which we find reflects accumulation of Heterochromatin Protein 1a (HP1a).
Testing various features of 1360, we found that 1360-sensitive PEV at site 1198 does not require the inverted repeats at the ends of the element, nor internal transcription start sites, but is impacted by small deletions over sites homologous to PIWI-interacting RNAs (piRNAs), a small RNA population initially generated in germ-line tissue and loaded into the zygote. This observation was extended to the retrotransposon Invader4, suggesting that the mechanism is broadly applicable. Thus, sites with homology to piRNAs may be cis-acting targets for heterochromatin assembly, likely in the early embryo (a stage when piRNA pathway components are abundant), with this decision subsequently propagated in somatic tissue by chromatin-modifying factors.
Results
1360-Sensitive Silencing Is Observed in a Subset of Euchromatic Domains.
We generated a collection of landing pad lines by mobilizing a P element construct containing attP sites flanking a Flippase FLP-excisable 1360, cloned upstream of an hsp70-white reporter (Fig. 1A). Mobilization led to the recovery of 38 variegating landing pad lines (∼5% recovery) with inserts mapping to unique genomic sites (Fig. 1B). Many (34%) mapped to the telomere-associated repeats (TASs) present on 2R and 3R. The subtelomeric repeats are known targets for P element insertion (6). These genomic regions are enriched for Polycomb-group proteins (PcG) and the associated trimethylation of histone H3 lysine 27 (H3K27me3) mark (7, 8). In 60% of the variegating lines (including those in TASs), the construct mapped to regions with a high repeat density (>30%), a property of canonical heterochromatin (9). (See Dataset S1 for a list of all lines.) In many of these cases, the construct insertion sites lie in the pericentric heterochromatin of 2L and 2R, in the fourth or the Y chromosome. A subset of lines have insertion sites in regions of low repeat density (<10%) that are enriched for chromatin marks associated with transcriptional activity, such as H3K4me2 and H3K9ac, in both S2 (undifferentiated male embryo cell line) and BG3 (differentiated neuronal cell line) cells (Dataset S1) (7). The distribution of variegating insertions within transcriptionally active domains is not random; all lie within two divisions (or numerical cytological positions) of pericentric or telomeric heterochromatin. These lines represent a unique resource with which to study position effects.
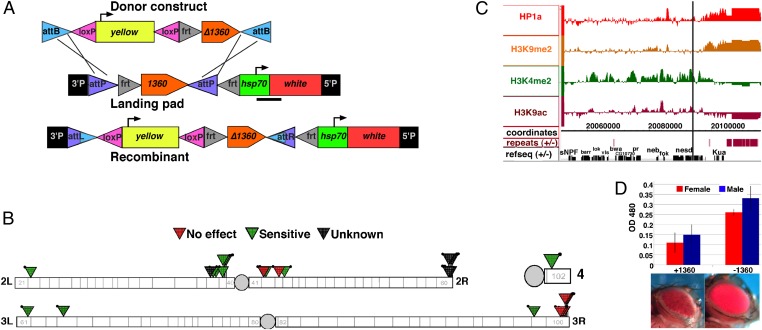
Fig. 1.
Sensitivity of PEV to 1360 can occur at euchromatic sites. (A) Schematic of phiC31-mediated cassette exchange between the donor construct and a landing pad, resulting in the recombinant product at the same genomic site. The yellow gene, used as a marker to recover recombinants, was removed by loxP excision before all assays. The bar below hsp70-white denotes the region assayed by ChIP-qPCR in Fig. 2. (B) Map of the landing pad insertions exhibiting variegation. Those showing suppressed variegation on excision of 1360 are marked by green triangles; those with no change, red triangles (see also Dataset S1). (Black triangles indicate that the line was not assayed.) (C) Chromatin state of the 1198 insertion site based on modENCODE ChIP-array data from third instar larvae (active marks H3K9ac and H3K4me2 and silent marks H3K9me2 and HP1a are shown). A vertical line denotes the P element insertion site within nesd. (D) Pigment assays +/−1360 for females (red) and males (blue), with representative eye pictures (see also Fig. S1). Bars represent the mean from two biological replicates (± SEM). Excision of the 1360 element results in a loss of silencing.
To determine which variegating landing pad reporters are 1360-sensitive, eye pigment assays were performed comparing 3- to 5-d-old adults with the 1360 element removed by FLP-mediated excision with their sibs with the 1360 element present, i.e., +/−1360. Fourteen lines exhibited pigment levels that were increased in the absence of 1360, a suppression of PEV (Fig. 1B; also see Dataset S1 and Fig. S1). Some of these lines had a reporter inserted within or close to genes, in repeat-poor regions enriched for euchromatic marks in S2 and BG3 cells (7). Thus, 1360 can support heterochromatin formation at a wider variety of sites than previously recognized (4, 10). We chose the 1360-sensitive landing pad line 1198, with an insert at the base of 2L, distal to a block of heterochromatin (cytological position 38B6, 2L:20094149) for further investigation. This line exhibits >twofold change +/−1360 in eye pigment, a range sufficient to detect partial suppression of 1360-sensitive silencing (Fig. 1D).
The construct is inserted within the gene nessun dorma (nesd). A survey of the chromatin environment at the insertion site (in the absence of the P element insert) shows that the region is enriched for marks associated with transcription (H3K4me2 and H3K9ac) in S2 and BG3 cells, as well as in third instar larvae (11, 12) (Fig. 1C). The 20-kb region surrounding the reporter has a repeat density of 2%, consistent with the notion that the reporter is not in canonical heterochromatin. Notably, the eye phenotype in the absence of 1360 corresponds with that observed on insertion of the reporter into a euchromatic environment, displaying a solid red eye; variegation is only observed in the presence of the 1360. However, in the presence of the 1360 element, pigment levels become comparable to some lines present in annotated heterochromatin (e.g., line 1310; compare line 1198 with 1360 to line 1310 without 1360; see Fig. S1), indicating that in the presence of a 1360, the site may be more representative of heterochromatin. The degree of silencing is greater in the female than the male, as is typical of PEV (Fig. 1D). This line gives us an opportunity to study 1360-sensitive ectopic silencing of a reporter.
1360 Supports Ectopic Heterochromatin Assembly.
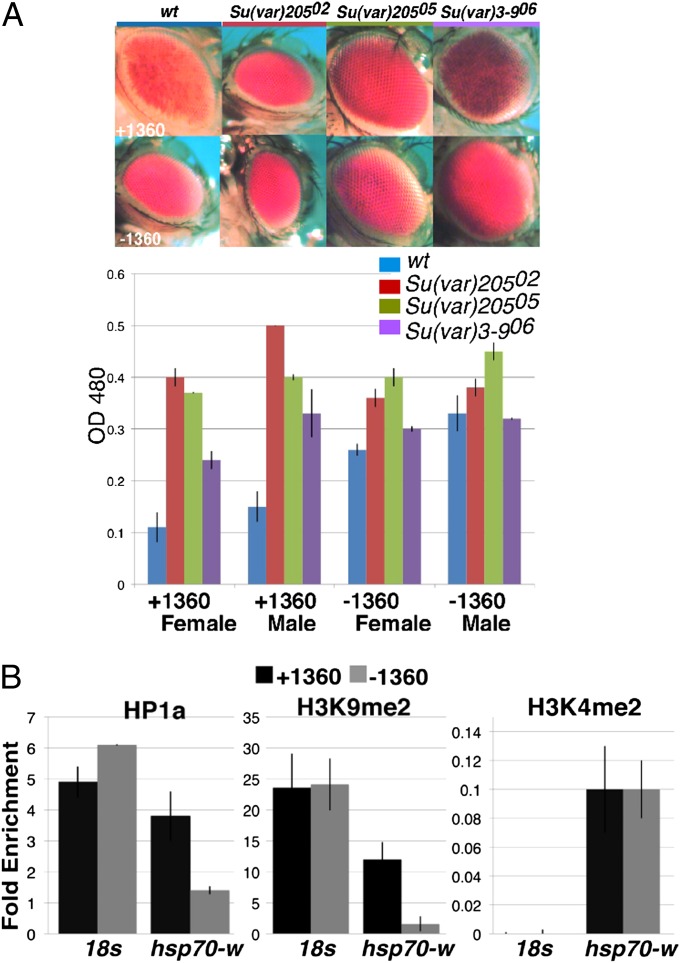
Heterochromatin assembly is associated with the presence of H3K9me2/3, deposited by a histone methyltransferase (HMT), typically (but not always) SU(VAR)3-9, and the chromo domain protein HP1a, encoded by Su(var)205 in flies. We assessed eye pigment levels for the 1198 reporter +/−1360 when mutant for either Su(var)3-9 or Su(var)205 to test whether these components are required for 1360-senseitive PEV. In the presence of 1360, a >twofold increase in pigment is detected with each mutant in both sexes, whereas no significant change is observed in the absence of 1360 in males (a minor quantitative change is seen in females), corroborating that 1360-sensitive PEV is dependent on heterochromatin components (Fig. 2A). Importantly, this also indicates that in the absence of 1360, HP1a-dependent silencing is not supported at this site.
Fig. 2.
1360 promotes heterochromatin assembly. (A) Eye phenotypes (Upper) of line 1198 +/−1360, comparing the starting line (wt) (Left) with those mutant for Su(var)205 (HP1a) or the HMT Su(var)3-9. (Lower) Corresponding pigment assay readings (OD480). Bars give the mean from two biological replicas (± SEM). (B) ChIP assessment of levels of HP1a, H3K9me2, and H3K4me2 at the hsp70-white promoter for line 1198 +1360 (black bars) and −1360 (gray bars). qPCR primers bracket the hsp70-white promoter (see Fig. 1A). All data were normalized to input; fold enrichment over the α-actinin locus is shown (± SEM).
To determine whether 1360-sensitive PEV is a consequence of a change in the chromatin landscape, we performed chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) using antibodies for HP1a and H3K9me2, assaying the promoter region of hsp70-white (as marked in Fig. 1A). This demonstrated that the local chromatin environment is substantially enriched for HP1a and H3K9me2 in the presence of 1360, whereas in the absence of 1360, levels were low (Fig. 2B). No significant difference in the euchromatic mark H3K4me2 was detectable +/−1360. Thus, the 1360-sensitive PEV exhibited by line 1198 reflects HP1a-dependent heterochromatin assembly at a normally euchromatic site in the genome.
Cis-Acting Sequence Element in the Right Half of 1360 Is Necessary and Sufficient for 1360-Sensitive PEV.
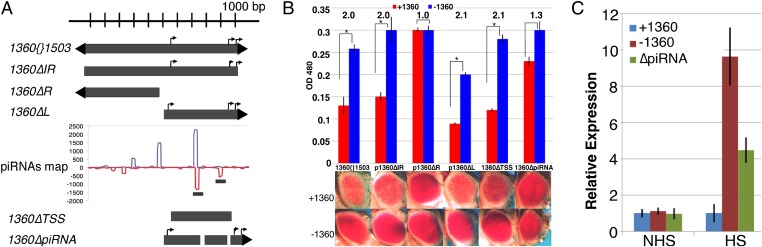
Several features present in 1360 could be used as a signal for heterochromatin assembly. Possibilities include the terminally inverted repeats; putative transcription start sites (TSSs) found within the right half of the element; and regions with similarity to piRNA reads. Terminally inverted repeats can contribute secondary structure or serve as binding sites for transposon-derived proteins, both mechanisms used in plants (13, 14). Present within the right half of 1360 are three putative TSSs, with similarity to the 1360 transcription initiation sites suggested to produce antisense Suppressor of stellate transcripts in the D. melanogaster male germ line (15). If the integrated copy of 1360 is a target for the transcription machinery, this might cause inappropriate local transcript production, targeting the site for silencing by a similar mechanism. The right half also contains sites that map to piRNA reads with an antisense bias; these sites could be targets for antisense piRNAs bound to Piwi, possibly targeting sense transcripts of the integrated 1360 (Fig. 3A).
Fig. 3.
piRNA sequence elements in the right half of 1360 support 1360-sensitive silencing. (A) The 1360 variants used in donor constructs for phiC31-mediated cassette exchange. Bent arrows indicate putative transcription start sites. Frequencies of sense (blue line) and antisense (red line) piRNA reads (20) that map to 1360{}1503 are shown. The bars below illustrate the deletions for the 1360ΔpiRNA constructs. (B) Representative eye pictures for each 1360Δ recombinant line are shown under the pigment assay data (from male flies) using OD480 (± SEM). Asterisks indicate a significant change +1360 (red) vs. −1360 (blue) with a P value of <0.05 (Student’s t test); numbers indicate fold change. Loss of the right half of 1360{}1503 or loss of the piRNA hotspots results in a significant loss of silencing (no or reduced change on excision of the 1360 remnant). (C) RT-qPCR of hsp70-w transcript levels in non–heat-shocked (NHS) and heat-shocked (HS) adult flies heterozygous for the P element. Shown is fold expression (normalized to RpL32) relative to full-length 1360 recombinant lines (± SEM).
To identify which sequence features in 1360 contribute to 1360-sensitive PEV, we tested 1360 fragments lacking the terminal inverted repeats (1360ΔIR), left half (1360ΔL), or right half (1360ΔR) for induction of silencing, as measured by pigment assays (Fig. 3A). Deletion of the right half of 1360 has the most pronounced effect on 1360-sensitive variegation, resulting in a twofold increase in eye pigment (Fig. 3B; compare red bars). Lines lacking the inverted repeats or the left half of 1360 retained 1360-sensitive silencing, indicating that these sequence elements are dispensable for 1360-sensitive PEV.
Because each 1360 element is flanked by flippase recognition target (FRT) sites (Fig. 1A), we can test whether silencing for each line is dependent on the presence of the 1360Δ construct. FLP-mediated excision of 1360ΔIR and 1360ΔL leads to a loss of silencing comparable to the loss of the full-length 1360 donor, whereas excision of 1360ΔR produced no change in pigment levels (Fig. 3B). Thus, deletion of the left half or inverted repeats of 1360 does not impact 1360-sensitive PEV, whereas deletion of the right half does, suggesting that sequence feature(s) within the right half of the element, independent of the terminally inverted repeat, are necessary and sufficient for 1360-sensitive PEV.
Putative Transcription Start Sites Present in 1360 Are Not Required for 1360-Sensitive PEV.
Although there are no universal core promoter elements in D. melanogaster, there are some canonical elements, spanning ∼80 bp, that are common targets for the RNA Pol II transcription machinery, including the TATA box and the Initiator (Inr) motif (16). The 1360 element in our construct contains one TATA box with a downstream Inr motif close to one of the mapped Su(ste) promoter sites, whereas the other two sites have nearby Inr motifs. To test whether these putative promoters are required, we made three 80-bp deletions in 1360ΔL, each spanning one of these sequence elements, to make 1360ΔTSS (Fig. 3A). The resulting construct maintains 1360-sensitive PEV, indicating that the putative TSSs are dispensable for silencing (Fig. 3B). FLP-mediated excision of 1360ΔTSS led to a ∼twofold increase in pigment levels, confirming that the sequences in 1360ΔTSS are sufficient to impart silencing (Fig. 3B). Thus, an alternative sequence element(s) must be responsible.
Disruption of Sites Matching Antisense piRNA Reads Compromises TE-Dependent Variegation.
The piRNA pathway, previously implicated in 1360-sensitive PEV (4), suppresses TEs in the germ line through posttranscriptional targeting of sense and antisense transcripts by the slicer-mediated activities of Aubergine (AUB) and Argonaute (AGO)3, using a ping-pong mechanism (17). The repertoire of defective and active TE copies present in the genome can skew the piRNA population to bias against active TEs, necessary to prevent further TE colonization of the genome (18, 19). A third component, Piwi, is a nuclear protein that binds mostly antisense piRNAs, a subset of which are generated by ping-pong cycle–dependent piRNA biogenesis (17, 20). Piwi interacts with HP1a and has been implicated in a transcriptional silencing role in the germ line (21–23). These piRNA components are loaded into the oocyte (17, 24); their mode of TE repression in the early embryo has been relatively unexplored but could include a role in establishing heterochromatin domains. Heterochromatin formation, and the boundary between heterochromatic and euchromatic domains, is established in the syncytium, at nuclear cycle 10–14, and must be propagated by chromatin-modifying mechanisms present in somatic tissue (25).
Large-scale sequencing of piRNA populations has revealed that individual TE families have distinct patterns of sense and antisense piRNAs (20, 26). To determine whether the right half of the 1360 element has a distinguishing piRNA distribution that could indicate key sites for silencing, we mapped the piRNA read density from wild-type (wt) ovary piRNA data generated by Li et al. (20), against the 1360 copy present in our construct, 1360{}1503. Both halves of the element exhibit matches to piRNA reads (Fig. 3A), but antisense peaks (one with a 10-nt overlapping sense and antisense peaks indicative of ping-pong cycle biogenesis) were present at two positions only, in the right half of the element (Fig. 3A). We tested whether these regions contributed to 1360-sensitive PEV by removing the signature ping-pong overlapping peaks (36-bp deletion) and the neighboring shorter antisense peak (10-bp deletion) of 1360ΔL to make 1360ΔpiRNA. The new junctions formed by the deletions did not result in new piRNA matches. Indeed, these relatively small deletions, 46 bp in total, significantly compromised the 1360 effect, although a complete loss of 1360-sensitive silencing was not observed (Fig. 3B).
Whether the observed loss of silencing was directly associated with the 1360ΔpiRNA construct was tested by assaying PEV after FLP excision of this 1360 fragment. Indeed, a 1.3-fold change was observed, indicating that much of the capacity to silence had been lost and that the residual silencing was attributable to the 1360 remnant (Fig. 3B). The results indicate that this 46-bp sequence component contributes significantly to 1360-dependent silencing but is not the sole sequence responsible for the effect. To verify that the PEV assay data were truly a reflection of a change in silencing (from less capacity for transcription +1360 to more capacity for transcription −1360), we performed RT-qPCR for hsp70-driven white transcripts in 3- to 5-d-old non–heat-shocked (NHS) and heat-shocked (HS) adults, comparing the full-length and 1360ΔpiRNA recombinant lines. Transcript levels for hsp70-w at 25 °C are sufficient to generate the eye phenotype. Heat shock increases the level of the appropriate transcription factor and provides a more robust test of the accessibility of the promoter region, producing levels of transcript suitable for a quantitative assay (3, 4). Indeed, the presence of 1360 significantly decreases the capacity for expression under heat shock; this capacity is restored on loss of 1360 and partially restored with the 1360ΔpiRNA construct (Fig. 3C). These findings indicate that piRNA sequence elements present in 1360 contribute to 1360-sensitive heterochromatin assembly.
To explore whether piRNA pathway components could impact 1360-sensitive PEV at this site, we tested reporter line 1198 both +/−1360 in the presence of piwi and aub mutant alleles. Mutations in both genes dominantly suppressed 1360-sensitive PEV (Fig. S2). This finding implicates an RNAi mechanism in establishing this 1360-sensitive silencing.
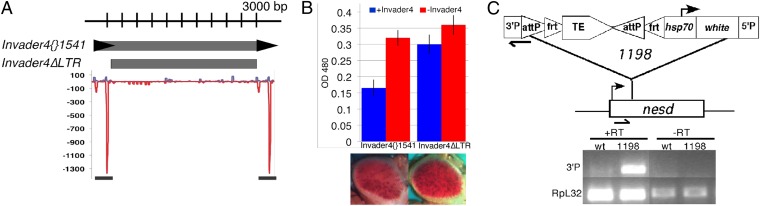
To determine whether this phenomenon is unique to element 1360, we swapped 1360 with the long terminal repeat (LTR) element Invader4 (Fig. 4A). Invader4 was chosen for two reasons. First, many variegating reporters generated for this study (Fig. 1B) are located within or in close proximity to an Invader4 element. [Similar criteria were used initially to select 1360 as a potential target for heterochromatin assembly (10).] Second, Invader4 is regulated similarly to 1360 by piRNA pathway components in that transcription from both elements is up-regulated in aub, ago3, and piwi ovaries (17, 20). When Invader4 replaces 1360 in the test construct, we found an equivalent level of variegation (Fig. 4B); this loss of expression was also suppressed by mutations in heterochromatin and piRNA components (Fig. S3). Deletion of sites with homology to piRNA reads (Invader4ΔLTR; Fig. 4A) resulted in an increase in pigment levels, similar to the loss of full-length Invader4 [compare silencing by the full-length element (Left) with that by the truncated element (Right) in Fig. 4B]. This evidence supports the suggestion that piRNA sequence elements can promote silencing; importantly, the phenomenon is not exclusive to 1360 and can be observed using either a DNA transposon- or a retrotransposon-derived element.
Fig. 4.
Invader4 piRNA sequence elements support PEV. (A) Invader4 variants used in donor constructs for phiC31-mediated cassette exchange are shown. Frequencies of sense (blue line) and antisense (red line) piRNA reads mapping to Invader4{}1541 are shown. The bar below denotes regions of Invader4 deleted in Invader4ΔLTR. (B) Pigment assay data using OD480 (± SEM) is shown for +/−Invader4 test constructs. Loss of regions with homology to piRNA reads (Invader4ΔLTR) results in a loss of Invader4-sensitive PEV. (C) Diagram of the P element insertion site for line 1198 in the 5′ end of nesd. Black bars represent primers used for RT-PCR (Upper). Read-through transcripts spanning the 3′P region of the P element (3′P) and control RpL32 products are shown (Lower). Lines used were wt control (no P element insert) and 1198, where +/−RT indicates the addition or omission of reverse transcriptase.
Work from plants and fungi argues that such a recognition event would likely be based on RNA-RNA base pairing (27), suggesting a requirement for transcription over the transposable element to generate a target template. Although the putative TSSs in 1360 are dispensable for 1360-sensitive PEV at this insertion site, read-through transcription could occur from a nearby promoter. In this case, the P element is inserted downstream of the transcription start site for nesd. This gene is expressed at low levels in the female ovary and at moderately high levels in the early embryo (28). To determine whether read-through transcription of the P element is occurring, we looked for transcripts across the P element junction by RT-PCR in 0- to 10-h embryos; such transcripts are detected (Fig. 4C). The combined results support a model in which a small RNA-targeting event using this transcript participates in the HP1a-dependent assembly of heterochromatin at this site in the early embryo, with the consequences of that event being evident in the tissues that go on to form the eye (see model in Fig. S4).
Discussion
Our screen to identify a 1360-sensitive phiC31 landing pad site generated many landing pad lines with inserts in unique chromatin domains, including several on the fourth chromosome and in repetitious elements such as the telomere-associated sequences on chromosomes 2R and 3R (Fig. 1). This study adds useful resources to the collection of phiC31 lines currently available (29). We find that reporters in a wide range of chromatin domains are sensitive to 1360; thus, the influence of 1360 is not limited to repeat-rich, heterochromatic sites. However, proximity to repeat-rich regions appears to be important, because most 1360-sensitive reporters are close to the base of the euchromatic arms or are telomere-proximal (Fig. 1B). Genetic analysis and ChIP experiments show that 1360-sensitive PEV is representative of HP1a-dependent heterochromatin assembly (Fig. 2). The use of phiC31 recombineering technology allowed us to show that a unique sequence element within the right half of 1360, independent of the inverted repeat terminal sequences and of putative transcription start sites, is required for optimal 1360 impact in line 1198 (Fig. 3). Importantly, an unrelated transposable element (Invader4) has the same effect as 1360, implicating a mechanism that can be applied to other TEs (Fig. 4 A and B). In both cases, the deletion of sites with homology to antisense-oriented piRNAs compromised silencing at this ectopic site. This suggests that a small RNA–directed, RNA-RNA targeting event is contributing to transposon-sensitive heterochromatin assembly, potentially using a mechanism similar to that documented in Schizosaccharomyces pombe and plants (Fig. S4) (30–33).
The ability to trigger ectopic HP1a assembly appears to be limited to genomic sites close to heterochromatic masses, because the distribution of 1360-sensitive sites is limited (Fig. 1B). Indeed, half of 1360-sensitive sites are in annotated heterochromatin (chromatin states 7/8, BG3 cells), whereas most sites inducing variegation but lacking 1360 sensitivity (without a change in pigment levels +/−1360) are in PcG-enriched regions, frequently in telomere-associated repeats (Dataset S1). Thus, 1360-sensitive PEV appears to be an HP1a-dependent phenomenon. Prior reports, as well as this study, find that single repetitious elements within the euchromatic arms do not trigger detectable reporter silencing (4). The high density of repetitious elements in heterochromatic domains is a fundamental characteristic of these regions, suggesting that these repeats may cooperatively participate in stabilizing heterochromatin factors. Cooperative function of interspersed signals has been reported for other domain-wide chromatin structures, for example that developed to provide dosage compensation (34). Many of the 1360-sensitive sites identified here are surrounded by repeats of different types; thus at different insertion sites, additional mechanisms could be at work. In the future it will be of interest to determine if our observations extend to other 1360-sensitive sites, both those adjacent to and within annotated heterochromatin. It is probably no coincidence that our experimentally recovered test site (Fig. 1C) resembles the classical rearrangements that result in PEV, having a reporter in a normally euchromatic domain close to a heterochromatic mass.
Evidence for piRNA-mediated effects on chromatin assembly in flies is mixed, with data suggesting both silencing and activating roles, depending on reporter location (4, 35, 36). Mutations in piRNA pathway components are weak suppressors of PEV for a reporter in a repeat-rich environment, with and without 1360 present, suggesting that both local heterochromatin and 1360-sensitive silencing are impacted (4). Other TEs with piRNA signals include HeT-A, I element, and copia (20). All three transposon families are silenced by HP1a and H3K9me2/3 assembly over their promoter elements in the female germ line by a mechanism dependent on RNAi components (22, 37). piRNA biogenesis is most active at this stage, and it has been argued that its effects may be limited to germ cells. For example, the RNA helicase Spindle E, required for piRNA biogenesis, is necessary for TE repression in the female germ line but not in mature somatic tissues (no-ovary carcasses) (37). Mutations in many of the RNAi components and chromosomal proteins result in female sterility, making the issue difficult to study.
Both piRNA and chromatin structural proteins (and/or their mRNAs) are synthesized during oogenesis, loaded into the ovary, and present in the early embryo (0–6 h) (24). Their presence overlaps the early stages of heterochromatin formation (cycles 11–14) and of zygotic transcription (nuclear cycle 14) (25). The latter coincides with the appearance of the histone H3K4 demethylase SU(VAR)3-3, without which HP1a and H3K9me2 levels in the heterochromatin become substantially reduced, whereas levels for the activating H3K4me2 mark become high (immunofluorescent staining of nuclei in cycle 14 embryos), resulting in suppression of PEV. Generation of wt clones from heterozygous Su(var)3-9 lines suggests that the chromatin patterns established in the late embryo persist during differentiation (25). Thus, piRNA components could play a role in initiation of heterochromatin formation in the early embryo at select sites (e.g., a subset of TEs) in the genome, with the structure maintained during development by chromosomal protein interactions. However, whether or not depletion of these products in the female germline/early embryo alone is sufficient to see an impact in the larvae/adult remains to be tested.
Although the impact on silencing of deleting the piRNA sites in 1360 and Invader4 is striking, full suppression of the PEV induced by 1360 was not observed (Figs. 3B and 4B), which suggests that additional sites may be critical. Such sequences could be missing from the small RNA library used for our analysis. Additional piRNA libraries (17) examined did not identify additional sites within the right half of the 1360 element. The piRNA libraries reported to date do not achieve saturation, so it is possible that additional piRNA targets remain.
Alternatively, a different mode of targeting could also be in effect, unrelated to a piRNA-mediated event. Such dual mechanisms have been documented for heterochromatin formation in S. pombe, a system in which RNAi-mediated heterochromatin targeting is well-established (38, 39). In many eukaryotes, transposases have been found to be sources of DNA binding domains with chromatin-modifying abilities (1). For example, Drosophila BEAF-32, derived from the hAT transposase, is a chromatin insulator that binds the scs chromatin boundary element (40). Similar mechanisms may have evolved in Drosophila to specifically target heterochromatin factors to TEs. However, no candidate proteins have emerged to date.
Evidence for an RNAi-based mechanism that contributes to heterochromatin assembly has been found in several model systems (worms, plants, fission yeast). In S. pombe, targeting of the HP1 homolog Swi6 and the H3K9 HMT Clr4 depends on the processing of RNA Pol II transcripts (generated from heterochromatic loci) by RNAi components. The RNAi-induced transcriptional silencing complex (RITS) contains the chromo domain protein Chp1, as well as the RNAi component Ago1, which binds small RNAs with homology to target sites (e.g., dg/dh repeats) in pericentric heterochromatin (38). In the system described here, the read-through transcripts of the nesd gene detected in early embryo samples (Fig. 4C) are plausible targets for Piwi associated with 1360 or Invader4 piRNAs, using a base-pairing interaction. Analysis of EST libraries from Drosophila embryos indicates that most TE families are transcribed (41). However, determining the origin of these transcripts is a challenging task given the high sequence similarity among TE family members. The current experiments have not allowed us to determine whether the 1360-sensitive silencing observed is transcription-dependent, as would be required for an RNA-RNA recognition event for targeted silencing; such a test will be critical for future work to establish the mechanism involved. The present results support a mechanism that uses piRNAs, possibly ping-pong cycle–derived [as are 1360 and Invader4 piRNAs (20)], for transposon-sensitive targeting of HP1a, likely early in development, with persistent effects observed using reporters in adult tissues.
Materials and Methods
Fly Stocks.
All Drosophila stocks and crosses were maintained at 25 °C on cornmeal sucrose-based media (42). Fly stocks were from the Bloomington Drosophila Stock Center unless otherwise indicated. Transgenesis of the landing pad construct into the starting stock yw67c12 was carried out by Genetic Services. Stocks and constructs used, plus the mobilization and mapping procedures, are described in detail in SI Materials and Methods.
Chromatin Immunoprecipitation.
Chromatin isolation and immunoprecipitation from third-instar larvae were carried out as described previously (12). The antibodies used were HP1a W191, Abcam 1220 H3K9me2, and Millipore 07-030 H3K4me2. Antibodies were validated by us and by others: see the Antibody Validation Database (http://compbio.med.harvard.edu/antibodies/about) (43). Quantitaive PCR (see Table S1 for the list of primers) was performed using Bio-Rad iQ SYBR Green Supermix in a Cepheid SmartCycler. Two biological replicates, each consisting of two technical replicates, were assayed for each immunoprecipitation assay.
Mapping piRNA Reads and Assessing RNA Products.
Direct-sequence mapping was carried out using small RNA sequence reads derived from wt Oregon R ovaries (20) retrieved from the National Center for Biotechnology Information (NCBI) trace archives with accession number SRP000458 (Figs. 3A and 4A). For RT-PCR, RNA was isolated from 0- to 10-h embryos (Fig. 4B) or 3- to 5-d adult flies, with or without heat shock (Fig. 3C), DNase I–treated, and reverse-transcribed using random hexamer primers (Fig. 4B) or oligo dT (Fig. 3C). See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Jack Bateman for providing clones and Wilson Leung for small RNA–mapping analysis. We thank Micaela Blank, Gregory Gandenberger, Patrick Ng, Anita Khoong, and Hao Yang for assistance in screening phiC31 transgenic lines. This work was supported by National Institutes of Health Grant GM068388 (to S.C.R.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207036109/-/DCSupplemental.
References
- 1.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: The lessons of PEV in Drosophila. Adv Genet. 2008;61:1–43. doi: 10.1016/S0065-2660(07)00001-6. [DOI] [PubMed] [Google Scholar]
- 3.Wallrath LL, Elgin SCR. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 4.Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- 9.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun F-L, et al. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol. 2004;24:8210–8220. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy S, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddle NC, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21:147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 16.Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 18.Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendez DL, et al. The HP1a disordered C terminus and chromo shadow domain cooperate to select target peptide partners. ChemBioChem. 2011;12:1084–1096. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenov MS, et al. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Klattenhoff C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slotkin RK, Martienssen RA. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 28.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venken KJ, et al. MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 32.Kloc A, Zaratiegui M, Nora E, Martienssen RA. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 34.Straub T, Becker PB. DNA sequence and the organization of chromosomal domains. Curr Opin Genet Dev. 2008;18:175–180. doi: 10.1016/j.gde.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 37.Klenov MS, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloc A, Martienssen RA. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–517. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 40.Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–423. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- 41.Deloger M, et al. Identification of expressed transposable element insertions in the sequenced genome of Drosophila melanogaster. Gene. 2009;439:55–62. doi: 10.1016/j.gene.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Shaffer CD, Wuller JM, Elgin SC. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 1994;44:99–108. doi: 10.1016/s0091-679x(08)60908-5. [DOI] [PubMed] [Google Scholar]
- 43.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.