Abstract
Glioblastoma multiforme (GBM) is the most aggressive of the astrocytic malignancies and the most common intracranial tumor in adults. Although the epidermal growth factor receptor (EGFR) is overexpressed and/or mutated in at least 50% of GBM cases and is required for tumor maintenance in animal models, EGFR inhibitors have thus far failed to deliver significant responses in GBM patients. One inherent resistance mechanism in GBM is the coactivation of multiple receptor tyrosine kinases, which generates redundancy in activation of phosphoinositide-3′-kinase (PI3K) signaling. Here we demonstrate that the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) tumor suppressor is frequently phosphorylated at a conserved tyrosine residue, Y240, in GBM clinical samples. Phosphorylation of Y240 is associated with shortened overall survival and resistance to EGFR inhibitor therapy in GBM patients and plays an active role in mediating resistance to EGFR inhibition in vitro. Y240 phosphorylation can be mediated by both fibroblast growth factor receptors and SRC family kinases (SFKs) but does not affect the ability of PTEN to antagonize PI3K signaling. These findings show that, in addition to genetic loss and mutation of PTEN, its modulation by tyrosine phosphorylation has important implications for the development and treatment of GBM.
Keywords: glioma, phosphatase, erlotinib, gefitinib
The PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor gene encodes a phosphatase responsible for the removal of phosphate from the 3′ position of the phospholipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3), thus opposing mitogenic signaling mediated by the class 1 phosphoinositide-3′-kinases (PI3Ks) (1). In recent years it has become clear that PTEN also performs a number of tumor suppressor functions independent of its lipid phosphatase activity, including the suppression of cell migration, maintenance of genomic stability, and inhibition of cell cycle progression (2–5). The PTEN gene is lost or mutated in ∼40% of glioblastoma multiforme (GBM) (6, 7), and retention of PTEN protein expression has been linked with responses to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in GBM patients (8, 9), suggesting that the detection of functional PTEN may inform the successful deployment of targeted therapeutics in this currently intractable disease. Even in cancer cells harboring wild-type PTEN genes, however, its protein function can be commandeered by a number of posttranslational modifications, including oxidation, phosphorylation, acetylation, and ubiquitination (10). Moreover, the requirement for precise regulation of PTEN function is underlined by the observation that even a 20% reduction in PTEN gene dosage can predispose to malignant transformation (11). Given these functionally relevant modifications, mere detection of wild-type PTEN in tumor cells may not convey the status of PTEN function, which can be critically associated with responsiveness to targeted therapeutics in GBM (8, 9). Thus, dissection of PTEN modifications in the context of human cancer biology and defining the molecules mediating these changes is essential in determining prognosis and guiding optimal treatment regimens.
Tyrosine phosphorylation of PTEN by SRC-family kinases (SFKs) has been proposed to modulate its function in a number of ways, including loss of membrane interaction and altered protein stability (12–15). PTEN function is compromised in cells displaying high SFK activity, and in particular, inhibition of ErbB2-driven SRC activity by trastuzumab allows PTEN to suppress PI3K signaling in breast cancer cell lines (13, 14). However, from these studies, it is unclear as to whether the observed effects on PTEN function are attributable to its tyrosine phosphorylation or whether they are mediated indirectly through the phosphorylation of other SRC substrates. Recently, the RAK nonreceptor tyrosine kinase has been shown to enhance PTEN tumor suppressor function by phosphorylating tyrosine 336, thus preventing its degradation by the proteasome (15). This finding highlights the importance of accurately mapping the specific PTEN tyrosine residues phosphorylated in vivo and determining whether they have any role in clinical behavior or as predictors of relapse or indicators of prognosis. To this end, we used mass spectrometry to identify PTEN tyrosine phosphorylation sites and developed phospho-specific antibodies against one such site (Y240), which we used to study PTEN tyrosine phosphorylation in cell lines and primary tumor samples from GBM patients. This analysis led to the discovery that a significant SFK-independent Y240 kinase activity in GBM cells is attributable to fibroblast growth factor receptors (FGFRs), which interact with and phosphorylate PTEN in vitro and in vivo. From analyzing clinical samples, we also found that in addition to loss or mutation of PTEN, its phosphorylation at Y240 is linked to shortened survival and EGFR TKI resistance in GBM patients. Finally, we show that substitution of tyrosine 240 for phenylalanine enhances the ability of PTEN to sensitize cells to the EGFR TKI erlotinib and that activation of FGFR signaling can protect cells from erlotinib, concordant with induction of Y240 phosphorylation.
Results
Mapping of PTEN Tyrosine Phosphorylation Sites by Mass Spectrometry.
To identify tyrosine residues in PTEN that are phosphorylated by c-SRC, mass spectrometric profiling was performed with cells coexpressing PTEN and c-SRC. Four sites of tyrosine phosphorylation were identified in the PTEN phosphatase domain (Y46, Y68, Y155, and Y174), two sites in the PTEN C2 domain (Y240 and Y315), and one site, Y377, in the C-terminal tail (SI Appendix, Table S1). To study the dynamics of PTEN phosphorylation in cells and tissues, antibodies specific for phosphorylated tyrosine 240 (pY240) were generated. Y240 has been proposed to play an important role in PTEN function (12), and correspondingly, this residue is highly conserved among vertebrate PTEN orthologs (SI Appendix, Fig. S1).
Upon confirming the specificity of the pY240 antibody for Western blotting (SI Appendix, Fig. S2), we then explored Y240 phosphorylation in GBM cell lines, a tumor type in which aberrant SFK activation is frequently observed (13, 16, 17). Phosphorylation of endogenous PTEN at Y240 was readily detectable, both in the serial xenograft line, GBM39, which harbors the constitutively active EGFR deletion mutant ΔEGFR (also known as EGFRvIII) commonly expressed in GBM (18) (Fig. 1A), and in primary neurosphere cultures derived from human GBM samples (Fig. 1B). Interestingly, although SFKs were potently inhibited by the SRC/ABL inhibitor dasatinib, as revealed by a loss of autophosphorylation at a conserved tyrosine residue (Y419 in human c-SRC), there remained a significant dasatinib-resistant phosphorylation of PTEN at Y240, suggesting the existence of one or more additional Y240 kinases in GBM cells (Fig. 1 B and C).
Fig. 1.
SFK and FGFR-dependent phosphorylation of PTEN at tyrosine 240 in GBM cells. (A) GBM39 cells cultured ex vivo were treated with 100 nM dasatinib for 24 h, as shown before analysis by Western blotting. (B) Neurosphere cultures from GBM specimens were treated with dasatinib as indicated for 24 h before analysis by Western blotting. (C) GBM39 neurospheres were treated with 100 nM dasatinib and 1 μM PD173074 alone or in combination as shown. (D) HEK 293T cells were transfected with wild-type or Y240F PTEN and c-SRC constructs as shown. Inhibitors (Das, 100 nM dasatinib; PD, 50 nM PD173074) were added to cells 24 h after transfection, and cells were harvested after a further 24 h. PTEN was immunoprecipitated from lysates, and pan-phosphotyrosine or phosphorylation of Y240 were detected by Western blotting. (E) HEK 293T cells were transfected with PTEN and FGFR2 expression constructs as shown, and samples were processed as in D. (F) NIH 3T3 cells were serum-starved for 24 h before stimulation with 20 ng/mL bFGF and harvested over a time course after stimulation as indicated. (G) HEK 293T cells were cotransfected with constructs expressing FGFR2 and either empty vector control or FLAG-HA–tagged PTEN. Lysates were subject to immunoprecipitation with anti-HA antibodies, and coprecipitation of FGFR2 was detected by Western blotting.
FGFRs Phosphorylate PTEN at Tyrosine 240.
Because we observed phosphorylation of Y240 in cells growing under neurosphere conditions that included EGF and basic fibroblast growth factor (bFGF), we tested whether FGF–FGFR signaling may be involved in the phosphorylation of PTEN. This possibility was tested by treatment of GBM39 cells with PD173074, an FGFR family inhibitor (19). Although ΔEGFR and SFK activation remained high in PD173074-treated GBM39 cells, PTEN Y240 phosphorylation was reduced by PD173074 alone, and the combination of dasatinib and PD173074 resulted in a complete extinction of Y240 phosphorylation (Fig. 1C). To study further the phosphorylation of PTEN by SFKs and FGFRs, 293T cells were cotransfected with PTEN and either SRC, FGFR2, or FGFR3—two FGFR family members known to be activated in GBM samples (20). Although expression of SRC induced Y240 phosphorylation of PTEN, as shown by pY240 blotting, we observed tyrosine phosphorylation of both wild-type and Y240F PTEN when blots were probed with pan-phosphotyrosine antibodies (Fig. 1D). Thus, consistent with the mass spectrometry findings above, SRC phosphorylates multiple tyrosine residues on PTEN, including Y240. As expected from our studies of GBM39, Y240 phosphorylation driven by SRC was highly sensitive to dasatinib but resistant to inhibitors of FGFR or EGFR, suggesting direct phosphorylation of PTEN by SRC. In contrast, enforced expression of FGFR2 (Fig. 1E) resulted in specific phosphorylation of Y240, as evidenced by almost complete loss of the pan-phosphotyrosine signal in the Y240F mutant. Consistent with our findings with GBM39, whereas the FGFR-driven phosphorylation of Y240 was sensitive to PD173074, it was unaffected by dasatinib treatment, confirming that FGFRs can phosphorylate Y240 independently of SFKs. This was also the case upon coexpression of PTEN with the activated FGFR3 mutant K650E (21) (SI Appendix, Fig. S3). Because overexpression of kinases can lead to phosphorylation of substrates not necessarily targeted under physiological conditions, we next investigated whether PTEN becomes phosphorylated at Y240 after bFGF stimulation of cells in the absence of any transfected proteins. Consistent with a physiological role for this modification, Y240 phosphorylation was undetectable in starved NIH-3T3 fibroblasts and was induced after addition of bFGF, peaking at 6–8 h after stimulation (Fig. 1F). Y240 phosphorylation was also induced after bFGF stimulation of astrocytes isolated from ink4a/arf−/− mice (SI Appendix, Fig. S4).
In coimmunoprecipitation studies, the physical association of PTEN with FGFR2 and with FGFR3 (Fig. 1G and SI Appendix, Fig. S5) reinforced a role for FGFRs in PTEN modification via phosphorylation. To secure evidence of a more direct role for FGFR in effecting these events, in vitro kinase assays were conducted using recombinant purified proteins. In these experiments, SFKs (SRC and LYN) and FGFR family kinases (FGFR2 and FGFR3) were able to directly phosphorylate GST-PTEN in vitro (SI Appendix, Fig. S6). These data suggest that FGFRs are highly specific Y240 kinases that may modulate PTEN function primarily through phosphorylation at this site, in contrast to SFKs, which regulate PTEN in a number of ways, presumably as a result of their ability to target multiple sites (13–15). Although PTEN has been shown to interact with platelet-derived growth factor receptors (PDGFRs) (22, 23), which are closely related to FGFRs, neither PDGFRα nor EGFR drove robust phosphorylation of Y240 in our in vitro assays (SI Appendix, Fig. S6). Our finding that SFKs and FGFRs both phosphorylate PTEN at Y240 is reminiscent of the capacity of these two classes of kinases to target a common site on the 90kDa ribosomal S6 kinase, RSK-2 (24, 25).
Phosphorylation of Tyrosine 240 Does Not Affect the Ability of PTEN to Antagonize PI3K/AKT Signaling.
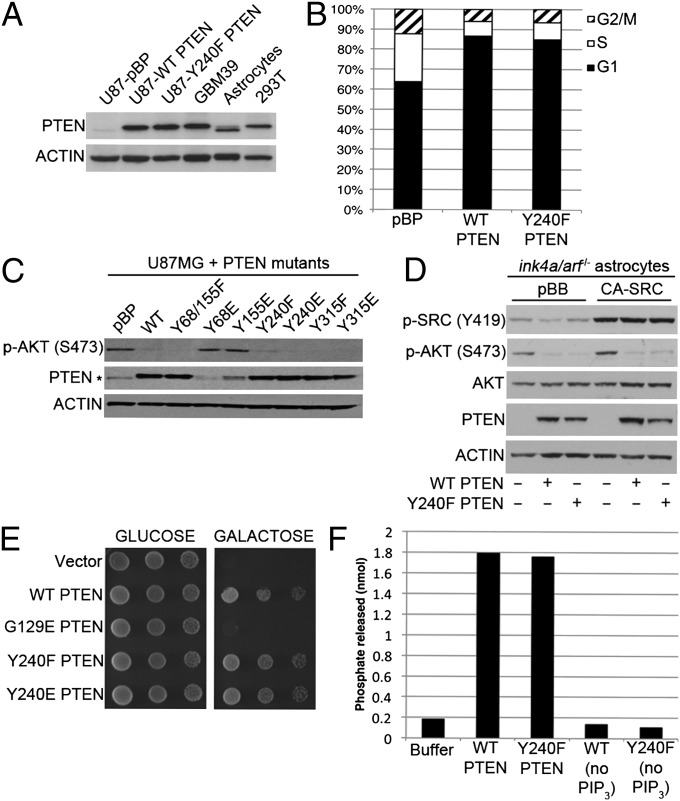
Both SFKs and FGFRs activate PI3K signaling by recruitment of the p85 regulatory subunit. The activity of PTEN to dephosphorylate PIP3 in membranes is also reduced upon coexpression with the SFK LCK (13), and inhibition of ErbB2-SRC signaling by trastuzumab treatment of breast cancer cells coincides with an increase in membrane-associated (and therefore potentially active) PTEN (14). These observations suggest that tyrosine phosphorylation of PTEN may inhibit its interaction with the plasma membrane, thus preventing access to PIP3. Conversely, it was previously reported that mutation of Y240, or of another SFK phosphorylation site, Y315, results in a loss of PTEN activity against PIP3 and concordant loss of tumor suppressor activity (12). These observations would suggest that phosphorylation of PTEN, at least at Y240 and Y315, may serve to activate the enzyme and enhance its ability to antagonize PI3K signaling. In an attempt to reconcile these observations, we compared the activity of wild-type PTEN with the Y240F mutant in several assays of PTEN function. We first established conditions by which we could reconstitute PTEN-mutant U87MG cells with physiological levels of PTEN, as demonstrated by Western blot showing expression levels equivalent to the endogenous levels of PTEN found in GBM39, mouse astrocytes, and 293T cells (Fig. 2A). We then analyzed the ability of wild-type and Y240F PTEN to cause G1 arrest in U87MG cells, a function dependent upon PTEN lipid phosphatase activity (26). Mutation of Y240 did not affect the G1 arrest activity of PTEN (Fig. 2B), suggesting that this allele retains lipid phosphatase activity in vivo. Furthermore, we assayed the ability of PTEN to antagonize AKT phosphorylation after mutation of a number of tyrosine phosphorylation sites (Fig. 2C and SI Appendix, Table S1) to either phenylalanine, to block phosphorylation, or to glutamate, which in some instances is able to mimic the effects of phosphorylation. Significantly, neither mutation of Y240 nor Y315 to phenylalanine prevented PTEN from suppressing AKT phosphorylation, nor did mutation of either residue to glutamate (Fig. 2C). Mutation of Y68 or Y155 to glutamate resulted in PTEN destabilization and increased AKT phosphorylation. Although it is possible that the inhibitory effects of SFK expression on PTEN activity and stability previously observed (13) could be due to phosphorylation at these sites, we found no evidence for Y68 or Y155 phosphorylation at endogenous levels of PTEN. Notably, point mutations resulting in substitution of Y68 and Y155 with cysteine or histidine and loss of PTEN function occur both in sporadic tumors and in the germline of patients with Cowden syndrome (27). Thus, the destabilization of PTEN is likely attributable to a loss of secondary structure caused by mutation of these residues, both of which reside in core α-helices (28, 29), rather than evidence for an effect of phosphorylation per se. To establish whether Y240 phosphorylation affects PTEN activity, we reconstituted PTEN-null astrocytes with either wild-type or Y240F PTEN. Even in the presence of constitutively active SRC, conditions under which PTEN tyrosine phosphorylation is readily detectable, the abilities of wild-type and Y240F PTEN to suppress AKT activation were indistinguishable (Fig. 2D and SI Appendix, Fig. S7). Next, we used a heterologous assay in which the ability of PTEN to rescue growth of yeast expressing a membrane-targeted mammalian PIK3CA allele (p110α-CAAX) is dependent upon its interaction with the membrane and dephosphorylation of PIP3 (30–32). Both the Y240F and Y240E alleles were equally capable of rescuing yeast expressing p110α-CAAX to viability as wild-type PTEN. In contrast, the lipid phosphatase-defective G129E mutant was unable to restore growth in the presence of p110α-CAAX (Fig. 2E). Finally, we directly compared the activities of recombinant purified wild-type and Y240F PTEN against soluble PIP3 in vitro and found them to be indistinguishable (Fig. 2F). Taken together, our data suggest that neither mutation of Y240 to phenylalanine nor phosphorylation of Y240 affect the ability of PTEN to dephosphorylate PIP3 and suppress PI3K signaling. It remains possible that a discrete pool of PTEN is modified by Y240 phosphorylation and that changes in the function of this specific fraction are masked in our assays of total cellular PTEN function. We are actively pursuing this possibility, together with our finding that Y240 phosphorylation peaks several hours after bFGF stimulation (Figs. 1F and 5D), thus with kinetics distinct from those of PI3K/AKT pathway activation.
Fig. 2.
Phosphorylation of Y240 does not affect PTEN control of PI3K/AKT signaling. (A) PTEN expression levels in U87MG cells after retroviral infection and selection compared with endogenous PTEN expression in GBM39, murine ink4a/arf−/− astrocytes, and HEK 293T cells. (B) Cell cycle profiles of U87MG cells reconstituted with PTEN wild-type or Y240F as in A, measured by propidium iodide staining and flow cytometry to assess DNA content. (C) AKT phosphorylation in U87MG cells reconstituted with wild-type PTEN or alleles bearing tyrosine phosphorylation site mutations as shown. Asterisk denotes the endogenous mutant PTEN. (D) Astrocytes derived from ink4a/arf−/−/ptenfl/fl mice after deletion of pten, expressing empty vector [pBabe-blasticidin (pBB)] or constitutively active SCR (pBB-SRC Y530F), were reconstituted with empty vector [pBabe-puro (pBP)] or pBP-wild-type PTEN or pBP-Y240F PTEN as shown. Cells were serum-starved for 24 h before harvesting. (E) Yeast expressing galactose-inducible p110α-CAAX were transformed with empty vector or PTEN alleles as shown and assessed for their ability to grow on galactose plates by spot assay as described in SI Appendix, SI Materials and Methods. (F) The catalytic activity of recombinant purified GST-tagged PTEN (wild-type or Y240F) proteins against soluble PI-3,4,5-P3 was measured by Biomol Green assay as described in SI Appendix, SI Materials and Methods.
Fig. 5.
Implication of PTEN Y240 phosphorylation in resistance to erlotinib in vitro. (A) Ink4a/Arf/PTEN−/− astrocytes expressing ΔEGFR were reconstituted with wild-type or Y240F PTEN alleles and after blasticidin selection were exposed to erlotinib for 72 h before measurement of cell viability by WST-1 assay. Error bars indicate SEM. (B) GBM39 cells were treated with erlotinib for 48 h in basal medium or in the presence of 20 ng/mL bFGF or 20 ng/mL EGF as shown, and relative cell viability was measured by WST1 assay. Decrease in viability is shown as the percentage change relative to untreated cells. (C) GBM39 cells collected after 48 h of erlotinib treatment were analyzed for cleaved PARP by Western blotting. (D) Acute changes in RTK signaling pathway components upon erlotinib treatment in the absence or presence of bFGF stimulation were assessed by Western blotting. Where indicated, 5 μM erlotinib was added to GBM39.
Detection of PTEN Y240 Phosphorylation in GBM Patient Samples.
Although PTEN tyrosine phosphorylation has been proposed to occur in human tumors, including GBM and breast cancer (12, 14, 15), it has not previously been possible to audit this modification in primary patient samples. To test whether our antibodies were potentially suitable for detection of pY240 by immunohistochemistry, we conducted peptide competition experiments using formalin-fixed, paraffin-embedded (FFPE) material from PTEN-positive human GBM samples (SI Appendix, Fig. S8) and staining of FFPE sections from xenograft tumors of U87-ΔEGFR cells reconstituted with either wild-type PTEN or Y240F PTEN (Fig. 3A). Whereas pY240 staining was evident in the tumors expressing wild-type PTEN, no reactivity was seen in tumor cells expressing the Y240F PTEN mutant, thus confirming the specificity of the antibodies for PTEN phosphorylated at Y240. Using the phospho-specific Y240 antibodies, we examined Y240 phosphorylation in a panel of GBM samples from two cohorts of patients at different institutions. As expected, GBM samples displayed heterogeneous staining for total PTEN, with a fraction of tumors exhibiting complete loss of PTEN expression. Of the PTEN-positive tumors analyzed at the University of São Paulo, 56 of 75 (75%) also displayed phosphorylation of Y240, suggesting that this modification is a common event in GBM (Fig. 3B). Because the available antibodies for monitoring FGFR activation are not suitable for immunohistochemistry of tissue sections, we analyzed FGFR expression levels by quantitative real-time PCR (qPCR). Consistent with our finding that FGFRs mediate PTEN phosphorylation in vitro, upon determining mRNA levels for the FGFR family members 1–4 by qPCR, we observed an increased expression (P < 0.05) of FGFR3 in those samples that displayed Y240 phosphorylation in the São Paulo cohort (Fig. 3C).
Fig. 3.
Phosphorylation of PTEN in GBM patient samples is correlated with FGFR3 expression and associated with shortened survival in EGFR-positive GBM. (A) Adjacent FFPE sections from s.c. xenograft tumors established from U87MG-ΔEGFR cells expressing either wild-type or Y240F PTEN were stained with antibodies specific for total PTEN and p-PTEN (Y240). Representative images from each tumor type are shown. (B) Paraffin-embedded tissue samples obtained after surgical resection of GBM patients were stained with antibodies specific for total and phospho-Y240 PTEN as indicated. Arrowheads indicate strong positivity for total PTEN in the vascular endothelium of a PTEN-negative tumor, characteristic of GBM (40× magnification). (C) FGFR3 transcript levels were measured by quantitative reverse transcriptase PCR using RNA isolated from GBM samples. (D) Kaplan-Meier survival curves showing overall survival of EGFR-positive GBM patients as a function of PTEN Y240 phosphorylation (SI Appendix, Table S2). (E) Representative images from GBM tissue microarray samples showing p-SFK, p-PTEN, and total PTEN staining.
When we analyzed those GBM samples that displayed elevated expression of EGFR (mRNA levels greater than median; SI Appendix, Table S2), we observed a striking association between Y240 phosphorylation and decreased overall survival (Fig. 3D) (median survival pY240-negative, 12 mo; median survival pY240-positive, 6 mo; P = 0.006). Sequencing revealed heterozygous PTEN mutations in several tumors, suggesting that mutant PTEN may also be targeted by Y240 phosphorylation (SI Appendix, Table S3). Staining of a GBM tissue microarray (TMA) from an independent validation set at the University of California, Los Angeles (UCLA) (Fig. 3E) revealed a similar frequency (82%) of p-PTEN staining among PTEN-positive samples (SI Appendix, Tables S2 and S4). We also observed pY240 staining in regions of residual PTEN expression in GBMs that showed intratumoral heterogeneity for expression of PTEN, exemplified in case 2, which interestingly also displays SFK activation restricted to the PTEN-positive region (Fig. 3E). Thus, PTEN function may be compromised by multiple genetic and posttranslational mechanisms within a single tumor—in the present case, loss of PTEN expression or modulation of PTEN function through Y240 phosphorylation. Examination of multiple slides from the São Paulo and UCLA studies revealed a largely nuclear localization for p-PTEN in the São Paulo samples, not apparent in slides from the UCLA patients or our xenograft material (compare Fig. 3 A and E with Fig. 3B). Staining of multiple slides from São Paulo at UCLA recapitulated this nuclear staining pattern (SI Appendix, Fig. S9), leading us to conclude that differences in the apparent localization of the pY240 signal are due to differences in the fixation and processing of the samples at the two institutions.
PTEN Y240 Phosphorylation Is Increased in EGFR TKI-Resistant GBM.
Because the presence of functional PTEN, together with expression of ΔEGFR, has been associated with response to the EGFR TKIs gefitinib and erlotinib in GBM patients (8, 9), we next analyzed PTEN phosphorylation in tumor tissue sections from GBM patients from an EGFR kinase inhibitor clinical trial (9). Significantly, tumors with high PTEN Y240 phosphorylation (P = 0.028) were those that failed to respond to EGFR inhibitors, despite expressing ΔEGFR and wild-type PTEN (Fig. 4A). Furthermore, we discovered that in two patients whose tumors initially responded to treatment but later acquired resistance, the PTEN pY240 levels were initially low but in the resistant tumors had increased to levels similar to those seen in the tumors that displayed resistance at the initiation of therapy (Fig. 4B). We also interrogated both the GBM TMA and TKI clinical trial samples for SFK activation by staining for Y419 phosphorylation and observed tight correlations with PTEN phosphorylation and TKI resistance (Fig. 3E and SI Appendix, Figs. S10 and S11 and Table S4), suggesting that SFKs may play important roles in TKI resistance in GBM, including but likely not restricted to, their contribution to PTEN phosphorylation. Although there are only a relatively small number of patients available from the EGFR TKI trial, all of which we examined, when taken together our findings suggest a link between PTEN phosphorylation and resistance to EGFR inhibitors in GBM. Furthermore, the persistence of PTEN and SFK phosphorylation observed in resistant tumors is consistent with a role for alternative receptor tyrosine kinases, such as FGFRs, in driving resistance by maintaining mitogenic signaling in the face of EGFR inhibition (20).
Fig. 4.
Phosphorylation of PTEN is increased in EGFR TKI-resistant GBM. (A) Quantification and representative staining patterns of PTEN Y240 phosphorylation observed in patients as a function of response to EGFR TKIs. (B) Quantification and representative staining of biopsies from two GBMs that initially responded to EGFR TKI therapy but later acquired resistance coincident with increased phosphorylation of PTEN.
PTEN Y240 Phosphorylation Plays a Causative Role in EGFR TKI Resistance.
To test for specific effects of PTEN Y240 phosphorylation on sensitivity to EGFR inhibitors in a genetically defined system, we reconstituted PTEN-null astrocytes expressing ΔEGFR (33) with either wild-type or Y240F PTEN. Introduction of the PTEN Y240F mutant sensitized these cells to erlotinib, whereas the wild-type PTEN allele lacked this activity (Fig. 5A; P < 0.01 at 500 nM/P < 0.001 at 2.5 μM). These data suggest that in addition to being a marker for EGFR TKI resistance, phosphorylation of PTEN at Y240 plays a causative role in driving resistance. Finally, on the basis of our findings that FGF signaling can drive phosphorylation of PTEN, we tested the potential for FGF to mediate resistance to EGFR inhibition. Consistent with the observed sensitivity of GBM39 to erlotinib in vivo (34), in the absence of exogenous growth factors treatment of GBM39 cells with erlotinib for 48 h resulted in cell rounding and detachment from the laminin substrate, induction of poly (ADP-ribose) polymerase (PARP) cleavage and loss of viability as measured by WST1 assay, all of which were blocked by the inclusion of bFGF in the medium (Fig. 5 B and C and SI Appendix, Fig. S12). Analysis of acute signaling changes in response to erlotinib treatment revealed a loss of AKT activation and PTEN Y240 phosphorylation, both of which were restored after addition of bFGF, although as we previously observed after bFGF stimulation of fibroblasts (Fig. 1F), Y240 phosphorylation was induced with delayed kinetics and only recovered to levels seen in untreated cells at 4 h after stimulation (Fig. 5D). Taken together, these data indicate that phosphorylation of PTEN at Y240 compromises its ability to mediate responses to EGFR TKIs, consistent with our observations that Y240 is highly phosphorylated in nonresponder tumors that retained wild-type PTEN expression. Furthermore, stimulation of FGFR signaling by addition of exogenous bFGF, which induces PTEN Y240 phosphorylation, is able to render the EGFR TKI-sensitive GBM cell line GBM39 resistant to erlotinib, demonstrating the potential for FGFR signaling to drive resistance in vivo.
Discussion
Although aberrant SFK activation has been linked to the suppression of PTEN function (13, 14), our study demonstrates that PTEN is tyrosine-phosphorylated in human tumors and provides a therapeutic rationale for attempting to inhibit this phosphorylation event in GBM. We have established that in addition to SFKs, the FGFRs also function as PTEN kinases, thus blocking PTEN phosphorylation in GBM will likely require combinations of tyrosine kinase inhibitors. Furthermore, the use of SFK inhibitors and/or FGFR inhibitors may sensitize resistant tumors to EGFR inhibition, at least in part, by suppressing PTEN phosphorylation. The finding that FGFRs phosphorylate PTEN is interesting in the context of recent observations that coactivation of multiple receptor tyrosine kinases (RTKs) is an important mechanism of resistance to TKIs in GBM cells, and that treatment of breast and lung cancer cell lines with PI3K or EGFR inhibitors induces transcription of several RTKs, including FGFR2 and -3 (20, 35–37). The signaling connection between FGFRs and PTEN may be of particular interest in those malignancies that frequently harbor activating mutations in FGFR, such as cancers of the breast, bladder, and endometrium (38).
In addition to phosphorylation of PTEN by SFKs, PTEN also dephosphorylates SFKs on their conserved autophosphorylation site (Y419 in c-SRC) (2). This activity is associated with inhibition of invasion in glioma cells and more recently has been shown to increase sensitivity to ErbB2 inhibition by trastuzumab in breast cancer cells (2, 39), but it is unclear as to which signals may determine the outcome of the PTEN–SFK interaction. By invoking FGFR as an additional input into PTEN phosphorylation, our data may help to explain how this reciprocal interaction can be regulated. Indeed, in our analysis the GBM samples displaying resistance to EGFR TKIs and high PTEN Y240 phosphorylation also displayed increased SFK phosphorylation at Y419 (SI Appendix, Figs. S10 and S11), again supporting our hypothesis that PTEN function is compromised in these tumors.
Our data suggest that phosphorylation of PTEN at Y240 does not affect its ability to attenuate PI3K signaling on a whole-cell level. We cannot rule out the possibility, however, that a specific pool of PTEN is modified in this way and that PIP3 levels at a particular subcellular location are regulated by PTEN tyrosine phosphorylation. In this regard, spatially restricted control of PTEN activity and the existence of discrete PIP3 pools within the cell have been documented (40–44). Alternatively, it is possible that Y240 phosphorylation affects a lipid phosphatase-independent function of PTEN, such as the control of cell migration or invasion, although we have not observed evidence for this, and it is less clear as to how this would impact on EGFR TKI sensitivity in vitro. We have not found evidence for an effect of Y240 phosphorylation on PTEN protein stability but are actively pursuing a potential role in directing PTEN to specific subcellular compartments. Our studies with the PTEN Y240F and Y315F mutants suggest that in contrast to a previous report (12), these tyrosines are not required for PTEN tumor suppressor function. The previous study was conducted in the U251MG cell line, which harbors mutant p53 (45), whereas the U87MG cells and murine astrocytes used in our study express wild-type p53. It is possible, therefore, that Y240 and Y315 become important for PTEN in the absence of functional p53, and we are investigating this possibility.
The contribution of suppressed PTEN function to a diverse range of malignancies suggests that our findings in GBM may be broadly applicable and that assessing the phosphorylation status of PTEN in patient biopsies will be a useful approach in guiding the treatment of multiple cancers and potentially other diseases associated with aberrant FGFR signaling.
Materials and Methods
Detailed descriptions of cell lines, antibodies and reagents, phosphorylation site mapping by mass spectrometry, PTEN phosphatase assays, yeast-based PTEN activity assays, U87MG xenografts, in vitro kinase assays, cell cycle analysis, drug sensitivity assays, and all immunohistochemical staining, qPCR, and statistical analysis of clinical samples are provided in SI Appendix, SI Materials and Methods. Experiments using animals were performed according to a protocol approved by the UCSD Institutional Animal Care and Use Committee. Human tumor specimens were obtained according to a protocol approved by the Institutional Review Board of UCLA.
Supplementary Material
Acknowledgments
We thank Cameron Brennan (Memorial Sloan-Kettering Cancer Center) for the TS543 GBM tumor sphere line, Daniel Donoghue (University of California, San Diego) for the plasmid encoding FGFR3 K650E, Richard Kolodner (University of California, San Diego) and Rob de Bruin [Medical Research Council (MRC) Laboratory for Molecular Cell Biology, United Kingdom] for yeast reagents, and Stephen Henderson (University College London, United Kingdom) for assistance with bioinformatics. This work was supported by an award from the Goldhirsh Foundation (to F.B.F.), National Institutes of Health Grants P01-CA95616 (to R.A.D., W.K.C., and F.B.F.) and P50-CA097257 (to C.D.J. and P.S.M.), and Fundaçao de Amparo à Pesquisa do Estado de São Paulo Grant 04/12433-6 (to S.K.N.M). W.K.C. is a fellow of the National Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211962109/-/DCSupplemental.
References
- 1.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Dey N, et al. The protein phosphatase activity of PTEN regulates SRC family kinases and controls glioma migration. Cancer Res. 2008;68:1862–1871. doi: 10.1158/0008-5472.CAN-07-1182. [DOI] [PubMed] [Google Scholar]
- 3.Okumura K, Zhao M, Depinho RA, Furnari FB, Cavenee WK. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc Natl Acad Sci USA. 2005;102:2703–2706. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 5.Song MS, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furnari FB, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 8.Haas-Kogan DA, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 9.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Jiang X. Post-translational regulation of PTEN. Oncogene. 2008;27:5454–5463. doi: 10.1038/onc.2008.242. [DOI] [PubMed] [Google Scholar]
- 11.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koul D, et al. Motif analysis of the tumor suppressor gene MMAC/PTEN identifies tyrosines critical for tumor suppression and lipid phosphatase activity. Oncogene. 2002;21:2357–2364. doi: 10.1038/sj.onc.1205296. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, et al. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J Biol Chem. 2003;278:40057–40066. doi: 10.1074/jbc.M303621200. [DOI] [PubMed] [Google Scholar]
- 14.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 15.Yim EK, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stettner MR, et al. Lyn kinase activity is the predominant cellular SRC kinase activity in glioblastoma tumor cells. Cancer Res. 2005;65:5535–5543. doi: 10.1158/0008-5472.CAN-04-3688. [DOI] [PubMed] [Google Scholar]
- 18.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 19.Mohammadi M, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 21.Webster MK, D’Avis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahimainathan L, Choudhury GG. Inactivation of platelet-derived growth factor receptor by the tumor suppressor PTEN provides a novel mechanism of action of the phosphatase. J Biol Chem. 2004;279:15258–15268. doi: 10.1074/jbc.M314328200. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, Morales FC, Kreimann EL, Georgescu M-M. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S, et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S, et al. Epidermal growth factor stimulates RSK2 activation through activation of the MEK/ERK pathway and src-dependent tyrosine phosphorylation of RSK2 at Tyr-529. J Biol Chem. 2008;283:4652–4657. doi: 10.1074/jbc.M709673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furnari FB, Huang HJ, Cavenee WK. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 27.Han SY, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60:3147–3151. [PubMed] [Google Scholar]
- 28.Georgescu MM, et al. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000;60:7033–7038. [PubMed] [Google Scholar]
- 29.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 30.Andrés-Pons A, et al. In vivo functional analysis of the counterbalance of hyperactive phosphatidylinositol 3-kinase p110 catalytic oncoproteins by the tumor suppressor PTEN. Cancer Res. 2007;67:9731–9739. doi: 10.1158/0008-5472.CAN-07-1278. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Escudero I, et al. A comprehensive functional analysis of PTEN mutations: Implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Escudero I, et al. Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem J. 2005;390:613–623. doi: 10.1042/BJ20050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Iglesia N, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkaria JN, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 35.Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci USA. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandarlapaty S, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ware KE, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS ONE. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner N, Grose R. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 41.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Lindsay Y, et al. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J Cell Sci. 2006;119:5160–5168. doi: 10.1242/jcs.000133. [DOI] [PubMed] [Google Scholar]
- 43.Maccario H, Perera NM, Gray A, Downes CP, Leslie NR. Ubiquitination of PTEN (phosphatase and tensin homolog) inhibits phosphatase activity and is enhanced by membrane targeting and hyperosmotic stress. J Biol Chem. 2010;285:12620–12628. doi: 10.1074/jbc.M109.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tibarewal P, et al. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci Signal. 2012;5:ra18. doi: 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- 45.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.