Abstract
We report the high-resolution (1.9 Å) crystal structure of oligomycin bound to the subunit c10 ring of the yeast mitochondrial ATP synthase. Oligomycin binds to the surface of the c10 ring making contact with two neighboring molecules at a position that explains the inhibitory effect on ATP synthesis. The carboxyl side chain of Glu59, which is essential for proton translocation, forms an H-bond with oligomycin via a bridging water molecule but is otherwise shielded from the aqueous environment. The remaining contacts between oligomycin and subunit c are primarily hydrophobic. The amino acid residues that form the oligomycin-binding site are 100% conserved between human and yeast but are widely different from those in bacterial homologs, thus explaining the differential sensitivity to oligomycin. Prior genetics studies suggest that the oligomycin-binding site overlaps with the binding site of other antibiotics, including those effective against Mycobacterium tuberculosis, and thereby frames a common “drug-binding site.” We anticipate that this drug-binding site will serve as an effective target for new antibiotics developed by rational design.
Keywords: F1Fo ATP synthase, proton pore
Oligomycin has been recognized as a potent inhibitor of the mitochondrial ATP synthase since 1958 when it was reported by Henry Lardy et al. (1). Studies in the 1960s from the laboratory of Efraim Racker demonstrated that the mitochondrial ATP synthase can be separated into two parts, coupling factor 1, or F1, which contains the catalytic site for ATP synthesis, and coupling factor o, or Fo, which is able to confer sensitivity to oligomycin (2–4). Despite more than 50 y of studies on mitochondrial F1Fo ATP synthase, the binding site of oligomycin on Fo has been elusive. Here we report the oligomycin-binding site on subunit-c of the Fo portion of the ATP synthase.
Subunit-c of the ATP synthase is an integral membrane protein consisting of two helices, 1 and 2, which span the inner mitochondrial membrane (Fig. 1). Subunit-c assembles as a homomeric ring consisting of 10 subunits in the yeast ATP synthase and eight subunits in the bovine ATP synthase (5, 6). The c-ring forms an essential component of the proton turbine of the ATP synthase, which spins coupled to the movement of protons down a potential gradient. The essential carboxylate of Glu59 in helix 2 of the yeast subunit-c is postulated to participate directly in the movement of protons from the cytosol to the mitochondrial matrix during ATP synthesis. The side-chain carboxyl of Glu59 is nearly in the middle of helix 2, positioning it in the lipid bilayer in the protonated, “closed” conformation (7). Subunit-a is postulated to form two aqueous half-channels that allow protons to gain access to the carboxylate of Glu59 in the “open” conformation, allowing protonation and deprotonation reaction (7).
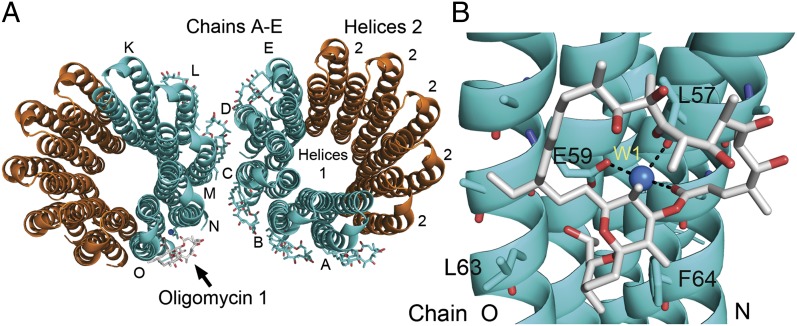
Fig. 1.
Oligomycin bound to c10 ring. (A) Shown is a representation of two c10 rings with bound oligomycin. The rings colored in cyan represent the copies in the asymmetric unit; the rings colored in orange are symmetrically related rings. The oligomycin molecules are shown only in the asymmetric unit. Oligomycin shown in cyan and gray represents the molecules that were modeled into the electron density map. Helix 1 is the first helix in subunit-c and is packed in the interior of the c-ring. Helix 2 is packed on the outer portion of the c-ring. The subunits of the asymmetric unit are designated A–E and K–O. Oligomycin 1 (gray) is bound to chains O and N and had the best corresponding electron density. (B) View of the oligomycin-binding site. The atoms in oligomycin are colored coded: gray, carbon; red, oxygen; blue, nitrogen. The water molecule (W1, blue) forms a bridging H-bond between Glu59 on chain O, the carbonyl of Leu57 on chain N, and the ester carbonyl of oligomycin (O36). The atoms of the c-ring are color coded: cyan, carbon; red, oxygen; blue, nitrogen.
Based on the results presented here, we propose that in the intact ATP synthase, oligomycin binds at the c-ring positioned at the proton channel and blocks proton translocation by blocking access to the essential carboxyl. Furthermore, we propose that the binding site framed out by oligomycin is a common drug-binding site for inhibitors that bind to the bacterial c-ring and likely for similar inhibitors that bind to Vo (analogous to Fo) of the vacuolar ATPase. Because of the widespread use of these inhibitors and the potential for drug discovery, the oligomycin-binding site constitutes an important basis for future drug discovery.
Results and Discussion
We recently reported the high-resolution crystal structure of the yeast c10-ring (8). The crystals, which formed in a solution containing 2-methyl-2,4-pentanediol (MPD), were tightly packed, containing only 47% solvent. The crystallographic space group of the crystals was P4222, with 40 c-subunits in the unit cell and five subunits per asymmetric unit. The five subunits in the asymmetric unit form half of a complete c-ring. The average B-factors for all the atoms were about 25 Å2 including the water molecules, indicating that the atoms are tightly constrained. The essential side-chain carboxylate of Glu59 was in an open and extended conformation independent of pH, suggesting that it is the structure involved in protonation and deprotonation during the reaction cycle.
To obtain the oligomycin-bound structure, the crystals were formed at pH 6.1 and incubated in a solution containing oligomycin. To mimic the biological conditions more closely, the MPD concentration was lowered in a stepwise fashion to a final concentration of 5% (vol/vol). The crystallographic characteristics remained the same until the MPD concentration was lowered to 20% (vol/vol), at which point the space group changed to I4122 with 80 c-subunits in the unit cell and 10 subunits per asymmetric unit (Table S1). In the absence of oligomycin, the diffraction quality diminished when the MPD concentration was lowered to 15% (vol/vol), whereas in the presence of oligomycin the diffraction quality was unchanged to the final MPD concentration of 5% (vol/vol). This result suggested that oligomycin was binding to the c-ring and aiding in the crystal integrity.
The structure was solved by molecular replacement using half a c-ring from the structure of yeast c10 (PDB ID 3UD0) (8). The resulting electron density map was clearly traceable and yielded an unambiguous model of the 10 c-subunits, each containing 74 or 75 amino acids without any outliers in the Ramachandran plot. The refinement did not use noncrystallographic constraints or restraints, and thus each of the 10 subunit-c structures is unique, although they are nearly identical, as evidenced by an average pair-wise rmsd of 0.18 Å (Cα atoms of residues 3–73). As in prior structures of the yeast c10 ring, the average B-factor for all of the atoms, including 150 assigned water molecules, was low (26.8 Å2), suggesting that the atoms were in a rather fixed position.
We could model seven molecules of oligomycin A reliably into the asymmetric unit, which has 10 c-subunits (Fig. 1). The electron density map of the bound oligomycin (Fig. S1 and Movie S1) was not equivalent for all seven molecules, possibly because crystal contacts and differences in the accessibility of the binding site to the solvent channels altered the occupancy. These factors also may explain why only seven molecules of oligomycin, not 10 molecules, are present in the asymmetric unit. However, this variance provided an internal control to assess the conformational changes elicited by the binding of oligomycin (Fig. S2). Although we used a mixture of oligomycin A, B, and C, the electron density map fit the model of oligomycin A; oligomycin B and C were likely bound also, but at a much lower occupancy. Even though this structure is that of the c-ring alone, that the mode of binding is likely to be identical to that in the intact ATP synthase.
Oligomycin binds to helix 2 of two adjacent subunit-c molecules (Fig. 1). The hydrophobic face of the oligomycin molecule covers the hydrophobic face of subunit-c, and the hydrophilic face of oligomycin is predominantly exposed to the bulk solvent. These atomic arrangements contribute both enthalpic and entropic energy to the binding energy. The oligomycin molecule covers Glu59 and obscures access of the side-chain carboxyl to the bulk solvent. The side-chain carboxyl of Glu59 forms an H-bond with a water molecule, which forms an H-bond with the carbonyl oxygen of Leu57 and an ester carbonyl oxygen (O36) in oligomycin. Thus, Glu59 forms an H-bond to oligomycin via a bridging water molecule. Compared with the ligand-free structures, the conformation of the side-chain carboxylate of Glu59 moves inward in the oligomycin-bound structure to accommodate the H-bond with the water (Fig. S2). Based on the pH and the atoms involved, it is likely that the side-chain carboxyl of Glu59 is in the protonated state. We propose that oligomycin inhibits proton translocation in the F1Fo ATP synthase by locking the essential carboxyl in a semiclosed conformation and shielding its access to the aqueous environment of the proton half-channel.
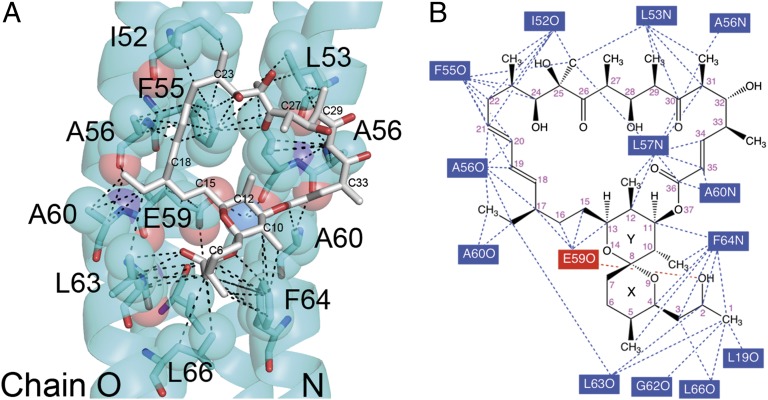
The remaining interactions between oligomycin and subunit-c occur primarily through van der Waals interactions (Fig. 2 and Movie S2). There are numerous instances in which the interatomic distance between the atoms of subunit-c and oligomycin are 3.7–4.9 Å. Phe64 forms a number of interactions with six carbon atoms of oligomycin, including C2 and C3, which have been reported to have significant double-bonding characteristics in oligomycin A (9), and thus also may interact via π–π interactions. Two residues, Ala56 and Ala60, each interact with opposite ends of the oligomycin molecule by forming contacts with oligomycin from both c-ring molecules, in this case on chains N and O. The remaining interacting residues form contacts to oligomycin from only one of the c-subunits in the pair forming the binding site.
Fig. 2.
Atomic interactions with oligomycin. (A) The atoms shown as spheres and residues shown as sticks are within 5 Å of an atom in the oligomycin molecule. The dashed lines indicate atomic distances in the range of 2.5–2.8 Å for the three H-bonds and 3.7–4.8 Å for the putative van der Waals interactions. Select carbon atoms of oligomycin are labeled. Oligomycin 1 is modeled here as oligomycin A, although the crystals were soaked with a mixture of oligomycin A, B, and C with 60% being oligomycin A. The water molecule, W1, is shown as a blue sphere. (B) A map of the interactions between oligomycin 1 and the c-ring. The residues for which an atom is within 4.8 Å are shown with dashed lines identifying the putative interactions. The single red dashed line shows a polar contact between E59 and the propanol group of oligomycin. As noted in the text, this distance is 3.2 Å, but the geometry does not favor a strong H-bond.
The backbone conformation of the c-ring does not change with oligomycin bound as shown by an average rmsd of 0.21 Å (Cα atoms). However, in addition to Glu59, the side chains of Leu63 and Phe64 form new rotamer conformations with the binding of oligomycin (Fig. S2). The rotation of the Leu63 and Phe64 side chains opens up a crevice between helix 2 of adjacent c-subunits, allowing the propanol group linked to the pyranose ring of oligomycin A to fit neatly. The oxygen atom (O2) of the propanol group is positioned within 3.6 Å of the amide carbonyl and 3.2 Å of the side-chain carboxyl of Glu59, but except for the distance, the geometries are poor for a strong H-bond. These rather minor changes that occur with the binding of oligomycin suggest that little binding energy is lost from entropic changes because of conformational changes in subunit-c.
The conformation of oligomycin 1 bound to the c-ring is nearly identical to the structure of oligomycin A as determined by X-ray crystallography (Fig. S3) (9). Comparative analysis indicates few conformational changes in the macrocyclic ring of oligomycin A upon binding subunit-c. The largest changes were associated with the conformation of the spiro-linked pyranose rings in which the propanol group inserts between the pair of subunit-c helices.
Overall, a single H-bond and numerous van der Waals interactions determine the enthalpy of the binding energy. Because the enthalpy of van der Waals interactions is strongly distance dependent (10), small changes in the backbone of subunit-c would be sufficient to affect the binding of oligomycin. Entropic losses caused by conformational changes are minimized by the highly constrained geometries of subunit-c, as indicated by the low B-factors, by the limited conformations the oligomycin molecule, and by minimal conformational changes in subunit-c caused by oligomycin binding. However, the hydrophobic effect (11) is likely the largest contributor to the binding energy, because the binding of oligomycin buries a calculated 345 Å2 of the hydrophobic surface of subunit-c, which is replaced with the hydrophilic face of the oligomycin molecule (12). Based on the mode of binding, we propose that the oligomycin binds only to subunit-c chains that are in contact with the proton half-channels formed by subunit-a. Thus, although we see seven oligomycin molecules bound to the c10-ring in the crystal structure, we believe that the number bound in the intact ATP synthase would be limited to the number of binding sites exposed to the proton half-channels.
Paradoxically, Lee and Ernster (13, 14) showed that at low concentrations oligomycin increased the phosphorylation capacity of EDTA/alkali-treated nonphosphorylating submitochondrial particles, and this finding was confirmed by studies in other laboratories (15). The most reasonable explanation for this finding is that oligomycin binds preferentially to the c-ring of the ATP synthases that are damaged and uncoupled, thereby preventing proton leak and allowing ATP synthesis by the undamaged synthase complexes. If correct, this notion suggests that the oligomycin binding site is more accessible in the damaged ATP synthase, but this suggestion is inconsistent with free access to the oligomycin binding site in the coupled ATP synthase.
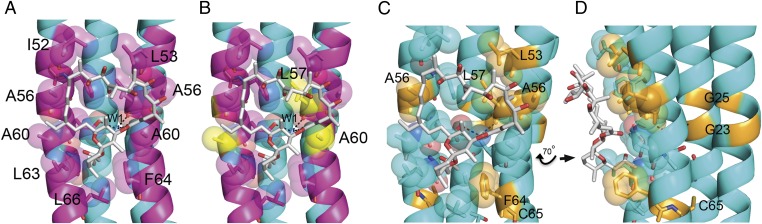
Oligomycin is a potent inhibitor of the mitochondrial, but not bacterial, enzyme. The residues involved in the oligomycin-binding site are conserved from yeast to human (Fig. 3A). In contrast, only two of the corresponding residues in the Escherichia coli subunit-c are conserved, Leu57 and Ala60 (Fig. 3B). Furthermore, the residue in E. coli subunit-c that corresponds to Glu59 is an aspartic acid residue, and this difference would affect the H-bonding network between subunit-c, the water molecule, and oligomycin. This analysis thus provides an explanation for, and is consistent with, the specificity of oligomycin on the ATP synthase from mitochondria and bacteria.
Fig. 3.
Conservation of the oligomycin-binding site. (A) The residues that are conserved from yeast to human are colored magenta. All the residues of the c-ring involved in binding oligomycin are conserved. Otherwise, this image is nearly identical to that in Fig. 2. (B) The residues colored in yellow are the only residues that are conserved from yeast to human (i.e., colored magenta in A) that are conserved in E. coli subunit-c. Thus, the oligomycin-binding site is not conserved in the bacterial subunit-c. Water (W1) is colored blue. (C and D) Residues that are critical for oligomycin binding as determined by genetic studies. The residues colored orange represent the residues identified in yeast that, when changed, confer resistance to oligomycin and a subset with cross-resistance to venturicidin and ossamycin. In D the molecules shown in C have been rotated 70° clockwise around the y axis.
A number of mutations in yeast have been shown to confer resistance to oligomycin and, in some cases, cross-resistance to the related antibiotics such as ossamycin and venturicidin (16–19). Unlike oligomycin and ossamycin, venturicidin is a potent inhibitor of mitochondrial and bacterial ATP synthase (20–22). The residues identified in yeast subunit-c that, when mutated, confer resistance to oligomycin are shown in Fig. 3 C and D. Four of the residues, Leu53, Ala56, Leu57, and Phe64 interact directly with oligomycin. The mechanism of resistance caused by mutations in these residues is direct and understandable. In contrast, mutations in Gly23 and Gly25 also confer resistance to oligomycin, but these residues are located in helix 1, which contains only one residue that forms the drug-binding site. However, Gly23 and Gly25 are adjacent to residues in helix 2 that are largely responsible for binding oligomycin. We propose that mutations in these residues alter the backbone conformation of helix 1, which in turn alters the backbone conformation of helix 2, thereby distorting the drug-binding site. Interestingly, mutations in Gly23 and Gly25 can confer resistance to multiple drugs, suggesting a general disruption of the drug-binding site (17, 18). Last, the side-chain of Cys65 is packed between the helices, and the oligomycin-resistance mutation, Cys65Ser (19), likely alters the interhelical packing and thus disrupts the oligomycin-binding site. The proximity of the residues that, when mutated, confer resistance to oligomycin, ossamycin, and venturicidin suggests that these antibiotics share a common binding region, although the exact molecular interactions almost certainly vary.
A seeming inconsistency is that resistance mutations also have been mapped to the gene encoding subunit-a of the ATP synthase (19, 20, 23, 24). There are two apparent explanations. First, the oligomycin binding site may be shared between subunits-a and -c. We do not favor this explanation, because oligomycin binds neatly to the face of subunit-c and does not seem to require additional interactions to form a stable complex. However, oligomycin derivatives that are considerably larger molecules, such as ossamycin, may bind to subunit-c and, by default, also interact with residues in subunit-a. The second explanation is that substitutions in subunit-a that confer resistance to oligomycin act by blocking access to the binding site. The occurrence of mutations in subunit-a that confer resistance to oligomycin thus provides supporting evidence that the oligomycin-binding site in the intact ATP synthase is at the face of the c-ring that is positioned at the proton access channel formed by subunit-a and that oligomycin does not bind at the c subunit facing the lipid bilayer.
Recently, the drug R207910 (TMC207) has been identified that is effective against Mycobacterium tuberculosis (25, 26). Strains resistant against R207910 were isolated, and three independent mutations in the gene encoding subunit-c, Asp26Val, Ala61Pro, Ile64Met (yeast numbering system), were identified as being responsible for conferring resistance to the drug (25, 27). In the model of the yeast subunit-c, Asp26 is in helix 1 and would be directly adjacent to yeast Gly25, which, when mutated, confers resistance to oligomycin and cross-resistance to related drugs (17, 18). Ala61 is adjacent to yeast Ala60, which is one of the two residues that interacts with oligomycin in both subunit-c molecules forming the binding site. The Ala61Pro mutation would be expected to cause a kink in the α-helix and to disrupt the drug-binding site. Last, Ile64 corresponds to Phe64 in yeast, which forms critical contacts with oligomycin. Although the structure of R207910 is quite distinct from that of oligomycin, ossamycin, and venturicidin, it shares some of their chemical properties. Thus, we propose that the binding site framed by oligomycin is a common drug-binding site for both bacterial and mitochondrial subunit-c. If this proposal is correct, then crystal structure analysis of the c-ring from pathogenic bacteria will provide a scaffold on which to build novel antibiotics by rational design. Finally, the V-type ATPases are similar in structure and are related in function to the F-type ATP synthases. Inhibitors that bind to Vo, such as bafilomycin and concanamycin, may bind to the corresponding region in the corresponding subunit of the vacuolar ATPase (28) and thus provide another potential drug target.
Materials and Methods
The yeast ATP synthase was purified, and the c-ring was crystallized as described (8), except that the crystallization buffer contained 50 mM Na citrate (pH 5.5) and 100 mM Na malonate (pH 7.0). The crystals were soaked in solution containing 0.5 mg/mL oligomycin (catalog no. 04876; Sigma), which is a mixture of oligomycin A, B, and C (60% oligomycin A). Crystal soaking to reduce the concentration of MPD to 5% was performed in a stepwise fashion, essentially as described previously (8), except that the buffer contained 50 mM Na citrate (pH 5.5), 0.4 M NaCl, 2 mM MgCl2, and 8% (vol/vol) propylene glycol and 1,2-dimyristoyl-sn-glycero-3-phosphocholine:1,2-dihexanoyl-sn-glycero-3-phosphocholine (3:1 molar ratio) lipid bicelles at 1% (wt/vol). On day 1, the crystal was soaked in buffer solutions containing 68% (vol/vol) MPD, 58% MPD, 48% MPD, and 38% MPD (0.1 mL); each soaking lasted 1–1.5 h at 21 °C. The final soaking was overnight at 21 °C in 28% MPD containing 0.5 mg/mL oligomycin. On day 2, the MPD concentration was reduced sequentially to 20%, 15%, 10%, and 5% with oligomycin (0.5 mg/mL). Glycerol was added as a cryoprotectant, so that the sum of the concentration of MPD (vol/vol) and glycerol (wt/vol) was equal to 30%. The crystals were soaked in each MPD concentration for 1–1.5 h. At the end of the final soaking period, the crystals were frozen in liquid nitrogen.
Diffraction data were collected at Advanced Photon Source (21ID-G, LS-CAT) at 100 K using a wavelength of 0.97856 Å. The data were processed using HKL2000 (29) and the CCP4 suite (30). Molecular replacement solution was obtained by Molrep (31) with the search model consisting of chains K, L, M, N, and O from the structure 3UD0 (8). The solution represented 10 chains (two half c-rings; see Fig. 1) in the asymmetric unit. Rigid body refinement was performed in Refmac (32), and the corresponding electron density maps revealed seven oligomycin molecules bound in a similar fashion to two neighboring c-subunits near Glu59. Coot (33) was used to display electron density maps, to make small adjustments in the model, and to add water molecules. The initial coordinates for oligomycin A were obtained from Cambridge Structural Database (CCDC 636099) (34), and the parameter file/library for oligomycin was generated by PRODRG (35). Final refinement procedures were performed in Refmac using restrained and TLS (Translation Libration Screw-motion) refinement. The model was validated by Procheck (36).
Accession Codes.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, under PDB ID code 4F4S.
Supplementary Material
Acknowledgments
Diffraction data were collected at Life Sciences Collaborative Access Team (LS-CAT, 21-ID-G) at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This work was supported by Nationals Institutes of Health Grant R01GM66223 (to D.M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID code 4F4S).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207912109/-/DCSupplemental.
References
- 1.Lardy HA, Johnson D, McMurray WC. Antibiotics as tools for metabolic studies. I. A survey of toxic antibiotics in respiratory, phosphorylative and glycolytic systems. Arch Biochem Biophys. 1958;78:587–597. doi: 10.1016/0003-9861(58)90383-7. [DOI] [PubMed] [Google Scholar]
- 2.Racker E. A mitochondrial factor conferring oligomycin sensitivity on soluble mitochondrial ATPase. Biochem Biophys Res Commun. 1963;10:435–439. doi: 10.1016/0006-291x(63)90375-9. [DOI] [PubMed] [Google Scholar]
- 3.Racker E. A reconstituted system of oxidative phosphorylation. Biochem Biophys Res Commun. 1964;14:75–78. doi: 10.1016/0006-291x(63)90214-6. [DOI] [PubMed] [Google Scholar]
- 4.Kagawa Y, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase. J Biol Chem. 1966;241:2461–2466. [PubMed] [Google Scholar]
- 5.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 7.Pogoryelov D, et al. Microscopic rotary mechanism of ion translocation in the F(o) complex of ATP synthases. Nat Chem Biol. 2010;6:891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- 8.Symersky J, et al. Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol. 2012;19:485–491, S1. doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer RA, Potter BS. X-ray structures and absolute configurations of the antibiotics oligomycins A, B, and C: Inhibitors of ATP synthase. J Chem Crystallogr. 2008;38(4):243–253. [Google Scholar]
- 10.Moore WJ. Physical Chemistry. Englewood Cliffs, NJ: Prentice-Hall; 1972. pp. 913–918. [Google Scholar]
- 11.Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 12.Tuñón I, Silla E, Pascual-Ahuir JL. Molecular surface area and hydrophobic effect. Protein Eng. 1992;5:715–716. doi: 10.1093/protein/5.8.715. [DOI] [PubMed] [Google Scholar]
- 13.Lee CP, Azzone GF, Ernster L. Evidence for Energy-Coupling in Non-Phosphorylating Electron Transport Particles from Beef-Heart Mitochondria. Nature. 1964;201:152–155. doi: 10.1038/201152a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee CP, Ernster L. Restoration of Oxidative Phosphorylation in Non-Phosphorylating Submitochondrial Particles by Oligomycin. Biochem Biophys Res Commun. 1965;18:523–529. doi: 10.1016/0006-291x(65)90785-0. [DOI] [PubMed] [Google Scholar]
- 15.Fessenden JM, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XI. Stimulation of oxidative phosphorylation by coupling factors and oligomycin; inhibition by an antibody against coupling factor 1. J Biol Chem. 1966;241:2483–2489. [PubMed] [Google Scholar]
- 16.Ooi BG, Novitski CE, Nagley P. DNA sequence analysis of the oli1 gene reveals amino acid changes in mitochondrial ATPase subunit 9 from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1985;152:709–714. doi: 10.1111/j.1432-1033.1985.tb09251.x. [DOI] [PubMed] [Google Scholar]
- 17.Galanis M, Mattoon JR, Nagley P. Amino acid substitutions in mitochondrial ATP synthase subunit 9 of Saccharomyces cerevisiae leading to venturicidin or ossamycin resistance. FEBS Lett. 1989;249:333–336. doi: 10.1016/0014-5793(89)80653-2. [DOI] [PubMed] [Google Scholar]
- 18.Nagley P, Hall RM, Ooi BG. Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance. FEBS Lett. 1986;195:159–163. doi: 10.1016/0014-5793(86)80152-1. [DOI] [PubMed] [Google Scholar]
- 19.Sebald W, Wachter E, Tzagoloff A. Identification of amino acid substitutions in the dicyclohexylcarbodiimide-binding subunit of the mitochondrial ATPase complex from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1979;100:599–607. doi: 10.1111/j.1432-1033.1979.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths DE, Houghton RL. Studies on energy-linked reactions: Modified mitochondrial ATPase of oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1974;46:157–167. doi: 10.1111/j.1432-1033.1974.tb03608.x. [DOI] [PubMed] [Google Scholar]
- 21.Perlin DS, Latchney LR, Senior AE. Inhibition of Escherichia coli H+-ATPase by venturicidin, oligomycin and ossamycin. Biochim Biophys Acta. 1985;807:238–244. doi: 10.1016/0005-2728(85)90254-3. [DOI] [PubMed] [Google Scholar]
- 22.Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol Mol Biol Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foury F, Tzagoloff A. Localization on mitochondrial DNA of mutations leading to a loss of rutamycin-sensitive adenosine triphosphatase. Eur J Biochem. 1976;68:113–119. doi: 10.1111/j.1432-1033.1976.tb10769.x. [DOI] [PubMed] [Google Scholar]
- 24.John UP, Nagley P. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 1986;207:79–83. doi: 10.1016/0014-5793(86)80016-3. [DOI] [PubMed] [Google Scholar]
- 25.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 26.Koul A, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- 27.Petrella S, et al. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob Agents Chemother. 2006;50:2853–2856. doi: 10.1128/AAC.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dröse S, Altendorf K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J Exp Biol. 1997;200:1–8. doi: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Allen FH. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr B. 2002;58(Pt 3 Pt 1):380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 35.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS, Thorton JM. PROCHECK: A program software suite for macromolecular structure determination. J Appl Cryst. 1993;26(Part 2):283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.