Abstract
Somatic mtDNA mutations have been reported in some human tumors, but their spectrum in different malignancies and their role in cancer development remain incompletely understood. Here, we describe the breadth of somatic and inherited mutations across the mitochondrial genome by sequence analyses of paired tumor and normal tissue samples from 226 individuals with five types of cancer using whole-genome data generated by The Cancer Genome Atlas Research Network. The frequencies of deleterious tumor-specific somatic mutations found in mtDNA varied across tumor types, ranging from 13% of glioblastomas to 63% of rectal adenocarcinomas. Compared with inherited mtDNA variants, somatic mtDNA mutations were enriched for nonsynonymous vs. synonymous changes (93 vs. 15; P < 2.2E−16) and were predicted to functionally impact the encoded protein. Somatic missense mutations in tumors were distributed uniformly among the mitochondrial protein genes, but 65% of somatic truncating mutations occurred in NADH dehydrogenase 5. Analysis of staging data in colon and rectal cancers revealed that the frequency of damaging mitochondrial mutations is the same in stages I and IV tumors. In summary, these data suggest that damaging somatic mtDNA mutations occur frequently (13–63%) in these five tumor types and likely confer a selective advantage in oncogenesis.
Keywords: deleterious mutations, colon adenocarcinoma, rectal adenomcarcinoma, acute myeloid leukemia, ovarian serous cystadenocarcinoma
Mutations in tumor suppressor genes and oncogenes are fundamental to the process of malignant transformation. Tumor cells also adapt to the dynamic requirements of unrestrained growth (1, 2), in part by shifting from oxidative phosphorylation to glycolysis, or the Warburg effect (3), which can be enabled by somatic mutations that impact the hypoxia activation-inducible factor (4–8). This metabolic shift results in less efficient energy production per molecule of glucose but also confers a selective advantage to tumors carrying these mutations through other mechanisms: increased availability of substrates needed by rapidly proliferating cells (8, 9), increased hypoxia activation-inducible factor levels that promote tumor growth, invasion, and metastasis (4, 5), and reduced reactive oxygen species levels that can induce cellular senescence (reviewed in ref. 10).
Mutations in nuclear genes encoding proteins of the citric acid cycle (11) that foster these adaptive changes occur in several human cancers. Some renal cell cancer and hereditary paragangliomas and pheochromocytomas carry inactivating mutations in genes encoding fumarate hydratase or succinate dehydrogenase subunits (12, 13). Somatic missense mutations in genes encoding isocitrate dehydrogenase 1 (IDH1) or IDH2 occur in adult-onset glioblastomas and acute myelogenous leukemia (11); these activating mutations attenuate enzyme activity and also induce epigenetic changes that promote tumor survival (8).
Variants in the mitochondrial genome, encoding 22 tRNAs, 2 rRNAs, and 13 proteins that comprise electron transport chain complexes critical for oxidative phosphorylation (14), can also affect tumorigenesis. mtDNA variants are matrilinearly inherited or arise as de novo somatic mutations in a fraction (heteroplasmic) or all (homoplasmic) of the mitochondrial genomes within each cell. Although somatic mtDNA mutations that induce metabolic reprogramming and oncogenic signaling have been previously reported (15–18), robust estimates of their frequency and types in different cancers remain limited because of analyses of few tumors, incomplete analyses of the entire mitochondrial genome, or inadequate ascertainment of insertions and deletions.
The Cancer Genome Atlas (TCGA) (19, 20) consortium has performed whole-genome massively parallel sequencing of select tumor and nontumor tissue pairs to identify somatic variations in the nuclear genome of cancer cells. Given the abundance of the 16,569-bp mitochondrial genomes in these cells, even low coverage of the nuclear genome results in deep coverage of mtDNA (median coverage > 800×) (SI Appendix, Table S1), which allows precise quantification of the heteroplasmic levels of mtDNA variants (15, 21). We therefore examined mtDNA sequence variants from tumor and nontumor tissue pairs obtained from 226 individuals with five different types of cancer. We identified highly significant enrichment of mtDNA variants in some tumor types that occurred with high levels of heteroplasmy. Thus, deleterious mtDNA mutations seem to be a common mechanism for altering metabolic pathways in tumorigenesis.
Results
mtDNA sequences were extracted from whole-genome sequences of paired tumor and normal tissues from 226 TCGA subjects. Available tumor types included colon adenocarcinoma (COAD; n = 86), rectal adenocarcinoma (READ; n = 43), acute myeloid leukemia (AML; n = 37), glioblastoma (GBM; n = 32), and ovarian serous cystadenocarcinoma (OV; n = 28).
Identification and Characterization of Somatic and Inherited mtDNA Variants.
Pairwise comparison of normal and tumor mtDNA sequences was used to identify inherited mtDNA variants or variants shared by both tissues (n = 2,440 in all 226 subjects) (Dataset S1). An average of 11 inherited mtDNA variants were found per individual, with no significant differences between tumor types.
Somatic or tumor-specific mtDNA variants were then identified in tumor mtDNA genomes (n = 233 in 130 of 226 tumors) (SI Appendix, Table S2). Although 94% of inherited mtDNA variants were common sequence polymorphisms found in the mitochondrial sequence database (annotated from >2,700 individuals) (22), only 17% of somatic mtDNA mutations had been described previously (P < 2.2E−16).
Both inherited and somatic mtDNA variants were found in similar proportions (24% and 29%, respectively) within nonprotein coding sequence regions, which include 22 tRNAs, 2 rRNAs, and the displacement loop control region (P = 0.13) (Dataset S1). Because the consequences of variants in these regions are not easily assessed, we focused subsequent analyses on mtDNA variants in protein coding regions, which include NADH dehydrogenase subunits (complex I), cytochrome b subunits (complex III), cytochrome c oxidase subunits (complex IV), and ATP synthase subunits (complex IV).
Functional Annotation of Inherited and Somatic mtDNA Variants.
Within protein coding regions, the distribution of nonsynonymous and synonymous mutations differed significantly between inherited variants and somatic mutations, a trend observed in all tumor types. Nonsynonymous changes occurred in 31% of 1,853 inherited coding variants but 86% of 166 somatic coding mutations (P < 2.2E−16) (Table 1). Somatic mtDNA mutations were, therefore, 6.3-fold more likely to alter protein amino acid sequence (Table 1). The distribution of heteroplasmic levels of synonymous and nonsynonymous variants (65.6% and 63.1%, respectively) was not significantly different; 94% (98/104) of tumors harboring somatic mtDNA mutations carried at least one nonsynonymous mutation (SI Appendix, Table S3), and 84 tumors (37%) carried somatic nonsynonymous mutations only, whereas only 6 tumors had synonymous mutations but no nonsynonymous mutations (P = 0.0006). These observations suggest that some deleterious mtDNA mutations could act as drivers of malignancy by contributing to metabolic dysregulation and associated oncogenic progression (2).
Table 1.
Distribution of mtDNA variants in 226 tumors
| Inherited variants |
Somatic mutations |
||||||||||
| Nonsyn |
Nonsyn |
||||||||||
| Tumor type | Number of tumor/nontumor pairs | Missense | Truncating | Syn | Nonsyn (%) | Number of tumors with nonsynonymous somatic mutations* | Missense | Truncating | Syn | Nonsyn (%) | P |
| COAD | 86 | 230 | 0 | 520 | 31 | 46 (53%) | 52 | 23 | 9 | 89 | <2.2E−16 |
| READ | 43 | 109 | 0 | 195 | 36 | 27 (63%) | 23 | 13 | 9 | 80 | 2.39E−08 |
| OV | 28 | 74 | 0 | 162 | 31 | 10 (36%) | 12 | 1 | 1 | 93 | 5.84E−06 |
| AML | 37 | 87 | 0 | 184 | 32 | 11 (30%) | 12 | 2 | 4 | 78 | 0.00016 |
| GBM | 32 | 91 | 0 | 201 | 31 | 4 (13%) | 4 | 0 | 1 | 80 | 0.03766 |
| Combined | 226 | 591 | 0 | 1,262 | 32 | 98 (43%) | 103 | 39 | 24 | 86 | <2.2E−16 |
Nonsyn, the number of nonsynonymous variants found per category; Nonsyn (%), nonsynonymous variants of the total number of coding variants; P, probability that the nonsynonymous variant frequency is different from the synonymous variant frequency (two-sided Fisher exact test); Syn, the number of synonymous variants found per category.
*P values comparing the number of tumors with and without nonsynonymous somatic mtDNA mutations were significant between COAD and GBM (4.93E−05), COAD and AML (0.02), READ and OV (0.03), READ and GBM (1.30E−05), and READ and AML (0.004).
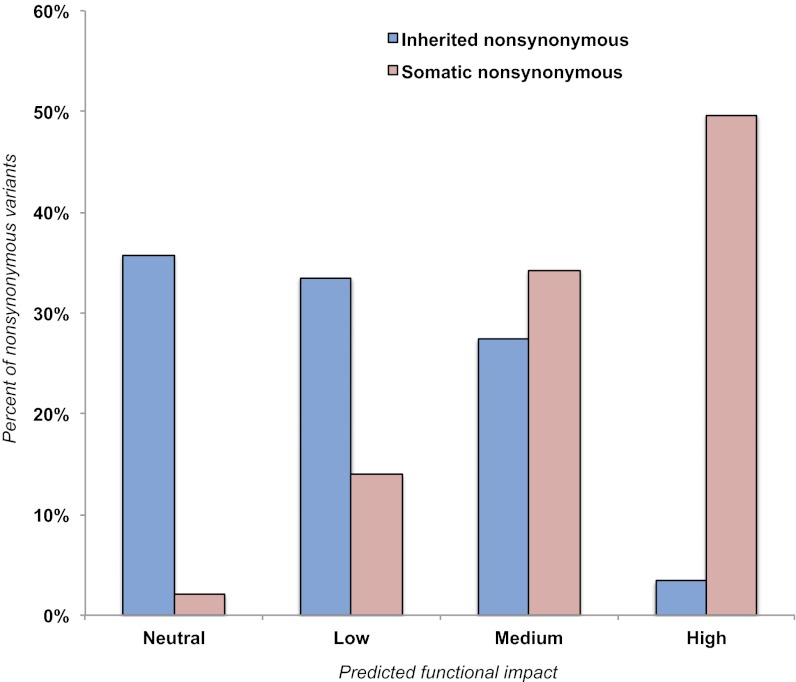
We considered the potential deleterious consequences of 142 somatic and 591 inherited nonsynonymous mutations (including missense and truncating) using MutationAssessor (v1) (23), which assigns high-, medium-, low-, or neutral-impact scores to mutations. We observed a significant difference in the distribution of impact scores between groups: 3% of inherited missense variants (20/591) vs. 50% (71/142) of somatic mtDNA mutations were predicted to have high impact on protein function (P < 2.2E−16) (Fig. 1). High-impact mutations occurred at higher levels of heteroplasmy than medium-, low-, or neutral-impact somatic mutations combined (P = 0.002, one-tailed t test), suggesting that cells carrying these mtDNA variants could have a selective advantage.
Fig. 1.
Impact of inherited and somatic nonsynonymous mitochondrial variants as assessed by MutationAssessor. Percent of inherited (blue; n = 591) and somatic (red; n = 142) nonsynonymous mtDNA mutations found in 226 individuals binned by predicted functional impact.
The capacity for massively parallel sequencing to detect small insertions and deletions, or indels (<10 bp), allowed us to identify 12 small (1–6 bp) inherited indels, all of which were homoplasmic and within noncoding regions (Dataset S1). We also observed 37 small (1–4 bp) somatic indels, 89% of which occurred in coding regions (SI Appendix, Table S2). Among these indels, 32 indels produced frameshift truncations and almost uniformly occurred within homopolymer tracts. In addition, we identified 7 somatic nonsense mutations, totaling 39 truncating mutations in tumor mtDNA.
These truncating mutations accounted for 27% (39/142) of all nonsynonymous mtDNA mutations (SI Appendix, Table S4). They occurred in 37 tumors, and 11 of these tumors had no other somatic mitochondrial variant. Truncating mutations were most frequently found in colorectal tumors, occurring in 26% of COAD (22/86) and 28% of READ (12/43). Strikingly, the majority of truncating mutations (25/39 or 64%) occurred in the NADH dehydrogenase 5 gene (ND5), which encodes a subunit of complex I of the electron transport chain. A single 8-bp homopolymer tract at position 12,418 of ND5 showed recurrent mutation: 17 tumors carried an indel within this longest homopolymer tract of the mitochondrial genome (SI Appendix, Table S7), and occurred in all tumor types except GBM. Using Sanger sequencing, we validated this indel in four COAD tumors and its absence from the corresponding normal tissue pairs. Levels of heteroplasmy for this indel ranged from 36% to 94% heteroplasmy among 11 COAD (13%), 3 READ (7%), 2 AML (5%), and 1 OV (4%) tumors.
Distribution of Somatic mtDNA Mutations by Tumor Type.
The frequency of somatic mtDNA mutations was significantly different between tumor types. Colon and rectal adenocarcinomas harbored significantly more nonsynonymous mtDNA mutations than each of the other three tumor types (57% versus 26%; P = 3.7E−06) (Table 1). Whether somatic mtDNA mutational frequency in colorectal epithelium is increased or whether there is an enhanced oncogenic advantage for tumors versus other cell types is unclear.
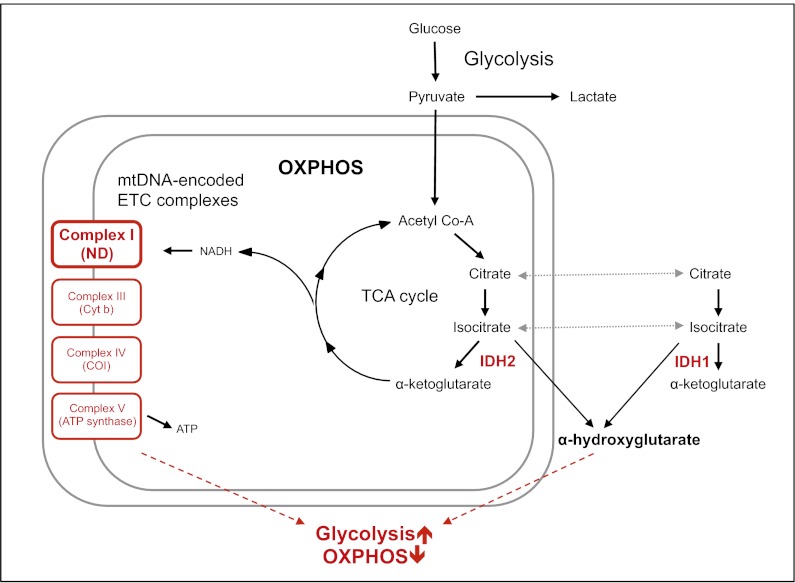
Prior studies have identified recurrent missense mutations in GBM and AML that impact specific residues of the nuclear genes IDH1 and IDH2, which encode isocitrate dehydrogenase 1 and 2 (24–26). Because somatic mtDNA mutations were significantly less prevalent in tumor types known to harbor these recurrent IDH mutations, we screened all GBM and AML nuclear genomes for three characteristic IDH mutations (R172 and R140 of IDH2 and R132 of IDH1) (11); 6 of 37 AML cases had IDH2 mutations (R140Q and R140L), and 2 AML cases had IDH1 mutations (R132H and R132C). Among 32 GBM cases, 2 cases had IDH1 mutations (R132H). Notably, none of the individuals with IDH mutations had somatic mtDNA mutations (SI Appendix, Table S5). The likelihood for mutual exclusivity of IDH and somatic mtDNA mutations in AML is low (P = 0.028; one-tailed Fisher exact test). This observation suggests that compound mutations in IDH and mitochondrial proteins are, at least to some extent, functionally redundant (Fig. 2).
Fig. 2.
A proposed common pathway by which somatic IDH mutations (red) and damaging mtDNA mutations (red; affecting complexes I, III, IV, and V of the electron transport chain) lead to metabolic deregulation, altering the relative amounts of oxidative phosphorylation and glycolysis (magenta) in early tumorigenesis. Nonsynonymous mtDNA mutations were identified in NADH-dehydrogenase (ND; complex I), cytochrome b (Cyt b; complex III), cytochrome c oxidase (COI; complex IV) and ATP synthase (ATP synth; complex V).
Analysis of Mutation Heteroplasmy.
In tumors carrying multiple somatic mtDNA mutations, assessment of heteroplasmy can offer preliminary insight into whether these mutations arose on independent mtDNA molecules or the same molecule. Of 64 tumors with multiple somatic mtDNA mutations, we found 35 tumors with examples of mutations at similar levels of heteroplasmy. For example, sample 2996 (AML) had ND1 mutation F270L in 56.3% (read depth = 2,901) of mtDNAs as well ND6 mutation I68V in 57.3% of mtDNAs (read depth = 2,320), suggesting that these mutations arose on the same mtDNA genome and underwent clonal expansion (SI Appendix, Table S2). By contrast, 29 tumors had two or more somatic mutations with different levels of heteroplasmy. An example is sample A016 (READ), which has two somatic mutations found in ∼93% of mtDNAs and two additional somatic mutations found in ∼43% of mtDNAs, suggesting that these mutations arose independently on different mtDNA molecules.
We also investigated tumors with multiple somatic mtDNA mutations located within a few hundred base pairs of one another. Because paired end sequencing was performed, we could use reads that spanned both mutations to determine if mutations arose in the same or different mitochondrial genomes. COAD samples 2672 and A00W carried examples of such mutations. Sample 2672 had two nonsynonymous substitutions in cytochrome oxidase III with ∼80% heteroplasmy (SI Appendix, Table S2). Each of six sequence reads that spanned these regions carried both mutations, indicating that they occurred in the same mitochondrial genomes. By contrast, sample A00W had two ND5 nonsynonymous mutations, an insertion with 13% heteroplasmy and a substitution with 76% heteroplasmy. All sequence reads containing the insertion lacked the substitution, implying that either these ND5 mutations arose in independent mitochondrial genomes or an initial mitochondrial mutation expanded in tumor cells followed a second mutation in a distinct subset of tumor cells.
Finally, we considered whether somatic mtDNA mutations were stable during clinically detectable tumor growth by assessing the frequency and heteroplasmy levels of somatic mtDNA mutations stratified by tumor stage in 127 colon and rectal adenocarcinomas (Table 2) (27, 28). There was no association between the presence of somatic mtDNA mutation and tumor stage. Similarly, the average heteroplasmy of somatic mtDNA mutations in early- and late-stage tumors did not differ. These observations suggest that these mutations likely provided advantages early in tumorigenesis and were clonally expanded; subsequently, their number and heteroplasmy remained constant in tumor mtDNA.
Table 2.
Distribution of nonsynonymous (NS) mtDNA mutations in 127 colon and rectal tumors
| Category (stage) | No somatic NS mtDNA mutations | Somatic NS mtDNA mutations | Tumors with ≥1 somatic NS mtDNA mutation (%) |
| I* | 7 | 18 | 72 |
| II | 21 | 27 | 56 |
| III | 19 | 16 | 46 |
| IV | 8 | 11 | 58 |
*P values comparing pairwise distributions of tumors with and without nonsynonymous somatic mtDNA mutations per stage (two-sided Fisher exact test) were not significant.
Discussion
Recent studies have greatly expanded understanding of the complex mechanisms by which altered metabolism promotes tumor progression (4, 5, 8, 9, 29). For example, mutations in IDH1 and IDH2, which encode isocitrate dehydrogenases, are prevalent in GBM and AML (11, 30), whereas mutations in encoding succinate dehydrogenase subunits, which encode succinate dehydrogenase, have been found to cause hereditary paragangliomas and pheochromocytomas (4, 5). These mutations impact tumor use of different pathways for energy production.
The advantages conveyed by metabolic deregulation and associated consequences provide a strong selective pressure on tumor cells. Acquisition of somatic mitochondrial mutations that impact oxidative phosphorylation seems to be an alternate mechanism for enhancing tumor growth. Capitalizing on next generation sequencing technologies that provided very high levels of sequence coverage for mitochondrial genomes, we quantified somatic mtDNA mutations in five types of cancer. We found high-impact somatic nonsynonymous mitochondrial mutations in several tumor types (Table 1) but most frequently, colon and rectal tumors. Among 226 tumors, somatic mtDNA mutations resulted in 39 truncations of proteins that comprise subunits of complex I (NADH dehydrogenase; n = 34), complex III (cytochrome b; n = 2), and complex IV (cytochrome c oxidase; n = 3) of the electron transport chain. The burden of these mutations is particularly remarkable given evidence that the mitochondrial genome is subjected to purifying selection (31). The work by Fan et al. (32) found that mice carrying a truncating ND6 mutation were eliminated from the female germ line within four generations, whereas a relatively milder missense mutation in cytochrome c oxidase subunit 1 was maintained.
Among somatic truncating mitochondrial mutations found in tumors, we identified a single base insertion or deletion at position 12,418 changing the length of an 8-bp homopolymer tract in ND5 in 17 different tumors (SI Appendix, Table S2). This ND5 truncating mutation was identified previously in a single tumor in each of six different studies involving 10 colorectal cell lines (33), 44 hepatocellular carcinomas (34), 45 colorectal tumors (35), 9 renal cell tumors (36), 58 breast cancers (37), and 31 gastric cancers (38). Collectively, these data indicate that truncating mutations caused by indels in the ND5 homopolymer tract occur in 3% (previous studies) to 7.5% (this study) of tumors. Other recurrent functional somatic mtDNA mutations, similar to the ND5 truncating mutations, have not been found in tumors.
The study by Hofhaus and Attardi (39) hinted at the metabolic consequences of this ND5 mutation. This work found this same ND5 truncation to arise with varying levels of heteroplasmy in cultured cell lines that spontaneously developed resistance to rotenone, a potent inhibitor of oxidative phosphorylation (39). Because transfer of mutant mitochondria into WT cells maintained a reliance on glycolysis for energy, the work by Hofhaus and Attardi (39) concluded that the ND5 mutation promoted respiration independence (39). More recently, the work by Park et al. (16) found that, compared with WT cells, cells carrying the ND5 truncation exhibited enhanced colony formation on soft agar and increased tumor growth when implanted into nude mice. Taken together, these functional studies imply that this ND5 mutation recapitulates the Warburg effect and provides early advantages for anchorage-independent growth. Consistent with this model, we observed no tumors with both an IDH mutation (n = 10) and somatic mtDNA mutations, raising the possibility that these tumors are functionally redundant (8). In summary, we suggest that this somatic ND5 truncating mutation is found at high heteroplasmy in tumors caused by the selective advantage that it conveys to tumor cells.
The distribution of somatic mutations in tumors provides some insights into the mutation-causing mechanisms; 82% of single-base somatic mtDNA mutations in tumors were transitions (G to A or C to T changes) (SI Appendix, Table S6), which can arise from abundant reactive oxygen species in mitochondria (17), and 31 of 32 tumor-specific indels detected in tumors occurred within homopolymer tracts (SI Appendix, Table S7). Because multiple mitochondrial mutations were found in some tumors, we speculate that these mutations arose simultaneously in different mitochondrial genomes or successive steps, resulting in an evolution of metabolic dysfunction.
Two lines of evidence suggest that damaging mtDNA mutations confer a selective advantage early in oncogenesis. First, the frequency of damaging somatic mtDNA mutations in colon and rectal tumors was the same in early (stage I) and late (stage IV) tumors (Table 2). Second, the average heteroplasmy of mitochondrial mutations was similar in all stages of colorectal tumors.
These data expand evidence for altered metabolism as a critical hallmark of many cancers, particularly colon and rectal adenocarcinomas, in which 26–28% of samples carried deleterious mtDNA mutations. By disrupting electron transport chain proteins, we predict that these mitochondrial mutations inhibit oxidative phosphorylation and provide selective growth advantages early in oncogenesis. Additional studies are needed to characterize the molecular pathways activated by mitochondrial mutations, define the clinical use of mitochondrial mutations as tumor biomarkers (18, 40), and address the therapeutic potential for targeting mutation-induced metabolic dysregulation in cancer.
Methods
Whole-genome sequence data generated from several centers of TCGA on either Illumina Genome Analyzer II or HiSeq (SI Appendix, Table S1) were acquired from the online repository of the consortium (dbGaP). Samples were selected based on sequencing depth and uniformity of coverage across the mitochondrial genome. All tumor tissue, with the exception of AML, was primary solid tumor tissue; AML samples were primary blood-derived cancer samples from peripheral blood. Normal samples were collected from either solid tissue or blood (SI Appendix, Table S1). Sequence reads were aligned to mtDNA (hg18 using bwa [21] or maq) (SI Appendix, Table S1). We then identified mtDNA variants from aligned reads using the ANNOVAR variant-calling pipeline (41), which was customized to adopt mtDNA alternative codon use, report heteroplasmy of variants, and convert the hg18 reference to Revised Cambridge Reference Sequence (rCRS). We conservatively set a minimum of 5% heteroplasmy as a threshold for variant calling, although the lowest level of heteroplasmy found in a somatic mtDNA variant was 11.1% (SI Appendix, Table S2). Somatic variants and all indels were visually inspected using the Integrative Genomics Viewer (42). The recurrent ND5 truncation was validated in tumor and nontumor tissue genomic DNA using the Beckman Coulter Genomics QuickLane Express Sequencing service. Where possible, paired end sequences were evaluated to define phase of neighboring variants. Unless otherwise specified, P values were generated using two-sided Fisher exact tests.
Supplementary Material
Acknowledgments
We thank the genome sequencing centers at the Broad Institute, Washington University, and Baylor College of Medicine for sequencing data production. We also thank Kai Wang (Children’s Hospital, Philadelphia) and Yevgeniy Antipin (Memorial Sloan-Kettering Cancer Center) for assistance with variant calling and computational analyses. Studies were supported by The Howard Hughes Medical Institute (T.C.L. and C.E.S.), National Cancer Institute, National Institutes of Health (T.C.L., R.K., and J.G.S.), LeDucq Foundation (C.E.S. and J.G.S.), National Heart, Lung and Blood Institute, National Institutes of Health (J.G.S.), and SysCODE (J.G.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211502109/-/DCSupplemental.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Greaves LC, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin WG., Jr Cancer and altered metabolism: Potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochachka PW, Rupert JL, Goldenberg L, Gleave M, Kozlowski P. Going malignant: The hypoxia-cancer connection in the prostate. Bioessays. 2002;24:749–757. doi: 10.1002/bies.10131. [DOI] [PubMed] [Google Scholar]
- 7.Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol. 2011;4:62–77. doi: 10.2174/1874467211104010062. [DOI] [PubMed] [Google Scholar]
- 8.Ichimura K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012 doi: 10.1007/s10014-012-0090-4. in press. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG, Jr, Thompson CB. Q&A: Cancer: Clues from cell metabolism. Nature. 2010;465:562–564. doi: 10.1038/465562a. [DOI] [PubMed] [Google Scholar]
- 10.Gupta-Elera G, Garrett AR, Robison RA, O’Neill KL. The role of oxidative stress in prostate cancer. Eur J Cancer Prev. 2012;21:155–162. doi: 10.1097/CEJ.0b013e32834a8002. [DOI] [PubMed] [Google Scholar]
- 11.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 12.Burnichon N, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frezza C, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 14.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 15.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JS, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ralph SJ, Rodríguez-Enríquez S, Neuzil J, Saavedra E, Moreno-Sánchez R. The causes of cancer revisited: “Mitochondrial malignancy” and ROS-induced oncogenic transformation—why mitochondria are targets for cancer therapy. Mol Aspects Med. 2010;31:145–170. doi: 10.1016/j.mam.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Fliss MS, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 19.Group TT. 2011. The Cancer Genome Atlas Web Page.
- 20.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott-Conner CE, Christie DW. Cancer staging using the American Joint Committee on Cancer TNM System. J Am Coll Surg. 1995;181:182–188. [PubMed] [Google Scholar]
- 28.Edge SB, et al., editors. AJCC Cancer Staging Manual (American Joint Committee on Cancer) 7th Ed. New York: Springer; 2010. [DOI] [PubMed] [Google Scholar]
- 29.Garber K. Oncology’s energetic pipeline. Nat Biotechnol. 2010;28:888–891. doi: 10.1038/nbt0910-888. [DOI] [PubMed] [Google Scholar]
- 30.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 32.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 34.Yin PH, et al. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion. 2010;10:174–182. doi: 10.1016/j.mito.2009.12.147. [DOI] [PubMed] [Google Scholar]
- 35.Habano W, Sugai T, Yoshida T, Nakamura S. Mitochondrial gene mutation, but not large-scale deletion, is a feature of colorectal carcinomas with mitochondrial microsatellite instability. Int J Cancer. 1999;83:625–629. doi: 10.1002/(sici)1097-0215(19991126)83:5<625::aid-ijc10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 36.Nagy A, Wilhelm M, Kovacs G. Mutations of mtDNA in renal cell tumours arising in end-stage renal disease. J Pathol. 2003;199:237–242. doi: 10.1002/path.1273. [DOI] [PubMed] [Google Scholar]
- 37.Tseng LM, et al. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer. 2011;50:800–811. doi: 10.1002/gcc.20901. [DOI] [PubMed] [Google Scholar]
- 38.Hung WY, et al. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta. 2010;1800:264–270. doi: 10.1016/j.bbagen.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Hofhaus G, Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol Cell Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahlquist DA, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.