Abstract
The human genome, like other mammalian genomes, encodes numerous natural antisense transcripts (NATs) that have been classified into head-to-head, tail-to-tail, or fully overlapped categories in reference to their sense transcripts. Evidence for NAT-mediated epigenetic silencing of sense transcription remains scanty. The DHRS4 gene encodes a metabolic enzyme and forms a gene cluster with its two immediately downstream homologous genes, DHRS4L2 and DHRS4L1, generated by gene duplication. We identified a head-to-head NAT of DHRS4, designated AS1DHRS4, which markedly regulates the expression of these three genes in the DHRS4 gene cluster. By pairing with ongoing sense transcripts, AS1DHRS4 not only mediates deacetylation of histone H3 and demethylation of H3K4 in cis for the DHRS4 gene, but also interacts physically in trans with the epigenetic modifiers H3K9- and H3K27-specific histone methyltransferases G9a and EZH2, targeting the promoters of the downstream DHRS4L2 and DHRS4L1 genes to induce local repressive H3K9me2 and H3K27me3 histone modifications. Furthermore, AS1DHRS4 induces DNA methylation in the promoter regions of DHRS4L2 by recruiting DNA methyltransferases. This study demonstrates that AS1DHRS4, as a long noncoding RNA, simultaneously controls the chromatin state of each gene within the DHRS4 gene cluster in a discriminative manner. This finding provides an example of transcriptional control over the multiple and highly homologous genes in a tight gene cluster, and may help explain the role of antisense RNAs in the regulation of duplicated genes as the result of genomic evolution.
Keywords: epigenetic regulation, long noncoding antisense RNA
The human DHRS4 gene cluster, located on chromosome 14q11.2, consists of three genes in tandem, among which DHRS4L1 and DHRS4L2 are believed to arise from the parental DHRS4 gene via gene duplication. DHRS4 (short-chain dehydrogenase/reductase family member 4) encodes the NADPH-dependent retinol dehydrogenase/reductase (NRDR), which was first identified in rabbit liver tissues and reported to have strong retinal reductase activity in the synthesis of retinoic acid (1, 2). In different species, this enzyme serves to reduce endogenous products, such as biogenic aldehydes, steroids, prostaglandins, and reactive lipid peroxidation products, and xenobiotic compounds, such as pharmacologic drugs, carcinogens, and toxicants, suggesting that NRDR may play an important role in the metabolism and detoxification of carbonyl compounds, especially in the liver (3–5).
The DHRS4 gene cluster is localized to an area of segmental duplication (6) and thus is suspected to contain clues for genomic evolution in primates. DHRS4 is highly conserved among mammals and aquatic animals, whereas DHRS4L1 and DHRS4L2 are present in primates. Mammalian genomes generate a wide variety of regulatory RNAs that are either small noncoding RNAs or long noncoding RNAs (lncRNAs) (7, 8). Based on their locations relative to protein-coding genes, lncRNAs are roughly classified into intergenic (between genes), intragenic/intronic (within genes), and antisense groups (9). Natural antisense transcripts (NATs) have previously been divided into head-to-head, tail-to-tail, and fully overlapped categories in reference to their sense transcripts (10).
Recent reports demonstrate that lncRNAs play an important role in epigenetic gene regulation, and that their dysfunction leads to disease (11, 12). Several models have been proposed to explain how lncRNAs epigenetically silence genes. Examples include fully overlapped NATs, such as the Xist–Tsix pair, Kcnq1ot1, and intergenic HOTAIR, which function in either a cis or a trans manner to regulate target gene transcription through different epigenetic mechanisms (9, 13, 14).
In the present study, we identified and characterized a head-to-head NAT of the DHRS4 gene, AS1DHRS4, in normal human hepatic (HL7702) and hepatocellular carcinoma (HepG2) cells. AS1DHRS4 is a 1,865-bp spliced and polyadenylated RNA transcribed from intron 1 of the DHRS4 gene in the antisense direction that controls the local epigenetic status of each gene promoter within the DHRS4 gene cluster in a discriminative manner. Because AS1DHRS4 transcription is initiated within the DHRS4 gene to regulate DHSR4 gene expression, AS1DHRS4 antisense regulates DHRS4 in cis; that is, AS1DHRS4 and DHRS4 exist within the same allele and form a regulatory unit. In contrast, AS1DHRS4 antisense must translocate to the DHRS4L2 and DHRS4L1 alleles, and thus regulates the DHRS4L2 and DHRS4L1 genes in trans.
Results
AS1DHRS4, a Natural Antisense Long Noncoding Transcript of the DHRS4 Gene.
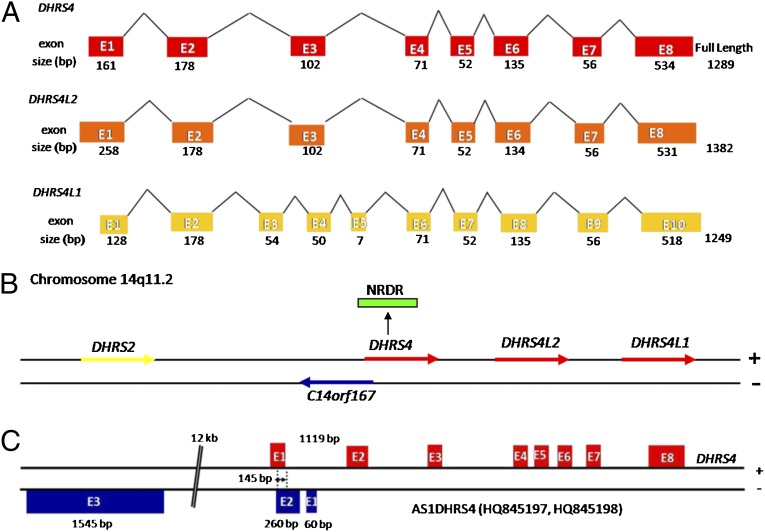
The human DHRS4 gene cluster comprises three genes: DHRS4, DHRS4L2, and DHRS4L1 (Fig. 1A). Sequence analysis revealed that the DHRS4L2 and DHRS4L1 mRNA sequences share high homology with DHRS4 (>80%), and that the DNA sequence homology of all three genes exceeds 77% (SI Appendix, Table S1). A GenBank search identified the gene C14ORF167 in a head-to-head orientation relative to the DHRS4 gene, on the opposite strand of the DHRS4 gene cluster. The transcription start site of C14ORF167 was inside the first intron of DHRS4, and the 3′end of C14ORF167 was completely upstream of DHRS4 (Fig. 1B).
Fig. 1.
AS1DHRS4 is a cis-antisense transcript to the DHRS4 gene. (A) Exon composition and homologous relationship between the three coding genes within the DHRS4 gene cluster. (B) Schematic representation of the DHRS4 gene cluster and C14ORF167 and DHRS2 gene loci on human chromosome 14q11.2. (C) Exon composition of the AS1DHRS4 transcript and organization of the overlapping exon with DHRS4.
Based on the sequence of C14ORF167, by performing 5′ and 3′ RACE, we cloned a 1,865-bp antisense transcript of DHRS4, designated AS1DHRS4, consisting of three exons with a 3′ polyadenylation tail (Fig. 1C). Exon 2 of AS1DHRS4 overlaps with the first exon of DHRS4 by 145 bp in an antisense fashion. The sequences of AS1DHRS4 from HepG2 and HL7702 cell lines have been submitted to GenBank (accession nos. HQ845197 and HQ845198, respectively). AS1DHRS4 contains a unique ORF spanning 222 bp and encoding 73 amino acids. Using the Coding Potential Calculator (15) and a manual approach (16) for annotating human long noncoding RNA genes, we classified AS1DHRS4 as a long noncoding RNA (SI Appendix, Fig. S1).
Antisense transcripts have been implicated in regulation of their overlapping sense transcripts (17, 18). As a first step in determining whether AS1DHRS4 regulates the expression of DHRS4 or the DHRS4 gene cluster, we analyzed the expression of AS1DHRS4 in human normal and cancer cell lines and compared it with DHRS4 gene cluster expression. AS1DHRS4 was detected in all cell lines examined. Quantitative RT-PCR (qPCR) showed relatively high AS1DHRS4 expression and low DHRS4 expression in PLC/PRF/5, EC109, and HeLa cell lines, and high expression of DHRS4 but relatively low expression of AS1DHRS4 in HepG2, HL7702, and BEL 7402 cell lines (SI Appendix, Fig. S2A).
The expression of NRDR protein was high in HL7702 and HepG2 cells, coinciding with relatively higher DHRS4 mRNA levels (SI Appendix, Fig. S2B). We did not detect protein expression of either DHRS4L2 or DHRS4L1, using anti-DHRS4L2 and anti-DHRS4L1, in any of the cell lines tested, possibly because of inadequate RNA levels.
To determine the stability of AS1DHRS4, we used actinomycin D to inhibit total cellular transcription, with c-MYC as a control for a short half-life mRNA species. Results from qPCR analysis in HL7702 and HepG2 cells treated between 30 min and 4 h with actinomycin D show that AS1DHRS4 was stable in HL7702 cells (SI Appendix, Fig. S3A). It is also possible that AS1DHRS4 is stabilized when it binds to a specific RNA-binding protein in a ribonucleoprotein complex. The splicing factor SF2/ASF also shuttles between the nucleus and cytoplasm, and participates in postsplicing RNA processing, including mRNA export, stability, and translation (19). SF2/ASF has been reported to recognize a functionally diverse landscape of RNA transcripts, including DHRS4 mRNA and DHRS4L2 mRNA in HEK293 cells (20). Using RNA-immunoprecipitation (RNA-IP) in HL7702 cells, we confirmed the binding of SF2/ASF to DHRS4 mRNA, but not to mRNAs for DHRS4L2 or DHRS4L1, or to AS1DHRS4. In parallel, we also tested other known specific RNA-binding proteins, including hnRNP K, upf2, and upf3x, for potential AS1DHRS4-binding activities, and found no positive results (SI Appendix, Fig. S3B). qPCR analysis of nuclear and cytoplasmic RNA extracts from HL7702 and HepG2 cells found that AS1DHRS4 was distributed more in the cytoplasm (60.47%) and less in the nucleus (39.53%) (SI Appendix, Fig. S3C).
AS1DHRS4 Regulates DHRS4 Gene Cluster Transcription.
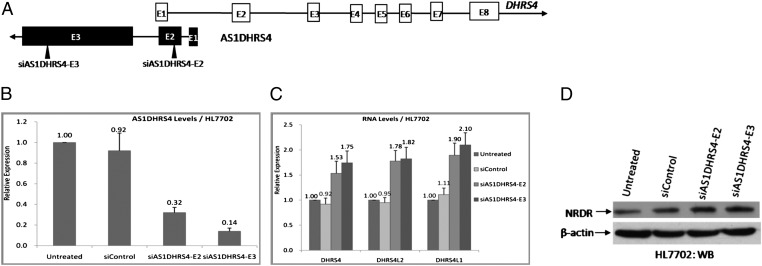
To check whether the antisense RNA plays a regulatory role in gene expression at the DHRS4 gene cluster, we analyzed the changes in mRNAs for DHRS4, DHRS4L2, and DHRS4L1, as well as NRDR protein levels, after silencing of AS1DHRS4 using two transcript-specific siRNAs that specifically targeted exon 2 of AS1DHRS4 (siAS1DHRS4-E2) and exon 3 of AS1DHRS4 (siAS1DHRS4-E3) (Fig. 2A). We first measured AS1DHRS4 levels in HepG2 and HL7702 cells treated with siAS1DHRS4-E2 or siAS1DHRS4-E3. The results indicated that both siAS1DHRS4-E2 and siAS1DHRS4-E3 caused visible reductions in AS1DHRS4 levels, with a greater inhibitory effect of siAS1DHRS4-E3 compared with siAS1DHRS4-E2 in both cell lines (Fig. 2B and SI Appendix, Fig, S4A).
Fig. 2.
AS1DHRS4 regulates DHRS4, DHRS4L2, and DHRS4L1 RNA levels and NRDR protein levels in HL7702 cells. (A) Positions of the siAS1DHRS4 oligonucleotides, indicated by black triangles. (B) qPCR analysis of AS1DHRS4 RNA levels at 36 h after siAS1DHRS4 treatment, with the GAPDH gene as an internal control. Error bars represent the SEs of three independent experiments.The siControl was a scrambled sequence with no homology to any known gene. (C) qPCR analysis of the RNA levels of the DHRS4 gene cluster after siAS1DHRS4 treatment for 36 h, with the GAPDH gene as an internal control. Error bars represent the SEs of three independent experiments. (D) Western blot (WB) analysis of total cell lysates, from siAS1DHRS4-transfected HL7702 cells, using anti-NRDR antibodies, with the β-actin protein as a loading control.
Introduction of siAS1DHRS4-E2 and siAS1DHRS4-E3 resulted in increases in DHRS4 gene cluster mRNA and NRDR protein levels (Fig. 2 C and D and SI Appendix, Fig. S4 B and C). Unexpectedly, DHRS4L2 and DHRS4L1 RNA expression increased significantly after the knockdown of AS1DHRS4, although the increased RNA levels of DHRS4L2 and DHRS4L1 were still much lower than DHRS4 RNA levels, and their encoded proteins remained undetected. This result indicates that the antisense transcript of DHRS4 regulates DHRS4 gene expression in cis, because AS1DHRS4 antisense transcription is initiated within the DHRS4 allele, but regulates DHRS4L2 and DHRS4L1 expressions in trans within the the DHRS4 gene cluster.
Nuclear-localized ncRNAs are less amenable to siRNA-based knockdown. However, the highly efficient knockdown of AS1DHRS4 with siAS1DHRS4-E2 and siAS1DHRS4-E3 in our system was sufficient to affect the nuclear AS1DHRS4. One possibility was that some of our siAS1DHRS4s entered the nucleus for action, consistent with a previous report (17). These results suggest that the ∼40% minor fraction of nuclear AS1DHRS4 was sufficient for epigenetic regulation of the three DHRS4 genes, but that after knockdown, insufficient AS1DHRS4 remained to exert regulation.
To examine whether knockdown of AS1DHRS4 affected expression of the nearby DHRS2 (short-chain dehydrogenase/reductase family member 2) gene, we analyzed RNA levels in parallel. DHRS2 is located 308.12 kb upstream of the DHRS4 gene and is the closest protein-coding gene to DHRS4. The cDNA sequence of DHRS2 has a homology of 44.7% compared with DHRS4, and the promoter sequences share less than 10% homology. Knockdown of AS1DHRS4 had no effect on DHRS2 gene expression at the transcriptional level (SI Appendix, Fig. S5A), suggesting that AS1DHRS4 specifically regulates the DHRS4 gene cluster.
Knockdown of AS1DHRS4 increased both mature mRNAs and pre-mRNAs of the DHRS4 gene cluster (SI Appendix, Fig. S5B). The fact that AS1DHRS4 regulates the nascent RNA form supports the notion that AS1DHRS4 regulates DHRS4 gene cluster expression at the transcriptional level.
AS1DHRS4 Mediates Gene-Suppressive Histone Modifications in the Promoter Regions of the DHRS4 Gene Cluster.
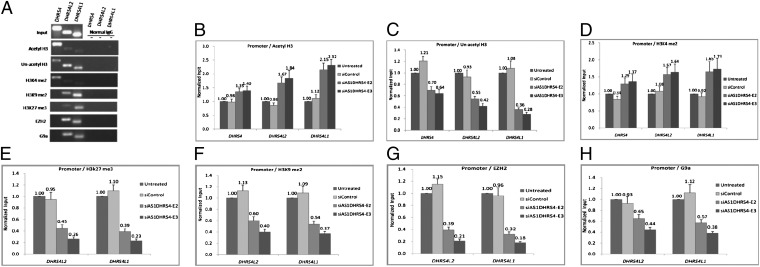
To identify a possible mechanism of ASDHRS4-mediated transcriptional inhibition, we analyzed histone modification status in the promoter regions of the DHRS4 gene cluster using ChIP assays. In our first ChIP assays, using antibodies against acetyl-H3, unacetylated H3, H3K4me2, H3K9me2, and H3K27me3, we found acetyl-H3 and H3K4me2 enrichment in the DHRS4 promoter and unacetylated H3, H3K9me2, and H3K27me3 enrichment in the DHRS4L2 and DHRS4L1 promoters. We next confirmed the interaction of histone H3K9me2 methyltransferase EZH2 and H3K27me3 methyltransferase G9a with the promoter regions of DHRS4L2 and DHRS4L1 (Fig. 3A). H3K4me2 and acetyl-H3 have been reported to associate with active genes (21). EZH2 is an H3K27-specific histone methyltransferase that is a component of PRC2 (22). G9a is responsible for monomethylation and dimethylation of H3K9 (23), and H3K9me2 and H3K27me3 are important modifications for gene silencing.
Fig. 3.
AS1DHRS4 mediates repressive histone modifications in the promoter regions of the DHRS4 gene cluster loci in HL7702 cells. (A) Agarose gel electrophoresis showing PCR analysis of acetyl-H3, unacetyl H3, H3K4me2, H3K27me3, H3K9me2, EZH2, and G9a enrichment in the DHRS4 gene cluster promoters in ChIP assays. (B–D) ChIP analysis of acetyl-H3 (B), unacetyl H3 (C), and H3K4me2 (D) enrichment in the DHRS4 gene cluster promoters. Knockdown of AS1DHRS4 induced an increase in acetyl-H3 and H3K4me2 and a decrease in unacetyl H3. (E and F) ChIP analysis of H3K27me3 (E) and H3K9me2 (F) enrichment in the DHRS4L2 and DHRS4L1 gene promoters. Knockdown of AS1DHRS4 induced a decrease in H3K27me3 and H3K9me2. (G and H) ChIP analysis of EZH2 (G) and G9a (H) binding to the DHRS4L2 and DHRS4L1 promoter regions. Knockdown of AS1DHRS4 induced a decrease in EZH2 and G9a. ChIP enrichment was measured at 36 h after siAS1DHRS4 treatment using qPCR, normalized by the input DNA. Error bars represent the SE of three independent experiments.
We performed identical ChIP assays after knockdown of AS1DHRS4. Active gene indicators, such as acetyl-H3 and H3K4me2, were increased, whereas inactive gene indicators, such as unacetylated H3, were decreased in the promoter region of the DHRS4 gene cluster. H3K9me2, G9a, H3K27me3, and EZH2 were decreased in the DHRS4L2 and DHRS4L1 promoter regions in HL7702 (Fig. 3 B–H) and HepG2 cells (SI Appendix, Fig. S6). These results indicate that silencing of AS1DHRS4 changed the chromatin status from a closed and inactive state to an open and active state, thereby increasing mRNA expression of the DHRS4 gene cluster.
AS1DHRS4 Interacts with H3K9 and H3K27 Methyltransferases to Maintain Epigenetic Silencing of the DHRS4L2 and DHRS4L1 Genes.
Previous studies (9, 24) have suggested that lncRNAs may directly or indirectly attract epigenetic modifiers, such as histone methyltransferases (G9a or EZH2), to bring about repressive epigenetic modifications in the gene cluster. Our ChIP assay results confirm that AS1DHRS4 mediated gene silencing through repressive H3K9me2 and H3K27me3 chromatin modifications. Moreover, G9a and EZH2 were also enriched in the promoter regions of DHRS4L2 and DHRS4L1.
To analyze the mechanism of AS1DHRS4, we first tested whether AS1DHRS4 interacts with G9a and EZH2 by performing RNA-IP assays using antibodies against G9a and EZH2. In HL7702 cells, AS1DHRS4 was pulled down by either antibody (SI Appendix, Fig. S7A), suggesting that AS1DHRS4 formed a complex with G9a and EZH2. This finding is consistent with the resulting histone modifications of H3K9me2 and H3K27me3 in the DHRS4L2 and DHRS4L1 gene domains. Furthermore, RNA-ChIP assays, performed using antibodies against H3K9me2 or H3K27me3, revealed that AS1DHRS4 was bound to histone H3K9me2 and H3K27me3, possibly at the DHRS4L2 and DHRS4L1 gene loci (SI Appendix, Fig. S7B).
We used two pairs of primers to detect AS1DHRS4 in immunoprecipitated total RNAs by qPCR. One primer was designed based on the sequence at an exon junction, and the other was located in the 3′ end. This indicated that AS1DHRS4, bound to histone methyltransferases, was long and had undergone splicing.
AS1DHRS4 Induces DNA Methylation in the Promoter Region of the DHRS4L2 Gene by Recruiting DNA Methyltransferases.
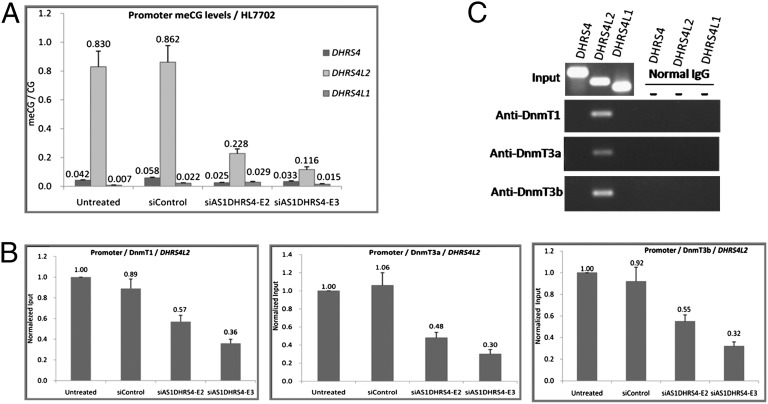
Antisense transcripts have been proposed to cause DNA methylation (25, 26). Tsix, a long antisense transcript, induces DNA methylation at the promoter of the sense transcript Xist (27). We examined the methylation status of the DHRS4 gene cluster in the promoter CpG islands of HepG2 and HL7702 cells. The CpG sites of the DHRS4L2 gene promoter were methylated, correlating with the promoter’s transcriptionally repressed status, whereas the DHRS4 and DHRS4L1 promoters were unmethylated, with DHRS4 transcriptionally active and DHRS4L1 transcriptionally inactive (SI Appendix, Fig. S8). To analyze DNA methylation levels, we measured the ratio of methylated CpG to total CpG sites in these promoter regions. After knockdown of AS1DHRS4, DNA methylation of DHRS4L2 was significantly decreased, changing the DHRS4L2 promoter from a hypermethylated state to a hypomethylated state but causing no changes in DHRS4 or DHRS4L1 promoters (Fig. 4A and SI Appendix, Fig. S9).
Fig. 4.
A1SDHRS4 induces CpG island methylation in the promoter region of the DHRS4L2 gene by recruiting DNA methyltransferases. (A) The ratio of methylated CpG to total CpG sites in the promoter regions of the DHRS4 gene cluster was measured to analyze DNA methylation status in HL7702 cells. Error bars represent the SE of three independent experiments, and each experiment detected eight clones for each promoter region. (B) ChIP analysis of DNA methyltransferase binding in the DHRS4L2 promoter region in HL7702 cells. siAS1DHRS4 treatment for 36 h induced decreases in DnmT1, DnmT3a, and DnmT3b, as measured by qPCR, normalized to the input DNA. Error bars represent the SEs of three independent experiments. (C) Agarose gel electrophoresis showing PCR analysis of DnmT1, DnmT3a, and DnmT3b enrichment in the DHRS4 gene cluster promoters in ChIP assays in HL7702 cells.
We then performed ChIP assays to detect the potential interaction between DnmTs and the DHRS4L2 gene promoter after AS1DHRS4-mediated knockdown. Enrichment of DnmT1, DnmT3a, and DnmT3b in the promoter region of DHRS4L2 gene was reduced by siAS1DHRS4-E2 and siAS1DHRS4-E3 in both HL7702 (Fig. 4B) and HepG2 cells (SI Appendix, Fig. S10). DnmT1, DnmT3a, and DnmT3b were not detected in the promoter regions of DHRS4 and DHRS4L1 (Fig. 4C). Given that hypomethylated DNA is associated with active genes, whereas hypermethylated genes are silent (28), we conclude that the transcriptional silencing of the DHRS4L2 gene is regulated in part by AS1DHRS4-directed DNA methylation.
AS1DHRS4 Mediates Suppressive Histone and DNA Modifications in the DHRS4 Gene Cluster Through RNA–RNA Interaction.
To clarify whether AS1DHRS4 functions at the target loci in a sequence-specific fashion, we performed RNA-ChIP assays using antibody against H3, along with DNase I digestion and RNase protection experiments, to identify any RNA–RNA or RNA–DNA interactions between the overlapped segments of AS1DHRS4 and the sense transcripts or cDNA strands of the DHRS4 gene cluster (29). Because DHRS4L2 and DHRS4L1 sequences shared ∼93% similarity with DHRS4 in the overlapped region, RNA–RNA or RNA–DNA pairs between AS1DHRS4 and DHRS4L2 or DHRS4L1 would have less than perfect complementarity. RNase A digests single-stranded RNA at C and U residues, whereas RNase T1 digests at G residues. During RNase digestion, total or partial cleavage of the duplexes may occur at the position of the mismatch. Cleavage at mismatch positions often can be minimized by decreasing the RNase A concentration or using RNase T1. We tested a series of RNase A concentrations (10, 20, 40, 60, and 80 ng/μL) and found that a concentration of 20 ng/μL was sufficient to digest single-strand RNA but could not cleave fully or partially complementary double-stranded RNAs. After RNase T1 treatment, primers were designed to avoid any G mismatch and to detect the partially complementary segments without G mismatches. No PCR products derived from single DNA template strands were detected with RNase H treatment. Normal rabbit IgG antibody was used as the negative control for nonspecific immunopositive targets, and it did not result in PCR products. The results indicate RNA–RNA interaction between AS1DHRS4 and the ongoing pre-mRNAs of the DHRS4 gene cluster, rather than RNA–DNA pairing in the immunoprecipitated nuclear extracts (SI Appendix, Figs. S11 and S12). All amplified PCR segments were confirmed by DNA sequencing.
To determine whether cytoplasmic duplexes between AS1DHRS4 and mRNAs of the DHRS4 gene cluster are also present, we extracted the cytoplasmic RNAs for RNase A and RNase T1 digestion, and performed qPCR. We did not detect the RNA pairs (Fig. 5E).
To elucidate whether RNA–RNA interactions were involved in the gene regulation, we performed blocking experiments with antisense phosphorothioate oligodeoxynucleotides (ODNs). Blocking of RNA–RNA interactions with ODNs targeting the overlapped exon 1 or intron 1 segment sequence, but not the junction between exon 7 and intron 7, could abrogate the effects of AS1DHRS4-mediated modifications in the promoters of the DHRS4 gene cluster (SI Appendix, Fig. S13 A–E). Nonetheless, all of the sense transcripts in the DHRS4 gene cluster were decreased by the blocking ODNs (SI Appendix, Fig. S13F). This might be because phosphorothioate ODNs operate by Watson–Crick hydrogen bonding to inhibit RNA expression by activating the RNase H pathway, which cleaves the targeted RNA at the heteroduplex site (29). These results indicate that the overlapping sequence between AS1DHRS4 and the DHRS4 gene cluster is responsible for AS1DHRS4-mediated and target-specific transcriptional regulation of the DHRS4 gene cluster.
Discussion
DHRS4 is an important metabolic enzyme in the human body, expressed predominantly in the liver. DHRS4L1 and DHRS4L2 appeared in succession through DHRS4 gene duplication, whereas the latter is a recent member in genomic evolution. The DHRS4 gene cluster is located in an area of segmental duplications, and its expression is down-regulated in some common human cancers (30). In the cell lines that we tested, there was no detectable protein expression of either DHRS4L2 or DHRSL1; however, a search for their expression in vivo is still needed. The possibility also exists that these two proteins are never translated. Thus, at this time we cannot conclude that the transregulation by AS1DHRS4 is totally irrelevant to metabolism or liver function. It is conceivable that AS1DHRS4 diminishes the transcription of DHRS4L2 and DHRS4L1 to affect DHRS4 expression at the transcriptional or translational level.
AS1DHRS4 is also a specific product of primate evolution; the NAT of the Dhrs4 gene in mice has a substantially different configuration and has no similar function (SI Appendix, Fig. S14). The transcription start site of AS1DHRS4 resides in the first intron of DHRS4. The entire gene including its introns was duplicated in mammals relatively recently. There are predicted potential promoters in the antisense strands of human DHRS4, DHRS4L2, and DHRS4L1. The finding that DHRS4L1 and DHRS4L2 do not give rise to their own, local, head-to-head antisense transcription similar to AS1DHRS4 is puzzling, but conceivable for two possible reasons: (i) the homology identity is ∼92% among the first intron of each gene, and these specific sequence differences between the duplicated paralogs could have simply abrogated transcription factor binding sites necessary for the antisense transcription of DHRS4L2 or DHRS4L1; and (ii) there also might be unknown distant enhancer sequences that promote only AS1DHRS4 transcription, not the antisense transcription from sites of DHRS4L2 and DHRS4L1.
Sequence complementarity to the target transcript is a requirement for a NAT to cause an effect. Our results demonstrate RNA–RNA pairing between exon 2 of AS1DHRS4 and all exon 1 segments of the DHRS4 gene cluster transcripts. The formation of RNA–RNA pairing may guide AS1DHRS4-associated modifiers or the resulting local higher-ordered RNA–RNA complexes, directly recruiting modifiers to the respective promoters of the DHRS4 gene cluster for epigenetic regulation. This supports the possible model proposed by Hekimoglu and Ringrose (31) and Guttman and Rinn (32) in which a free RNA strand binds to modifier proteins and then anneals back to a nascent transcript of the complementary sequence, transcribed from the same site.
Only a few examples of DNA methylation mediated by lncRNA have been documented (33). G9a- and EZH2-containing complexes could recruit DnmTs, with DnmT3a and DnmT3b initiating and DnmT1 maintaining DNA methylation at the promoter (34). Our results support the view that lncRNA is a major player in epigenetic regulation.
Why the modifications are different for each promoter of the DHRS4 gene cluster remains to be elucidated. For the transcriptionally active DHRS4 gene locus, bidirectional transcription may be a factor, with the steps of AS1DHRS4 transcription possibly interfering with or removing some chromatin-bound modifier proteins by passing through the promoter of the DHRS4 gene (31). Selective AS1DHRS4-mediated promoter methylation of the DHRS4L2, but not of DHRS4L1, may result from the different lengths of 5′ UTRs between the two sense transcripts. Sequence analysis revealed that the 5′ end of exon 1 of DHRS4L2 is 130 bp longer than that of DHRS4L1 (Fig. 1A). This makes an extended 5′ UTR that partially overlaps with the gene promoter, which may be required for some RNA-directed epigenetic silencing through recruitment of chromatin remodeling complexes, including DnmT3a (29, 35). Another possible explanation is that RNA polymerase II could act in RNA-directed DNA methylation. The closer position of the CpG islands relative to the transcription start site also might be responsible for the specific hypermethylation of DHRS4L2 (36–38).
Although most sense–antisense pairs are reported to exhibit direct or synergistic relationships in the levels of the two transcripts (17), there are also some experimentally validated examples in which the two transcripts are expressed inversely, as in the case of AS1DHRS4 (9, 39). A number of models have been proposed to explain the mechanism of NATs and other lncRNAs in epigenetic gene silencing, including Kcnq1ot1 and Air for parental imprinting (40), the Xist–Tsix pair for mammalian X-chromosome inactivation (41), and HOTAIR for long-range control of HOX genes (42). Kcnq1ot1, an unspliced and overlapping NAT of the Kcnq1 gene, is required for epigenetic silencing of neighboring genes upstream or downstream of the Kcnq1 locus, an activity dependent on interaction of the Kcnq1ot1 transcript with both G9a and PRC2, as well as DnmTs at the loci of silenced genes (43). Air is a 108-kb unspliced NAT expressed from a promoter located in intron 2 of the Igf2r gene that mediates bidirectional silencing of three genes, Slc22a2 and Slc22a3, located upstream, and Igf2r, located downstream (40, 44).
HOTAIR is transcribed from an intergenic sequence of HOXC genes on chromosome 12, and recruits EZH2 in trans to regulate gene expression of the HOXD gene cluster on chromosome 2 (42). Examples in which NATs function in trans have been reported as well. A p15AS expression construct induced p15 silencing in cis and in trans through heterochromatin formation, but not DNA methylation (45). When an lncRNA antisense to Oct4 pseudogene 5 was suppressed, transcription of Oct4 and Oct4 pseudogenes 4 and 5 was observed to increase, in correlation with a loss of silent-state epigenetic marks and histone methyltransferase Ezh2 at the Oct4 promoter (46).
Based on our present results, we present a schematic model to illustrate the regulatory roles of AS1DHRS4 in the expression of the DHRS4 gene cluster (SI Appendix, Fig. S15). This model involves substantially different patterns of epigenetic modification from reported NATs and lncRNA regulatory mechanisms in that, except for the overlapping configuration, AS1DHRS4 has both specific cis and trans activity on inducing different repressive enzyme complexes in the individual promoters within the DHRS4 gene cluster locus. The hierarchical order of events and dependencies leading to gene silencing remain largely unknown. Some previous studies have suggested that DNA methylation patterns guide histone modifications (including histone acetylation and methylation) during gene silencing, whereas other studies have argued that DNA methylation takes its cues primarily from histone modification states (47).
Gene duplications are quite common phenomena. DHRS4L2 is the most recently reported member of the gene cluster resulting from DHRS4 gene duplication. Genome-wide analysis suggests that the vast majority of newly duplicated genes (functional genes or pseudogenes) are silenced within a few million years, because degenerative mutations are much more frequent than advantageous ones (48–50). Thus, we presume that the methylation of histones and CpG islands in the DHRS4L2 promoter reflects a dual strategy for the cell to limit expression of newly duplicated genes to avoid any potential dramatic damages to the organism.
Between 5% and 30% of transcriptional units in diverse eukaryotes have been found to harbor a cis-NAT (51). Head-to-head overlapping is the most common of the three different NAT configurations, and the first exon and the 5′ end of the first intron are hotspots of antisense transcription (10). Thus, we speculate that the regulatory pattern of AS1DHRS4 may be common among other duplicated genes. Structurally different RNAs engage in diverse mechanisms that lead to different regulatory outcomes. This study provides insight into how multiple and highly homologous genes in a very tight gene cluster are differentially regulated by NATs in an epigenetic manner, and explains the precise transcriptional control of newly duplicated genes by NATs as the result of genomic evolution.
Materials and Methods
RACE, RNA interference, RT-PCR and qPCR, Western blotting, bisulfite sequencing for DNA methylation analysis, and ChIP were performed according to routine protocols. RNA-IP and RNA-ChIP assays, DNase I and RNase treatments, ODN blockade experiments, and the sources of specialized materials are described in SI Appendix, SI Materials and Methods. Primer sequences, siRNA oligonucleotides, and ODNs are listed in SI Appendix, Tables S1–S10.
Supplementary Material
Acknowledgments
We thank Junhui Bian and Stanley L. Lin for their critical reading of the manuscript. This work was supported by National Basic Research Project of China Program 973 Grant 2010CD912503 and National Natural Science Foundation of China Grants 30970626 and 31071153.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.T.L. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. HQ845197 (AS1DHRS4 sequence in the HepG2 cell line) and HQ845198 (AS1DHRS4 RNA sequence in the HL7702 cell line)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116597109/-/DCSupplemental.
References
- 1.Huang DY, Ichikawa Y. Purification and characterization of a novel cytosolic NADP(H)-dependent retinol oxidoreductase from rabbit liver. Biochim Biophys Acta. 1997;1338:47–59. doi: 10.1016/s0167-4838(96)00183-5. [DOI] [PubMed] [Google Scholar]
- 2.Endo S, et al. Molecular determinants for the stereospecific reduction of 3-ketosteroids and reactivity towards all-trans-retinal of a short-chain dehydrogenase/reductase (DHRS4) Arch Biochem Biophys. 2009;481:183–190. doi: 10.1016/j.abb.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Matsunaga T, et al. Characterization of human DHRS4: An inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch Biochem Biophys. 2008;477:339–347. doi: 10.1016/j.abb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Oppermann U. Carbonyl reductases: The complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann F, Maser E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab Rev. 2007;39:87–144. doi: 10.1080/03602530600969440. [DOI] [PubMed] [Google Scholar]
- 6.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 8.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Regha K, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malecová B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- 15.Kong LY, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345-9. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia H, et al. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama S, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 18.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 19.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders G, et al. doRiNA: A database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012;40(Database issue):D180–D186. doi: 10.1093/nar/gkr1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 22.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 23.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 24.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matzke M, Matzke AJ, Kooter JM. RNA: Guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 26.Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 27.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 28.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, et al. Common human cancer genes discovered by integrated gene-expression analysis. PLoS ONE. 2007;2:e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6:129–137. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 32.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 34.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 35.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He XJ, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law JA, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano T, Fraser P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm Genome. 2009;20:557–562. doi: 10.1007/s00335-009-9218-1. [DOI] [PubMed] [Google Scholar]
- 41.Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 44.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 45.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 49.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 50.Rodin SN, Riggs AD. Epigenetic silencing may aid evolution by gene duplication. J Mol Evol. 2003;56:718–729. doi: 10.1007/s00239-002-2446-6. [DOI] [PubMed] [Google Scholar]
- 51.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.