Abstract
Although tyrosine-phosphorylated or activated STAT3 (pY-STAT3) is a well-described mediator of tumorigenesis, its role in thyroid cancer has not been investigated. We observed that 63 of 110 (57%) human primary papillary thyroid carcinoma (PTC) cases expressed nuclear pY-STAT3 in tumor cells, preferentially in association with the tumor stroma. An inverse relationship between pY-STAT3 expression with tumor size and the presence of distant metastases was observed. Using human thyroid cancer-derived cell lines [harboring rearranged during transfection (RET)/PTC, v-RAF murine sarcoma viral oncogene homolog B (BRAF), or rat sarcoma virus oncogene (RAS) alterations], we determined that IL-6/gp130/JAK signaling is responsible for STAT3 activation. STAT3 knockdown by shRNA in representative thyroid cancer cell lines that express high levels of pY-STAT3 had no effect on in vitro growth. However, xenografted short hairpin STAT3 cells generated larger tumors than control cells. Similarly, STAT3 deficiency in a murine model of BRAFV600E-induced PTC led to thyroid tumors that were more proliferative and larger than those tumors expressing STAT3wt. Genome expression analysis revealed that STAT3 knockdown resulted in the down-regulation of multiple transcripts, including the tumor suppressor insulin-like growth factor binding protein 7. Furthermore, STAT3 knockdown led to an increase in glucose consumption, lactate production, and expression of Hypoxia-inducible factor 1 (HIF1α) target genes, suggesting that STAT3 is a negative regulator of aerobic glycolysis. Our studies show that, in the context of thyroid cancer, STAT3 is paradoxically a negative regulator of tumor growth. These findings suggest that targeting STAT3 in these cancers could enhance tumor size and highlight the complexities of the role of STAT3 in tumorigenesis.
Keywords: cytokine, tumor microenvironment, metabolic reprogramming
Papillary thyroid carcinoma (PTC) is the most common endocrine malignancy in humans (1); 70% of PTCs harbor alterations in receptor tyrosine kinases or their downstream effectors that activate the ERK/MAPK pathway (2), namely v-RAF murine sarcoma viral oncogene homolog B (BRAF) V600E mutations, rearranged during transfection (RET)/PTC rearrangements, and rat sarcoma virus oncogene (RAS) mutations (3). Although these oncogenic alterations have been extensively studied, other factors, particularly those factors mediating the interaction between the tumor and the surrounding microenvironment, have not been fully characterized (4). PTCs often display an immune cell and desmoplastic stromal infiltrate associated with increased IL-6 and IFNγ, although the precise role of the inflammatory mediators is not yet known (5).
IL-6 plays an important part in immune cell development and behavior (6). This cytokine signals through a dual receptor system comprising a membrane or soluble receptor for IL-6 (IL-6R) and the signal transducing component, gp130, which is common to the IL-6 family of cytokines (7). Engagement of IL-6 with the IL-6R and gp130 leads to the phosphorylation of associated JAKs, which in turn, mediate tyrosine phosphorylation of STATs (particularly STAT3) as well as activation of the ERK/MAPK pathway (6). Several reports have shown that oncogenic RET can promote STAT3 Y705 phosphorylation (pY-STAT3) and transcriptional activation (8). pY-STAT3 is persistently expressed in a wide variety of solid tumors (9, 10), and IL-6 has been shown to be its major activator (11, 12). Several strategies have been developed to target pY-STAT3, including IL-6 and IL-6R blocking antibodies and JAK inhibitors (13). However, the canonical tumor-promoting function of STAT3 has been challenged by recent studies. For instance, STAT3 behaves as a tumor suppressor in phosphatase and tensin homolog-deficient glioblastomas (14). Moreover, STAT3 deficiency in the adenomatosis polyposis coli (APCMin) model of intestinal cancer led to enhanced tumor progression characterized by the development of invasive, more proliferative carcinomas (15). In models of hepatocellular carcinoma, p19 alternate open reading frame (ARF) dictated the tumor-suppressive vs. -promoting behavior of STAT3 (16).
Until now, the role of pY-STAT3 has not been examined in thyroid cancer. We determined that pY-STAT3 is expressed in benign follicular thyroid adenomas (FTAs) and a subset of human PTC and thyroid cancer-derived cell lines (TCCs). Knockdown of STAT3 in TCCs as well as its targeted deletion in a murine model of BRAFV600E-induced PTC led to enhanced tumor growth. Gene expression profiling revealed the differential expression of transcripts, such as insulin-like growth factor binding protein 7 (IGFBP7), which may also participate in the growth inhibitory activity of STAT3. Furthermore, using both standard conditions and those conditions mimicking an in vivo metabolic state, we determined that STAT3-deficient cells are more glycolytic, express higher levels of hypoxia-inducible transcription factor 1α (HIF1α) targets, and are less sensitive to the growth inhibitory effects of cobalt chloride (hypoxia-mimetic) treatment than their short hairpin control (shCT) counterparts. These studies underlie a role for STAT3 in thyroid cancer, adding to the growing number of examples describing STAT3 as a tumor suppressor in cancer.
Results
pY-STAT3 Is Expressed in Human Primary PTC.
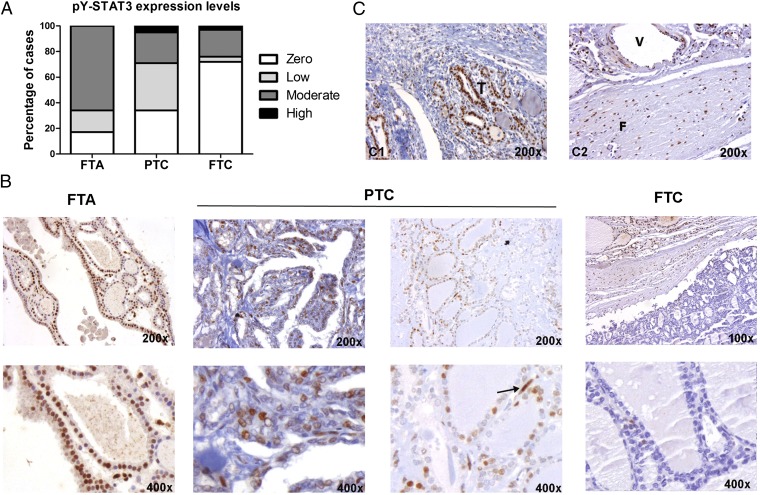
The expression of activated pY-STAT3 in human thyroid tumors has been poorly characterized. We analyzed nuclear pY-STAT3 levels by immunohistochemistry (IHC) in a panel of 146 primary thyroid lesions. pY-STAT3 expression was seen in 10 of 12 (83%) benign FTAs, 63 of 110 (57%) PTCs, and 6 of 24 (25%) follicular thyroid carcinomas (FTCs) (Fig. 1 A and B and Table 1). We also investigated whether pY-STAT3 expression was associated with clinical–pathological parameters in PTC, such as tumor size, vascular invasion, and presence of distant metastases. The average PTC tumor size was ∼4 cm, and tumors were grouped into ≥4- and <4-cm size categories. We found that PTCs larger than 4 cm expressed lower levels of pY-STAT3 in tumor cells [score: 0.7 ± 0.3 (≥4 cm) vs. 2.9 ± 0.6 (<4 cm); Fisher P = 0.04]. No significant association was found between pY-STAT3 and vascular invasion. However, an inverse association between pY-STAT3 expression and the presence of distant metastases was determined (Fisher P = 0.03). Specifically, in a subset of aggressive PTC cases, only 7 of 20 (35%) were pY-STAT3–positive, whereas 56 of 90 (62%) of the remaining more indolent PTCs expressed pY-STAT3. Approximately one-half of PTCs harbor BRAFV600E mutations (17). Here, the BRAF mutational status was determined in 98 of 110 of the PTCs (17); 42% harbored the BRAFV600E mutation and expressed higher levels of pY-STAT3 relative to BRAFwt tumors (Mann–Whitney P = 0.04) (Table 1). In ∼70% of the PTCs (as well as in the FTAs and FTCs), we detected the highest levels of pY-STAT3 within tumor cells at the leading edge of the tumor in association with stromal cells (fibroblasts and endothelial and immune cells), which were also strongly positive for pY-STAT3 (Fig. 1C).
Fig. 1.
pY-STAT3 expression in human thyroid lesions. (A) Quantification of pY-STAT3 expression levels in thyroid tumors. (B) Representative pY-STAT3 staining in benign FTA and malignant PTC and FTC. Arrow points at a pY-STAT3–positive endothelial cell in PTC. (C, 1) Tumor cells (T) within the stroma are strongly pY-STAT3–positive. (C, 1) Stromal cells, namely fibroblasts (F) and endothelial cells lining up vessels (V), highly express pY-STAT3.
Table 1.
STAT3 and pY705-STAT3 (pY-STAT3) expression in primary thyroid lesions
| pY-STAT3 | BRAF (exon 15) |
| FTA | |
| pY-STAT3pos cases = 10 of 12 | 100% WT |
| Percentage of positive cells: 3.8% ± 0.8% | |
| Mean intensity of staining: moderate (2) | |
| Score: 2.7 ± 0.3 | |
| FTC | |
| pY-STAT3pos = 6 of 24 | 100% WT |
| Percentage of positive cells: 1.5% ± 0.5% | |
| Mean intensity of staining: weak (1) | |
| Score: 1.4 ± 0.6 | |
| PTC | |
| pY-STAT3pos = 63 of 110 | 41/98 V600E+ |
| Percentage of positive cells: 4.3% ± 0.9% | [1.5 ± 0.3 (wt) vs. |
| Mean intensity of staining: moderate (2) | 3.1 ± 0.6 (V600E)] |
| Score: 2.4 ± 0.5 | P = 0.013 |
Percentages and scores represent mean ± SEM.
STAT3 Is Regulated by the IL-6/gp130/JAK Signaling in TCCs.
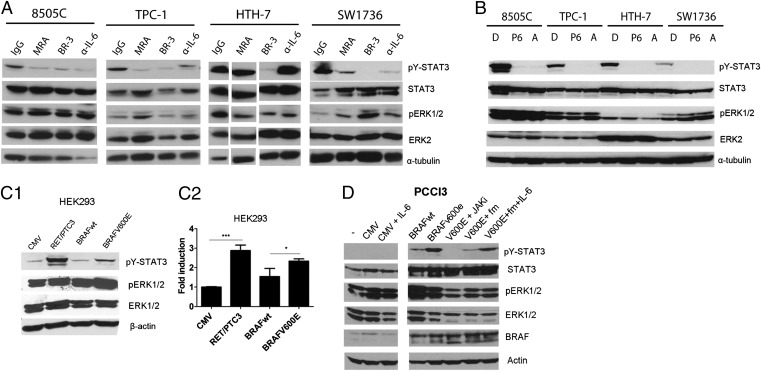
Aberrant IL-6/gp130/JAK signaling has been shown to be a principle regulator of STAT3 activation in many cancers (11, 12, 18). We analyzed pY-STAT3 expression in several TCCs harboring RET, BRAF, and RAS alterations (Table S1) (19, 20). As in primary tumors, pY-STAT3 was heterogeneously expressed in these cell lines (Fig. S1A). We determined the role of IL-6 signaling in mediating STAT3 activation in the pY-STAT3–expressing cell lines (8505C, TPC-1, HTH-7, and SW1736) with inhibitors of the IL-6 pathway, namely anti-human IL-6 (α-IL6), IL-6R [MRA (also known as tocilizumab)], and gp130 (clone BR3) antibodies, (Fig. 2A) and JAK inhibitors P6 and Astrazeneca (AZD) 1480 (Fig. 2B). BR3, P6, and AZD1480 effectively blocked STAT3 phosphorylation (Fig. 2 A and B). IL-6 was secreted by all of the cell lines to variable levels (Fig. S1B). In all cell lines, except HTH-7, α-IL-6 and MRA reduced pY-STAT3 (Fig. 2A). Additionally, 8505C, TPC-1, and SW1736 cells expressed mRNA for IL-6 and the membrane form of the IL-6R (mIL-6R), whereas none expressed the soluble form of the receptor (sIL-6R) (Fig. S1C). We also examined the mRNA levels of other members of the IL-6/gp130 family of cytokines, including leukemia inhibitory factor (LIF) and oncostatin M (OSM) ligands and receptors (Fig. S1C). HTH-7 did not express mIL-6R but did express OSM and OSM receptor (Fig. S1C). However, because of technical limitations, attempts to block OSM signaling were unsuccessful.
Fig. 2.
pY-STAT3 is regulated by the IL-6/gp130/JAK signaling pathway in TCCs. (A) Extracts from TCCs treated for 4 h with control IgG (at 10 μg/mL) as well as blocking antibodies targeting IL-6R (MRA; at 50 μg/mL), gp130 (BR3; at 10 μg/mL), and IL-6 ligand (α-IL-6; at 1 μg/mL) were immunoblotted with the indicated antibodies. HTH-7 lanes were noncontiguous in the same gel. (B) Extracts from TCCs were treated for 16 h with JAK inhibitors P6 (1 μM) and AZD1480 (A; 1 μM) and immunoblotted with the indicated antibodies. α-tubulin was the loading control. (C) RET/PTC and BRAFV600E induce STAT3 Y-phosphorylation and transcriptional activation in HEK293 cells. (C, 1) Cells were transiently transfected with RET/PTC3, BRAFwt, and BRAFV600E expression vectors. Protein extracts were immunoblotted with the indicated antibodies. (C, 2) HEK293 cells were transiently transfected with the indicated expression vectors combined with Interleukin-6 response element (IRE)-Luciferase (Luc) reporter plasmid. Luciferase activity was measured after 48 h (mean ± SD; n = 3). (D) IL-6/JAK signaling mediates BRAFV600E-induced pY-STAT3 in PCCl3 cells. Cells were transfected with pCMV (control vector) or pCMV-BRAFV600E. After 48 h, the medium from BRAFV600E cells was replaced with fresh medium RPMI (column 7) or fresh RPMI supplemented with 50 ng/mL IL-6 (column 8) for 30 min. Cells were lysed, and protein extracts were incubated with the indicated antibodies.
In addition to the IL-6 family of cytokines, STAT3 activation can be regulated by growth factors and nonreceptor tyrosine kinases (11, 21). In thyroid cells, RET/PTC (8) and mutant RET (22, 23) mediate STAT3 phosphorylation, but no studies have examined the role of oncogenic BRAF in STAT3 activation. Given the association between BRAFV600E and pY-STAT3 positivity in our series of human PTC (Table 1), we investigated whether oncogenic BRAF could also lead to STAT3 Y705 phosphorylation and transcriptional activation. We transfected HEK293 cells with exogenous pCMV-RET/PTC3, pCMV-BRAFwt, and pCMV-BRAFV600E expression vectors. Both RET/PTC3 and mutant BRAF (but not control CMV or BRAFwt) induced pY-STAT3 expression (Fig. 2C, 1) and transcriptional activity (Fig. 2C, 2). BRAFV600E has been shown to up-regulate IL-6 in melanoma cells (24). To determine whether BRAFV600E-mediated STAT3 activation was through increased IL-6 expression, we transfected a rat thyroid cell line, PCCl3, with either pCMV-BRAFV600E or pCMV expression vectors. The addition of exogenous IL-6 on control (pCMV) transfected PCCl3 cells did not increase pY-STAT3 levels (Fig. 2D). In contrast, BRAFV600E expression (and BRAFwt, albeit at lower levels) markedly increased pY-STAT3 levels, which were abrogated by the JAK inhibitor, AZD1480 (Fig. 2D). Similarly, removal of conditioned media from the BRAFV600E-PCCl3 cells led to a reduction in pY-STAT3 levels, which could be reversed through the addition of IL-6 (Fig. 2D). These data suggest that BRAFV600E can up-regulate downstream IL-6 signaling, resulting in STAT3 activation.
STAT3 Knockdown Increases Tumorigenicity of TCC.
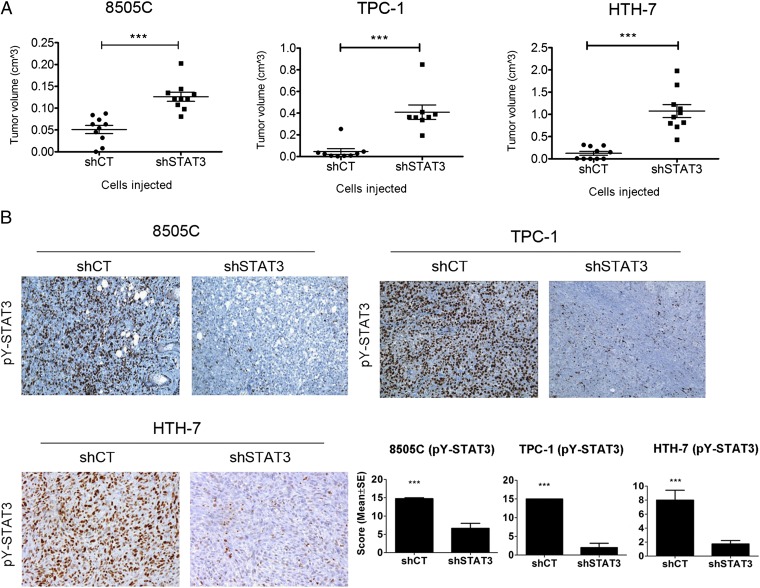
We evaluated the effects of STAT3 knockdown in pY-STAT3–expressing cell lines (8505C, TPC-1, HTH7, and SW1736) by transduction of shSTAT3 and scrambled control (shCT) vectors. Decreases in pY-STAT3 protein expression were confirmed by Western blot analysis (Fig. S2A). pERK1/2 levels remained unchanged, with the exception of TPC-1, where shSTAT3 displayed lower pERK1/2 levels than their respective shCT (Fig. S2A). The proliferation of shSTAT3 cells was compared with shCT cells, and viable cell numbers were determined daily, revealing no differences in growth between matched cell lines (Fig. S2B). The in vivo tumorigenic capacity of 8505C, TPC-1, and HTH7 shCT and shSTAT3 cells was assessed by s.c. injection of cells in athymic nude mice. The SW1736 cell line in our hands did not form tumors and was not further analyzed. Tumors were allowed to grow, and mice were killed 5 (8505C and TPC-1) and 3 (HTH-7) wk after injection. For all matched xenografts, shSTAT3 tumors were significantly larger than the shCT (Fig. 3A). Down-regulation of pY-STAT3 in the shSTAT3 tumors was confirmed by IHC analysis of tumor sections (Fig. 3B). No differences were found in tumor vasculature (Meca-32) or apoptosis (TUNEL) between shCT and shSTAT3 tumors (Fig. S3). Notably, pERK1/2 levels were down-regulated in 8505C- and TPC-1–shSTAT3 tumors compared with shCT. pS6 (S235/236) and pSTAT1 (Y701) levels remained unchanged in all tumors (Fig. S3).
Fig. 3.
STAT3 knockdown in thyroid cancer cell lines enhances their tumorigenic capacity. (A) 8505C (106 cells), TPC-1 (106 cells), and HTH-7 (107 cells) were s.c. coinjected with Matrigel in female athymic nude mice (n = 10 tumors/cell line), and tumor volume was estimated at 5 (8505C and TPC-1) or 3 (HTH-7) wk (mean ± SEM). (B) Representative sections from shCT and shSTAT3 tumor sections stained for pY-STAT3. Quantification of tumor pY-STAT3 levels (n ≥ 3) is on the right. (Magnification: 100×.) ***P < 0.0001.
Given the recently described roles for unphosphorylated STAT3 in tumorigenesis (25, 26), several experimental controls were used. First, STAT3 levels were reduced in the K1 cell line, which expresses total STAT3 but very low levels of the phosphorylated protein (Figs. S1A and S4A, 1). Injection of these cells in nude mice generated tumors with similar sizes independently of their STAT3 status (Figs. S4A, 2). Notably, all pY-STAT3–positive cells were stromal in origin (Figs. S4A, 3). Second, we introduced either a tyrosine-mutant form of STAT3 (STAT3Y705F) or WT murine-STAT3 (to test for off-target effects of the shSTAT3) into the 8505C-shSTAT3 cells (Figs. S4B, 1). These cell lines were injected s.c., and tumor volumes were determined. The expression of the tyrosine mutant did not rescue the tumor-suppressive effects of STAT3, whereas reexpression of WT STAT3 reduced tumor growth (Figs. S4B, 2). Expression of both pY-STAT3 and total STAT3 was confirmed in xenografts by IHC (Figs. S4B, 3).
Thyrocyte-Specific Deletion of STAT3 in a Murine Model of BRAFV600E-Induced PTC Leads to Increased Thyrocyte Proliferation and Tumor Growth.
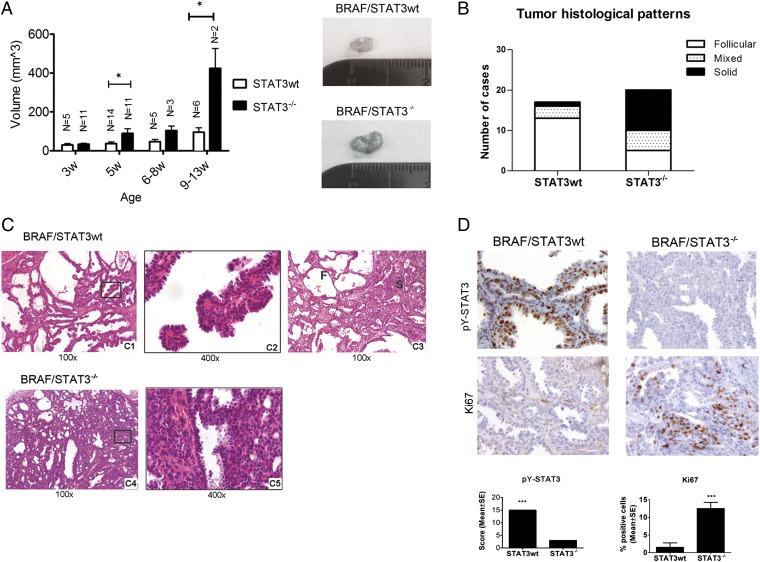
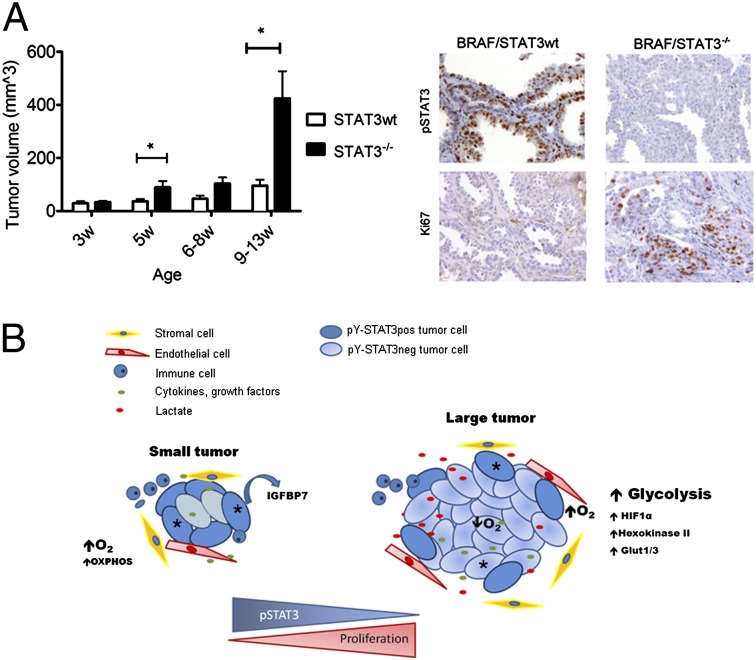
The thyroid peroxidase (TPO) -Cre/lox-stop-lox (LSL)-BRAFV600E murine model of thyroid cancer has been recently characterized (27). These tumors express high levels of pY-STAT3 throughout the tumor (Fig. 4). To examine the role of STAT3 in this model, we deleted STAT3 in BRAFV600E-expressing thyrocytes by crossing STAT3flox/flox mice (28) with TPO-Cre mice (Fig. S5A). Importantly, STAT3 deficiency in thyrocytes from BRAFwt mice (TPO-Cre/STAT3−/−) did not alter the phenotype and histological appearance of the thyroids compared with STAT3 deficiency in thyrocytes from C57BL/6 WT mice (Fig. S5B). TPO-Cre/STAT3−/− mice were crossed with BRAF/STAT3flox/flox mice to generate mice that expressed BRAFV600E in thyrocytes with or without STAT3 (Fig. S5A). BRAF/STAT3−/− mice were phenotypically similar to BRAF/STAT3wt mice. However, by 5 wk of age and at later time points (6–8 and 9–13 wk), the BRAF/STAT3−/− mice exhibited significantly larger thyroid tumors than those tumors from BRAF/STAT3wt mice (Fig. 4A). Histologically, by 5 wk of age, tumors from mice with either genotype had similar histological features, including signs of local invasion to the skeletal muscle and blood vessels (Fig. 4C and Fig. S5B, 2). In age-matched mice, the tissue architecture of the PTC from BRAF/STAT3wt mice was homogenous: neoplastic tissue was composed of follicles and areas of papilliferous hyperplasia (Fig. 4 B and C). In contrast, the BRAF/STAT3−/− tumors were more heterogeneous, displaying early signs of solid growth (∼75% by 5 wk of age) that were only apparent in older BRAF/STAT3wt mice (∼24% by 8 wk of age) (Fig. 4 B and C). IHC confirmed reduction of pY-STAT3 levels in BRAF/STAT3−/− thyrocytes (Fig. 4D). Protein Ki67 levels were higher in the BRAF/STAT3−/− tumors compared with the BRAF/STAT3wt tumors (Fig. 4D), and no differences were detected in the apoptotic index (TUNEL and cleaved-caspase 3) between the two groups (Fig. S5C). We tested whether compensatory up-regulation of several pathways, known to be important in thyroid pathogenesis [PI3K/mammalian target of rapamycin (mTOR), MAPK, and TGF-β], was occurring in BRAF/STAT3−/− tumors. Comparative IHC for pS6, p-p38/MAPK, pJNK/MAPK, and pSmad2/3 revealed no significant differences in the expression levels of any of these markers between the tumor groups. A slight reduction in pERK1/2 levels was observed in BRAF/STAT3−/− thyrocytes compared with BRAF/STAT3wt, similar to what we observed in the 8505C and TPC-1 xenografts (Figs. S3 and S5C).
Fig. 4.
STAT3 deficiency in thyrocytes of LSL-BRAFV600E mice increases thyroid tumor size and proliferation. (A) BRAF/STAT3wt and BRAF/STAT3−/− mice were killed at distinct time points and thyroid volume was determined (mean ± SEM). Images represent thyroids from 5-wk-old mice. (B) Distribution of mouse genotype by histological pattern. (C) Histological sections of thyroids from BRAF/STAT3wt and BRAF/STAT3−/− mice. (C, 1 and 2) BRAF/STAT3wt thyroids revealing hobnail PTC with follicular architecture at 5 wk and (C, 3) mixed follicular (F) and solid (S) architecture at 9 wk. (C, 4 and 5) Thyroid section of a 5-wk-old BRAF/STAT3−/− mouse revealing predominantly solid areas. (D) IHC analysis of pY-STAT3 and Ki67 in the thyroids of 5-wk-old BRAF/STAT3wt and BRAF/STAT3−/− mice. *P < 0.05, ***P < 0.0001.
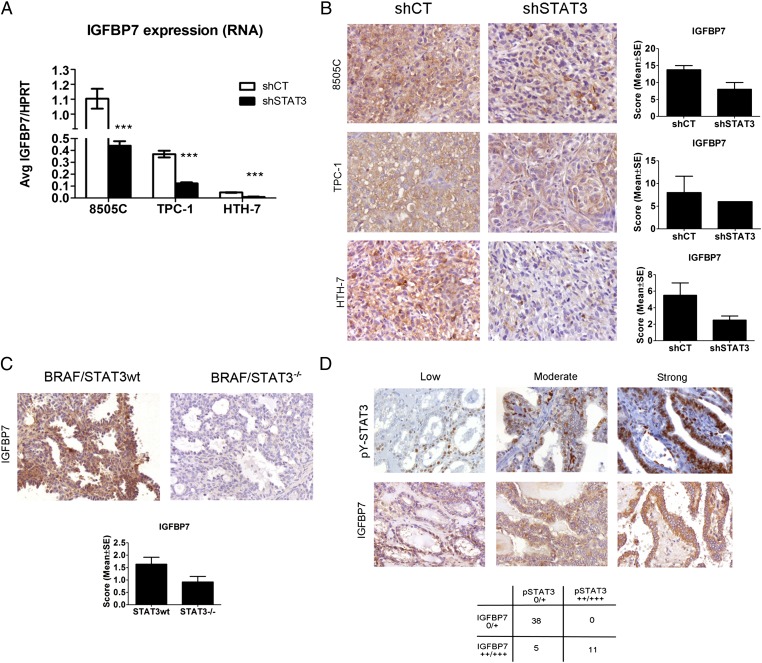
STAT3 Knockdown in TCCs and Transgenic Mice Leads to IGFBP7 Down-Regulation.
We performed genome-wide expression analysis in 8505C, TPC-1, and HTH-7 shCT and shSTAT3 cell lines to identify potential molecules or signaling pathways explaining the increased growth of shSTAT3 tumors. A total of 235 genes were down-regulated, and 146 genes were up-regulated in the three shSTAT3 cell lines compared with their respective shCT (Dataset S1). We focused on those genes with differential regulation that was common to at least two of the cells lines (Table S2). Of the seven commonly down-regulated genes, IGFBP7 was decreased in 8505C and HTH-7 shSTAT3 cells compared with shCT (Table S2). IGFBP7 has been shown to have a tumor-suppressive activity in models of melanoma, colorectal cancer, and thyroid cancer (29–31). We determined that IGFBP7 RNA (Fig. 5A) [by quantitative real-time RT-PCR (Q-PCR)] and protein levels (by IHC) (Fig. 5 B and C) were decreased in 8505C-, HTH-7– and TPC-1–shSTAT3 cells, xenografts, and transgenic tumor models compared with control mice. In the xenografts and transgenic tumors, a positive association between pY-STAT3 and IGFBP7 levels was observed (Fisher exact P = 0.06) (Fig. 5 B and C). A similar relationship between pY-STAT3 and IGFBP7 was detected in 47 primary human PTC samples by IHC (Fig. 5D). IGFBP7 was heterogeneously expressed in all 47 cases: 10 cases (21%) displayed low levels of IGFBP7, 17 cases (36%) had moderate levels of IGFBP7, and 20 cases (43%) had high levels of IGFBP7. A significant positive correlation between pY-STAT3 and IGFBP7 expression was observed (Spearman correlation P = 0.03) (Fig. 5D).
Fig. 5.
IGFBP7 levels are reduced in STAT3-deficient thyroid cancer models. (A) Q-PCR of IGFBP7 RNA levels in shCT and shSTAT3 8505C, TPC-1, and HTH-7 cell lines. Bars represent average IGFBP7/HRPT expression ± SEM of three independent experiments. (B) Representative immunostaining of IGFBP7 in sections of shCT and shSTAT3 tumor xenografts and (C) BRAF/STAT3wt vs. BRAF/STAT3−/− transgenic tumors. (D) pY-STAT3 expression positively correlates with IGFBP7 expression in human PTC. Representative serial sections of 54 human PTC specimens were stained for nuclear pY-STAT3 and cytoplasmic IGFBP7. Examples of low, moderate, and strong pY-STAT3 and IGFBP7 staining are shown. Of 54 cases, 38 cases (70%) displayed none or low pY-STAT3 IGFBP7 (0/+), and 11 cases displayed equally moderate/high (++/+++) levels of both proteins (20%). Fisher exact P < 0.001; Spearman correlation P = 0.03. (Magnification: 200×.)
Promoter methylation has been described as the most frequent event accounting for IGFBP7 down-regulation in cancer models (30, 32). We analyzed the −237- to +10-bp region of the IGFBP7 promoter, corresponding to a CpG island associated with variations in IGFBP7 expression (30, 32), in shCT and shSTAT3 8505C and TPC-1 cell lines. We detected additional partially methylated CpG sites in 8505C and TPC-1 shSTAT3 cell lines at positions −100, −169 (8505C), and −100 (TPC-1) bp (Fig. S6).
Increased Glycolysis of STAT3-Deficient TCCs.
STAT3 down-regulation in 8505C, TPC-1, and HTH-7 cell lines led to increased tumor growth with no apparent effects in vitro (Fig. S2). We investigated whether the tumor microenvironment might explain such differences in cellular behavior. IHC characterization of xenografts and transgenic mice revealed no differences in tumor vasculature (Figs. S3 and S5C). Moreover, no significant differences were detected in T-cell numbers (CD3 staining) and activated macrophages (Iba-1) in BRAF/STAT3−/− tumors compared with STAT3wt tumors from transgenic mice (Fig. S5C).
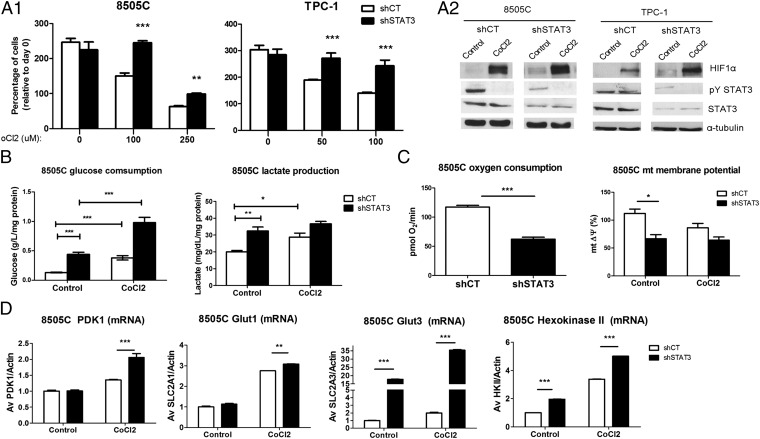
The metabolic switch from oxidative phosphorylation (OXPHOS) to aerobic glycolysis is a hallmark of many cancers (33). STAT3 has been shown to mediate metabolic changes in cells through the regulation of energy metabolism and oxidative stress through canonical (transcriptional) and noncanonical activities (34, 35). We hypothesized that STAT3 may function primarily as a positive regulator of OXPHOS in thyroid cancer. Thus, a reduction in STAT3 levels might shift the balance to enhanced glycolysis for energy production, leading to a selective growth advantage in a hypoxic in vivo tumor microenvironment. To test this hypothesis, we determined the growth of 8505C and TPC-1 shCT and shSTAT3 cell lines under different concentrations of cobalt chloride (CoCl2), a commonly used hypoxia-mimetic; 8505C and TPC-1 shSTAT3 cells grew more efficiently under CoCl2 treatment than their respective shCT cells (P < 0.0001) (Fig. 6A, 1). CoCl2 stabilizes the HIF1α in normoxia, impeding its proteasomal-dependent degradation (36). STAT3 has been shown to both transcriptionally regulate HIF1α and impede its degradation through the sequestration of the von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase (VHL-E3) (37, 38). We observed that CoCl2 induced HIF1α accumulation at similar levels in both shCT and shSTAT3 cells (Fig. 6A, 2). Surprisingly, HIF1α protein levels were higher in shSTAT3 cells compared with shCT at basal levels (Fig. 6A, 2). Notably, HIF1a mRNA levels were lower in shSTAT3 compared with shCT cells (Fig. S7A). Finally, CoCl2 treatment led to a reduction in pY-STAT3 levels (Fig. 6A, 2). These observations suggest that STAT3 is a negative regulator of HIF1α protein expression/stability in these TCCs. Response to hypoxia through HIF1α leads to the up-regulation of glycolytic enzymes, increased glucose consumption and lactate production, and negative regulation of OXPHOS (33). Both under normoxic conditions and after treatment with CoCl2, shSTAT3 cells consumed larger amounts of glucose and produced more lactate than their respective shCT cells (Fig. 6B and Fig. S7B). Consistently, in shSTAT3 cells, significant drops in oxygen consumption rate (P < 0.001) as well as mitochondrial membrane potential (mtΔψ), which reflects the pumping of hydrogen ions across the inner membrane during OXPHOS (P = 0.03), were detected (Fig. 6C).
Fig. 6.
STAT deficiency induces a glycolytic metabolism in TCCs. (A, 1) shCT and shSTAT3 TCCs (8505C and TPC-1) were grown for 96 and 48 h, respectively, with RPMI (1% serum) and different concentrations of the hypoxia-mimetic CoCl2. Growth was determined by the sulpharhodamine B assay. (A, 2) shCT and shSTAT3 cell lines (8505C and TPC-1) were treated with CoCl2 (100 μM) for 24 h. Total protein was probed with the indicated antibodies. (B and C) Cells were grown in plain RPMI for 48 h with or without CoCl2 (100 μM). (B) Glucose uptake was determined by subtracting the glucose concentration in the supernatant from the initial concentration in fresh RPMI. Lactate was measured in the supernatant of the cell. Results were normalized to protein amount (milligrams). Bars represent mean ± SEM (n = 3). (C) The oxygen consumption rate and mitochondrial membrane potential (JC-1) were measured in equal numbers of 8505C shCT and shSTAT3 cells (mean ± SEM; n = 3). (D) 8505C shCT and shSTAT3 cell lines were treated with CoCl2 (100 μM) for 24 h, and the RNA levels of the indicated glycolytic enzymes (PDK1 and GLUT1/3) were determined and normalized to β-actin. Bars are mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.0001.
The glycolysis regulator, pyruvate dehydrogenase kinase (PDK), inactivates the oxidation of pyruvate by pyruvate dehydrogenase in the mitochondria, resulting in increased lactate production. CoCl2 induced an expected increase in PDK1 mRNA levels in shCT cells (1.4-fold for 8505C and twofold for TPC-1), which was more pronounced in the 8505shSTAT3 (twofold) and TPC-1shSTAT3 (sixfold) cell lines (Fig. 6D and Fig. S7C). In parallel, glucose transporter 3 (GLUT3) and hexokinase II (HK2) RNA levels were up-regulated in 8505C shSTAT3 cells compared with shCT. GLUT3, HK2, and GLUT1 levels were all increased to a greater degree in the shSTAT3 cells after CoCl2 treatment (Fig. 6D). Altogether, these data suggest that glycolysis is increased in STAT3-deficient TCCs.
Discussion
The present study aimed to define the role and mechanism of STAT3 regulation in PTC. A growing number of studies have shown that high nuclear pY-STAT3 expression in tumors, including breast, head and neck, and lung, is correlated with improved survival, smaller tumors, or less aggressive histology, suggesting a growth-suppressive function for pY-STAT3 rather than its accepted role as an oncogene (12, 39, 40). We analyzed the presence of Tyr705-phosphorylated STAT3 in primary human thyroid lesions and observed the highest levels of pY-STAT3 in benign FTAs and PTCs without evidence of distant metastasis. Conversely, the lowest levels of pY-STAT3 were found in a subset of PTC cases presenting with lung and bone metastases. Furthermore, we found a positive association between the expression of pY-STAT3 and smaller PTC tumor sizes. Similar observations have been described in the recent work by Kim et al. (41), which showed an analogous inverse relationship between pY-STAT3, STAT3 DNA binding activity, and PTC tumor size, suggesting a growth-suppressive role for pY-STAT3 in PTC.
Interestingly, pY-STAT3–positive tumor cells were preferentially found on the edge of tumors within malignant epithelial cells, stromal cells, and adjacent nonmalignant thyrocytes. Because STAT3 is ubiquitously expressed in PTC (42), this pattern of activation suggests that paracrine factors (cytokines and growth factors) produced by the stroma may be mediating STAT3 activation in tumor cells, similar to what we recently described in breast cancers (43).
Several studies have shown that, in models of RAS and EGF receptor-driven tumorigenesis, ERK/MAPK signaling can enhance STAT3 phosphorylation through autocrine/paracrine production of IL-6 or LIF (12, 44, 45). Studies have also shown that oncogenic RET can increase proinflammatory mediators, such as IL-6, in thyroid cells and tumors (46–48). In addition, BRAFV600E has been shown to induce the secretion of IL-6 in melanoma cell lines (24). In our PTC series, we observed that BRAFV600E-harboring cases expressed higher levels of pY-STAT3 compared with BRAFwt, suggesting a role for dysregulated BRAF signaling in STAT3 activation. We also showed that BRAFV600E expression led to STAT3 phosphorylation and transcriptional activation in HEK293 and PCCl3 cell lines dependent on IL-6/JAK signaling. Furthermore, we showed that blocking members of the IL-6/gp130 family in TCCs led to a potent reduction in pY-STAT3 levels. Our data show a principle role for the IL-6/gp130/JAK signaling pathway in regulating STAT3 activation in thyroid cancer, similar to what has been observed in breast (11), lung (12), colorectal (49), and prostate (18, 50) cancers.
We investigated the role of STAT3 in cell lines and in vivo models (xenografts and transgenics) of thyroid cancer. Stable knockdown of STAT3 in TCCs did not alter in vitro growth, whereas in vivo, shSTAT3 tumors grew significantly faster than matched controls. In our transgenic murine model of BRAFV600E-induced PTC, thyrocyte-specific ablation of STAT3 led to larger and more proliferative tumors, with increased areas of solid growth compared with age-matched BRAF/STAT3wt mice. A similar scenario has been described in p19 null RAS-transformed hepatocytes, in which STAT3 deficiency did not cause differences in proliferation in vitro but gave rise to larger tumors in nude mice (16). Furthermore, the introduction of a nontyrosine phosphorylateable form of STAT3 in shSTAT3 cells could not suppress tumor growth, showing that the Y705 residue is necessary for the in vivo growth-restraining activity of STAT3.
We observed decreased activation of the MAPK signaling pathway (pERK1/2) in STAT3-deficient tumors. Although this pathway is considered a driver of thyroid tumorigenesis, studies have shown that it can suppress the growth of human medullary thyroid carcinoma cell lines and xenografts (51, 52).
Gene expression profiling led us to the identification of IGFBP7 as a commonly down-regulated target in the shSTAT3 cell lines, xenografts, and BRAF/STAT3−/− murine PTC. In addition, we detected a positive correlation between pY-STAT3 and IGFBP7 in primary human PTC. In cancer, IGFBP7 has been shown to induce apoptosis and senescence and block proliferation in an autocrine/paracrine fashion (53). Furthermore, IGFBP7 has also been described as a tumor suppressor that is down-regulated in thyroid cancer (31), melanoma (54), and colorectal cancer through epigenetic silencing (hypermethylation) (29, 32, 55). Interestingly, we found that STAT3-deficient 8505C and TPC-1 cell lines had increased IGFBP7 promoter methylation compared with shCTs. Additionally, the human IGFBP7 promoter sequence has a number of optimal STAT3 binding sites, suggesting that STAT3 may be a direct transcriptional activator of IGFBP7. Although showing the functional consequences of IGFBP7 expression to the mediated growth restraint of STAT3 would be of interest, a single protein is unlikely to be regulating in vivo growth.
We hypothesized that the microenvironment might account for the differential growth capacity of STAT3-deficient tumors. Surprisingly, we did not observe differences in the number of blood vessels or immune cell infiltration.
STAT3 has been implicated as a modulator of cellular metabolism, including glycolysis and mitochondrial respiration. Phospho-S727 STAT3 has been shown to localize in the mitochondria, where it positively regulates the activity of complex I/II of OXPHOS (26, 56). In contrast, pY-STAT3 was shown to up-regulate glycolysis in fibroblasts and STAT3-dependent cancer cell lines (57, 58). Given the hypoxic nature of tumors, we investigated whether STAT3 deficiency could alter the metabolic function of TCCs. Knockdown of STAT3 led to a growth advantage under hypoxic stress because of a metabolic reprogramming characterized by increased glucose consumption, lactate production, and reduced rate of oxygen consumption. Consistent with this phenotype, the absence of STAT3 increased the expression of the genes encoding glycolytic enzymes (PDK1, GLUT3, and HK2). We also observed the up-regulation of additional glycolysis regulators (PDK3 and enolase-2) in 8505C shSTAT3 cells compared with shCT cells through gene profiling analysis (Dataset S1).
STAT3 has been shown to positively regulate HIF1a expression through transcriptional and posttranscriptional mechanisms (37, 38, 59). However, we showed that STAT3-deficient TCCs expressed slightly higher levels of HIF1α and its transcriptional targets compared with controls, suggesting a paradoxical role for STAT3 as a negative regulator of HIF1α. Although the reduction in STAT3 led to a decrease in HIF1a mRNA levels, HIF1α protein levels were slightly elevated, suggesting that STAT3 may negatively regulate HIF1α at the posttranscriptional level. CoCl2 led to the reduction of pY-STAT3, consistent with a recent study showing hypoxia-induced reduction of pY-STAT3 through increased SOCS-3 expression (60). In summary, these results suggest that, in the absence of STAT3, TCCs undergo a metabolic reprogramming, resulting in enhanced glycolysis under hypoxic stress.
Serine- but not tyrosine-phosphorylated STAT3 was identified in the mitochondria of cells (mSTAT3), where it was shown to modulate OXPHOS chain activity (26, 56). Given that a serine-phosphorylated but tyrosine-mutant form of STAT3 was unable to rescue the STAT3 knockdown phenotype in our studies, the regulation of OXPHOS activity by mSTAT3 does not seem to be at play in our models. However, work in the laboratory of Valeria Poli has shown that transcripts encoding for mitochondrial proteins belonging to OXPHOS complexes were reduced in cells expressing constitutively activated pY-STAT3, which was associated with reduced complexes IV/V activity (57, 58). Adding to the complexity of the roles of STAT3 in regulating metabolism, recent work showed that nuclear STAT3 can be tyrosine-phosphorylated by the dimeric M2 isoform of pyruvate kinase (PKM2), and together, they form a transcriptional activating complex participating in a PKM2/HIF1α-positive feedback loop (61, 62). Thus, in the models of thyroid cancer studied here, it remains to be determined whether STAT3 is the transcriptional regulator of genes encoding proteins involved in OXPHOS activity and/or STAT3 can influence the activity of PKM2. Future studies will also be needed to determine the mechanisms by which STAT3 negatively regulates HIF1α protein synthesis and/or turnover in thyroid cancer models and the additional STAT3 targets that participate in the regulation of glycolysis and oxidative phosphorylation.
In conclusion, our data show that STAT3 is a suppressor of thyroid tumor growth in preclinical models, although it remains to be determined whether this finding is the case for all histological subtypes and stages of thyroid cancer, including distant metastatic progression. Nevertheless, our findings suggest that targeting this transcription factor should be used with caution. Importantly, a better understanding of the mechanisms and contexts that predict the dual-edged function of STAT3 in tumorigenesis must be found.
Materials and Methods
Human Tumor Samples.
Human PTC samples (formalin-fixed and paraffin-embedded) were retrieved from Hospital de S. João/Medical Faculty of the University of Porto, Porto, Portugal; Institute of Pathology and Molecular Immunology of the University of Porto, Porto, Portugal; Portuguese Institute of Oncology (IPO), Porto, Portugal; Hospital de S. Marcos, Braga, Portugal; and Hospital Clínico Universitario, Santiago de Compostela, Spain. All of the procedures were in accordance with institutional ethical standards. The histological classification of the samples was done by an experienced thyroid pathologist (M.S.-S.).
IHC, Reagents, and Antibodies.
IHC was carried out using the streptavidin-biotin-HRP technique. The results were independently evaluated by two observers and scored either semiquantitatively (pY-STAT3, STAT3, pY-STAT1, pS6, pERK1/2, p-p38, p-JNK, pSmad2/3, vimentin, E-cadherin, and IGFBP7) or quantitatively (Ki67, cyclin D1, Meca-32, TUNEL, cleaved-caspase 3, iba1, and CD3). Semiquantitative analysis was performed by considering both the intensity (0, negative; 1, low; 2, moderate; 3, strong) and the percentage of cells displaying identical staining (0, negative; 1, 0–5%; 2, 5–25%; 3, 25–50%; 4, 50–75%; 5, 75–100%), and the respective indexes were multiplied by each other to calculate a score value. The score values of pY-STAT3 and IGFBP7 were grouped into categories: low reactivity (score < 4), moderate reactivity (score = 4–7), and high reactivity (score ≥ 8).
Cell Lines and Reagents.
8505C, TPC-1, HTH-7, and SW1736, as well as their derived shCT and shSTAT3 cell lines, were maintained in RPMI, 10% FBS, and 1% PenStrep. PCCl3 was maintained as described (48). 293T and HEK293T cells were maintained in DMEM, 10% FBS, and 1% PenStrep. STAT3 shRNA lentiviral construct and scrambled control construct were previously described (63). Retroviral vectors pBabe-STAT3 and pBabe-Y705F are described in ref. 21.
Protein Extracts and Western Blotting.
Cells were lysed in radioimmunoprecipitation assay buffer (RIPA) buffer supplemented with proteinase and phosphatase inhibitors. For subcellular protein fractionation, the Standard Fractionation Kit from Mitosciences (Abcam) was used according to the manufacturer’s instructions. Protein extracts were run in SDS gels and transferred to Hybond ECL membrane (Amersham Biosciences). The primary antibodies were the same as used for IHC at different dilutions. Secondary antibodies (anti-rabbit, anti-mouse, and anti-goat) conjugated with peroxidase were from Santa Cruz BT, and bands were visualized by the ECL detection system (Perkin-Elmer).
ELISA.
Cells were plated at similar densities and grown until subconfluence in serum-free medium. The supernatants were collected, and IL-6 levels were assessed using an IL-6 ELISA kit (Cell Sciences) according to the manufacturer’s instructions. IL-6 levels were normalized to total protein extracted from the cells.
Plasmids and Luciferase Reporter Assay.
The pIRE-ti-LUC and pDM2-LacZ reporter plasmids were provided by Robert Hofstra (University of Groningen, Groningen, The Netherlands) (23). The BRAF and RET/PTC expression vectors are described in ref. 64.
In Vitro Cell Growth.
Cell growth was assessed by Calcein AM (calcein acetoxymethylester; Invitrogen) or sulforhodamine (SRB) staining as previously described (65).
In Vivo Tumorigenicity Studies.
All of the procedures were included in a protocol approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee. Cells were harvested, mixed with an equal volume of matrigel (BD Biosciences), and injected s.c. into the flank of female homozygous athymic nu/nu mice (Harlan Laboratories). Tumor dimensions length, width, and height were determined, and tumor volume was calculated by the expression (1/2 × length × width × height) (66).
Transgenics.
All of the procedures were included in a protocol approved by the MSKCC Institutional Animal Care and Use Committee. All of the genotypes were in C57BL/6 background. LSL-BRAFV600E mice are described in ref. 27. STAT3flox/flox mice were a gift from Shizuo Akira (Hyogo College of Medicine, Nishinomiya, Japan) (28). These mice were backcrossed to C57BL/6 (a gift from Hua Yu, City of Hope, El Duarte, CA).
Gene Expression Profiling Analysis.
RNA samples were hybridized to a Human HT12 Illumina array according to the manufacturer’s instructions. Data analysis was done in Partek. A gene was considered differentially expressed according to an arbitrary cutoff of twofold change with P < 0.05.
IGFBP7 Promoter Methylation Analysis.
Genomic DNA was extracted using phenol–chloroform and converted using sodium bisulfite with the EpiTect Bisulfite Kit (Qiagen). The converted DNA was amplified using primers specific for the −237- to +40-bp region of the IGFBP7 promoter, and the amplification products were subjected to direct sequencing.
RT-PCR/Q-PCR.
Total RNA was extracted from cells using the TRIzol method (Invitrogen) followed by cDNA conversion (BioRad) using the manufacturer’s instructions. For Q-PCR, the Quantitect Sybr Green (Qiagen) was used for human IGFBP7 RNA quantification, and hypoxanthine phosphoribosyltransferase 1 (HPRT) was used as the housekeeping control. Primer sequences are available on request. For human HIF1a, PDK1, GLUT1 (SLC2A1), GLUT3 (SLC2A3), and HK2 RNA quantification, Taqman gene expression assays (Applied Biosystems) were used.
Glucose and Lactate Quantification.
Identical cell numbers were plated, and after 24 h, the medium was replaced with fresh RPMI supplemented with 1% serum and the indicated drugs. After 24 h, the medium was collected. Glucose present in the conditioned medium was quantified using the Glucose GOD/PAP Kit (Roche) and subtracted from the initial concentration present in fresh RPMI. Lactate was quantified with the LO-POD enzymatic colorimetric assay (Spinreact).
Oxygen Consumption.
Oxygen consumption in 8505C-derived cells was measured using the Seahorse 24XF instrument (Seahorse Biosciences) according to the manufacturer’s protocols.
Membrane Potential.
Mitochondrial membrane potential was measured using the JC1 Mitochondrial Membrane Potential Assay Kit (Mitosciences; Abcam) following the manufacturer’s instructions.
Statistical Analysis.
Statistical analysis was done in StatView (SAS Institute) using Student t, Mann–Whitney u, χ2, and Fisher correlation tests. Results were considered statistically significant when P < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Supplementary Material
Acknowledgments
We thank Vítor Trovisco for the RET/PTC and BRAF expression vectors, Hugo Prazeres and Joana Nunes for their helpful comments, and the Molecular Cytology Core at Sloan–Kettering Institute, especially Alex Baldys and Afsar Barlas, for help with immunostainings. pIRE-Luc and pDM2-LacZ were provided by Dr. Robert Hofstra (University of Groningen, Groningen, The Netherlands). Astra Zeneca generously provided AZD1480. This study was supported in part by a Portuguese Foundation for Science and Technology (FCT) Project Grant. The Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP) is an Associate Laboratory of the Portuguese Ministry of Science, Technology, and Higher Education that is partially supported by the FCT. J.P.C. was supported by FCT Grant SFRH/BD/40260/2007. This project was supported by grants from the NIH-CA87637, U54: CA148967, Astra Zeneca, Sussman Family Fund, Marjorie and Charles Holloway Foundation and Lerner Awards (to J.F.B.). D.L. was supported by grants from the Children's Cancer and Blood Foundation, The Manning Foundation, The Hartwell Foundation, Pediatric Oncology Experimental Therapeutics Investigators Consortium, Stavros S. Niarchos Foundation, Champalimaud Foundation, The Nancy C. and Daniel P. Paduano Foundation, The Mary Kay Foundation, American Hellenic Educational Progressive Association 5th District, The Malcolm Hewitt Wiener Foundation, The George Best Costacos Foundation, NCI-R01CA 098234-01, NCI UO1 TMEN, Susan G. Komen for the Cure, and NCI-U54-CA143836 PSOC training grant. D.L. and J.F.B. were supported by The Beth C. Tortolani Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.G. is a guest editor invited by the Editorial Board.
See Author Summary on page 13904 (volume 109, number 35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201232109/-/DCSupplemental.
References
- 1.DeLellis RA, Lloyd RV, Heitz PU, Eng C. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Endocrine Glands. Lyon, France: IARC Press; 2004. [Google Scholar]
- 2.Fagin JA, Mitsiades N. Molecular pathology of thyroid cancer: Diagnostic and clinical implications. Best Pract Res Clin Endocrinol Metab. 2008;22:955–969. doi: 10.1016/j.beem.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trovisco V, et al. Molecular genetics of papillary thyroid carcinoma: Great expectations. Arq Bras Endocrinol Metabol. 2007;51:643–653. doi: 10.1590/s0004-27302007000500002. [DOI] [PubMed] [Google Scholar]
- 4.Ryder M, Ghossein RA, Ricarte-Filho JC, Knauf JA, Fagin JA. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr Relat Cancer. 2008;15:1069–1074. doi: 10.1677/ERC-08-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koperek O, Asari R, Niederle B, Kaserer K. Desmoplastic stromal reaction in papillary thyroid microcarcinoma. Histopathology. 2011;58:919–924. doi: 10.1111/j.1365-2559.2011.03791.x. [DOI] [PubMed] [Google Scholar]
- 6.Kishimoto T. Interleukin-6: From basic science to medicine—40 years in immunology. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 7.Taga T, et al. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell. 1989;58:573–581. doi: 10.1016/0092-8674(89)90438-8. [DOI] [PubMed] [Google Scholar]
- 8.Hwang JH, et al. Activation of signal transducer and activator of transcription 3 by oncogenic RET/PTC (rearranged in transformation/papillary thyroid carcinoma) tyrosine kinase: Roles in specific gene regulation and cellular transformation. Mol Endocrinol. 2003;17:1155–1166. doi: 10.1210/me.2002-0401. [DOI] [PubMed] [Google Scholar]
- 9.Mora LB, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: Direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 10.Pedranzini L, Leitch A, Bromberg J. Stat3 is required for the development of skin cancer. J Clin Invest. 2004;114:619–622. doi: 10.1172/JCI22800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berishaj M, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sansone P, Bromberg J. Environment, inflammation, and cancer. Curr Opin Genet Dev. 2011;21:80–85. doi: 10.1016/j.gde.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 14.de la Iglesia N, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musteanu M, et al. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 2010;138:1003–1011. doi: 10.1053/j.gastro.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 16.Schneller D, et al. p19(ARF) /p14(ARF) controls oncogenic functions of signal transducer and activator of transcription 3 in hepatocellular carcinoma. Hepatology. 2011;54:164–172. doi: 10.1002/hep.24329. [DOI] [PubMed] [Google Scholar]
- 17.Soares P, et al. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003;22:4578–4580. doi: 10.1038/sj.onc.1206706. [DOI] [PubMed] [Google Scholar]
- 18.Hedvat M, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meireles AM, et al. Molecular and genotypic characterization of human thyroid follicular cell carcinoma-derived cell lines. Thyroid. 2007;17:707–715. doi: 10.1089/thy.2007.0097. [DOI] [PubMed] [Google Scholar]
- 20.Ricarte-Filho JC, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69:4885–4893. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plaza Menacho I, et al. RET-familial medullary thyroid carcinoma mutants Y791F and S891A activate a Src/JAK/STAT3 pathway, independent of glial cell line-derived neurotrophic factor. Cancer Res. 2005;65:1729–1737. doi: 10.1158/0008-5472.CAN-04-2363. [DOI] [PubMed] [Google Scholar]
- 23.Schuringa JJ, et al. MEN2A-RET-induced cellular transformation by activation of STAT3. Oncogene. 2001;20:5350–5358. doi: 10.1038/sj.onc.1204715. [DOI] [PubMed] [Google Scholar]
- 24.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 26.Gough DJ, et al. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franco AT, et al. Thyrotrophin receptor signaling dependence of Braf-induced thyroid tumor initiation in mice. Proc Natl Acad Sci USA. 2011;108:1615–1620. doi: 10.1073/pnas.1015557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda K, et al. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 29.Wajapeyee N, Kapoor V, Mahalingam M, Green MR. Efficacy of IGFBP7 for treatment of metastatic melanoma and other cancers in mouse models and human cell lines. Mol Cancer Ther. 2009;8:3009–3014. doi: 10.1158/1535-7163.MCT-09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki H, et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- 31.Vizioli MG, et al. IGFBP7: An oncosuppressor gene in thyroid carcinogenesis. Oncogene. 2010;29:3835–3844. doi: 10.1038/onc.2010.136. [DOI] [PubMed] [Google Scholar]
- 32.Lin J, et al. Reactivation of IGFBP7 by DNA demethylation inhibits human colon cancer cell growth in vitro. Cancer Biol Ther. 2008;7:1896–1900. doi: 10.4161/cbt.7.12.6937. [DOI] [PubMed] [Google Scholar]
- 33.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Demaria M, Poli V. From the nucleus to the mitochondria and back: The odyssey of a multitask STAT3. Cell Cycle. 2011;10:3221–3222. doi: 10.4161/cc.10.19.17379. [DOI] [PubMed] [Google Scholar]
- 35.Szczepanek K, Chen Q, Larner AC, Lesnefsky EJ. Cytoprotection by the modulation of mitochondrial electron transport chain: The emerging role of mitochondrial STAT3. Mitochondrion. 2012;12:180–189. doi: 10.1016/j.mito.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Hilliard G, Ferguson T, Millhorn DE. Cobalt inhibits the interaction between hypoxia-inducible factor-alpha and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-alpha. J Biol Chem. 2003;278:15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- 37.Jung JE, et al. STAT3 inhibits the degradation of HIF-1alpha by pVHL-mediated ubiquitination. Exp Mol Med. 2008;40:479–485. doi: 10.3858/emm.2008.40.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu G, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–1105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolled-Filhart M, Camp RL, Kowalski DP, Smith BL, Rimm DL. Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin Cancer Res. 2003;9:594–600. [PubMed] [Google Scholar]
- 40.Pectasides E, et al. Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin Cancer Res. 2010;16:2427–2434. doi: 10.1158/1078-0432.CCR-09-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim WG, et al. Basal STAT3 activities are negatively correlated with tumor size in papillary thyroid carcinomas. J Endocrinol Invest. 2012;35:413–418. doi: 10.3275/7907. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, et al. Upregulation of the signal transducers and activators of transcription 3 (STAT3) pathway in lymphatic metastases of papillary thyroid cancer. Int J Clin Exp Pathol. 2011;4:356–362. [PMC free article] [PubMed] [Google Scholar]
- 43.Studebaker AW, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- 44.Leslie K, et al. Differential interleukin-6/Stat3 signaling as a function of cellular context mediates Ras-induced transformation. Breast Cancer Res. 2010;12:R80. doi: 10.1186/bcr2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JI, Strock CJ, Ball DW, Nelkin BD. The Ras/Raf/MEK/extracellular signal-regulated kinase pathway induces autocrine-paracrine growth inhibition via the leukemia inhibitory factor/JAK/STAT pathway. Mol Cell Biol. 2003;23:543–554. doi: 10.1128/MCB.23.2.543-554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borrello MG, et al. Induction of a proinflammatory program in normal human thyrocytes by the RET/PTC1 oncogene. Proc Natl Acad Sci USA. 2005;102:14825–14830. doi: 10.1073/pnas.0503039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engelmann D, et al. Transcriptome analysis in mouse tumors induced by Ret-MEN2/FMTC mutations reveals subtype-specific role in survival and interference with immune surveillance. Endocr Relat Cancer. 2009;16:211–224. doi: 10.1677/ERC-08-0158. [DOI] [PubMed] [Google Scholar]
- 48.Puxeddu E, et al. RET/PTC-induced gene expression in thyroid PCCL3 cells reveals early activation of genes involved in regulation of the immune response. Endocr Relat Cancer. 2005;12:319–334. doi: 10.1677/erc.1.00947. [DOI] [PubMed] [Google Scholar]
- 49.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flowers LO, Subramaniam PS, Johnson HM. A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene. 2005;24:2114–2120. doi: 10.1038/sj.onc.1208437. [DOI] [PubMed] [Google Scholar]
- 51.Vaccaro A, Chen H, Kunnimalaiyaan M. In-vivo activation of Raf-1 inhibits tumor growth and development in a xenograft model of human medullary thyroid cancer. Anticancer Drugs. 2006;17:849–853. doi: 10.1097/01.cad.0000217424.36961.47. [DOI] [PubMed] [Google Scholar]
- 52.Kim EJ, Park JI, Nelkin BD. IFI16 is an essential mediator of growth inhibition, but not differentiation, induced by the leukemia inhibitory factor/JAK/STAT pathway in medullary thyroid carcinoma cells. J Biol Chem. 2005;280:4913–4920. doi: 10.1074/jbc.M410542200. [DOI] [PubMed] [Google Scholar]
- 53.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 54.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinoue T, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wegrzyn J, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demaria M, et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demaria M, et al. STAT3 can serve as a hit in the process of malignant transformation of primary cells. Cell Death Differ. 2012;19:1390–1397. doi: 10.1038/cdd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Q, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 60.Gu Q, Kong Y, Yu ZB, Bai L, Xiao YB. Hypoxia-induced SOCS3 is limiting STAT3 phosphorylation and NF-κB activation in congenital heart disease. Biochimie. 2011;93:909–920. doi: 10.1016/j.biochi.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 61.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- 64.Faustino A, et al. mTOR pathway overactivation in BRAF mutated papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97:E1139–E1149. doi: 10.1210/jc.2011-2748. [DOI] [PubMed] [Google Scholar]
- 65.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 66.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]