Abstract
Myostatin and activin A are structurally related secreted proteins that act to limit skeletal muscle growth. The cellular targets for myostatin and activin A in muscle and the role of satellite cells in mediating muscle hypertrophy induced by inhibition of this signaling pathway have not been fully elucidated. Here we show that myostatin/activin A inhibition can cause muscle hypertrophy in mice lacking either syndecan4 or Pax7, both of which are important for satellite cell function and development. Moreover, we show that muscle hypertrophy after pharmacological blockade of this pathway occurs without significant satellite cell proliferation and fusion to myofibers and without an increase in the number of myonuclei per myofiber. Finally, we show that genetic ablation of Acvr2b, which encodes a high-affinity receptor for myostatin and activin A specifically in myofibers is sufficient to induce muscle hypertrophy. All of these findings are consistent with satellite cells playing little or no role in myostatin/activin A signaling in vivo and render support that inhibition of this signaling pathway can be an effective therapeutic approach for increasing muscle growth even in disease settings characterized by satellite cell dysfunction.
Keywords: activin receptors, GDF-8, follistatin
Myostatin (MSTN) is a transforming growth factor-β family member that acts as a negative regulator of skeletal muscle mass. Mstn−/− mice exhibit an approximate doubling of skeletal muscle mass throughout the body as a result of a combination of increased numbers of muscle fibers and increased muscle fiber sizes (1). The function of myostatin is highly conserved among mammals: naturally occurring mutations in the MSTN gene resulting in increased muscling have been identified in cattle (2–5), sheep (6), dogs (7), and humans (8).
The identification of myostatin and its biological function immediately suggested the possibility that inhibitors of this pathway may have clinical applications for treating patients with muscle loss. Indeed, there has been considerable focus on elucidating the molecular mechanisms underlying myostatin activity, with the goal of identifying strategies for pharmacological intervention. A number of key regulatory components of this signaling system have been identified, including inhibitory extracellular binding proteins, such as follistatin (9), FSTL-3 (10), GASP-1/GASP-2 (11), and the myostatin propeptide (9, 12), as well as myostatin receptors, which include both the type II receptors, ACVR2 and ACVR2B (9, 13), and the type I receptors, most likely ALK4 and ALK5 (14). The elucidation of the myostatin regulatory system has led to the development of a wide range of myostatin inhibitors that are active in vivo, and postnatal elimination of myostatin activity in mice either by systemic administration of these inhibitors (13, 15–18) or by induced muscle-specific deletion of the Mstn gene (19) has been shown to cause significant growth of muscle fibers, demonstrating the critical importance of this pathway in limiting muscle growth in adult animals. Finally, the function of myostatin in muscle seems to be redundant with that of at least one other TGF-β family member (13, 20), including activin A (21).
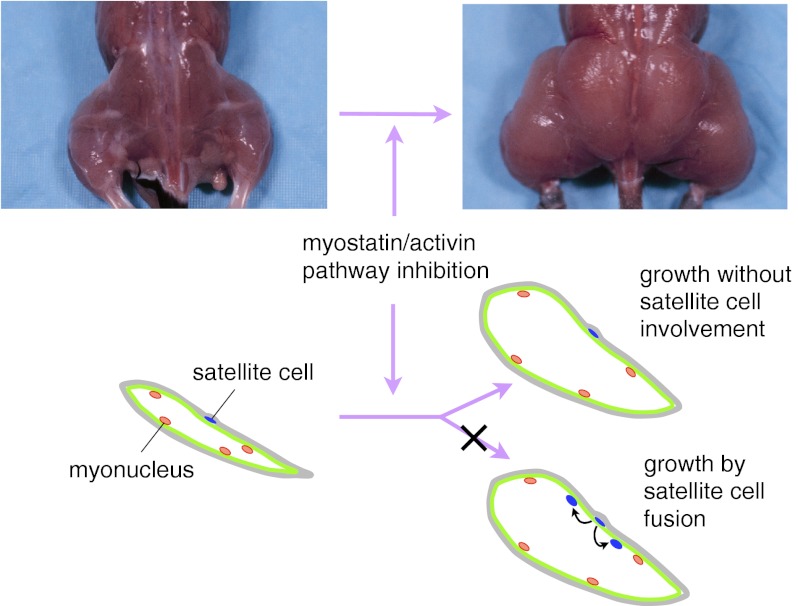
Although the fundamental role of myostatin and activin A in negatively regulating muscle growth has been firmly established, there is considerable debate as to the identities of their cellular targets. A critical question is whether these ligands act in vivo by signaling to satellite cells, which are the stem cells resident in adult muscle, or directly to myofibers. Defining the role of satellite cells in mediating myostatin/activin A signaling and the effects of myostatin/activin A inhibition is important not only for understanding the basic biology of skeletal muscle growth but also for pursuing clinical applications based on targeting this pathway. A major question has been whether therapies based on myostatin/activin A inhibition will have beneficial effects in clinical settings in which satellite cells are largely dormant or exhausted, such as in muscular dystrophy or age-related sarcopenia. In this regard, conflicting results have been reported in a number of studies that have examined the effect of myostatin/activin A loss or inhibition on satellite cells in vivo (22–27). Here, using various combinations of genetic and pharmacological approaches in mice, we examine the contribution of satellite cells to muscle hypertrophy induced by myostatin/activin A inhibition.
Results
If inhibition of myostatin/activin A activity results in muscle hypertrophy by causing activation and fusion of satellite cells to myofibers, one prediction is that the effect of blocking this pathway would be attenuated in mice in which satellite cells are defective. To test this prediction, we examined two mutant strains that have previously been reported to have defects in muscle regeneration.
We first examined the effect of blocking myostatin/activin A signaling in mice lacking syndecan4 (Sdc4). Sdc4 is expressed by satellite cells (28), and Sdc4−/− mice are relatively healthy with normal body weights (29) but exhibit a severe defect in muscle regeneration after chemical injury owing to a failure of satellite cell activation (30). We used both genetic and pharmacological approaches to determine the effect of blocking myostatin/activin A signaling in Sdc4−/− mice. For the genetic approach, we examined the effect of overexpressing the myostatin/activin A inhibitor follistatin by crossing Sdc4 mutants to F66 transgenic mice, which express follistatin under the control of a myosin light chain promoter and enhancer (20). We elected to use the F66 transgene rather than the Mstn deletion mutation in these studies for two reasons. First, we showed previously that whereas increases in both fiber numbers and fiber sizes contribute significantly to increased muscling in Mstn−/− mice (1), the increases in muscle mass seen in F66 transgenic mice result almost entirely from muscle fiber hypertrophy (20). Second, because follistatin has a relatively broad range of specificity in that it is capable of blocking multiple ligands, including both myostatin and activin A, we reasoned that the F66 transgenic approach would allow us to observe effects of inhibiting the general signaling pathway rather than inhibiting just myostatin.
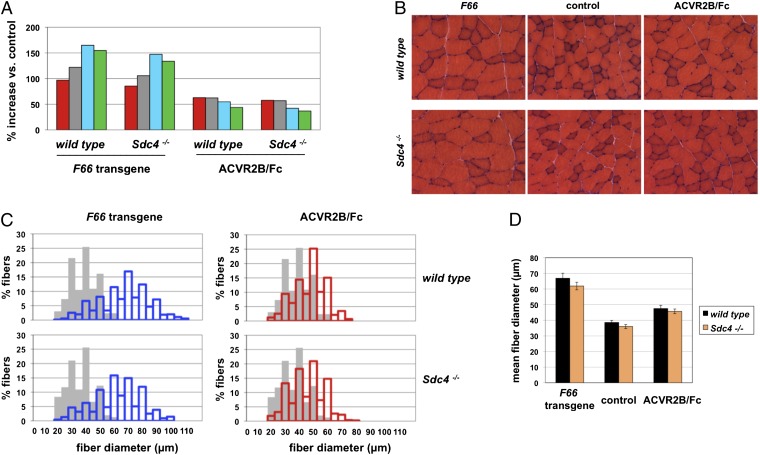
Consistent with our previous report, the F66 transgene in a WT background caused increases in muscle mass ranging from 97% to 165%, depending on the specific muscle group (Fig. 1A and Table 1), which resulted from dramatic increases in muscle fiber sizes (Fig. 1 B and C). Morphometric analysis of the gastrocnemius muscle showed that the mean fiber diameter in F66 transgenic mice was increased by 73% compared with that in WT mice (Fig. 1D). We observed similar effects of the F66 transgene in an Sdc4−/− background (Fig. 1A and Table 1). The magnitude of the increases in muscle weights caused by the F66 transgene in the Sdc4−/− background was comparable to that seen in a WT background. Moreover, the extent of muscle fiber hypertrophy induced by the F66 transgene in Sdc4−/− mice (Fig. 1B) was similar to that seen in a WT background in terms of the distribution of fiber sizes (Fig. 1C) and in terms of mean fiber diameter (Fig. 1D).
Fig. 1.
Effect of blocking the myostatin/activin A pathway in Sdc4−/− mice. (A) Percentage increase in weights of the pectoralis (red), triceps (gray), quadriceps (blue), and gastrocnemius/plantaris (green) muscles of WT and Sdc4−/− mice resulting either from the presence of the F66 transgene or from administration of ACVR2B/Fc (10 mg/kg for 4 wk). Actual weights, SEs, and P values are shown in Table 1. (B) Hematoxylin and eosin-stained sections prepared from gastrocnemius muscles showing the hypertrophy induced by the F66 transgene and by ACVR2B/Fc. (C) Distribution of fiber sizes in gastrocnemius muscles of mice carrying the F66 transgene (blue bars) or injected with ACVR2B/Fc (red bars) compared with control mice (gray bars) either lacking the transgene or injected with vehicle. (D) Mean fiber diameters in the gastrocnemius muscles.
Table 1.
Effect of the follistatin transgene (F66) and the soluble ACVR2B receptor (ACVR2B/Fc) on muscle weights (mg) in Sdc4 mutant mice
| Variable | Pectoralis | Triceps | Quadriceps | Gastrocnemius |
| Sdc4+/+ (n = 11) | 72.6 ± 1.6 | 94.2 ± 2.0 | 190.6 ± 3.7 | 134.9 ± 2.3 |
| Sdc4+/+ + F66 (n = 13) | 143.0 ± 4.3* | 209.1 ± 9.5* | 504.9 ± 17.6* | 343.7 ± 9.6* |
| Effect of F66 (%) | +96.9 | +122.0 | +164.9 | +154.7 |
| Sdc4−/− (n = 12) | 71.2 ± 1.9 | 92.3 ± 2.1 | 182.4 ± 3.7 | 128.2 ± 2.2 |
| Sdc4−/− + F66 (n = 14) | 131.9 ± 4.8* | 189.9 ± 8.4* | 450.9 ± 18.5*,† | 299.5 ± 9.3*,‡ |
| Effect of F66 (%) | +85.4 | +105.7 | +147.2 | +133.7 |
| Sdc4+/+ + PBS (n = 3) | 65.3 ± 2.2 | 86.0 ± 3.1 | 171.7 ± 8.0 | 128.3 ± 4.5 |
| Sdc4+/+ + ACVR2B/Fc (n = 3) | 106.3 ± 7.6§ | 139.7 ± 10.4§ | 265.7 ± 17.0§ | 184.3 ± 14.9§ |
| Effect of ACVR2B/Fc (%) | +62.8 | +62.4 | +54.8 | +43.6 |
| Sdc4−/− + PBS (n = 3) | 69.3 ± 2.1 | 91.0 ± 3.2 | 188.7 ± 5.9 | 136.3 ± 2.9 |
| Sdc4−/− + ACVR2B/Fc (n = 3) | 109.3 ± 2.9¶ | 143.0 ± 1.4¶ | 267.7 ± 3.9¶ | 186.3 ± 7.1‖ |
| Effect of ACVR2B/Fc (%) | +57.7 | +57.1 | +41.9 | +36.7 |
*P < 0.001 vs. Sdc4+/+;†P < 0.05 vs. Sdc4+/+ + F66;‡P < 0.01 vs. Sdc4+/+ + F66;§P < 0.05 vs. Sdc4+/+ + PBS;¶P < 0.001 vs. Sdc4−/− + PBS;‖P < 0.01 vs. Sdc4−/− + PBS.
For the pharmacological approach, we examined the effect of blocking myostatin/activin A in adult Sdc4−/− mice using a soluble form of the activin type IIB (ACVR2B) receptor in which the extracellular ligand binding domain was fused to an Fc domain (ACVR2B/Fc; ref. 13). We showed previously that ACVR2B/Fc is a potent myostatin/activin A inhibitor capable of inducing significant muscle growth when administered systemically to adult mice. As shown in Fig. 1 and Table 1, the ACVR2B/Fc inhibitor induced significant muscle hypertrophy not only in WT mice but also in Sdc4−/− mice. As in the case of the F66 transgene, the magnitude of the increases in muscle weights seen in Sdc4−/− mice (37–58%, depending on the specific muscle group analyzed) and the shift in the distribution in fiber sizes resulting from ACVR2B/Fc administration were comparable to those observed in WT mice (Fig. 1 and Table 1). Hence, muscle hypertrophy induced by myostatin/activin A inhibition either by follistatin or by ACVR2B/Fc is Sdc4 independent.
It is possible that muscle hypertrophy induced by myostatin/activin A inhibition is dependent on satellite cell activity that is distinct from Sdc4-dependent activity required for muscle regeneration after chemical injury. For this reason, we examined the effect of myostatin/activin A inhibition in a mouse strain in which satellite cells are severely depleted, namely, Pax7−/− mice. Unlike Sdc4−/− mice, Pax7−/− mice are considerably smaller than WT mice and mostly die within the first 2 wk after birth (31). Skeletal muscles of Pax7−/− mice are significantly smaller overall and contain fibers with reduced diameters (32).
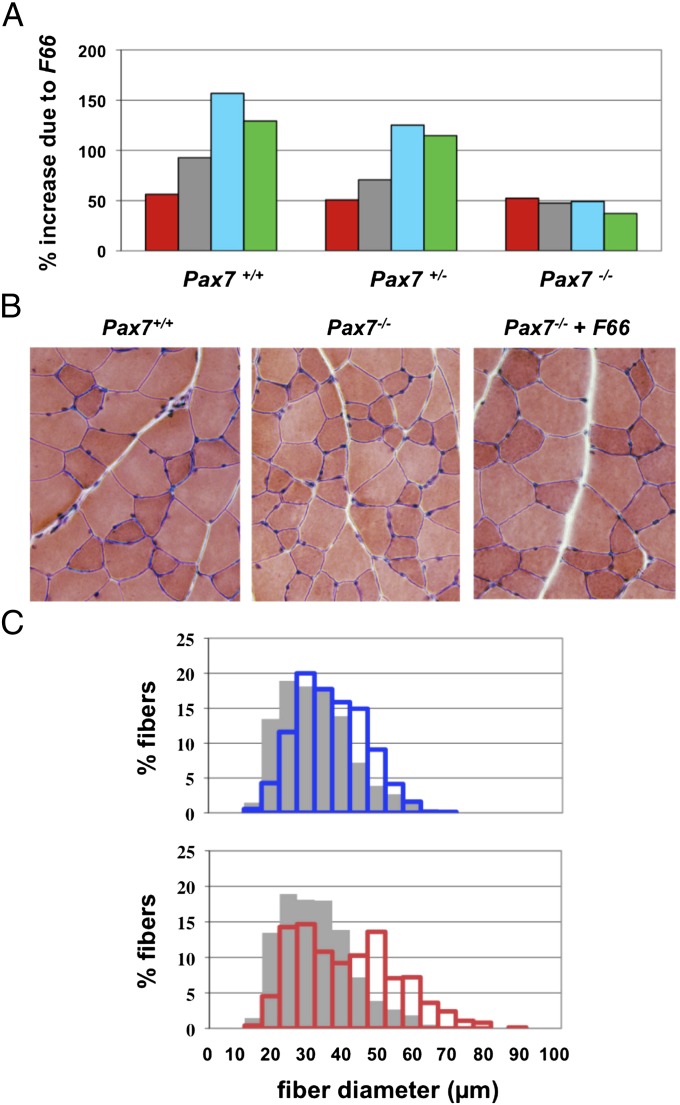
For our studies, we used a Pax7 mutant line that we generated independently by gene targeting. Specifically, we replaced most of the Pax7 coding sequence with an rtTA tetracycline activator cassette (Fig. S1). Consistent with previous reports, mice homozygous for our Pax7 mutant allele were severely wasted, and most died before weaning. Some mutant animals on a hybrid background, however, were viable to adulthood, and these mice were small and had muscle weights that were reduced by 43–55% compared with WT mice (Table 2). As in the Sdc4 studies, we examined the effect of overexpressing follistatin in Pax7−/− mice by crossing in the F66 transgene. As shown in Fig. 2A and Table 2, the F66 transgene had a significant effect even in the absence of Pax7, with muscle weights in F66, Pax7−/− mice being 37–52% higher than those of Pax7−/− mice. Analysis of muscle sections revealed significant hypertrophy induced by the F66 transgene (Fig. 2B), and morphometric analysis revealed a clear shift in the distribution of fiber sizes toward larger fibers (Fig. 2C). The F66 transgene affected fibers throughout the size spectrum. At the lower end of the spectrum, less than 5% of fibers in F66, Pax7−/− mice had diameters ≤ 20 μm compared with approximately 15% of fibers in Pax7−/− mice. At the higher end of the spectrum, nearly 36% of fibers in F66, Pax7−/− mice had diameters ≥50 μm, compared with less than 9% of fibers in Pax7−/− mice. Overall, the relative effect of the F66 transgene was lower in Pax7−/− mice than in WT mice, which was not unexpected given the severe muscle phenotype resulting from the absence of Pax7. Nevertheless, inhibition of myostatin/activin A by follistatin clearly caused significant increases in muscle growth in the mutant mice.
Table 2.
Effect of the follistatin transgene (F66) on muscle weights (mg) in Pax7 mutant mice
| Variable | Pectoralis | Triceps | Quadriceps | Gastrocnemius |
| Pax7+/+ (n = 12) | 81.8 ± 3.4 | 107.7 ± 4.5 | 212.3 ± 8.9 | 144.0 ± 5.7 |
| Pax7+/+ + F66 (n = 13) | 127.8 ± 5.5* | 207.6 ± 14.2* | 545.3 ± 35.0* | 330.2 ± 21.1* |
| Effect of F66 (%) | +56.2 | +92.8 | +156.8 | +129.3 |
| Pax7+/− (n = 15) | 79.5 ± 1.7 | 108.0 ± 1.9 | 208.6 ± 3.5 | 140.7 ± 2.7 |
| Pax7+/− + F66 (n = 22) | 119.9 ± 3.3† | 184.5 ± 7.4† | 469.5 ± 18.5† | 302.0 ± 10.5† |
| Effect of F66 (%) | +50.7 | +70.8 | +125.1 | +114.7 |
| Pax7−/− (n = 9) | 40.1 ± 2.1* | 48.3 ± 1.7* | 115.0 ± 6.6* | 81.7 ± 4.9* |
| Pax7−/−+ F66 (n = 13) | 61.1 ± 2.5‡ | 71.3 ± 2.6‡ | 171.4 ± 5.9‡ | 112.1 ± 4.0‡ |
| Effect of F66 (%) | +52.2 | +47.5 | +49.0 | +37.2 |
*P < 0.001 vs. Pax7+/+;†P < 0.001 vs. Pax7+/−;‡P < 0.001 vs. Pax7−/−.
Fig. 2.
Effect of the F66 transgene in Pax7−/− mice. (A) Percentage increase in weights of the pectoralis (red), triceps (gray), quadriceps (blue), and gastrocnemius/plantaris (green) muscles of WT, Pax7+/−, and Pax7−/− mice resulting from the presence of the F66 transgene. Actual weights, SEs, and P values are shown in Table 2. (B) Hematoxylin and eosin-stained sections prepared from gastrocnemius muscles showing the hypertrophy induced by the F66 transgene in Pax7−/− mice. (C) Distribution of fiber sizes in gastrocnemius muscles of WT mice (blue bars) and Pax7−/− mice without (gray bars) or with (red bars) the F66 transgene.
These studies demonstrated that inhibition of the myostatin/activin A pathway can lead to significant muscle hypertrophy even in mice severely compromised for satellite cell function (i.e., Sdc4 mutants) or number (i.e., Pax7 mutants). To complement these satellite cell deficiency studies, we next investigated the role of satellite cells in mediating the effect of myostatin/activin A inhibition by directly monitoring the behavior and fate of satellite cells after pharmacological blockade of this pathway. For these studies, we used an inducible cell marking-lineage tracing system for adult satellite cells (33). We have previously shown that combining a Pax7-CreERT2 knockin allele (Pax7CE) with a Cre reporter LacZ allele (R26RLacZ; ref. 33) allows tamoxifen-inducible permanent marking of PAX7-positive satellite cells, which can be easily identified by X-gal staining. Moreover, lineage tracing of these marked cells revealed that they are capable of fusing to myofibers either during the perinatal period or after injury in adult mice, which can be visualized by X-gal staining in myofibers.
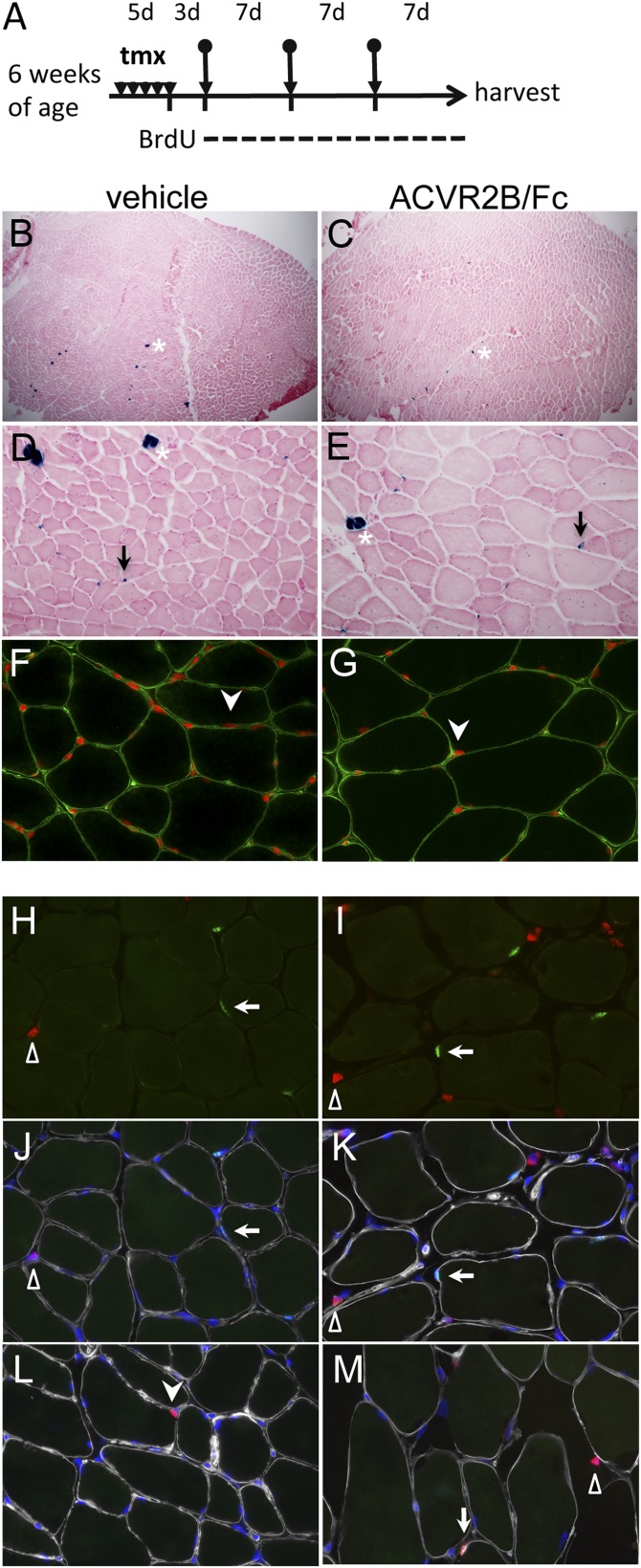
We examined the effect of myostatin/activin A inhibition by treating these mice with ACVR2B/Fc (scheme outlined in Fig. 3A). Three days after tamoxifen-induced cell marking, we initiated three weekly i.p. injections of ACVR2B/Fc and killed the mice 1 wk after the last injection. As expected, systemic administration of the soluble receptor to tamoxifen-treated Pax7CE, R26RLacZ mice caused significant muscle growth of all muscle groups examined (Table S1). We focused our analysis on the tibialis anterior (TA), where this marking-tracing system has been extensively characterized. TA muscles showed a 35% increase in wet weight upon treatment with the soluble receptor. Despite this dramatic muscle hypertrophy, however, we did not observe any significant fusion of satellite cells into the myofibers as assessed by X-gal staining (Fig. 3 B–E). Consistent with the very low number of X-gal–stained myofibers, the number of X-gal–stained satellite cells relative to total myofibers was similar in ACVR2B/Fc and PBS injected mice (Table 3).
Fig. 3.
No substantial myofiber incorporation from satellite cells after ACVR2B/Fc-induced muscle hypertrophy. (A) Schema of experimental time line. Bullet-arrows indicate the time of vehicle or ACVR2B/Fc administration; d, day; tmx, tamoxifen. Vehicle and ACVR2B/Fc-treated samples are indicated at top. (B–E) X-gal stained (blue) TA muscle sections from lineage-labeled Pax7CE/+, R26RLacZ mice treated with vehicle (B and D) and ACVR2B/Fc (C and E), counterstained with Nuclear Fast Red; B and C, low magnification; D and E, high magnification; asterisks, spindle myofibers; arrows, β-gal+ satellite cells. (F and G) Immunostaining of dystrophin (green) to delineate myofiber boundaries for identifying myonuclei (arrowheads) stained by DAPI (red). (H–M) Immunostaining of PAX7 (green), BrdU (red), and Laminin (white), counterstained with DAPI (blue); H and I, overlaid images of PAX7 and BrdU; J and K, same images further overlaid with Laminin and DAPI; L and M, rare examples of a PAX7+BrdU+ satellite cell and a PAX7−BrdU+ myonucleus, respectively. For H–M, arrows, filled arrowheads, and open triangles indicate satellite cells, myonuclei, and interstitial cells, respectively. Color scheme for antigens is the same as in H–K.
Table 3.
Myonuclei and satellite cells in ACVR2B/Fc-injected mice
| Variable | PBS | ACVR2B/Fc |
| X-gal stained sections* | ||
| Total myofibers | 3,893 ± 411 | 2,694 ± 480 |
| LacZ+ myofibers | 6.0 ± 2.9 | 5.0 ± 4.0 |
| LacZ+ mononuclear cells | 156.3 ± 6.3 | 96.5 ± 6.8 |
| LacZ+ mononuclear cells/myofiber | 0.041 ± 0.003 | 0.038 ± 0.007 |
| DAPI/dystrophin stained sections† | ||
| Total myofibers | 1,094 ± 132 | 823 ± 183 |
| Total myonuclei | 580 ± 75 | 425 ± 76 |
| Myonuclei/myofiber | 0.523 ± 0.22 | 0.511 ± 0.21 |
| % Central nucleated fibers | 0.28 ± 0.08 | 0.26 ± 0.10 |
*n = 4 mice per group; quantification of eight representative fields per muscle.
†n = 3 mice per group; quantification of 15 representative fields per muscle.
Thus, both the satellite cell functional deficiency mouse models and the forward satellite cell marking strategy were consistent with myostain/activin A inhibition-induced muscle hypertrophy requiring minimal input from satellite cells. To further confirm these findings, we used two additional methods to assess the extent of satellite cell fusion to myofibers in mice treated with ACVR2B/Fc. First, we quantified numbers of myonuclei and central nucleated fibers by staining sections with both DAPI and dystrophin. As shown in Fig. 3 F and G and Table 3, we found no significant differences in either the number of myonuclei (i.e., DAPI+ nuclei within the dystrophin boundary) per myofiber or the percentage of central nucleated fibers in mice injected with ACVR2B/Fc compared with mice injected with vehicle. Second, we modified the experimental scheme outlined in Fig. 3A by feeding mice with BrdU supplemented in drinking water after the first ACVR2B/Fc injection and throughout the duration of the study. As shown in Fig. 3 H and I, nuclei in which BrdU had been incorporated into the newly replicated DNA were readily detected. To identify newly divided satellite cells, we coimmunostained for PAX7, and to identify BrdU-positive myonuclei (Pax7-negative), we coimmunostained for laminin to outline the myofibers. As shown in Fig. 3 H–M and Table 4, we detected very few BrdU-positive satellite cells and very few BrdU-positive myonuclei in muscles of mice injected with ACVR2B/Fc. Hence, the results of all of these experiments indicate that the soluble receptor caused significant muscle hypertrophy in these mice, seemingly with very little satellite cell recruitment.
Table 4.
Numbers of Pax7- and BrdU-positive cells in ACVR2B/Fc-injected mice
| Mononuclear cells |
Myofibers |
||||
| Cells counted | Pax7+ | BrdU+ | Pax7+/BrdU+ | Total analyzed | BrdU+ |
| PBS sample 1 | 102 | 79 | 0 | 1,263 | 0 |
| PBS sample 2 | 104 | 61 | 0 | 1,358 | 1 |
| PBS sample 3 | 105 | 62 | 1 | 1,417 | 1 |
| ACVR2B/Fc sample 1 | 104 | 438 | 1 | 1,353 | 2 |
| ACVR2B/Fc sample 2 | 101 | 294 | 2 | 847 | 1 |
| ACVR2B/Fc sample 3 | 101 | 288 | 1 | 863 | 1 |
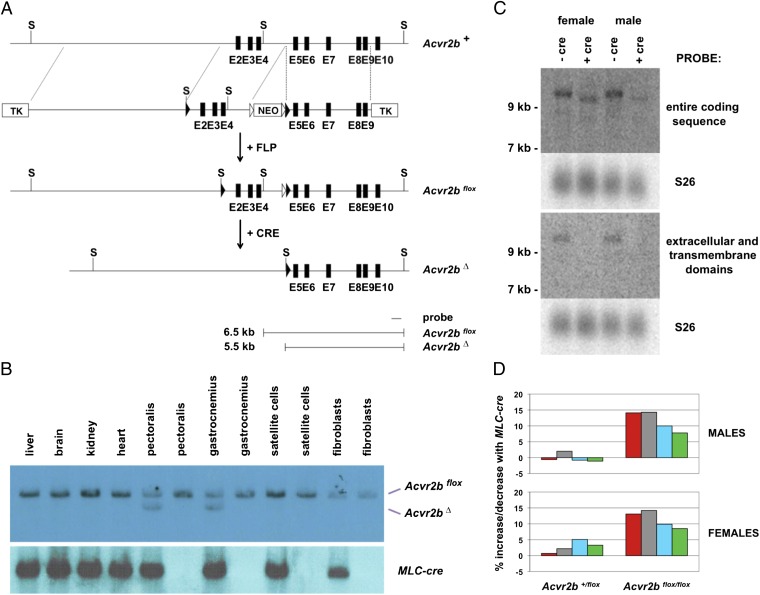
Given the lack of evidence for satellite cell contribution to muscle hypertrophy induced by myostatin/activin A inhibition, we tested whether blocking myostatin/activin A signaling in myofibers is sufficient to induce muscle hypertrophy. Specifically, we examined the effect of targeting myostatin/activin A receptors in myofibers. In previous studies we had shown that myostatin is capable of binding the two activin type II receptors, ACVR2 and ACVR2B, but has a higher affinity for ACVR2B (9). ACVR2B has also been shown to be a high-affinity receptor for activin A (34). To ablate ACVR2B function selectively in myofibers, we generated mice carrying a conditional deletion allele for Acvr2b, in which we flanked exons 2–4 with LoxP sites (Fig. 4A). Our rationale was that removal of exons 2–4 by cre-mediated recombination would delete the entire ligand-binding domain, the transmembrane domain, and part of the cytoplasmic domain and would put the remaining downstream coding sequence out of frame. Hence, deletion of exons 2–4 would almost certainly result in a null allele. We then targeted Acvr2b in myofibers by breeding mice carrying the floxed allele to transgenic mice expressing cre recombinase from a myosin light chain promoter and enhancer (MLC-cre; ref. 35).
Fig. 4.
Effect of targeting Acvr2b in myofibers. (A) Gene targeting strategy. Locations of exons 2–10 (E2–E10) are shown as black boxes, FRT (Flippase recognition target) sites are denoted by open triangles, and LoxP sites are denoted by filled triangles. The neo cassette was removed by crossing mice carrying the targeted allele with transgenic mice expressing FLP (Flippase) recombinase in the germline, resulting in an Acvr2bflox allele. Recombination between the LoxP sites results in a deletion allele (Acvr2bΔ) lacking exons 2–4. Southern blot analysis of genomic DNA digested with ScaI (S) and hybridized using the probe indicated is predicted to give 6.5-kb and 5.5-kb fragments for the Acvr2bflox and Acvr2bΔ alleles, respectively. (B) Southern blot analysis of genomic DNA isolated from various tissues or cell preparations (as indicated) using either the probe shown in A or a probe for the MLC-cre transgene. (C) Northern blot analysis of total RNA (25 μg) isolated from the gastrocnemius muscle using probes corresponding either to the entire coding sequence or to the extracellular and transmembrane domains of ACVR2B. Blots were reprobed with S26 (ribosomal protein) as a loading control. (D) Percentage increase or decrease in weights of the pectoralis (red), triceps (gray), quadriceps (blue), and gastrocnemius/plantaris (green) muscles of Acvr2b+/flox or Acvr2bflox/flox mice resulting from the presence of the MLC-cre transgene. Actual weights, SEs, and P values are shown in Table 5.
To determine the specificity of the MLC-cre transgene in directing recombination of the floxed Acvr2b allele, we carried out Southern analysis of DNA isolated from various tissues. Recombination between the LoxP sites was detected only in skeletal muscles of mice carrying the MLC-cre transgene (Fig. 4B); in these muscles, approximately half of the genomic DNA had undergone recombination, which is the maximal amount that would be predicted for myofiber-specific targeting, given the known fraction of myonuclei relative to total nuclei in muscle (36). As further confirmation that recombination in muscle was restricted to the myofibers, we freshly isolated satellite cells from these animals (Materials and Methods). Using this procedure, we obtained preparations of cells that were 88–89% Pax7-positive. We also harvested fibroblasts during the satellite cell isolation procedure for an additional negative control. As shown in Fig. 4B, no recombination was detected in either satellite cells or fibroblasts from MLC-cre mice, supporting that the MLC-cre transgene directs efficient recombination specifically in myofibers. We further examined the efficiency of recombination by Northern blot analysis of muscle RNA isolated from Acvr2bflox/flox mice either with or without the MLC-cre transgene. As shown in Fig. 4C, a probe corresponding to the entire coding sequence of ACVR2B detected a single RNA species with an apparent size of greater than 9 kb in Acvr2bflox/flox mice lacking cre. In Acvr2bflox/flox mice carrying the MLC-cre transgene, the probe detected a smaller RNA with an electrophoretic mobility consistent with cre-mediated deletion of exons 2–4. Moreover, unlike the full-length RNA, the smaller RNA species was not detected using a probe corresponding to the extracellular and transmembrane domains of the ACVR2B. These results demonstrate that the MLC-cre transgene was effective in inducing recombination of the Acvr2bflox allele, and the absence of the full-length transcript in muscles of Acvr2bflox/flox, MLC-cre mice further suggests that myofibers are the primary site of Acvr2b expression in muscle.
We then assessed the effect of eliminating ACVR2B in myofibers by comparing muscle weights in Acvr2b+/flox and Acvr2bflox/flox mice with and without the MLC-cre transgene. As shown in Fig. 4D and Table 5, mice that were homozygous for Acvr2bflox and that also carried the MLC-cre transgene exhibited statistically significant differences in muscle size, with muscle weights being increased by 8–14% in males and 9–14% in females, depending on the specific muscle group. Given that we have previously shown that ACVR2B is functionally redundant with the other type II activin receptor, namely ACVR2, in terms of regulation of muscle growth (13), we presume that targeting both receptors in myofibers would lead to an enhanced response. Nevertheless, our data show that eliminating even just ACVR2B function in myofibers is sufficient to induce muscle hypertrophy, consistent with myofibers being a target for myostatin/activin A signaling.
Table 5.
Effect of the targeting Acvr2b in myofibers on muscle weights (mg)
| Variable | Pectoralis | Triceps | Quadriceps | Gastrocnemius |
| Males | ||||
| Acvr2b+/flox (n = 23) | 71.3 ± 2.1 | 91.0 ± 2.3 | 189.1 ± 5.6 | 129.2 ± 4.1 |
| Acvr2b+/flox + MLC-cre (n = 28) | 70.9 ± 1.7 | 92.8 ± 2.1 | 187.6 ± 4.2 | 127.8 ± 2.7 |
| Effect of MLC-cre (%) | −0.6 | +2.0 | −0.8 | −1.1 |
| Acvr2bflox/flox (n = 27) | 70.2 ± 1.7 | 92.4 ± 1.9 | 191.9 ± 4.4 | 130.4 ± 2.6 |
| Acvr2bflox/flox + MLC-cre (n = 20) | 80.1 ± 1.9*,† | 105.6 ± 2.3†,‡ | 211.1 ± 4.8*,§ | 140.6 ± 3.2¶,‖ |
| Effect of MLC-cre (%) | +14.1 | +14.3 | +10.0 | +7.8 |
| Females | ||||
| Acvr2b+/flox (n = 15) | 44.2 ± 1.5 | 66.7 ± 1.7 | 128.5 ± 2.9 | 87.2 ± 2.3 |
| Acvr2b+/flox + MLC-cre (n = 25) | 44.5 ± 0.9 | 68.2 ± 1.1 | 135.0 ± 2.6 | 90.1 ± 1.7 |
| Effect of MLC-cre (%) | +0.7 | +2.2 | +5.1 | +3.3 |
| Acvr2bflox/flox (n = 21) | 42.7 ± 1.2 | 63.5 ± 1.0 | 129.6 ± 2.3 | 86.7 ± 1.4 |
| Acvr2bflox/flox + MLC-cre (n = 31) | 48.3 ± 0.9†,¶ | 72.5 ± 0.9*,† | 142.4 ± 2.2†,‡ | 94.1 ± 1.7§,¶ |
| Effect of MLC-cre (%) | +13.1 | +14.2 | +9.9 | +8.5 |
*P < 0.01 vs. Acvr2b+/flox;†P < 0.001 vs. Acvr2bflox/flox;‡P < 0.001 vs. Acvr2b+/flox;§P < 0.01 vs. Acvr2bflox/flox;¶P < 0.05 vs. Acvr2b+/flox;‖P < 0.05 vs. Acvr2bflox/flox.
Discussion
Although considerable progress has been made in elucidating the regulatory and signaling mechanisms of myostatin at the molecular level, there is considerable controversy as to the cell types in muscle that are direct targets of myostatin signaling in vivo. A variety of studies have demonstrated that myostatin can regulate both proliferation and differentiation of myoblasts and satellite cells in culture (22, 37–43), and these findings, taken together with the phenotype of Mstn−/− mice, have led to the model that a major role for myostatin is to maintain satellite cells in quiescence. Consistent with this model, muscle hypertrophy induced by electroporation of a follistatin expression construct was found to be blunted in mice that had been subjected to local γ irradiation, implying a role for satellite cells in this process (24). Indeed, one study reported that muscles of mice lacking myostatin have increased numbers of satellite cells, as well as an increased proportion of satellite cells in an activated state (22), and a second recent study reported that pharmacological inhibition of the myostatin pathway in mice could induce satellite cell proliferation (26). Several other studies, however, reported that the increased muscle mass seen either in Mstn−/− mice (23), in muscles electroporated with a dominant negative Acvr2b expression construct (25), or in mice treated with a myostatin inhibitor (27) occurs in the absence of satellite cell activation and fusion. Moreover, both in vitro and in vivo studies have suggested that myostatin is capable of acting directly on the myofibers themselves. In particular, purified myostatin has been shown to be capable of inhibiting protein synthesis (39) and reducing myotube diameter (44) when added to differentiated myotubes in culture, and overstimulation of this pathway in mice by implantation of myostatin-expressing cells (45) or by electroporation of constructs expressing either myostatin (46) or a constitutively active type I receptor (25) has been shown to induce muscle fiber atrophy.
Here, we have used complementary approaches to investigate further the role of satellite cells in mediating the effects of blocking this pathway in vivo. In previous studies, we had shown that muscle growth is regulated by not only myostatin but by multiple members of the TGF-β family, including activin A (13, 20, 21). For the studies presented here, we attempted to maximize the effect of blocking this pathway in muscle by using inhibitors and approaches capable of targeting both myostatin and activin A. Specifically, we used a pharmacological approach in which we administered a soluble form of the ACVR2B receptor to adult mice and genetic approaches in which we either overexpressed follistatin as a transgene or ablated the Acvr2b gene specifically in muscle. First, we show that muscle hypertrophy induced by myostatin/activin A inhibition can occur in mice that are null for either Sdc4 or Pax7, both of which are deficient in satellite cell activity. Second, we show that pharmacological blockade of the myostatin/activin A pathway using ACVR2B/Fc can cause significant muscle growth with little or no fusion of satellite cells to the growing myofibers and with little or no change in the number of myonuclei per fiber. Finally, we show that blockade of myostatin/activin A signaling specifically in myofibers by genetically targeting the high-affinity receptor, ACVR2B, is sufficient to induce muscle hypertrophy, consistent with myofibers being a direct target for myostatin/activin A signaling. The results of all of these studies suggest that satellite activation and fusion to myofibers do not play a significant role in muscle growth induced solely by inhibition of the myostatin/activin A pathway in adult mice. Additional studies will be required to determine whether the role of myostatin/activin A signaling may be more complex in physiologic settings in which satellite cells are activated by other stimuli, such as after exercise or muscle injury; in such settings, for example, it is possible that inhibition of myostatin/activin A signaling to myofibers may influence satellite cell function either by release of secondary signals that act directly on activated satellite cells or by making the myofibers more permissive for satellite cell fusion.
These findings have direct implications not only for understanding the cellular mechanisms underlying muscle growth but also for targeting the myostatin/activin A pathway for clinical applications. Although inhibitors of this pathway are being pursued as potential therapeutic agents for a variety of different diseases leading to muscle loss, a critical question is whether this strategy will be effective in disease states in which the satellite cell population may already be compromised. For example, for diseases like muscular dystrophy, a widely held view is that although satellite cell activation can compensate for the degenerative process in early stages of the disease, this pool of cells is eventually depleted, thereby leading to an accelerated rate of disease progression at later stages of the disease. Similarly, satellite cell exhaustion has been proposed to be a major contributor to the muscle loss that occurs during aging. Our data indicate that satellite cells are not a primary target for myostatin/activin A signaling and suggest that myostatin and activin A regulate muscle homeostasis predominantly by signaling directly to myofibers. If this is the manner in which these ligands function, many different diseases affecting muscle should potentially be responsive to inhibition of this pathway irrespective of satellite cell loss or dysfunction in these disease settings.
Materials and Methods
All animal experiments were carried out in accordance with protocols that were approved by the Institutional Animal Care and Use Committees at the Johns Hopkins University School of Medicine, the Carnegie Institution for Science, or Baylor College of Medicine. Pax7 and Acvr2b targeting constructs were generated from phage clones isolated from a 129 SvJ genomic library. Embryonic stem cell targeting was carried out using either R1 (kindly provided by A. Nagy, R. Nagy, and W. Abramow-Newerly, Samuel Lunenfeld Research Institute, Toronto, Canada) or E14Tg2A (BayGenomics) cells. Blastocyst injections of targeted clones were carried out either by the Johns Hopkins Transgenic Core Facility or by the Baylor Genetically Engineered Mouse Core. F66 transgenic mice and Sdc4 mutant mice were maintained on a C57BL/6 background, and Pax7 mutant and Acvr2b flox mice were maintained on a hybrid C57BL/6 and 129/SvJ background. The ACVR2B/Fc fusion protein was expressed in Chinese hamster ovary cells, purified from the conditioned medium using a protein A Sepharose column, and administered by weekly i.p. injections. Control mice were injected with vehicle (PBS). Tamoxifen preparation, storage, and injection regimen were as previously described (33). BrdU was supplemented in the drinking water as previously described (47) immediately after the first ACVR2B/Fc and vehicle injections and for the entire duration.
For measurement of muscle weights, individual muscles from both sides of 10-wk-old mice were dissected, and the average weight was used for each muscle. Morphometric analysis of the gastrocnemius muscle was carried out as previously described (20). For plotting the distribution of fiber sizes, all data for a given genotype were pooled. X-gal histochemistry and immunostaining were carried out as previously described (33). Images were taken under a Nikon E800 scope with a Canon EO5 camera. Immunostaining was carried out with mouse anti-PAX7 (1:10; DSHB), rabbit anti-Laminin (1:2,000; Sigma), mouse anti-dystrophin (1:1,000; Abcam), and sheep anti-BrdU (1:1,000; AbCam) primary antibodies, followed by host-specific secondary antibodies conjugated with Alexa 488, 568, and 647 (all at 1:1,000; Molecular Probes). DAPI (0.4 μg/mL; Sigma) was used to stain nuclei before mounting in Fluoromount G (Southern Biotech). Images were taken under an Axioskope with a monochrome Axiocam. Image processing and quantification were performed by using Photoshop or Metamorph.
Satellite cell isolation was performed essentially as described by Sherwood et al. (48) using collagenase and dispase digestion of hindlimb muscles. Dissociated cells were filtered through 40-μm restrainers and plated onto Matrigel (BD Biosciences) coated dishes for 48 h in culture. They were then trypsinized and consecutively preplated twice on noncoated dish for 30 min each. Attached cells were designated fibroblasts, and the unattached cell fraction was designated satellite cells, both of which were immediately harvested for DNA isolation. A small fraction of satellite cells was plated onto four-well chamber slides coated with Matrigel for 6 h, fixed, and subjected to Pax7 immunostaining as described above to evaluate the enrichment of satellite cells.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants R01AR059685 (to S.-J.L.), R01AR060636 (to S.-J.L.), P01NS0720027 (to S.-J.L.), DP5OD009208 (to C.L.), R01HD032067 (to M.M.M.), and R01AR060042 (to C.-M.F.) and a grant from the Jain Foundation (to S.-J.L.).
Footnotes
Conflict of interest statement: Under a licensing agreement between Pfizer, Inc. and Johns Hopkins University, S.-J.L. is entitled to a share of royalty received by the University on sales of products related to myostatin. The terms of this arrangement are being managed by the University in accordance with its conflicts of interest policies.
This article is a PNAS Direct Submission.
See Author Summary on page 13902 (volume 109, number 35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206410109/-/DCSupplemental.
References
- 1.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.Grobet L, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 3.Kambadur R, Sharma M, Smith TPL, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 4.McPherron AC, Lee S-J. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grobet L, et al. Molecular definition of an allelic series of mutations disrupting the myostatin function and causing double-muscling in cattle. Mamm Genome. 1998;9:210–213. doi: 10.1007/s003359900727. [DOI] [PubMed] [Google Scholar]
- 6.Clop A, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 7.Mosher DS, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuelke M, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 9.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill JJ, et al. The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem. 2002;277:40735–40741. doi: 10.1074/jbc.M206379200. [DOI] [PubMed] [Google Scholar]
- 11.Hill JJ, Qiu Y, Hewick RM, Wolfman NM. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: A novel protein with protease inhibitor and follistatin domains. Mol Endocrinol. 2003;17:1144–1154. doi: 10.1210/me.2002-0366. [DOI] [PubMed] [Google Scholar]
- 12.Thies RS, et al. GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors. 2001;18:251–259. doi: 10.3109/08977190109029114. [DOI] [PubMed] [Google Scholar]
- 13.Lee SJ, et al. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci USA. 2005;102:18117–18122. doi: 10.1073/pnas.0505996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor β-like signaling pathway to block adipogenesis. Mol Cell Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittemore L-A, et al. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun. 2003;300:965–971. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 16.Wolfman NM, et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeBrasseur NK, et al. Myostatin inhibition enhances the effects of exercise on performance and metabolic outcomes in aged mice. J Gerontol A Biol Sci Med Sci. 2009;64:940–948. doi: 10.1093/gerona/glp068. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 2011;25:1653–1663. doi: 10.1096/fj.10-176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welle S, Bhatt K, Pinkert CA, Tawil R, Thornton CA. Muscle growth after postdevelopmental myostatin gene knockout. Am J Physiol Endocrinol Metab. 2007;292:E985–E991. doi: 10.1152/ajpendo.00531.2006. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ. Quadrupling muscle mass in mice by targeting TGF-β signaling pathways. PLoS ONE. 2007;2:e789. doi: 10.1371/journal.pone.0000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SJ, et al. Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 2010;24:1998–2008. doi: 10.1210/me.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amthor H, et al. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc Natl Acad Sci USA. 2009;106:7479–7484. doi: 10.1073/pnas.0811129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilson H, et al. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. Am J Physiol Endocrinol Metab. 2009;297:E157–E164. doi: 10.1152/ajpendo.00193.2009. [DOI] [PubMed] [Google Scholar]
- 25.Sartori R, et al. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–C1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, et al. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, McPherron AC. Myostatin inhibition induces muscle fibre hypertrophy prior to satellite cell activation. J Physiol. 2012;590:2151–2165. doi: 10.1113/jphysiol.2011.226001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornelison DDW, Filla MS, Stanley HM, Rapraeger AC, Olwin BB. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 29.Echtermeyer F, et al. Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J Clin Invest. 2001;107:R9–R14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornelison DDW, et al. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–2236. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7-/- mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- 32.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 33.Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attisano L, Wrana JL, Cheifetz S, Massagué J. Novel activin receptors: Distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell. 1992;68:97–108. doi: 10.1016/0092-8674(92)90209-u. [DOI] [PubMed] [Google Scholar]
- 35.McPherron AC, Huynh TV, Lee S-J. Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev Biol. 2009;9:24–32. doi: 10.1186/1471-213X-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmalbruch H, Hellhammer U. The number of nuclei in adult rat muscles with special reference to satellite cells. Anat Rec. 1977;189:169–175. doi: 10.1002/ar.1091890204. [DOI] [PubMed] [Google Scholar]
- 37.Thomas M, et al. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 38.Ríos R, Carneiro I, Arce VM, Devesa J. Myostatin regulates cell survival during C2C12 myogenesis. Biochem Biophys Res Commun. 2001;280:561–566. doi: 10.1006/bbrc.2000.4159. [DOI] [PubMed] [Google Scholar]
- 39.Taylor WE, et al. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab. 2001;280:E221–E228. doi: 10.1152/ajpendo.2001.280.2.E221. [DOI] [PubMed] [Google Scholar]
- 40.Langley B, et al. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- 41.Ríos R, Carneiro I, Arce VM, Devesa J. Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol. 2002;282:C993–C999. doi: 10.1152/ajpcell.00372.2001. [DOI] [PubMed] [Google Scholar]
- 42.Joulia D, et al. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res. 2003;286:263–275. doi: 10.1016/s0014-4827(03)00074-0. [DOI] [PubMed] [Google Scholar]
- 43.Wagner KR, Liu X, Chang X, Allen RE. Muscle regeneration in the prolonged absence of myostatin. Proc Natl Acad Sci USA. 2005;102:2519–2524. doi: 10.1073/pnas.0408729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trendelenburg AU, et al. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296:C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 45.Zimmers TA, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- 46.Durieux A-C, et al. Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology. 2007;148:3140–3147. doi: 10.1210/en.2006-1500. [DOI] [PubMed] [Google Scholar]
- 47.Lepper C, Partridge TA, Fan C-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherwood RI, et al. Isolation of adult mouse myogenic progenitors: Functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]