Abstract
Background
Despite their wide occurrence, cryptosporidiosis and giardiasis are considered neglected diseases by the World Health Organization. The epidemiology of these diseases and microsporidiosis in humans in developing countries is poorly understood. The high concentration of pathogens in raw sewage makes the characterization of the transmission of these pathogens simple through the genotype and subtype analysis of a small number of samples.
Methodology/Principal Findings
The distribution of genotypes and subtypes of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in 386 samples of combined sewer systems from Shanghai, Nanjing and Wuhan and the sewer system in Qingdao in China was determined using PCR-sequencing tools. Eimeria spp. were also genotyped to assess the contribution of domestic animals to Cryptosporidium spp., G. duodenalis, and E. bieneusi in wastewater. The high occurrence of Cryptosporidium spp. (56.2%), G. duodenalis (82.6%), E. bieneusi (87.6%), and Eimeria/Cyclospora (80.3%) made the source attribution possible. As expected, several human-pathogenic species/genotypes, including Cryptosporidium hominis, Cryptosporidium meleagridis, G. duodenalis sub-assemblage A-II, and E. bieneusi genotype D, were the dominant parasites in wastewater. In addition to humans, the common presence of Cryptosporidium spp. and Eimeria spp. from rodents indicated that rodents might have contributed to the occurrence of E. bieneusi genotype D in samples. Likewise, the finding of Eimeria spp. and Cryptosporidium baileyi from birds indicated that C. meleagridis might be of both human and bird origins.
Conclusions/Significance
The distribution of Cryptosporidium species, G. duodenalis genotypes and subtypes, and E. bieneusi genotypes in urban wastewater indicates that anthroponotic transmission appeared to be important in epidemiology of cryptosporidiosis, giardiasis, and microsporidiosis in the study areas. The finding of different distributions of subtypes between Shanghai and Wuhan was indicative of possible differences in the source of C. hominis among different areas in China.
Introduction
Cryptosporidium, Giardia, and microsporidia infections are significant causes of diarrhea in humans worldwide [1], [2]. Despite their wide occurrence, cryptosporidiosis and giardiasis are considered neglected diseases by the World Health Organization, largely due to a lack of studies in developing countries [3]. In addition to infecting humans, these parasites are found in a wide range of animals including domestic and wild animals. Humans can acquire Cryptosporidium infections through several transmission routes, such as direct contact with infected persons (anthroponotic transmission) or animals (zoonotic transmission) and ingestion of contaminated water or food [4]. Although the epidemiology of Giardia and microsporidia is less clear, potential sources of infections can be identified by comparing the distribution of genotypes of each parasite among different hosts [5], [6].
The host-adaptive nature of different species/genotypes of these parasites helps us understand the potential infection sources and transmission routes of the disease. Among the over 70 species and genotype of Cryptosporidium, five species are responsible for most human infections, including C. hominis infecting humans, C. parvum infecting ruminants, C. meleagridis infecting birds, C. felis infecting cats, and C. canis infecting dogs [7]. In contrast, giardiasis in humans and most other mammals is caused by Giardia duodenalis (also known as Giardia lamblia or Giardia intestinalis) [6]. Giardia duodenalis has at least eight genotypes, assemblages A-H. Among them, assemblages A and B infect humans and a broad range of other hosts, including livestock, cats, dogs, and wild mammals. Within the assemblage A, there are three major subtypes groups (sub-assemblages), A-I, A-II and A-III, with A-I mainly infecting most animals, A-II mainly infecting humans, and A-III mainly infecting wild ruminants. Assemblages C-H appear generally to be mostly restricted to companion animals, livestock, rodents, and seals [6]. Enterocytozoon bieneusi is the most common microsporidian species infecting humans, domestic animals, wild mammals, and birds [5], [8]. Enterocytozoon bieneusi forms several phylogenetic groups of genotypes, with group 1 infecting humans and most animals and groups 2–5 infecting different animals [1], [9]. Groups 2–5 represent host-adapted genotypes that have no major public health significance [10], [11]. Therefore, using both an understanding of the host-specificity of various pathogen species/subtypes and describing the most common pathogen species/subtypes in wastewater can shed light into the potential infections sources and transmission routes of diseases. For example, a common occurrence of the human-specific C. hominis rather than potentially zoonotic C. parvum would indicate that anthroponotic transmission is important in cryptosporidiosis transmission.
The monitoring of raw wastewater for pathogens has been used in surveillance of a few bacterial, viral and parasitic pathogens in urban communities [12]–[16]. The identification of pathogens in human specimens is still the standard method for the elucidation of disease transmission. However, because of the low prevalence of most pathogens, the screening of a large number of specimens is frequently needed, which is time-consuming and expensive. In contrast, the high concentration of pathogens in raw sewage facilities the detection of these pathogens via the analysis of a small number of samples and can provide a quick overall picture of the disease transmission at the community level, although some zoonotic pathogens in wastewater may also come from domestic animals. Therefore, this approach is especially useful for emerging infectious pathogens, such as E. coli O157:H7 and H5N1. Because some emerging infectious diseases are also endemic in urban communities in addition to causing large-scale outbreaks of illnesses, simple detection of the pathogens in raw wastewater is not enough for molecular surveillance of the pathogens. For these endemic pathogens, molecular epidemiologic tracking (subtyping) is frequently needed to assess the endemic transmission at the community level and to identify the occurrence of outbreaks, which are characterized by the presence of one or a few subtypes at high frequency.
In two U.S. studies, genotyping and subtyping parasites in raw wastewater was used as a rapid and cost-effective tool for characterizing the transmission of cryptosporidiosis and giardiasis in an urban community [15], [17]. In one study, promising results were obtained in molecular surveillance of an outbreak strain of C. hominis in urban wastewater [15]. Our previous study on species and subtype distribution of Cryptosporidium spp. in domestic wastewater in Shanghai, China suggested that anthroponotic transmission might play an important role in cryptosporidiosis epidemiology, and the dominant C. hominis subtypes in this region were different from those in other countries [16]. The objectives of this study were to identify the species/genotype distribution of Cryptosporidium spp., G. duodenalis, and E. bieneusi in domestic wastewater in four large cities in China, to assess endemic transmission of cryptosporidiosis, giardiasis and microsporidiosis based on the species or genotype identified and our knowledge on their host sources and geographic distribution, and to determine whether the unique subtypes of C. hominis identified in Shanghai also existed in other areas in China. Because domestic animals may contribute to parasite occurrence in urban wastewater, Eimeria spp., which commonly infect domestic mammals and birds and have strong host specificity, were also analyzed to facilitate the animal source attribution of Cryptosporidium, G. duodenalis, and E. bieneusi genotypes found in wastewater.
Materials and Methods
Wastewater sample collection and processing
A total of 386 raw wastewater samples were collected from four cities in China from 2006 to 2009, including 90 from four wastewater treatment plants (WWTPs) in Shanghai (December 2006 to April 2007), 87 from five WWTPs in Nanjing (July 2007 to May 2009), 109 from five WWTPs in Qingdao (April–July 2008), and 100 from five WWTPs in Wuhan (April–July 2008). All WWTPs in each city were located at areas with high population density. The four cities were selected based on the following criteria: (1) different population densities, with 3,631 people/km2 in Shanghai, which is the highest in mainland China, and 815 to 890 people/km2 in the other three cities; (2) different pet densities, with the highest in Shanghai; (3) different conditions of the sewage system, with the best in Shanghai and more antique in Qingdao. Qingdao has separate storm water and domestic wastewater systems, and the other three cities use combined sewer systems in the inner city areas. Grab samples of 500 to 1,000 ml of raw wastewater were collected at the input of WWTPs. Cysts, oocysts and spores of parasites in samples were concentrated by centrifugation at 6,000×g for 10 min. Samples were stored in 2.5% potassium dichromate solution at 4°C prior to DNA extraction.
DNA extraction
After washing the samples twice in distilled water, genomic DNA was extracted from 0.5 ml of concentrate using a FastDNA SPIN Kit for Soil (BIO 101, Carlsbad, CA) and eluted in 100 µl of reagent-grade water as described previously [18]. DNA was stored at −80°C until analyzed by PCR. DNA from each sample was analyzed at least five times for each genetic target using 2 µl of the extraction DNA in PCR. Following analysis with a nested PCR described below, secondary PCR products were detected by gel electrophoresis in 1.5% agarose.
Cryptosporidium detection, genotyping and subtyping
A ∼830-bp fragment of the small subunit (SSU) rRNA gene was amplified by nested PCR as previously described [19]. Cryptosporidium species and genotypes were differentiated by restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products using restriction enzymes SspI and VspI [20]. Cryptosporidium muris and C. andersoni were further differentiated by RFLP analysis using restriction enzymes DdeI [21]. To identify C. parvum, C. hominis and C. cuniculus subtypes, a ∼850-bp fragment of the 60 kDa glycoprotein (gp60) gene was amplified by nested PCR [16]. The secondary PCR products were sequenced using the secondary PCR primers and an intermediary sequencing primer (5′-GAGATATATCTTGTTGCG-3′). The established subtype nomenclature [22] was used to classify gp60 subtypes.
Giardia duodenalis detection, genotyping and subtyping
To detect G. duodenalis, a 530-bp fragment of the triosephosphate isomerase (tpi) gene was amplified by nested PCR [23]. Genotypes and subtypes of G. duodenalis were determined by sequence analysis of the secondary PCR products.
Enterocytozoon bieneusi detection and genotyping
To detect E. bieneusi, a ∼392-bp fragment of the ribosomal internal transcribed spacer (ITS) was amplified by nested PCR [10]. Genotypes of E. bieneusi were determined by sequence analysis of the secondary PCR products, and named according to the established nomenclature [24].
Detection and identification of Eimeria spp. and Cyclospora cayetanensis
To detect Eimeria spp. and Cyclospora spp., a 294-bp fragment of the SSU rRNA gene was amplified by nested PCR [25]. The differentiation of Eimeria and Cyclospora species were done by sequence analysis of the secondary PCR products.
DNA sequencing and data analysis
After being purified using the Montage PCR filters (Millipore, Bedford, MA), the secondary PCR products were sequenced directly with the secondary PCR primers using an ABI BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and the manufacturer-suggested procedures. Sequences were read on an ABI3130 Genetic Analyzer (Applied Biosystems). To confirm the accuracy of the sequence and to detect multiple genotypes and subtypes in a sample, all positive PCR products from each sample were sequenced, except for C. muris-positive samples in Qingdao, which was identified mostly by SspI and DdeI RFLP analysis, and for Eimeria/Cyclospora-postive samples, for which only one PCR product per sample was sequenced. The nucleotide sequences of Cryptosporidium spp., G. duodenalis, E. bieneusi, and Eimeria/Cyclospora spp. were aligned with reference sequences downloaded from the GenBank using ClustalX (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) to determine their genotype or subtype identity. Each pathogen genotype or subtype was presented as the number of positive samples or PCR products, with its frequency calculated based on the total number of samples analyzed in each area.
Phylogenetic analysis
The genotypes of Enterocytozoon bieneusi obtained were compared to known E. bieneusi genotypes by phylogenetic analysis of ITS sequences. After eliminating sequences with ambiguous positions, 579 E. bieneusi ITS sequences were obtained from wastewater samples, representing 23 genotypes. They were used in a neighbour-joining analysis together with reference sequences downloaded from the GenBank, using 500 replicates in bootstrap assessment of the robustness of clusters. Likewise, 298 clean SSU rRNA sequences of Eimeria/Cyclospora representing 39 genotypes were assigned to host groups based on their phylogenetic positions in a similarly constructed neighbour-joining tree with reference sequences from known animal hosts.
Nucleotide sequence accession numbers
Unique nucleotide sequences generated from the study were deposited in GenBank under accession numbers JQ863254-JQ863307.
Results
Cryptosporidium species/genotypes in wastewater
Based on PCR analysis of the SSU rRNA gene, 63 of 90 (70.0%) wastewater samples from Shanghai, 32 of 87 (36.8%) samples from Nanjing, 75 of 109 (68.8%) samples from Qingdao, and 47 of 100 (47.0%) samples from Wuhan were positive for Cryptosporidium spp. RFLP analysis and DNA sequencing of PCR products revealed the presence of 14 species/genotypes of Cryptosporidium in these samples: C. hominis, C. parvum, C. andersoni, C. canis, C. suis, C. cuniculus, C. meleagridis, C. baileyi, C. muris, pig genotype II, Avian genotype III, rat genotype I, rat genotype IV, and a new genotype (Table 1).
Table 1. Cryptosporidium species and genotypes in wastewater samples from four cities in China during 2006–2009.
| Species/genotype or gp60 subtype | Ref No. | No. of positive samples (No. of positive PCR products)a | |||
| Shanghaib | Nanjing | Qingdao | Wuhan | ||
| Humans | |||||
| C. hominis | EF570922 or AY204230 | 59 (78) | 6 (9) | 1 (1) | 19 (21) |
| IaA18R4 | FJ153246 | 3 (3) | |||
| IaA19R4 | FJ153245 | 3 (3) | |||
| IaA27R3 | AF164502 | 1 (1) | |||
| IbA19G2 | FJ707312 | 5 (6)c | 1 (1) | ||
| IbA20G2 | FJ707313 | 3 (3) | |||
| IbA21G2 | FJ153239 | 22 (35)c | |||
| IdA14 | GU214350 | 2 (2) | |||
| IdA16 | HQ149034 | 2 (3) | 1 (1) | 1 (1)d | |
| IeA11G3T3 | GU214354 | 1 (1) | |||
| IeA12G3T3 | FJ153242 | 6 (7) | 1 (1) | 1 (1) | 9 (12)d |
| IfA20G1 | FJ153243 | 1 (1) | |||
| IfA22G1 | FJ153244 | 4 (5) | |||
| Cryptosporidium W13314 | HM191258 | 1 (1) | |||
| Livestock and companion animals | |||||
| C. parvum | AF093490 | 1 (1) | 2 (3) | ||
| IIdA19G1 | HQ009809 | 2 (3) | |||
| C. andersoni | FJ463187 | 4 (5) | |||
| C. canis | EU754833 | 1 (2) | 1 (1) | ||
| C. suis | AF115377 or GQ345008 | 1 (2) | 7 (7) | 6 (7) | 17 (20) |
| Pig genotype II | AB449825 | 1 (1) | 5 (5) | ||
| C. cuniculus | EU437413 | 3 (3) | 2 (2) | ||
| VaA31 | FJ262734 | 1 (1) | |||
| Birds | |||||
| C. meleagridis | AF112574 | 7 (7) | 2 (2) | 7 (7) | 6 (6) |
| C. baileyi | AF093495 or EU717825 | 3 (3) | 6 (6) | 2 (2) | 4 (4) |
| Avian genotype III | HM116386 | 1 (1) | |||
| Rodents | |||||
| C. muris | AF093498 | 1 (1) | 18 (24) | 2 (2)e | 10 (10) |
| Rat genotype I | AY268584 | 1 (1) | 1 (1) | 2 (2) | 1 (2) |
| Rat genotype IV | AY737585, AY737580 or AY737584 | 8 (8) | 2 (2) | ||
| Totalf | 63/90 (70.0%) | 32/87 (36.8%) | 75/109 (68.8%) | 47/100 (47.0%) | |
All data on specific genotypes and subtypes are based on sequence analysis.
Cryptosporidium data from Shanghai wastewater samples were previously reported in [16].
Concurrent presence of IbA19G2 and IbA21G2 was found in one sample.
Concurrent presence of IdA16 and IeA12G3T3 was found in one sample.
A total of 60 (109) were positive for C. muris by SspI and DdeI RFLP analysis.
No. of positive samples/No. of samples (positivity in percentage) based on PCR analysis of the SSU rRNA gene.
Among the five major human-pathogenic Cryptosporidium species (C. hominis, C. parvum, C. meleagridis, C. canis, and C. felis), both the human-specific C. hominis and bird-adapted C. meleagridis were found in samples from all four cities. The occurrence of C. hominis was more common than C. meleagridis in Shanghai (59 versus seven), Nanjing (six versus two), and Wuhan (19 versus six). The only exception was Qingdao where C. hominis was found in only one sample while C. meleagridis was found in seven samples (Table 1). Similar to the common existence of C. meleagridis (22 samples), other species from birds, such as C. baileyi was also detected in 15 samples from the four cities studied. Cryptosporidium parvum, which is a common parasite of cattle, was found only in one sample in Shanghai and two samples in Qingdao. The rare occurrence of C. parvum was consistent with the frequency of other species infecting cattle, such as C. andersoni (found in four samples only). Cryptosporidium canis, commonly found dogs, was only seen in one sample each in Qingdao and Wuhan, where the common species in cats, C. felis, was not detected in any of the samples.
Surprisingly, C. muris and other Cryptosporidium genotypes infecting rodents were the most commonly detected species/genotypes in Qingdao, being found in 70 samples, or 93.3% of all Cryptosporidium-positive samples. Within C. muris from Qingdao, the detection in 12 samples was confirmed by DNA sequencing and the other 58 diagnosed by RFLP analysis with restriction enzymes SspI and DdeI (Table 1). Although these species/genotypes were also found at a high frequency in Nanjing (19 samples, or 59.4% of Cryptosporidium-positive samples), they were less commonly seen in Wuhan (13 or 27.7% Cryptosporidium-positive samples), and were only rarely seen in Shanghai (two samples, or 3.2% Cryptosporidium-positive samples) (Table 1).
The partial SSU rRNA gene sequence of a novel genotype in this study (FJ153238) was identical to that isolated from a patient in the United Kingdom (HM191258) [26]. A recently identified zoonotic species, C. cuniculus [27], which was responsible for a waterborne outbreak in the United Kingdom in 2008 [28], was also found in five samples in this study. The concurrent presence of multiple Cryptosporidium species/genotypes was seen in many samples: two species/genotypes in 44 samples, three species/genotypes in 13 samples, and four species/genotypes in six samples.
Cryptosporidium hominis, C. parvum and C. cuniculus gp60 subtypes
Samples positive for C. hominis, C. parvum or C. cuniculus were subtyped by sequence analysis of the gp60 gene. For C. hominis-positive samples, 47 of 59 samples in Shanghai, four of six samples in Nanjing, one sample in Qingdao, and 10 of 19 samples in Wuhan were successfully subtyped. Altogether, there were 12 subtypes in five subtype families in the 62 C. hominis-positive wastewater samples: Ia (three subtypes in seven samples), Ib (three subtypes in 30 samples), Id (two subtypes in six samples), Ie (two subtypes in 18 samples), and If (two subtypes in five samples) (Table 1). Although Ib subtypes were commonly detected in Shanghai samples (30/48 C. hominis-positive samples subtyped successfully), only one sample in Nanjing was positive for one of the Ib subtypes (IbA19G2). In contrast, the Ie subtype IeA12G3T3 that previously thought to be unique to Shanghai, was found in 11 samples in the other three cities. Of the five C. cuniculus-positive samples in Nanjing and Wuhan, only one was successfully subtyped as VaA31 (Table 1). Two of the three C. parvum-positive samples were subtyped as IIdA19G1 (Table 1). Concurrent presence of two subtypes was found in five samples, including IbA19G2 and IbA21G2 in one sample from Shanghai, and IdA16 and IeA12G3T3 in one sample from Wuhan. Except for samples from Qingdao, all C. meleagridis-positive samples were also analyzed by gp60 PCR. Only four of the seven samples from Shanghai and one of the six samples from Wuhan produced the expected gp60 PCR products, all of which belonged to C. hominis subtypes indicating the concurrent presence of C. hominis.
Giardia duodenalis genotypes and subtypes
The PCR analysis of tpi gene identified a high occurrence of G. duodenalis in the four cities studied, with positive rates of 67.8% (59/87) in Nanjing, 69.0% (69/100) in Wuhan, 94.5% (85/90) in Shanghai and 97.2% in Qingdao (106/109). All of them belonged to two known genotypes of G. duodenalis (assemblages A and B) of humans. The prevalent genotype was assemblage A, which was detected in 76.1% (243 of 319) G. duodenalis-positive samples, including 51 samples in Shanghai, 38 samples in Nanjing, 97 samples in Qingdao, and 57 samples in Wuhan (Table 2). Assemblage B was found in 59 samples, but in 53 of them it was in concurrence with assemblage A (Table 2). In 27 samples, the presence of mixed genotypes was identified by the occurrence different assemblages in different replicates of analysis while in the remaining 36 samples the presence of mixed genotypes was demonstrated by underlying peaks in DNA sequencing trace files. All samples containing assemblage A had the A2 subtype. Sixteen assemblage B subtypes were found in this study, only one of which was identical to a sequence reported before (GenBank No. L02116).
Table 2. Giardia duodenalis assemblages and genotypes in wastewater samples of four cities in China during 2006–2009.
| Genotype or subtype | Ref No. | No. of positive samples (No. of positive PCR products)a | |||
| Shanghai | Nanjing | Qingdao | Wuhan | ||
| Humans | |||||
| A2 | U57897 | 51 (105) | 38 (76) | 97 (180) | 57 (86) |
| Mammals (including humans) | |||||
| B1 | L02116 | 1 (1) | |||
| B2 | 17238-1 | 1 (1) | |||
| B3 | 17257-2 | 1 (1) | |||
| B4 | 17239-1 | 1 (1) | |||
| B5 | 19068-2 | 1 (1) | |||
| A2+B1b | U57897, L02116 | 1 (1) | |||
| A2+B2b | 17238-1 | 2 (5) | |||
| A2+B3b | 17257-2 | 3 (6) | |||
| A2+B4b | 17239-1 | 1 (2) | |||
| A2+B6b | 17219-2 | 1 (2) | |||
| A2+B7b | 18930-2 | 1 (2) | |||
| A2+B8b | 17231-2 | 1 (5) | |||
| A2+B9b | 18933-2 | 1 (2) | |||
| A2+B10b | 17252-1 | 1 (4) | |||
| A2+B11b | 18972-2 | 1 (2) | |||
| A2+B12b | 17259-1 | 1 (2) | |||
| A2+B13b | 19088-1 | 1 (2) | |||
| A2+B14b | 17241-2 | 1 (2) | |||
| A2+B15b | 19121-2 | 1 (2) | |||
| A2+Bc | 30 (38) | 6 (10) | |||
| B2+B16 | 17238-1, 19108-2 | 1 (2) | |||
| Totald | 85/90 (94.5%) | 59/87 (67.8%) | 106/109 (97.2%) | 69/100 (69.0%) | |
All data on specific genotypes and subtypes are based on sequence analysis.
Different sequencing results in different PCR replicate analyses of each sample. B2–B16 are new subtypes identified in this study.
Based on the occurrence of underlying peaks in DNA sequencing trace files; the subtype identity of assemblage B could not be established under such a circumstance.
No. of positive samples/No. of samples (positivity in percentage) based on PCR analysis of the tpi gene.
Enterocytozoon bieneusi genotypes
Enterocytozoon bieneusi was detected in more than 90% of samples in Shanghai, Qingdao and Wuhan, with prevalence lower in Nanjing (62.1% or 54/87 samples). In the 338 E. bieneusi-positive samples, a total of 23 ITS genotypes were seen in 579 sequences obtained (Table 3). Genotype D was the most prevalent, being found in 279 of 338 (82.5%) E. bieneusi-positive samples or 469 of 592 (79.2%) sequences.
Table 3. Enterocytozoon bieneusi ITS genotypes in wastewater samples of four cities in China during 2006–2009a.
| Genotype | Ref No. | Reported hosts in the literature | No. of positive samples (No. of positive PCR products)a | |||
| Shanghai | Nanjing | Qingdao | Wuhan | |||
| Humans and/or animals | Group 1a–1hb (Group I)c | |||||
| Type IV | AY371277 | Humans, cats, cattle and dogs | 4 (4) | 5 (5) | 3 (3) | |
| EbpC | AY371279 | Humans, pigs, beaver, otter, muskrat, raccon and fox | 2 (2) | 1 (1) | 5 (5) | |
| EbpD | AF076043 | Pigs | 1 (1) | |||
| Peru6 | AY371281 | Humans, dog, cattle, birds | 1 (1) | 3 (4) | 1(1) | |
| Peru8 | AY371283 | Humans | 1 (1) | 1 (1) | 11 (13) | |
| Peru11 | AY371286 | Humans | 2 (2) | 2 (2) | 6 (6) | |
| C | AF101199 | Humans | 1 (1) | |||
| D | AY371284 | Humans, pigs, cattle, dogs, beaver, falcons, fox, macaque, muskrat, raccoon | 78 (138) | 50 (99) | 71 (99) | 80 (133) |
| EbpA | AF076040 | Cattle, pigs | 2 (2) | |||
| PtEb IV | DQ885580 | Cats | 1 (1) | |||
| BEB6 | EU153584 | Cattle | 1 (1) | 1 (1) | ||
| PigEBITS7 | AF348475 | Pigs | 6 (7) | 1 (1) | ||
| PigEBITS8 | AF348476 | Pigs | 1 (1) | |||
| WL12 | AY237220 | Beaver, otter | 1 (1) | |||
| WL14 | AY237222 | Muskrat | 2 (2) | |||
| Unknown | Group 1id | |||||
| WW1e | 19031-2 | 4 (5) | ||||
| WW2e | 19055-2 | 1 (1) | ||||
| WW3e | 19083-1 | 15 (17) | 5 (6) | |||
| WW4e | 19063-2 | 3 (3) | ||||
| WW5e | 19054-1 | 1 (1) | ||||
| Muskrat and cat | Group 3b (Group Ic) | |||||
| WL4 | AY237212 | Muskrat | 1 (1) | 1 (1) | ||
| Unknown | Group 6d | |||||
| WW6e | 19081-2 | 4 (4) | ||||
| WW7e | 19107-1 | 1 (1) | ||||
| Dog | Outliersb (Group IVc) | |||||
| PtEb IX | DQ885585 | Dogs | 4 (4) | 3 (3) | ||
| WW8e | 19029-1 | 4 (5) | ||||
| WW9e | 19079-1 | 1 (1) | ||||
| Totalf | 85/90 (94.5%) | 54/87 (62.1%) | 100/109 (91.7%) | 99/100 (99.0%) | ||
All data on specific genotypes are based on sequence analysis.
Group name given by Thellier and Breton (2008).
Group name given by Henriques-Gil et al. (2010).
Group or subgroup newly identified in this study.
New genotype in this study.
No. of positive samples/No. of samples (positivity in percentage) based on PCR analysis of the ITS.
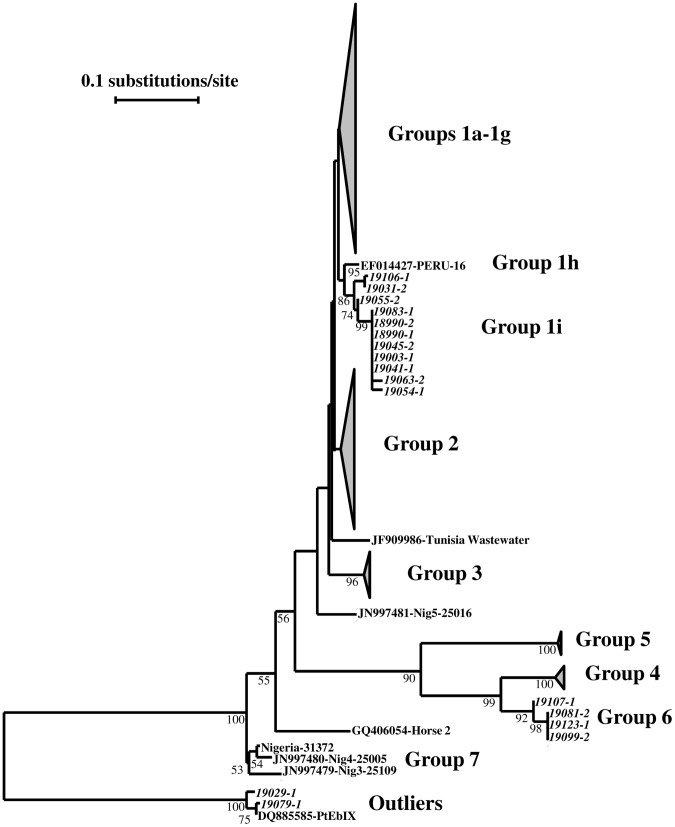
Two new groups of E. bieneusi genotypes were found in this study. A new group named 1i consisted of five genotypes seen in 33 sequences and formed a cluster sister to the previously designed subgroup 1 h [9]. This cluster was within the large group (group 1) of ITS sequences originated from humans and animals [1], [9], [29]. The other new group was named as group 6 in this study and consisted of two genotypes seen in five sequences (Fig. 1). In addition, we also found two new genotypes belonged to the so called Outliers group from dogs [1]; one was seen in five sequences and the other one in one sequence. Consistent with the high prevalence of E. bieneusi, 34 samples had concurrent presence of two genotypes.
Figure 1. Phylogenetic relationship of Enterocytozoon bieneusi genotype groups.
The relationship of new E. bieneusi genotype groups (1i and 6) identified in this study to established E. bieneusi genotype groups was inferred by a neighbor-joining analysis of nucleotide sequences of the ITS, based on Kimura 2-parameter distances. Some sequences generated in the study are italicized, while the remaining 538 sequences in Groups 1a–1g and one sequence in Group 3 are not shown (see Table 3). Several unique ITS sequences from AIDS patients in a recent Nigerian study [43] are designed as Group 7.
Eimeria spp. and Cyclospora cayetanensis
Among 386 samples analyzed, 310 samples (80.3%) were positive for Eimeria spp. or Cyclospora spp. The majority of the PCR products were from Eimeria spp., such as Eimeria alabamensis and its relative from cattle (167 samples), E. acervulina, E. tenella and their relatives from chicken (six samples), E. nieschulzi, E. falciformis, E. separata and their relatives from rodents (95 samples), E. flavecens from rabbits (one sample), and E. arnyi relatives from snakes (four samples). Cyclospora spp. were detected in 13 wastewater samples; in two samples, the sequence obtained had minor differences from C. cayetanensis. Nine Eimeria spp. of unknown animal sources were also found in 13 sequences from 13 samples in this study (Table 4).
Table 4. Eimeria species and related parasites in wastewater samples of four cities in China during 2006–2009a.
| Species or genotype | Ref No. | No. of positive samples (No. of positive PCR products)a | |||
| Shanghai | Nanjing | Qingdao | Wuhan | ||
| Eimeria spp. from cattle (?) | |||||
| E. alabamensis | AF291427 | 1 (1) | 12 (12) | 38 (38) | 43 (43) |
| Eimeria sp. | 18097 | 1 (1) | 7 (7) | 27 (27) | 37 (37) |
| Eimeria sp. | 22035 | 1 (1) | |||
| Eimeria spp. from chicken | |||||
| E. acervulina | EF210324 | 1 (1) | |||
| E. tenella | U40264 | 1 (1) | |||
| Eimeria sp. | 26221 | 2 (2) | |||
| Eimeria sp. | 17236 | 1 (1) | |||
| Eimeria sp. | 17242 | 1 (1) | |||
| Eimeria spp. from rabbits | |||||
| E. flavescens | EF694011 | 1 (1) | |||
| Eimeria spp. from snakes | |||||
| Eimeria sp. | 18110 | 2 (2) | |||
| Eimeria sp. | 18118 | 1 (1) | |||
| Eimeria sp. | 22024 | 1 (1) | |||
| Eimeria spp. from rodents | |||||
| E. nieschulzi | U40263 | 4 (4) | 8 (8) | 11 (11) | |
| E. separata | AF311643 | 5 (5) | 6 (6) | 2 (2) | 1 (1) |
| Eimeria sp. | AB447983 | 1 (1) | 5 (5) | 2 (2) | |
| E. falciformis | AF080614 | 1 (1) | 1 (1) | ||
| Eimeria sp. | 19027 | 6 (6) | 3 (3) | 13 (13) | 4 (4) |
| Eimeria sp. | 22032 | 3 (3) | 4 (4) | ||
| Eimeria sp. | 19118 | 3 (3) | 1 (1) | ||
| Eimeria sp. | 19110 | 2 (2) | 1 (1) | ||
| Eimeria sp. | 19108 | 2 (2) | |||
| Eimeria sp. | 17256 | 1 (1) | |||
| Eimeria sp. | 22029 | 1 (1) | |||
| Eimeria sp. | 19111 | 1 (1) | |||
| Eimeria sp. | 18928 | 1 (1) | |||
| Eimeria sp. | 18969 | 1 (1) | |||
| Eimeria sp. | 18934 | 1 (1) | |||
| Eimeria spp. from unknown source | |||||
| Eimeria sp. | 18109 | 3 (3) | |||
| Eimeria sp. | 18132 | 2 (2) | |||
| Eimeria sp. | 18079 | 2 (2) | |||
| Eimeria sp. | 18121 | 1 (1) | |||
| Eimeria sp. | 18076 | 1 (1) | |||
| Eimeria sp. | 18120 | 1 (1) | |||
| Eimeria sp. | 18137 | 1 (1) | |||
| Eimeria sp. | 18075 | 1 (1) | |||
| Eimeria sp. | 18100 | 1 (1) | |||
| Cyclospora spp. from humans and primates | |||||
| C. cayetanensis | AF111183 | 5 (5) | 6 (6) | ||
| Cyclospora sp. | 26218 | 1 (1) | |||
| Cyclospora sp. | 18979 | 1 (1) | |||
| Totalb | 42/90 (46.7%) | 60/87 (69.0%) | 109/109 (100%) | 99/100 (99.0%) | |
All data on specific species/genotypes are based on sequence analysis.
No. of positive samples/No. of samples (positivity in percentage) based on PCR analysis of the SSU rRNA gene.
Discussion
The transmission of Cryptosporidium, G. duodenalis and E. bieneusi in humans in China is poorly understood. There are only three studies that have genotyped and subtyped Cryptosporidium spp. in humans. All five specimens collected from diarrheic children in Tianjin had C. hominis [30]. Nine specimens were positive for C. hominis and one for C. felis in a small study of out-patients in Henan Province [31]. In a study conducted in in-patients in three pediatric hospitals in Shanghai, C. hominis was identified in 92, C. meleagridis in 6, C. felis in 2, and C. canis in 2 cases, but the majority of C. hominis-positive patients (74/92) were from a hospital-associated outbreak of cryptosporidiosis, which had made less samples available for the examination of transmission dynamics in the general population. Likewise, there are only three small-scale studies of G. duodenalis genotypes in humans in China. In one study in Anhui Province, assemblages A and B were each found in four patients [32]. In another study in Hebei Province, sub-assemblage A-II was found in three patients [33]. In a recent study in Henan Province, sub-assemblages A-I and A-II and assemblage B were was found in eight, four and six patients, respectively [31]. In the only study on E. bieneusi infection in China, genotypes I and J and four novel genotypes were found in nine diarrheic children in Jilin Province [34].
Data generated in this study suggest that anthroponotic transmission is important in the epidemiology of cryptosporidiosis in the study areas. This was supported by the common finding of C. hominis in the wastewater samples in Shanghai and Wuhan, where 93.7% and 40.4% of Cryptosporidium-positive samples had C. hominis, respectively. The proportion of samples positive for C. hominis was lower than Nanjing, where 18.8% had C. hominis. In contrast, only one sample from Qingdao was positive for C. hominis. In Nanjing and Qingdao, the occurrence of C. hominis was probably more common than the data suggested, as C. muris and other genotypes from rodents in the wastewater distribution system were the dominant Cryptosporidium spp. in samples from the two cities, which could have masked the concurrent presence of C. hominis. It is known that minor populations of other species or genotypes in specimens are frequently under-detected with Cryptosporidium genus-specific primers [35]. The higher occurrence of C. muris in samples from Nanjing and Qingdao could be the result of higher number of rodents in the wastewater distribution system. Therefore, the distribution of Cryptosporidium species in these cities may be affected by human populations in the city as well as rodents in the sewer system.
In agreement with this conclusion, other human-pathogenic species that are traditionally associated with zoonotic transmission of cryptosporidiosis were mostly detected in wastewater samples from the four cities at low frequencies. Thus, C. felis was not found in any samples, and C. parvum, C. canis, and C. cuniculus were each found in only 2–5 samples. The only exception was C. meleagridis, which was found in all four cities studied and in 22 of 210 (10.1%) Cryptosporidium-positive wastewater samples (Table 1). The isolated detection of C. parvum, C. canis, and C. cuniculus was probably largely the result of Cryptosporidium contamination from domestic animals, as rodents, dogs and rabbits are known hosts of these Cryptosporidium species, respectively, in addition to humans. Likewise, pet birds and other domestic birds like chickens and pigeons might have also contributed to the relative high occurrence of C. meleagridis, as the more common avian Cryptosporidium species, C. baileyi, was also found in 15 of the 217 (6.9%) Cryptosporidium-positive samples. Nevertheless, the higher occurrence of C. meleagridis than C. baileyi in wastewater samples indicated that some of the C. meleagridis probably originated from humans. In a recent study conducted in in-patients in three hospitals in Shanghai, C. meleagridis was detected in 6 of 28 (21.4%) sporadic cases of cryptosporidiosis [36]. Although some of the C. meleagridis could have been transmitted anthroponotically, we could not rule out the possibility that some zoonotic transmission of C. meleagridis may be occurring in the study areas. Previously, anthroponotic transmission has been identified as a major route in cryptosporidiosis epidemiology in most other developing countries [37].
There could be some differences in the transmission of C. hominis among the cities studied, at least between Shanghai and Wuhan. In Shanghai, 29 of the 47 (61.7%) C. hominis-positive samples subtyped had three unique subtypes (IbA19G2, IbA20G2, and IbA21G2) of the Ib subtype family. In Wuhan, however, none of the Ib subtypes were found in any of the 10 C. hominis-positive samples subtyped. There are not enough C. hominis-positive samples from Nanjing and Qingdao to infer C. hominis transmission, although IbA19G2 was found in one of the three C. hominis-positive samples from Nanjing. Interestingly, the Ie subtype IeA12G3T3 that was previously thought to be unique in Shanghai [16] was found in all other cities studied, and was the dominant C. hominis subtype in Wuhan, being found in 9/10 C. hominis-positive samples.
Anthroponotic transmission is probably also important in the epidemiology of giardiasis in China. The majority (296 of 319 or 92.8%) of samples genotyped and subtyped had the G. duodenalis sub-assemblage A-II. The predominance of sub-assemblage A-II, a parasite mainly found in humans, and the absence of sub-assemblage A-I, a parasite more often found in animals [6], suggests that zoonotic transmission might not play an important role in giardiasis transmission in the study cities. In some samples, several subtypes of assemblage B, which infects humans, ruminants, dogs and other mammals, were also found. Like the sub-assemblage A-II, humans are the likely source for these parasites. This was supported by the absence of animal-specific assemblages of G. duodenalis, such as C and D, which are common in dogs, E, which is common in livestock, F, which is common in cats, and G. which is common in rodents [6]. The absence of assemblage G was surprising considering the common presence of rodents in wastewater in cities studied and the high prevalence of Cryptosporidium spp. of rodents (Table 1). This was possibly because rodents were more commonly infected with G. muris than G. duodenalis assemblage G [38], [39]. The primers used in this study are largely G. duodenalis-specific, thus have a poor sensitivity in detecting G. muris. Only a single target was used in genotyping and subtyping G. duodenalis, which could be problematic for assemblage B, as it has been shown to exhibit heterozygosity at some genetic loci.
The transmission of E. bieneusi in China is less clear. The majority of E. bieneusi found in this study belonged to Group 1 [1], [9]. Parasites in this group infect both humans and animals. Within this group, genotype D was the dominant one, being found in 279/338 (82.5%) E. bieneusi-positive samples. The known hosts for genotype D include humans, cattle, pigs, dogs, and some wild mammals. In this study, the contribution of these animals to genotype D was probably limited, as host-adapted E. bieneusi genotypes of cattle (BEB6 of group 2), pigs (EbpD of 1e), and wild mammals (groups 3–5) were not found or only found in one sample each (Table 3). The only host-adapted genotypes found in wastewater samples were the called outlier genotype from dogs and two novel genogroups (group 1i and group 6). As they were mostly found in samples from Qingdao, rodents were the likely sources for the two new genogroups. We also cannot rule out the contribution of rodents to genotype D and other minor genotypes in group 1 found in wastewater, as these genotypes generally lack host specificity.
Genotyping data from Eimeria spp. and Cyclospora spp. generated from wastewater samples mostly support the interpretations on transmission of human-pathogenic protists and attribution of contamination sources. As Eimeria spp. and Cyclospora form host-specific clusters in phylogenetic analysis of the SSU rRNA sequences, they serve as good markers for contamination source attribution. Thus, the contribution of birds to C. meleagridis in wastewater was supported by the occasional finding of Eimeria spp. from chickens and other birds. The latter were probably from pet birds, which are known to be infected with C. meleagridis in China [40]. Likewise, the common occurrence of Eimeria spp. from rodents (seen in 97 samples) was also in agreement with the occurrence of C. muris in wastewater samples, especially those from Qingdao and Nanjing. The only discrepancy was the common occurrence of E. alabamensis (AF291427) or E. alabamensis-like sequences in 167 wastewater samples. Although E. alabamensis is a known parasite of cattle, C. parvum, C. andersoni, G. duodenalis assemblage E, and E. bieneusi group 2, all of which are common parasites of cattle, were largely absent in wastewater samples in this study. Whether the E. alabamensis sequence seen in this study was from cattle is still uncertain, as there are two very different SSU rRNA sequences of E. alabamensis in the GenBank (AF291427 and AY876926), with AY876926 forming a cluster with other bovine and ovine Eimeria spp. while AF291427 (the type seen in wastewater samples) was alone by itself in a different part of the phylogenetic tree.
In summary, results of this study indicated that molecular surveillance of enteric parasites in wastewater can be used to assess of the characteristics of endemic transmission of pathogens. The data acquired have shown a common occurrence of Cryptosporidium spp., G. duodenalis and E. bieneusi in urban communities in China, although traditional epidemiologic studies are few and routine surveillance of these pathogens is non-existent. These data have also shown a major role of anthroponotic transmission in the epidemiology of cryptosporidiosis, giardiasis, and microsporidiosis in four cities in China. They have also led to the identification of the occurrence of several rare parasites in China, including a new Cryptosporidium genotype recently seen in a patient in the United Kingdom [26], the newly identified human pathogen C. cuniculus, which was responsible for a recent waterborne outbreak of cryptosporidiosis in the United Kingdom [28], and C. cayetanensis, which was only recently identified in humans in China [41], [42]. Thus, genotyping and subtyping enteric parasites in wastewater can be an effective and economical supplement to conventional surveillance and epidemiologic tools. For example, giving the complexity of C. hominis population in wastewater in Shanghai [16], the finding of only two major subtypes from C. hominis in 86 of 3,245 in-patient in a pediatric hospital in Shanghai has led to the recent identification of a hospital-associated outbreak of cryptosporidiosis [36]. Nevertheless, caution should be exercised when interpreting transmission routes and dynamics using data from molecular surveillance of pathogens in urban wastewater. When some species are shed in larger numbers or longer durations than others, molecular techniques that can detect minor species should be adopted to avoid the misinterpretation of the data. More studies of wastewater samples from other areas are needed to validate the utility of the molecular surveillance system, perhaps in concurrence with case-control or longitudinal studies in humans.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Funding Statement
This work was supported in part by the National Natural Science Foundation of China (31110103901, 41001316); the National Basic Research Program of China (973 Project) (2011CB200903); Fundamental Research Funds for the Central Universities, China; Open Funding Projects of the State Key Laboratory of Veterinary Etiological Biology at the Lanzhou Veterinary Research Institute and State Key Laboratory of Bioreactor Engineering. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Henriques-Gil N, Haro M, Izquierdo F, Fenoy S, del Aguila C (2010) Phylogenetic approach to the variability of the microsporidian Enterocytozoon bieneusi and its implications for inter- and intrahost transmission. Appl Environ Microbiol 76: 3333–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiao L, Fayer R (2008) Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol 38: 1239–1255. [DOI] [PubMed] [Google Scholar]
- 3. Savioli L, Smith H, Thompson A (2006) Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends Parasitol 22: 203–208. [DOI] [PubMed] [Google Scholar]
- 4. Xiao L (2010) Molecular epidemiology of cryptosporidiosis: An update. Exp Parasitol 124: 80–89. [DOI] [PubMed] [Google Scholar]
- 5. Santin M, Fayer R (2011) Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci 90: 363–371. [DOI] [PubMed] [Google Scholar]
- 6. Feng Y, Xiao L (2011) Zoonotic potential and molecular epidemiology of Giardia and giardiasis. Clin Microbiol Rev 24: 110–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao L, Feng Y (2008) Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol 52: 309–323. [DOI] [PubMed] [Google Scholar]
- 8. Didier ES (1998) Microsporidiosis. Clin Infect Dis 27: 1–7; quiz 8. [DOI] [PubMed] [Google Scholar]
- 9. Thellier M, Breton J (2008) Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 15: 349–358. [DOI] [PubMed] [Google Scholar]
- 10. Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW 3rd, et al. (2003) Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi . Appl Environ Microbiol 69: 4495–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sulaiman IM, Fayer R, Yang C, Santin M, Matos O, et al. (2004) Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol Res 92: 328–334. [DOI] [PubMed] [Google Scholar]
- 12. Talebi M, Rahimi F, Katouli M, Mollby R, Pourshafie MR (2008) Epidemiological link between wastewater and human vancomycin-resistant Enterococcus faecium isolates. Curr Microbiol 56: 468–473. [DOI] [PubMed] [Google Scholar]
- 13. Vincent V, Scott HM, Harvey RB, Alali WQ, Hume ME (2007) Novel surveillance of Salmonella enterica serotype Heidelberg epidemics in a closed community. Foodborne Pathog Dis 4: 375–385. [DOI] [PubMed] [Google Scholar]
- 14. Carducci A, Verani M, Battistini R, Pizzi F, Rovini E, et al. (2006) Epidemiological surveillance of human enteric viruses by monitoring of different environmental matrices. Water Sci Technol 54: 239–244. [DOI] [PubMed] [Google Scholar]
- 15. Zhou L, Singh A, Jiang J, Xiao L (2003) Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: Implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol 41: 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Y, Li N, Duan L, Xiao L (2009) Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol 47: 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sulaiman IM, Jiang J, Singh A, Xiao L (2004) Distribution of Giardia duodenalis genotypes and subgenotypes in raw urban wastewater in Milwaukee, Wisconsin. Appl Environ Microbiol 70: 3776–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, et al. (2007) Cryptosporidium genotypes in wildlife from a new york watershed. Appl Environ Microbiol 73: 6475–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng Y, Zhao X, Chen J, Jin W, Zhou X, et al. (2011) Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in shanghai, china. Appl Environ Microbiol 77: 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang J, Alderisio KA, Xiao L (2005) Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol 71: 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao L, Alderisio K, Singh A (2006) Development and standardization of a Cryptosporidium genotyping tool for water samples. Awwa Research Foundation, Denver, CO., 39 p.
- 22. Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, et al. (2005) Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43: 2805–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng Y, Ortega Y, Cama V, Terrel J, Xiao L (2008) High intragenotypic diversity of Giardia duodenalis in dairy cattle on three farms. Parasitol Res 103: 87–92. [DOI] [PubMed] [Google Scholar]
- 24. Santin M, Fayer R (2009) Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol 56: 34–38. [DOI] [PubMed] [Google Scholar]
- 25. Ortega YR, Sterling CR, Gilman RH, Cama VA, Diaz F (1993) Cyclospora species–a new protozoan pathogen of humans. N Engl J Med 328: 1308–1312. [DOI] [PubMed] [Google Scholar]
- 26. Chalmers RM, Smith R, Elwin K, Clifton-Hadley FA, Giles M (2011) Epidemiology of anthroponotic and zoonotic human cryptosporidiosis in England and Wales, 2004–2006. Epidemiol Infect 139: 700–712. [DOI] [PubMed] [Google Scholar]
- 27. Robinson G, Wright S, Elwin K, Hadfield SJ, Katzer F, et al. (2010) Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int J Parasitol 40: 1539–1548. [DOI] [PubMed] [Google Scholar]
- 28. Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Xiao L, et al. (2009) Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg Infect Dis 15: 829–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widmer G, Akiyoshi DE (2010) Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi . Infect Genet Evol 10: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, et al. (2001) A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol Suppl: 28S–31S. [DOI] [PubMed] [Google Scholar]
- 31. Wang R, Zhang X, Zhu H, Zhang L, Feng Y, et al. (2011) Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp Parasitol 127: 42–45. [DOI] [PubMed] [Google Scholar]
- 32. Yong TS, Park SJ, Hwang UW, Yang HW, Lee KW, et al. (2000) Genotyping of Giardia lamblia isolates from humans in China and Korea using ribosomal DNA Sequences. J Parasitol 86: 887–891. [DOI] [PubMed] [Google Scholar]
- 33. Chen XN, Lu SQ, Li JH, Wang FY, Wang NX, et al. (2001) Isolation of Hebei isolates of Giardia lamblia and investigation on their genotypes. Chinese J Parasitic Dis Control 14: 100–102. [Google Scholar]
- 34. Zhang X, Wang Z, Su Y, Liang X, Sun X, et al. (2011) Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol 49: 2006–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cama V, Gilman RH, Vivar A, Ticona E, Ortega Y, et al. (2006) Mixed Cryptosporidium infections and HIV. Emerg Infect Dis 12: 1025–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, et al. (2012) Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis 18: 312–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao L, Ryan UM (2008) Molecular Epidemiology. In Fayer, R., Xiao, L. (eds). Cryptosporidium and Cryptosporidiosis, second edition. CRC Press, New York, USA. p119–172.
- 38. Sogayar MI, Yoshida EL (1995) Giardia survey in live-trapped small domestic and wild mammals in four regions in the southwest region of the state of Sao Paulo, Brazil. Mem Inst Oswaldo Cruz 90: 675–678. [DOI] [PubMed] [Google Scholar]
- 39. Franjola R, Soto G, Montefusco A (1995) [Prevalence of protozoa infections in synanthropic rodents in Valdivia City, Chile]. Bol Chil Parasitol 50: 66–72. [PubMed] [Google Scholar]
- 40. Qi M, Wang R, Ning C, Li X, Zhang L, et al. (2011) Cryptosporidium spp. in pet birds: Genetic diversity and potential public health significance. Exp Parasitol 128: 336–340. [DOI] [PubMed] [Google Scholar]
- 41. Wang KX, Li CP, Wang J, Tian Y (2002) Cyclospore cayetanensis in Anhui, China. World J Gastroenterol 8: 1144–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Y, Lv BA, Wang Q, Wang RJ, Jian FC, et al. (2011) Prevalence and Molecular Characterization of Cyclospora cayetanensis, Henan, China. Emerg Infect Dis 17: 1887–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, et al. (2012) Molecular epidemiologic characterization of Enterocytozoon bieneusi in HIV-infected persons in Benin City, Nigeria. Am J Trop Med Hyg 86: 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]