Abstract
Reduced DNA methylation has been reported in DICER1-deficient mouse ES cells. Reductions seen at pericentric satellite repeats have suggested that siRNAs are required for the proper assembly of heterochromatin. More recent studies have postulated that the reduced methylation is an indirect effect: the loss of Mir290 cluster miRNAs leads to upregulation of the transcriptional repressor RBL2 that targets the downregulation of DNA methyltransferase (Dnmt) genes. However, the observations have been inconsistent. We surmised that the inconsistency could be related to cell line “age,” given that DNA methylation is lost progressively with passage in DNMT-deficient ES cells. We therefore subjected Dicer1 −/− ES cells to two experimental regimes to rigorously test the level of functional DNMT activity. First, we cultured them for a prolonged period. If DNMT activity was reduced, further losses of methylation would occur. Second, we measured their DNMT activity in a rebound DNA methylation assay: DNA methylation was stripped from Cre/loxP conditionally mutant Dicer1 ES cells using a shRNA targeting Dnmt1 mRNA. Cre expression then converted these cells to Dicer1 −/−, allowing for DNMT1 recovery and forcing the cells to remethylate in the absence of RNAi. In both cases, we found functional DNMT activity to be normal. Finally, we also show that the level of RBL2 protein is not at excess levels in Dicer1 −/− ES cells as has been assumed. These studies reveal that reduced functional DNMT activity is not a salient feature of DICER1-deficient ES cells. We suggest that the reduced DNA methylation sometimes observed in these cells could be due to stochastic alterations in DNA methylation patterns that could offer growth or survival advantages in culture, or to the dysregulation of pathways acting in opposition to the DNMT pathway.
Author Summary
In mammalian cells, DNA methylation is required for the maintenance of genome stability. Recent studies have shown that the genome-wide levels of DNA methylation can be reduced in DICER1-deficient mouse embryonic stem (ES) cells, suggesting that the activity of DNA methylating enzymes (DNMTs) may be regulated by small RNA molecules. The enzyme DICER1 catalyses the production of these small RNAs that serve as sequence-specific guides for modifying chromatin or transcription. However, these observations of defective DNA methylation have been inconsistent. We surmised that this inconsistency could be due to cell line “age,” because it can take many cell divisions before reduced DNMT activity may result in loss of DNA methylation. To test this idea, we rigorously assayed the functional level of DNMT activity in DICER1-deficient ES cells. First, we tested their ability to maintain DNA methylation over prolonged culture. Second, we tested their ability to rebound in DNA methylation after first stripping it from the genome. In both cases functional DNMT activity was entirely normal. We suggest that losses of DNA methylation sometimes seen in DICER1-deficient ES cells is stochastic and could involve cell line adaptation.
Introduction
Most CpG dinucleotides in the mammalian genome are methylated [1], much of this being present at localized centric (minor satellite, MinS) and pericentric (major satellite, MajS) repeats, and dispersed repeats or transposable elements (TEs). Here DNA methylation plays a role in maintaining genome stability, being required for chromosome (Chr) stability [2], [3] and the silencing of TEs [4], [5]. At single copy sequences, the stabilization of transcriptional repression through promoter methylation is a common theme, although other functions for DNA methylation within the gene body and at regulatory regions are apparent [6]. For example, DNA methylation is central to the establishment and maintenance of the parental-specific expression of imprinted genes [7], and for the stabilization of X Chr inactivation [8]. Given the wide-ranging and essential functions of DNA methylation in mammalian cells, it is important to identify mechanisms that assist in its establishment and propagation.
A number of studies have reported a disruption in DNA methylation at repetitive sequences in mouse embryonic stem (ES) cells lacking activity of the key RNAi enzyme DICER1 (Dicer1, Dcr-1 homolog (Drosophila)). Two different mechanisms have been suggested for these methylation defects. In the first mechanism, it was suggested that reduced DNA methylation at MajS and MinS repeats is linked with a failure in RNAi-mediated heterochromatinization [9]. This mechanism has been extensively studied in regard to the pericentric region in fission yeast (Schizosaccharomyces pombe). Transcripts from this region are processed into siRNAs by Dicer, which then guide the Clr4 histone methyltransferase back to the repeats to catalyse the formation of histone H3 lysine 9 trimethylation (H3K9me3). This post-translational modification (PTM) is an essential waypoint in heterochromatin induction and recruits heterochromatin-inducing proteins such as Swi6 [10], [11]. The latter part of this pathway in fission yeast is conserved in mammals. Pericentric H3K9me3 is catalysed by SUV39H1 (suppressor of variegation 3–9 homolog 1 (Drosophila)) and SUV39H2, the mammalian homologues of Clr4. This PTM then recruits the heterochromatin-inducing CBX proteins, the mammalian homologues of Swi6. In addition, H3K9me3 recruits de novo DNA methyltransferase (DNMT) activity and DNA methylation, thereby adding an additional repressive layer [2], [3]. If pericentric DNA methylation is indeed defective in DICER1-deficient ES cells [9], then this would point to earlier parts of the pathway also being conserved, that is, mammalian pericentric H3K9me3 could also be guided by siRNAs that are processed from MajS transcripts by DICER1. However, defective DNA methylation at MajS repeats in ES cells has not been consistently observed. While one study observed losses [12], three others did not [13]–[15].
The second mechanism attributes losses of DNA methylation in DICER1-deficient ES cells, or their immediate differentiated derivatives, to an indirect effect of the loss of DICER1 pre-miRNA processing. More specifically, it has been proposed that Mir290 (microRNA 290) cluster miRNA deficiency results in an upregulation of a target transcript encoding the transcriptional suppressor RBL2 (retinoblastoma-like 2), with increased RBL2 in turn suppressing Dnmt gene transcription and lowered DNMT protein. Sequences reduced in DNA methylation were the promoters of pluripotency genes on ES cell differentiation [15], the whole genome [12], and the Xist (inactive X specific transcripts) promoter [16]. However, the effects observed on Dnmt expression and DNA methylation were again inconsistent. For example, a generalized reduction in genomic DNA was reported, including a loss of DNMT1 activity [12], that was not observed in another study [15]. Also, the degree of demethylation at the Xist promoter varied between sublines [16]. It should be noted that the relative level of RBL2 protein in DICER1-deficient ES cells was not measured in immunoblots [12], [15]. A summary of the various findings in DICER1-deficient ES cells published to date is provided (Table S1).
While RNAi plays important roles in establishing epigenetic states in fission yeast and in plants, the latter possessing a robust RNAi-mediated DNA methylation mechanism [17], it remains unclear that RNAi might play similar roles in mammalian cells. Because of the broad implications for our understanding of the maintenance of genome stability, differentiation, and X Chr inactivation, it is important to resolve if there does exist direct or indirect mechanistic links between RNAi and DNA methylation in ES cells or other mammalian cell types. We therefore decided to reinvestigate the effects of ablating Dicer1 on DNA methylation at various sequences in ES cells. We considered that, if DNMT activity was critically impaired in Dicer1 −/− ES cells, then the inconsistent losses of DNA methylation across the various studies could be related to the length of time the lines used had previously spent in culture. For example, impaired maintenance DNMT activity would lead to a rapid loss of global methylation with passage, while impaired de novo DNMT activity would lead to a gradual global loss. To rigorously test this possibility, we subjected Dicer1 −/− ES cells to two types of experimental manipulation designed to amplify the effect of reduced DNMT activity. These were (i) a prolonged culture regime, and (ii) a test of their ability to ‘rebound’ in DNA methylation after experimentally stripping them of DNA methylation. In neither case did we obtain evidence for defective functional DNMT activity. We conclude that the reductions of DNMT protein levels that can be observed in DICER1-deficient ES cells are generally insufficient to account for losses in DNA methylation. We propose that the losses of DNA methylation sometimes observed could involve an adaptive response of the cells to the lack of si- and miRNAs, or even the upregulation of pathways that work in opposition to the DNMT machinery.
Results
Effect of DICER1-deficiency on Dnmt and Rbl2 mRNA and encoded protein levels
A euploid Dicer1 c/− (c, Cre/loxP conditional mutant allele; -, mutant allele) mouse ES cell line of XY sex Chr constitution was derived. The Dicer1 mutation [18] involves deletion of the terminal three exons encoding 76 of 213 aa of the second RNase III catalytic domain and all 57 aa of the dsRNA binding domain. Also, the translation stop codon and poly(A) signal sequences are deleted, these being generally required for transcript stability and nuclear export, respectively. The Dicer1 c/− ES cells were transiently transfected with a Cre expression cassette to obtain Dicer1 −/− clones. These were distinguishable at picking by their small size, with genotypes confirmed by Southern blot. Picked clones were expanded and DNA and RNA harvested at passage 5.
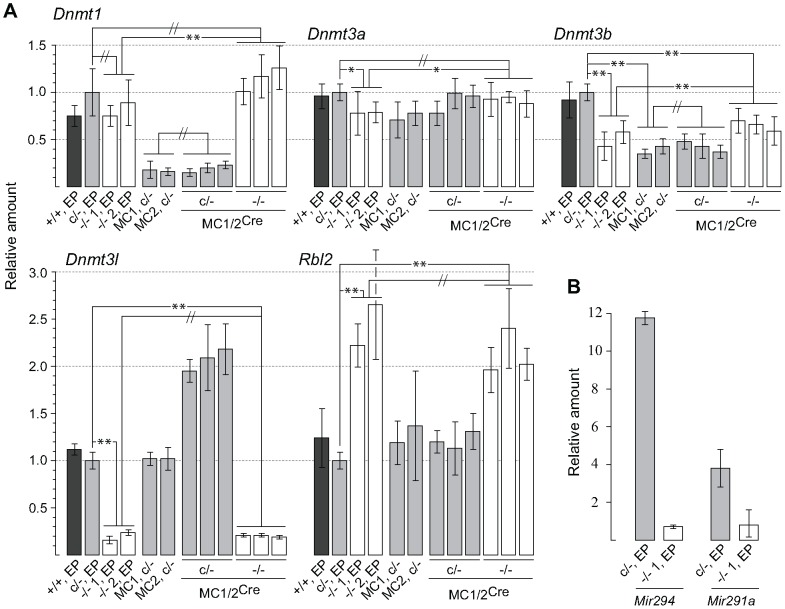
qPCR assays showed that, in the two independently derived Dicer1 −/− lines at early passage (EP), the amount of Dnmt1 (DNA methyltransferase (cytosine-5) 1) mRNA was unchanged relative to the parental Dicer1 c/− EP cell line (Figure 1A). By contrast, the amounts of Dnmt3a, Dnmt3b (DNA methyltransferase 3A and -B) and Dnmt3l (DNA (cytosine-5-)-methyltransferase 3-like) mRNAs were significantly reduced to ∼75%, 50% and 20% respectively, while the amount of Rbl2 mRNA was increased to ∼200%. Mir290 cluster miRNAs were essentially undetectable in our Dicer1 −/− lines (Figure 1B). Overall, these findings indicate that, in terms of the mRNA levels for these genes, our Dicer1 −/− EP ES cell lines were similar to those previously described [12], [15], [16].
Figure 1. qPCR assays.
(A) Values obtained for the parental Dicer1 c/− line were used to calibrate all other values, and are set at 1.0. The y axis is a linear scale. Bars are mean ± s.d. Probabilities obtained from Student's t-test, 2-tail, type 2: P<0.05 (*), P<0.01 (**), not significantly different (//). Bar 1; Dicer1 +/+ cell line 2A at early passage (EP). Bar 2; parental Dicer1 c/− EP line. Bars 3 and 4; Two independent Dicer1 −/− EP lines derived from Cre transfection of the parental Dicer1 c/− EP line. Bars 5 and 6; Two independent sublines (MC1 and MC2) derived from concatemeric targeting at Hprt with the Dnmt1 shRNA. Bars 7–9 and 10–12; three sublines derived from exposure of MC1 (two sublines) and MC2 (one subline) to Cre (MC1/2) that did not excise (c/−) and did excise (−/−) the floxed Dicer1 sequence, respectively. (B) Measurements were of the parental Dicer1 c/− line and one Dicer1 −/− subline. The y axis is a linear scale. Values are normalized to those obtained for Rps7, which are set at 1.0. Other details are contained in the Materials and Methods section.
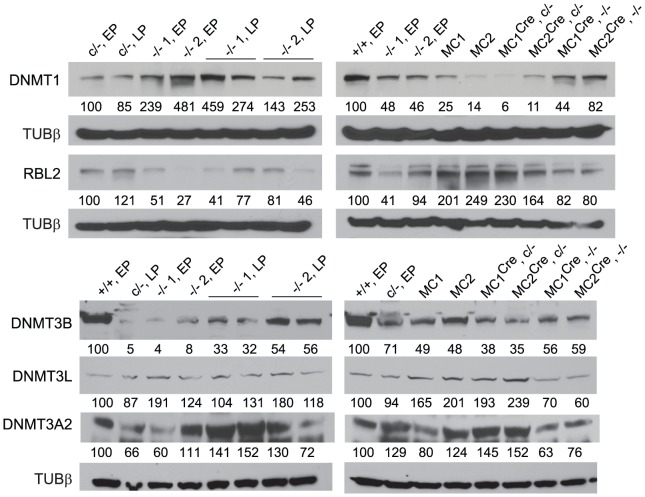
In the Dicer1 −/− EP cell lines, immunoblots (Figure 2) showed that there was no deficiency in DNMT1 relative to the parental Dicer1 c/− EP line (left blot). Indeed, DNMT1 levels were elevated. The levels of DNMT3A2 in the Dicer1 −/− EP lines were on average ∼85% of that present in the Dicer1 +/+ EP line (left blot). Surprisingly, the relative levels of the other proteins assayed were very different to their corresponding mRNAs (Figure 2). In the Dicer1 −/− EP lines, DNMT3B levels were less than 10% that in the Dicer1 +/+ EP line (left blot). However, they were probably not as deficient relative to the parental Dicer1 c/− EP line, as this line had ∼70% the amount of DNMTB in comparison to the Dicer1 +/+ EP line (right blot). Despite the low amount of Dnmt3l mRNA in Dicer1 −/− EP lines, DNMT3L protein content in these lines was similar if not higher relative to the Dicer1 +/+ EP line (left blot). Significantly, the amount of RBL2, for which elevated levels have been proposed to cause repression of Dnmt genes and consequent demethylation in ES cells, was lower in the Dicer1−/− EP cells compared to the parental Dicer1 c/− and Dicer1 +/+ EP lines (left and right blots).
Figure 2. Immunoblots.
Values below each band represent the densitometry reading taken for that band. These were normalized to the reading taken for lane 1, which was for the WT Dicer1 +/+ EP cell line (+/+, EP), or the parental Dicer1 c/− EP cell line (c/−, EP). The loading was consistent throughout as shown by the detection of Tubulin β (TUBβ).
DICER1-deficient ES cells retain DNA methylation over extended passage
The large majority of CpG dyads in the mammalian genome are symmetrically methylated. The stability of this high degree of DNA methylation, at least in ES cells, relies on the presence of de novo and maintenance DNMT activity. De novo activity is provided largely by DNMT3A and DNMTB working with the cofactor DNMT3L. These enzymes are able to hemimethylate unmethylated CpG dyads, or fully methylate hemimethylated dyads. However, the system is relatively inefficient, that is, the proportion of sites that can be de novo methylated per cell division is low. Maintenance activity is provided largely by DNMT1. With high efficiency, this enzyme fully methylates hemimethylated dyads produced at DNA replication. Loss of the de novo (DNMTs 3A, 3B and 3L) or maintenance (DNMT1) system results in the progressive loss of DNA methylation at virtually all sequences. On ablation of Dnmt1, global loss occurs over 5 passages (25 cell divisions), although a background of methylation is retained indefinitely due to continuing de novo DNMT activity. This ongoing de novo activity is crucial for the remethylation of the genome should DNMT1 activity be reintroduced, as DNMT1 alone has negligible de novo activity in ES cells. Ablating Dnmt3a and Dnmt3b, or Dnmt3l, results in a much slower loss of DNA methylation than does ablation of Dnmt1. Unmethylated dyads produced stochastically can no longer be remethylated with any efficiency so they gradually accumulate. In Dnmt3a-Dnmt3b double-mutant ES cells, virtually all methylation is eventually lost [1], [19]–[27].
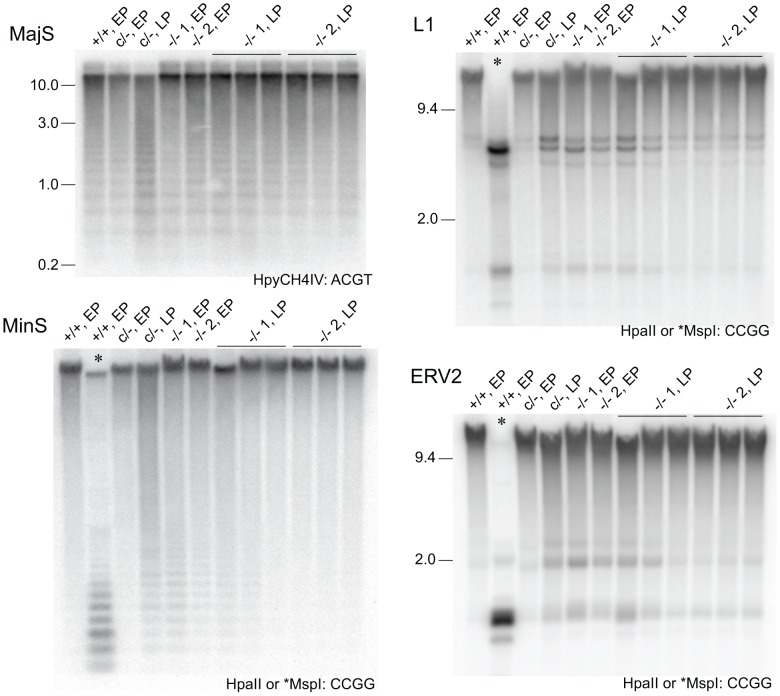
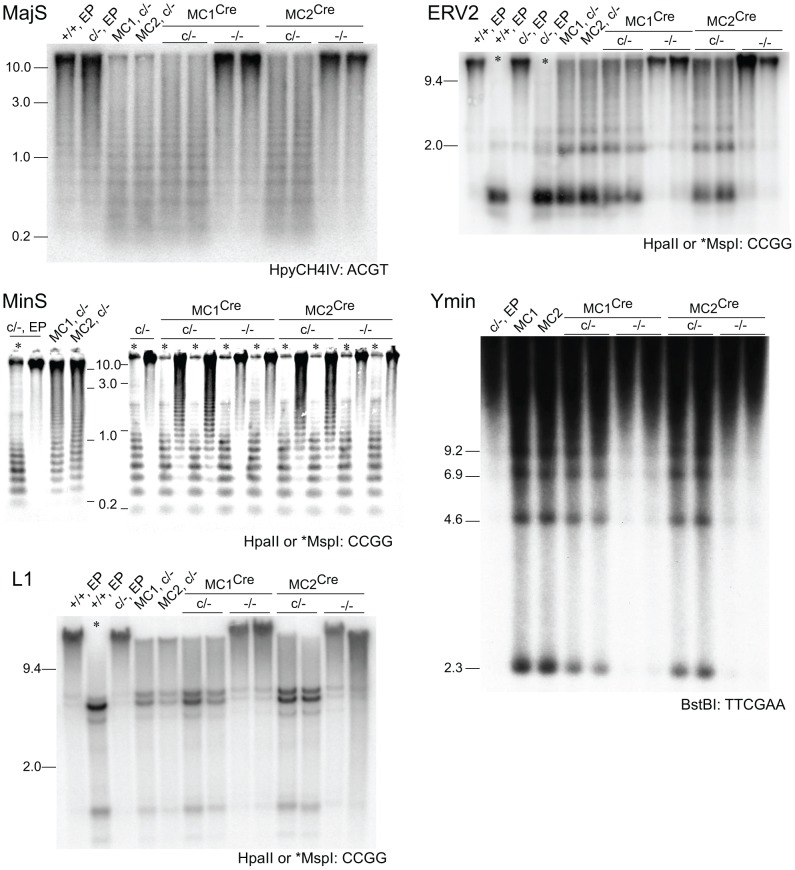
Given the dynamics of DNA methylation as summarized above, critically reduced DNMT activity in DICER1-deficient ES cells could result in variable DNA methylation content depending on the length of time the cells have spent in culture. It was not clear if the reduction in de novo Dnmt mRNA and DNMT protein in our Dicer1 −/− EP lines was sufficient to lead to reduced DNA methylation. Methylation-sensitive Southern blot analysis was therefore used to analyse the degree of methylation at MajS and MinS repeats, and two dispersed repetitive transposable elements (TEs)—long interspersed nuclear element type 1 (L1) and endogenous retrovirus type 2 (ERV2) or intracisternal A particle. No reduction in methylation was seen at the MajS repeats (Figure 3). By contrast, some reduction in DNA methylation was evident at the MinS repeats and L1 and ERV2 elements in the two Dicer1 −/− EP lines (Figure 3).
Figure 3. Stability of DNA methylation in DICER1-deficient ES cells.
Methylation-sensitive Southern blots. Details are provided in the legend to Figure 1, except that later passage (LP) cells were also used. Passage (p) numbers were: +/+, EP (p8), c/−, EP (p7), c/−, L (p24), −/− 1, EP and −/− 2, EP (p5), −/− 1, LP and −/− 2, LP (p20). Generally, a single passage for +/+ and c/− lines lasted 3 days, while for −/− cells a single passage lasted 5 days. The restriction enzyme and its recognition sequence are at the bottom right of each plate. At CCGG sites, HpaII cuts only when the CpG dyad is unmethylated. The isoschizomer MspI can cut when the CpG dyad is fully, hemi- or unmethylated.
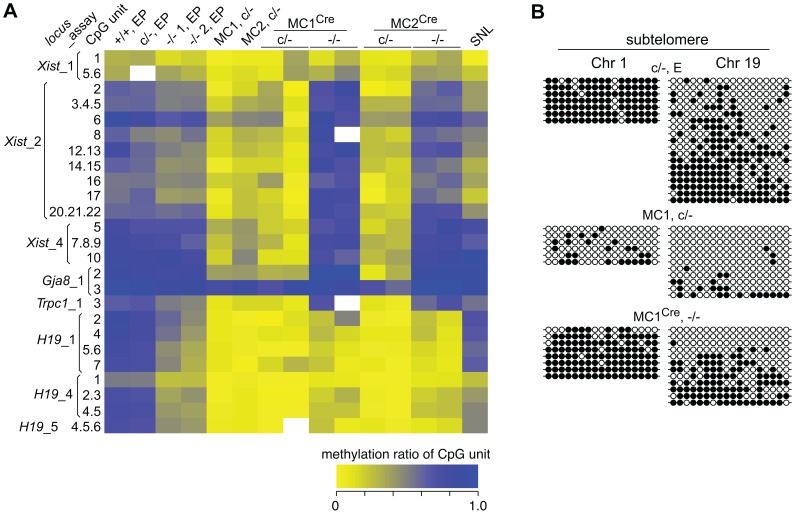
Selected single copy sequences in Dicer1 −/− EP lines were also analysed for DNA methylation using the EpiTYPER assay (Sequenom, San Diego, CA, USA). This assay is similar to bisulphite sequencing in that DNA is converted using sodium bisulphite and then amplified [28], but differs in that the PCR product is subjected to cleavage and analysis by mass spectroscopy [29]. Sequences assayed were the Xist promoter region, reported to show reduced DNA methylation in XY Dicer1 −/− ES cells [16], two regions reported to be heavily methylated in WT ES cells—the upstream regions of the Gja8 (gap junction protein, alpha 8) and Trpc1 (transient receptor potential cation channel, subfamily C, member 1) genes [30], and the promoter and imprinting control region of the imprinted H19 (H19 fetal liver RNA) gene. The latter imprinted sequences were included as a reference: differential DNA methylation and monoallelic expression at imprinted regions can be lost over passage in ES cells and cannot be regained, this being shown for the H19 promoter [31], [32]. This is presumably because the imprintable CpGs are permissive for de novo methylation only during germ cell development, and therefore gradually lose methylation in ES cells as would any other sequence if it were not a target for de novo methylation. In the Dicer1 −/− EP lines, some reduction in DNA methylation was evident at some CpG units in the Xist_2 region, while no reduction was seen for the Xist_1 and _4 regions (Figure 4A). In a previous study, substantial reductions were seen at all of these Xist regions using the EpiTYPER assay [16]. A reduction was also seen for the Trpc_1 CpG unit, and more noticeable demethylation at all of the CpG units in the H19_1, _4 and _5 regions (Figure 4A). For the H19 CpG units, the loss of methylation is expected as the EP clonally-derived Dicer1 −/− cell lines are effectively at much higher passage than the Dicer1 c/− EP parental cell line.
Figure 4. DNA methylation analysis of single copy sequences.
(A) Heat-map showing the degree of methylation at CpG units as assayed by EpiTYPER. For example, a yellow rectangle indicates a small fraction of the CpG unit was methylated. Actual values obtained are provided (Dataset S1). The Xist_1 and _2 assays correspond to CpG-rich region 2 within exon 1, while the Xist_4 assay corresponds to CpG-rich region 1 directly adjacent to the 5′ end of exon 1. These regions correspond to the Xist promoter [16]. The Gja8 and Trpc1 assays correspond to 5′ upstream regions of these genes. The H19_1 assay corresponds to the H19 promoter region, while the H19_4 and _5 assays correspond to the insulin-like growth factor 2 (Igf2)/H19 imprinting control region located 5′ upstream of H19. In EpiTYPER assays, CpGs are analysed as ‘units’, that is, the methylated fraction measured may correspond to one CpG, e.g. (Xist_2, 8), or be the average of more than one CpG, for example (Xist_2, 20.21.22). SNL, mitomycin C-inactivated SNL feeder cells. (B) Bisulphite sequencing assays. Each row represents a sequenced clone. Closed and open circles represent a methylated and non-methylated CpG, respectively.
To determine if the reduction in DNA methylation seen in the two Dicer1 −/− EP lines was the beginning of a generalized reduction of DNA methylation with passage—as would be expected if DNMT activity were compromised—they were propagated for a further 60 days involving at least 15 additional passages. All cell lines reached passage 20 or above, which is sufficient to result in substantial loss of global DNA methylation in the absence of de novo DNMT activity [27]. By passage 15, each of the two cultures of Dicer1 −/− lines had developed a strong tendency for differentiation, but this was eliminated by picking and expanding undifferentiated clones. The parental Dicer1 c/− line was passaged alongside the two Dicer1 −/− lines and was always phenotypically normal. After this passaging regime, or at later passage (LP), DNA was harvested from three sublines for each of the two Dicer1 −/− lines, and from the Dicer1 c/− line. In the Dicer1 −/− LP sublines, the DNA methylation levels had remained stable for the MajS repeats, or even increased slightly for the MinS repeats and the L1s and ERV2s (Figure 3). In the parental Dicer1 c/−LP line, a loss of methylation at the MinS and two dispersed repeats occurred (Figure 3). This effect seen in the Dicer1 c/− line—which would be expected to behave similarly to WT, may reflect a normal level of fluctuation. While, the degree of stability of DNA methylation in WT cells has not been studied extensively, we note that WT cell lines used in other studies exhibit small reductions of methylation at repetitive sequences [9], [13], [14]. The absence of any progressive DNA methylation loss at repetitive elements in Dicer1 −/− ES cells with passage indicated that longer-term DNMT activity was essentially intact in these si- and miRNA-deficient ES cells. However, the reductions in DNA methylation seen at the MinS repeats and TEs in Dicer1 −/− EP lines and in the parental Dicer1 c/− LP line (Figure 3 and Figure 4A) did correlate with a low level of DNMTB (Figure 2). To further examine this phenomenon, we devised a rebound DNA methylation assay to test the ability of EP Dicer1 −/− ES cells to methylate their DNA.
The rebound DNA methylation assay
The rebound DNA methylation assay is depicted (Figure 5A). Genomic DNA in Dicer1 c/− (c, Cre/loxP conditional null allele; -, null allele) ES cells is largely stripped of methylation using a shRNA complementary to Dnmt1 mRNA. These ES cells are then exposed to Cre recombinase, converting them to Dicer1 −/− while simultaneously abrogating shRNA processing and restoring Dnmt1 mRNA. Then, the ability of these cells to regain DNA methylation provides a readout of functional DNMT activity. This activity is the combined functional activity of DNMTs 1, 3A, 3B and 3L, in a si- and miRNA-deficient environment. Rebound DNA methylation could not occur unless all components of this DNMT system are essentially intact [21], [24].
Figure 5. Experimental strategy to test rebound DNA methylation after Dicer1 ablation.
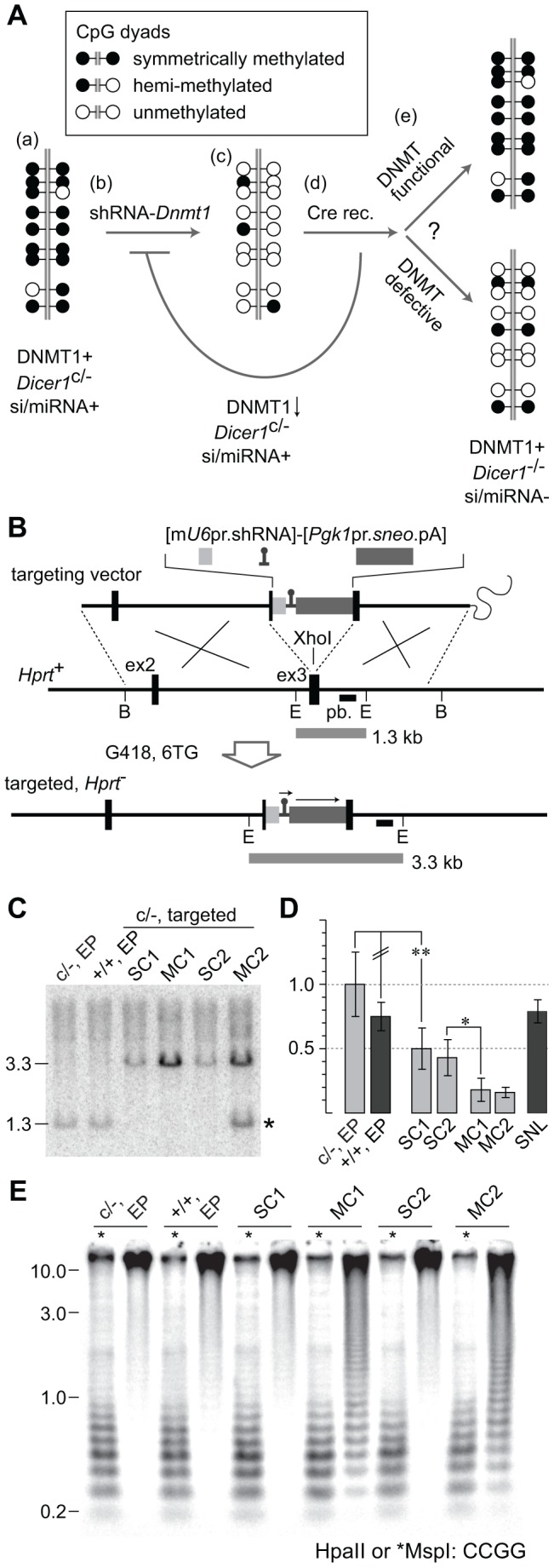
(A) (a) The parental Dicer1 c/− XY ES cell line is normally methylated. (b) A construct containing an shRNA targeting Dnmt1 mRNA (shRNA-Dnmt1) is homologously recombined into the X-linked Hprt locus. (c) The integrated shRNA effects substantial reduction in genome-wide DNA methylation content, although a background of DNA methylation is retained due to ongoing de novo DNMT activity. (d) Recombinant clones are transiently transfected with Cre to ablate the Dicer1 c allele. After ablation, clearing of residual DICER1 and si- and miRNAs, as well as restoration of functional DNMT1 activity, is expected to occur within a few cell divisions. (e) DNA methylation would potentially be re-established over ∼5 passages, corresponding to ∼25 cell divisions through the combined action of the DNMT system as a whole. The extent of rebound DNA methylation serves as an indicator of the dependency of this process on DICER1 activity and the presence of si- and miRNAs. (B) The targeting vector consisted of a 6.8 kb BamHI (B) genomic fragment of Hprt spanning exons 2 and 3 (ex2 and ex3). The mU6 promoter (pr) driving the shRNA and a sneo selection cassette were inserted into ex3. The sequence of the shRNA cassette is provided (Text S1). All clones surviving G418 and 6TG dual selection are expected to be targeted. pb, probe used in Southern blot, hybridizing to EcoRI (E) bands as indicated by horizontal bars. (C) Southern blot to detect gene targeting. WT bands are seen in the parental Dicer1 c/− line, and in a Dicer1 +/+ line. An increase in band size is occurs when the single Hprt sequence is replaced by a single copy (SC) or multiple copy concatemer (MC) of the targeting vector. In addition to the mutant band, clone MC2 clone shows a WT band of normal intensity (*). This results from recombination occurring in the 5′ arm, rather than in the 3′ arm, of the terminal vector in the concatemer. (D) qPCR of Dnmt1 mRNA. All values were calibrated to that obtained for the Dicer1 c/− line, adjusted to 1.0. SNL, mitomycin C-inactivated feeder cells. Other details as above. (E) Southern blot to detect methylation levels at MinS repeats. Details as above.
To strip DNA methylation, a mU6-shRNA complementary to Dnmt1 mRNA was targeted to the X-linked housekeeper Hprt (hypoxanthine guanine phosphoribosyl transferase) gene via sequential G418 and 6-thioguanine (6TG) selection (Figure 5B). This locus is expression-permissive for targeted cassettes, including shRNA cassettes [33], [34]. Following electroporation, ∼20 dual drug resistant clones derived from the parental Dicer1 c/− line were obtained per 10 cm plate. This was a ∼10-fold higher frequency of clone recovery compared to our previous experience using the same homology arms for Hprt [33], and could be related to the opposite transcriptional orientation of the positive selection cassette, or use of the sneo rather than the neo selection cassette. After two passages—with growth of the initial clone being the first passage, half of the cells were frozen while the other half were used for DNA extraction. Of 16 clones analysed by Southern blot, band intensity relative to DNA loading indicated that 12 clones were single copy (SC) recombinants, while four clones were multiple copy (MC) recombinants. The latter derive from concatemeric insertion of the complete targeting vector including vector sequences, this being expected when no negative selection cassette is present. Two SC and two MC clones were selected for further study. Relative band intensity indicates the number of vector copies integrated in MC clones was ∼four (Figure 5C).
Frozen cells were thawed and grown for another 2 passages, corresponding to 5 passages after DNA integration, then total RNA and genomic DNA extracted for qPCR and DNA methylation analysis. qPCR revealed that the level of reduction in Dnmt1 mRNA content correlated with SC or MC integration. Relative to the parental cell line, the two MC lines showed more than 80% reduction in mRNA while the former showed ∼50% reduction (Figure 5D). Immunoblots showed that DNMT1 was low in the two MC lines relative to the Dicer1 +/+ EP line (Figure 2). Also noticeable in these MC lines was a considerable reduction in the amount of Dnmt3b mRNA relative to the parental Dicer1 c/− EP line (Figure 1A). While this could have resulted from off-targeting activity of the introduced shRNA, this would not present a problem for our strategy as we were trying to achieve as much reduction of DNA methylation as possible before ablation of Dicer1 activity.
Reduction of genomic DNA methylation content in MC sublines
Genomic DNA isolated from the SC and MC lines was assayed for DNA methylation at MinS repeats. No reduction in methylation was seen in the two SC lines (Figure 5E), and these were not analysed further. By contrast, substantial loss of DNA methylation in the two MC lines was seen. Other sequences assayed in these MC lines were (i) localized repeats —MajS and Y Chr centric repeats (Ymin), using methylation-sensitive Southern blots, (ii) dispersed repeats—L1s and ERV2s, also using Southern blots, and (iii) selected single copy sequences—as described above, using bisulphite sequencing and EpiTYPER. All repeat and single copy sequences displayed a substantial loss of DNA methylation (Figure 4A, 4B and Figure 6), with the exception of CpG unit 3 within the Gja8_1 region (Figure 4A). This retention indicates that there is an unusually high frequency of maintenance methylation effected by DNMT3A or DNMT3B at this site. This atypical CpG unit was only 121 bp from another CpG unit, Gja8_1 unit 2, that displayed the typical DNA methylation dynamics associated with loss of DNMT1. In general, the degree of DNA methylation loss was similar to that seen in Dnmt1 −/− ES cells and in ES cells in which the same shRNA sequence was introduced by lentivirus [35].
Figure 6. Rebound DNA methylation at repetitive sequences after Dicer1 ablation.
Southern blots. MC1Cre and MC2Cre are sublines derived from Cre transfection of line MC1 and MC2 respectively. Other details are as provided in the legends to previous figures.
Effect of Dicer1 ablation on Dnmt mRNA and DNMT protein content
The two targeted Dicer1 c/− lines MC1 and MC2 were transiently transfected with a Cre expression vector then plated at low density to obtain clonal growth. For each of the MC1 and MC2 transfections, normally growing (presumptive Dicer1 c/−) and slow growing (presumptive Dicer1 −/−) sublines were picked, propagated until passage 5, then total RNA and genomic DNA extracted and genotype confirmed by Southern blot. Three each of control MCCre Dicer1 c/− and experimental MCCre Dicer1 −/− sublines were selected for qPCR. As expected, the amount of Dnmt1 mRNA remained low in the former and was restored in the latter to the level seen in the parental Dicer1 c/− cell line. This restoration was to a level higher than that seen in the two Dicer1 −/− EP lines (Figure 1A). In the three MCCre Dicer1 −/− sublines the amount of Dnmt3a mRNA was higher than in the Dicer1 −/− EP lines, although was not significantly different to that in the parental Dicer1 c/− cell line. Similarly, the amount of Dnmt3b mRNA in the three MCCre Dicer1 −/− sublines was higher than in the Dicer1 −/− EP lines. However, this remained lower than in the parental Dicer1 c/− cell line (Figure 1A). Dnmt3l and Rbl2 mRNA content was no different in MCCre Dicer1 −/− sublines compared to the Dicer1 −/− EP lines, being elevated ∼twofold and reduced ∼fivefold relative to the parental Dicer1 c/− EP line, respectively (Figure 1A).
Immunoblots (Figure 2) confirmed that there was a marked rebound in DNMT1 level in the MCCre Dicer1 −/− sublines, although this did not reach the level seen in the Dicer1 +/+ EP line. As expected, the amount of DNMT1 remained low in the MCCre Dicer1 c/− sublines (right blot). DNMT3A2 levels were lower in the MCCre Dicer1 −/− sublines compared to the MCCre Dicer1 c/− sublines and the parental Dicer1 c/− EP line (right blot). DNMT3B levels in MCCre Dicer1 −/− sublines were ∼80% of that seen in the parental Dicer1 c/− EP line (right blot). Also, there was a marked increase in the amount of DNMT3B in the Dicer1 −/− LP sublines compared to the Dicer1 −/− EP cells from which they were derived (left blot). Thus, DNMT3B levels had increased over passage, but only to ∼50% the level in the Dicer1 +/+ EP line. DNMT3L and RBL2 levels in the MCCre Dicer1 −/− sublines were ∼70% and ∼80% that detected in the Dicer1 +/+ EP line, respectively (right blot). Also, RBL2 was clearly lower in these MCCre Dicer1 −/− sublines compared to the MCCre Dicer1 c/− sublines (right blot). Thus, again, DNMT3L and RBL2 protein levels did not reflect the amounts of mRNA present.
Effect of Dicer1 ablation on rebound DNA methylation
In our experimental system, at the point of Cre-mediated conversion from Dicer1 c/− to Dicer1 −/−, ES cells were globally hypomethylated. From this stage, only a few cell divisions (one passage), would be required for depletion of si- and miRNAs [36]. This would include depletion of the introduced siRNA that targets Dnmt1 mRNA, therefore there would be a lag in the restoration of DNMT1 protein. Given these kinetics, and that complete restoration of global DNA methylation content requires 5 passages with full DNMT activity [37], a complete rebound in DNA methylation in the experimental Dicer1 −/− sublines must be realized largely in the absence of si- and miRNAs. In the four experimental Dicer1 −/− sublines, two each derived from lines MC1 and MC2, after a total of 5 passages, essentially full restoration of DNA methylation content occurred at all sequences examined—MajS, MinS, Ymin, L1 and ERV2 repeats (Figure 6). In addition, using immunofluorescence, the MajS-specific mark H3K9me3 and downstream heterochromatic marks CBX5 (chromobox homolog 5 (Drosophila HP1α)) and H4K20me3 appeared to be as prevalent in one Dicer1 −/− subline examined as in controls (Figure S1A–S1C). Indeed, the labelling of these marks appeared to be highest in the Dicer1 −/− cells and could be indicative of some alteration of chromatin structure in the form of a greater concentration or accessibility of marks. This could be related to the increased transcription seen at these sequences [9], [13]. Essentially complete rebound in DNA methylation was also seen for single copy sequences that had lost methylation due to DNMT1 deficiency, these being the three Xist regions, the Trpc1_1 region, and the two subtelomeric regions. The exceptions were the three H19 regions (Figure 4A and 4B), no rebound being expected given that the methylation of these sites is germ line-specific. Overall, the results indicate that functional DNMT activity is normal in Dicer1 −/− EP ES cells.
Discussion
DICER1-deficient mouse ES cells have often been found to be deficient in DNA methylation. These observations have been predominantly of localized and dispersed repeats which constitute the large majority of methylated CpG dyads in the mammalian genome [4]. While a loss in RNAi-mediated pathways of heterochromatin formation could be involved, particularly at MajS repeats, the predominant cause has been proposed to be a downregulation in Dnmt gene activity primarily due to the loss of Mir290 cluster miRNAs.
In re-examining these possibilities, we assayed functional DNMT activity in Dicer1 −/− ES cells using two experimental strategies. The first strategy was the continued passage of Dicer1 −/− cells in order to assay for the longer-term stability of functional DNMT activity. Dicer1 −/− EP lines had reduced DNA methylation at the MinS repeats and TEs. The significance of this effect in respect to DICER-deficiency is unclear, as similar reductions were also seen in the parental Dicer1 c/− LP cell line, and is sometimes evident in WT cell lines in other studies. The reductions were correlated with low amounts of DNMT3B, and this may have been causal. However, in one of these cell lines the DNMT3A2 level was not reduced and this enzyme alone is sufficient for normal methylation at dispersed repeats [21]. In any event, if the methylation reductions in the Dicer1 −/− EP lines were due to an irreversible and critical reduction in DNMT activity, then the levels of methylation would further decrease. Instead, under this regime, no line lost methylation at TE or MajS sequences on passage. Thus, over prolonged passage, almost certainly attaining higher passage numbers in Dicer1 −/− ES cells than have previously been investigated, fully functional DNMT activity was present.
The results obtained with LP cells did not clearly indicate if, specific to EP, there was some compromise in DNMT activity. There would seem no practical method by which to directly measure this activity in these cell lines per se. Therefore, a second experimental strategy was devised to determine the amount of functional DNMT activity that is generally present in Dicer1 −/− ES cells very soon after their origination. After stripping the genome of DNA methylation, then ablating Dicer1 function, we found that all methylation-permissive sequences were remethylated at normal kinetics to full capacity, these being satellites, TEs, and non-imprinted single copy sequences. These results show that full DNMT activity is generally present in Dicer1 −/− ES cells in the earliest passages after their origination.
Our examination of Dnmt and Rbl2 mRNA levels and their respective protein levels in Dicer1 −/− ES cells are not in support of previous studies which concluded that increased amounts of the transcriptional repressor RBL2 leads to downregulation of Dnmt gene activity [12], [15]. These conclusions were based predominantly on the finding that Rbl2 mRNA was increased, and on the presumption that RBL2 protein was also increased. While we also saw increased Rbl2 mRNA in our Dicer1 −/− lines, immunoblots revealed that RBL2 protein was actually relatively low. Thus, the cause of Dnmt transcriptional downregulation in Dicer1 −/− ES cells would appear to lie elsewhere. Reduced levels of mRNA for all three Dnmts involved in de novo DNA methylation were found in our Dicer1 −/− EP lines. At the protein level, only DNMT3B was reduced, which was inconsistent with other studies that saw only DNMT3A to be reduced in Dicer1 −/− ES cells [15], [16]. Despite these reductions, it should be noted that ES cells are able to retain high levels of DNA methylation at least at TEs despite a substantial reduction in total de novo DNMT activity: Dnmt3a −/− or Dnmt3b −/− ES cells are not generally DNA methylation deficient. Each is able to de novo methylate introduced retroviral sequences at normal rates, and each is stably methylated at TEs. However, they can be deficient at particular sites according to site preference. For example, DNMT3A preferentially methylates MajS repeats and Xist promoter sequences, while DNMT3B preferentially methylates MinS repeats [21], [38].
Our results suggest that H3K9me3, that at least in part directs DNA methylation to MajS repeats, remains intact and functional in ES cells in the absence of si- and miRNAs. While these results indicate that RNAi is not indispensable for the establishment and propagation of heterochromatin in ES cells, it remains possible that it plays some role. Relative to fission yeast, eukaryotes probably possess a higher degree of redundancy in pathways that lead to heterochromatin formation. Even in the former, histone deacetylase activity is able to establish constitutive heterochromatin independent of RNAi [39]. At early stages of mouse preimplantation development, heterochromatinization of pericentric repeats is dependent on their transcription and it has not been discounted that siRNAs contribute to the overall mechanism [40]. Indeed, deep sequencing in mouse ES cells has revealed the presence of satellite-specific small ncRNAs, although these are not DICER1-dependent [14]. Further, relative to somatic cells, which can show substantial defects in heterochromatin on loss of DICER [41], the mammalian POU5F1/OCT4-positive lineage may rely less on RNAi for heterochromatin establishment and propagation.
Previously, it was shown that the transfection of Dicer1 −/− ES cells with Dnmt expression cassettes restored a deficiency in genomic DNA methylation [12]. While this result is consistent with there being a critical reduction in DNMT activity in Dicer1 −/− cells, the transfected Dnmt cDNAs were substantially over-expressed relative to WT, therefore excess DNMT activity could have overridden other mechanisms acting to reduce DNA methylation. There is considerable evidence that the over-expression of DNMTs results in hypermethylation with oncogenic consequences, for example [42]–[44]. Further, production of stable transfectants requires considerable further propagation of the cells, and as demonstrated here, this alone can result in increased DNA methylation in Dicer1 −/− cells.
Defective DNA methylation is also a feature of XX ES cells [27], [45], thereby bearing a resemblance to the phenomenon being examined here. However, the effect in XX ES cells can be substantial, with up to 50% reduction in the total amount of DNA methylation relative to XY and XO ES cells. This reduction does appear attributable to lowered levels of DNMT3A and DNMT3B, in particular the former, as the restoration of physiological amounts of these proteins through stable transfection resulted in normalization of DNA methylation status [45]. The mechanism causing reduced Dnmt3a and Dnmt3b expression in XX ES cells is unknown, although it was speculated that the loss may be attributable to the excessive expression of an X-linked de novo Dnmt repressor [45]. This repressor could not be the Mir290 gene family, as this maps to Chr 7. Overall, the demethylation phenomenon in XX ES cells is more pronounced than that seen in Dicer1 −/− ES cells, and appears to involve different mechanisms.
We have obtained no convincing evidence that the reductions in DNA methylation sometimes seen in Dicer1−/− ES cells, and also seen here, are the result of a critical reduction in DNMT activity. Certainly, we show that reduced DNMT activity, and a reduced capacity for DNA methylation, are not consistent features of DICER1-deficient ES cell lines. It is conceivable that there could be other mechanisms by which DNA methylation could be lost in ES cells lacking DICER1. These ES cells are defective in the G1-S phase transition due to the loss of Mir290 family miRNAs, and consequently have a greatly extended cell cycle [46]. This extended time in phase G1 might result in a higher probability of stochastic changes in DNA methylation patterns that could then be propagated—particularly if they offered a growth or survival advantage in culture. Also, the loss of miRNAs might result in a dysregulation of gene expression that works against the stable retention of DNA methylation. For example, this could involve DNMT mislocalization, or even an enhancement of DNA demethylation pathways. While the latter are not clearly defined, there is strong evidence for their existence in mouse ova, primordial germ cells and some somatic cells [47]. Irrespective of the actual mechanism for the loss of DNA methylation seen in DICER1-deficient ES cells, the effect appears to be sporadic and transient, and is not a salient or major epigenetic alteration resulting from loss of the RNAi machinery.
Materials and Methods
Ethics statement
This work was performed in accordance with the ‘Australian Code of Practice for the Care and Use of Animals for Scientific Purposes’ published by the National Health and Medical Research Council (Australia) and as approved by the Animal Ethics Committee of The University of Melbourne.
Derivation of ES cells, gene targeting, and ablation of Dicer1
ES cell lines were derived from (Dicer1 c/c ♀×Dicer1 +/− ♂) matings in mouse strain 129S1/SvImJ and propagated using mitomycin C inactivated STO-neo-LIF (SNL) feeder cells [18], [48]. One euploid XY Dicer1 c/− ES cell line—named DC8—was selected for further experiments. Dicer1 c/− ES cells were mutated to Dicer1 −/− by electroporating 107 cells with 25 µg of the circular pCAGGS-Cre vector [49]. One thousand cells were seeded onto 6 cm plates and after 8 days of growth colonies were picked into wells and expanded. In Southern blots, Dicer1 +/+, Dicer1 c/− and Dicer1 −/− genotypes were identified after EcoRI digestion by a 7.7 kb, a 4.0 and 5.6 kb, and a 5.6 kb band, respectively, using a 0.75 kb probe amplified from genomic DNA using primers 5′-CCAA GAAC CCAT AGCT TCCC ATC-3′ and 5′-TCAG ACAA CTGT TACG GTGT CGTG-3′. The Dicer1 +/+ cell line used was 2A: euploid, XY, strain 129S1/SvImJ [50].
The Hprt targeting construct consisted of a 6.8 kb genomic fragment in which an expression cassette was cloned into a unique XhoI site in exon 3 [33], [51]. The mU6 promoter sequence identical to that in the pSilencer 1.0 vector, (Ambion), together with cloning sites, was synthesized (Integrated DNA Technologies, Coralville, IA, USA). The short hairpin sequence [35] was cloned into inverted BsmBI sites, and a G418 resistance cassette—mouse Pgk1 (phosphoglycerate kinase 1) promoter driving a synthetic neo coding sequence (sneo) [52] and the bovine growth hormone poly(A) signal sequence—was inserted downstream into a unique AscI site. These cassettes were inserted in the same transcriptional orientation as Hprt (Figure 4A). Ten million DC8 ES cells electroporated with 25 µg of the linearized targeting vector (Figure 3A) were plated per 10 cm plate containing SNL feeder cells that are G418 and 6TG resistant. G418 selection (175 µg/mL active weight) began 1 day after plating, while 6TG selection (10 µg/mL) was initiated after another 4 days with G418 selection being continued. Dual resistant clones were picked after another 3 days.
Isolation of genomic DNA and RNA
At the final passage before harvesting, all cells in a confluent 6 cm plate of ES cells were passaged into one gelatinized 10 cm plate without feeder cells and grown for 2 days. ES cells were then further enriched by differential adherence, resulting in <2% level of SNL feeder cell contamination [31]. For genomic DNA extraction, these cells were pelleted, resuspended in 4 mL of phosphate buffered saline, then four pellets of equal size derived in four 1.5 mL tubes. Each pellet was incubated in 0.5 mL of lysis buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 100 mM EDTA, 1% SDS; with no protease added) (37°C, 2 h or room temperature, 16 h or longer). To pellet protein and SDS, 0.17 mL of 10 M ammonium acetate was added, the tube vortexed then centrifuged (14,000× g,10 min). The supernatant was removed and mixed thoroughly with an equal volume of isopropanol to precipitate DNA, then a centrifuge pulse applied. The pellets were washed in 1 mL 70% ethanol. Air dried pellets were resuspended in 50 µL of TE buffer. RNA was isolated using TriReagent (Ambion).
qPCR
For assay of miRNA content, TaqMan MicroRNA assays (Applied Biosystems) were used. Ct values were normalized to values for Rps7 (ribosomal protein S7) obtained using a SYBR Green assay as described [53]. For assay of mRNA content, TaqMan qPCR assays were designed using PrimerExpress software (Applied Biosystems) for use in multiplex reactions: 4-plex (Rps7, Alpl, Dnmt1, Dnmt3a mRNAs), 4-plex (Rps7, Alpl, Dnmt3b, Dnmt3l mRNAs), and 3-plex (Rps7, Alpl, Rbl2 mRNAs). The Rps7 transcript is ubiquitous while the Alpl (alkaline phosphatase, liver/bone/kidney) gene is expressed at a relatively high level in ES cells and germ cells. For each sample, two reverse transcription (RT) reactions with oligo-dT and triplicate PCR reactions for each RT were performed, yielding six Ct values. These values were converted to the relative amount of mRNA by multiplying the fold increase in product per cycle (equivalent to amplification efficiency) to the power of ΔCt. Values were normalized using combined Rsp7 and Alpl values. The TaqMan qPCR primers and probe for Dnmt3b mRNA detected only the Dnmt3b1 and -2 isoforms, these being the only isoforms encoding active protein, while those for Dnmt3a mRNA detected the two existent isoforms, Dnmt3a1 and -2, each encoding an active protein [21]. Primer and probe (Integrated DNA Technologies) sequences are provided (Text S1).
DNA methylation analysis
For Southern blots, genomic DNA was digested for 6 h with a fivefold excess of restriction enzyme (New England Biolabs, Ipswich, MA, USA) in a minimum volume. This resulted in complete digestion as confirmed in blots hybridized with a probe specific for mitochondrial DNA. Digested DNA was loaded at 1 µg/well for repetitive targets, or 5 µg/well for single copy targets. Blots were performed with Hybond XL membrane according to the supplied protocol (GE Healthcare), except that the pre- and hybridization solution was 5× SSPE, 5× Denhardt's solution, 1% SDS. All gels were depurinated. Blots were washed twice (15 min, 65°C) with 0.2× SSC, 0.5% SDS (single copy targets) or 0.1× SSC, 0.5% SDS (repetitive targets). Probes: Detection of homologous recombination at Hprt—0.25 kb PCR product made using primers 5′-TCAC TGAT ACTT CATA TCAC A-3′ and 5′-GCCT AAGA ATTG CTAT TGAA T-3′. MajS and MinS—as previously described [54]. L1, pL1-ORF1—0.92 kb EcoRI fragment representing a conserved portion of open reading frame 1 (ORF1), amplified with primers 5′-AACA CTGC TAAA GAGT TACA AGTC C-3′ and 5′-CCGT CCTT GTAT TGGT TTTT TTCT G-3′. ERV2, pERV2-ORF1—0.52 kb EcoRI fragment representing a conserved portion of ORF1, amplified with primers 5′-TTCA GGAC AAGC TATC AGAA G-3′ and 5′-AATG AATG AGTC TGCG CACT G-3′. Sequences of pL1-ORF1 and pERV2-ORF1 inserts were validated by sequencing. Y Chr centromere, pYmin2.3a [55]. Mitochondrial DNA, pMIT1—0.66 kb EcoRI fragment amplified with primers 5′-GCAC ACAC CGCC CGTC AC-3′ and 5′-GGTT TTTT CCGT TCCA GAAG AGC-3′. All amplified fragments were cloned into vector pGEM-T Easy (Promega) and verified by sequencing.
For bisulphite sequencing analysis, genomic DNA isolated as above was converted with sodium bisulphite using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). Three separate PCR reactions were performed for each sample with a similar number of clones sequenced from each. Sequencing showed that the conversion rate of Cs to Ts in non-CpGs was complete. Primers were designed with MethPrimer <http://www.urogene.org/methprimer/index1.html>. EpiTYPER assays were performed as previously described [56]. Primers were according to the EpiPanel (Sequenom) or designed with EpiDesigner <www.mysequenom.com>.
Immunoblots and immunofluorescence
Immunoblots were performed with Hybond P membrane according to the supplied protocols (GE Healthcare). Primary antibodies: anti-DNMT1 (Imgenex, San Diego, CA, USA; IMG-261A), Anti-DNMT3A2 (Millipore; 07-2050), Anti-DNMT3B (Abcam, Cambridge, MA, USA; ab122932), Anti-DNMT3L (Millipore; ABD78), Anti-RBL2/p130 (Abcam; ab89457). Secondary antisera: Goat Anti-mouse IgG, HRP conjugate (Millipore; 12-349) or goat Anti-rabbit IgG, HRP conjugate (Millipore; 12-348). Immunofluorescence was performed as previously described [57], [58]. Primary antibodies: Anti-H3K9me3 (Millipore; 07-523), Anti-CBX5 (Millipore; MAB3584), Anti-H4K20me3 (Abcam; ab9053), and CREST (supplied by the Royal Melbourne Hospital, Australia). Secondary antisera: donkey anti-mouse or anti-rabbit Alexa Fluor 488 -594, or -647 (Life Technologies).
Supporting Information
DNA methylation analysis of single copy sequences. Numerical data graphically depicted in Figure 4A.
(XLSX)
Immunofluorescence for markers of pericentric heterochromatin. (A) Panels: First row; metaphase spreads were probed with an antibody specific for H3K9me3. Second row; CREST antibody is specific for the centromere. Third row; merge of the two images directly above, concomitant with detection of DNA with DAPI, reveals the pericentric localization of H3K9me3. (B) As for A, except that CBX5 (synonym HP1α) is shown to be localized to the pericentric region. (C) As for A, except that H4K20me3 is shown to be localized to the pericentric region. Dicer1 c/− panels at left are the EP parental cell line.
(PDF)
Characteristics of Dicer1 −/− ES cell lines. Characteristics surveyed are indicated at left. Green, yellow and blue cell background indicates normal, low and high levels or content, respectively. The letters inside the cells indicate the assay method used: BS, bisulphite sequencing assay; ChIP DB, chromatin immunoprecipitation followed by dot blot; COBRA, comined bisulphite restriction analysis; ET, EpiTYPER assay; IB, immunoblotting; IF, immunofluorescence; MA, microarray; NB, northern blot; nd, not done; PCR, polymerase chain reaction; RTPCR, reverse transcription PCR; SB, Southern blot; qPCR, quantitative PCR.
(XLSX)
Primers and sequences. Sequences of primers used in qPCR, bisulphite sequencing assays, EpiTYPER assays and stem-loop cloning are shown. For insertion of the shRNA sequence to be driven by the mU6 promoter, each of two pairs of oligos as shown were preannealed, phosphorylated, then combined and ligated into the inverted BsmBI sites (red highlight). This strategy avoided cloning with full-length oligos containing a stem-loop. The functional guide strand (green highlight), which forms one strand of the siRNA as produced by DICER1 processing of the shRNA, is exactly complementary to the Dnmt1 mRNA sequence. The G418 selection cassette was cloned into the unique AscI site in the orientation as indicated (>). To complete the targeting vector, the cassette was excised with XhoI and SalI and cloned into the unique XhoI site in exon 3 of the ∼6 kb Hprt genomic fragment. The transcription start site is indicated by the G.
(PDF)
Funding Statement
This work was supported by grants 509311 and 350217 awarded by the National Health and Medical Research Council, Australia, and by the Victorian Government's Operational Infrastructure Support Program, Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, et al. (1982) Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, et al. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337. [DOI] [PubMed] [Google Scholar]
- 3. Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, et al. (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200. [DOI] [PubMed] [Google Scholar]
- 4. Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340. [DOI] [PubMed] [Google Scholar]
- 5. Walsh CP, Chaillet JR, Bestor TH (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117. [DOI] [PubMed] [Google Scholar]
- 6. Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet doi:10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 7. Arnaud P (2010) Genomic imprinting in germ cells: imprints are under control. Reproduction 140: 411–423. [DOI] [PubMed] [Google Scholar]
- 8. Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, et al. (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669. [DOI] [PubMed] [Google Scholar]
- 9. Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, et al. (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grewal SI (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lejeune E, Allshire RC (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265. [DOI] [PubMed] [Google Scholar]
- 12. Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, et al. (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A 102: 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calabrese JM, Seila AC, Yeo GW, Sharp PA (2007) RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A 104: 18097–18102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, et al. (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267. [DOI] [PubMed] [Google Scholar]
- 16. Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, et al. (2008) Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ (2007) Targets of RNA-directed DNA methylation. Curr Opin Plant Biol 10: 512–519. [DOI] [PubMed] [Google Scholar]
- 18. Mattiske DM, Han L, Mann JR (2009) Meiotic maturation failure induced by DICER1 deficiency is derived from primary oocyte ooplasm. Reproduction 137: 625–632. [DOI] [PubMed] [Google Scholar]
- 19. Pfeifer GP, Steigerwald SD, Hansen RS, Gartler SM, Riggs AD (1990) Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A 87: 8252–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, et al. (2002) Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol 22: 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T, Ueda Y, Dodge JE, Wang Z, Li E (2003) Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23: 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, et al. (2004) Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci U S A 101: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S (2004) DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem 279: 27816–27823. [DOI] [PubMed] [Google Scholar]
- 24. Chen ZX, Riggs AD (2005) Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol 83: 438–448. [DOI] [PubMed] [Google Scholar]
- 25. Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chédin F (2005) Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem 95: 902–917. [DOI] [PubMed] [Google Scholar]
- 26. Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, et al. (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11: 805–814. [DOI] [PubMed] [Google Scholar]
- 27. Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, et al. (2010) Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, et al. (2005) Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A 102: 15785–15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, et al. (2008) Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szabó P, Mann JR (1994) Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120: 1651–1660. [DOI] [PubMed] [Google Scholar]
- 32. Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, et al. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev 10: 1008–1020. [DOI] [PubMed] [Google Scholar]
- 33. Tang SH, Silva FJ, Tsark WM, Mann JR (2002) A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32: 199–202. [DOI] [PubMed] [Google Scholar]
- 34. Zheng GD, Hidaka K, Morisaki T (2005) Stable and uniform gene suppression by site-specific integration of siRNA expression cassette in murine embryonic stem cells. Stem Cells 23: 1028–1034. [DOI] [PubMed] [Google Scholar]
- 35. Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, et al. (2004) Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A 101: 10380–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, et al. (2011) Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 39: 5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, et al. (2007) In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324. [DOI] [PubMed] [Google Scholar]
- 38. Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- 39. Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185. [DOI] [PubMed] [Google Scholar]
- 40. Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G (2010) A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 19: 625–638. [DOI] [PubMed] [Google Scholar]
- 41. Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, et al. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6: 784–791. [DOI] [PubMed] [Google Scholar]
- 42. Girault I, Tozlu S, Lidereau R, Bièche I (2003) Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 9: 4415–4422. [PubMed] [Google Scholar]
- 43. Arai E, Kanai Y, Ushijima S, Fujimoto H, Mukai K, Hirohashi S (2006) Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int J Cancer 119: 288–296. [DOI] [PubMed] [Google Scholar]
- 44. Roll JD, Rivenbark AG, Jones WD, Coleman WB (2008) DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer 7: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, et al. (2005) Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet 37: 1274–1279. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen ZX, Riggs AD (2011) DNA methylation and demethylation in mammals. J Biol Chem 286: 18347–18353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mann JR (2001) Deriving and propagating mouse embryonic stem cell lines for studying genomic imprinting. Methods Mol Biol 181: 21–39. [DOI] [PubMed] [Google Scholar]
- 49. Araki K, Imaizumi T, Okuyama K, Oike Y, Yamamura K (1997) Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. J Biochem 122: 977–982. [DOI] [PubMed] [Google Scholar]
- 50. Szabó PE, Han L, Hyo-Jung J, Mann JR (2006) Mutagenesis in mice of nuclear hormone receptor binding sites in the Igf2/H19 imprinting control region. Cytogenet Genome Res 113: 238–246. [DOI] [PubMed] [Google Scholar]
- 51. Deng C, Capecchi MR (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol 12: 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jin SG, Mann JR (2005) Synthetic neomycin-kanamycin phosphotransferase, type II coding sequence for gene targeting in mammalian cells. Genesis 42: 207–209. [DOI] [PubMed] [Google Scholar]
- 53. Lefèvre C, Mann JR (2008) RNA expression microarray analysis in mouse prospermatogonia: identification of candidate epigenetic modifiers. Dev Dyn 237: 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kalitsis P, Fowler KJ, Earle E, Griffiths B, Howman E, et al. (2003) Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res 11: 345–357. [DOI] [PubMed] [Google Scholar]
- 55. Pertile MD, Graham AN, Choo KH, Kalitsis P (2009) Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res 19: 2202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wong NC, Ashley D, Chatterton Z, Parkinson-Bates M, Ng HK, et al. (2012) A distinct DNA methylation signature defines pediatric pre-B cell acute lymphoblastic leukemia. Epigenetics 7: 535–541. [DOI] [PubMed] [Google Scholar]
- 57. Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, et al. (2000) Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 10: 1319–1328. [DOI] [PubMed] [Google Scholar]
- 58. Wong LH, Saffery R, Anderson MA, Earle E, Quach JM, et al. (2005) Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J Biol Chem 280: 3954–3962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA methylation analysis of single copy sequences. Numerical data graphically depicted in Figure 4A.
(XLSX)
Immunofluorescence for markers of pericentric heterochromatin. (A) Panels: First row; metaphase spreads were probed with an antibody specific for H3K9me3. Second row; CREST antibody is specific for the centromere. Third row; merge of the two images directly above, concomitant with detection of DNA with DAPI, reveals the pericentric localization of H3K9me3. (B) As for A, except that CBX5 (synonym HP1α) is shown to be localized to the pericentric region. (C) As for A, except that H4K20me3 is shown to be localized to the pericentric region. Dicer1 c/− panels at left are the EP parental cell line.
(PDF)
Characteristics of Dicer1 −/− ES cell lines. Characteristics surveyed are indicated at left. Green, yellow and blue cell background indicates normal, low and high levels or content, respectively. The letters inside the cells indicate the assay method used: BS, bisulphite sequencing assay; ChIP DB, chromatin immunoprecipitation followed by dot blot; COBRA, comined bisulphite restriction analysis; ET, EpiTYPER assay; IB, immunoblotting; IF, immunofluorescence; MA, microarray; NB, northern blot; nd, not done; PCR, polymerase chain reaction; RTPCR, reverse transcription PCR; SB, Southern blot; qPCR, quantitative PCR.
(XLSX)
Primers and sequences. Sequences of primers used in qPCR, bisulphite sequencing assays, EpiTYPER assays and stem-loop cloning are shown. For insertion of the shRNA sequence to be driven by the mU6 promoter, each of two pairs of oligos as shown were preannealed, phosphorylated, then combined and ligated into the inverted BsmBI sites (red highlight). This strategy avoided cloning with full-length oligos containing a stem-loop. The functional guide strand (green highlight), which forms one strand of the siRNA as produced by DICER1 processing of the shRNA, is exactly complementary to the Dnmt1 mRNA sequence. The G418 selection cassette was cloned into the unique AscI site in the orientation as indicated (>). To complete the targeting vector, the cassette was excised with XhoI and SalI and cloned into the unique XhoI site in exon 3 of the ∼6 kb Hprt genomic fragment. The transcription start site is indicated by the G.
(PDF)