Abstract
Analysis of microbial communities by high-throughput pyrosequencing of SSU rRNA gene PCR amplicons has transformed microbial ecology research and led to the observation that many communities contain a diverse assortment of rare taxa–a phenomenon termed the Rare Biosphere. Multiple studies have investigated the effect of pyrosequencing read quality on operational taxonomic unit (OTU) richness for contrived communities, yet there is limited information on the fidelity of community structure estimates obtained through this approach. Given that PCR biases are widely recognized, and further unknown biases may arise from the sequencing process itself, a priori assumptions about the neutrality of the data generation process are at best unvalidated. Furthermore, post-sequencing quality control algorithms have not been explicitly evaluated for the accuracy of recovered representative sequences and its impact on downstream analyses, reducing useful discussion on pyrosequencing reads to their diversity and abundances. Here we report on community structures and sequences recovered for in vitro-simulated communities consisting of twenty 16S rRNA gene clones tiered at known proportions. PCR amplicon libraries of the V3–V4 and V6 hypervariable regions from the in vitro-simulated communities were sequenced using the Roche 454 GS FLX Titanium platform. Commonly used quality control protocols resulted in the formation of OTUs with >1% abundance composed entirely of erroneous sequences, while over-aggressive clustering approaches obfuscated real, expected OTUs. The pyrosequencing process itself did not appear to impose significant biases on overall community structure estimates, although the detection limit for rare taxa may be affected by PCR amplicon size and quality control approach employed. Meanwhile, PCR biases associated with the initial amplicon generation may impose greater distortions in the observed community structure.
Introduction
High-throughput pyrosequencing of PCR amplicons has emerged as a valuable technique in microbial ecology and revealed, in unprecedented detail, the microbial diversities found in various marine and terrestrial environments [1]–[9] and the human microbiome [10]–[13]. The power of this approach lies in the read depth achieved, where tens to hundreds of thousands of individual sequencing reads are simultaneously generated and used to estimate the composition and abundance of microbial operational taxonomic units (OTUs) in a given community. However, this high read depth comes at a cost of relatively high error rates for individual reads obtained using commonly employed sequencing technology (i.e., Roche 454 GS FLX with Titanium chemistry, 454-Ti) [14]. In the context of genomic (re-) sequencing, low consensus error rates are achieved through sequence assembly; however, for PCR amplicons, redundancy is indistinguishable from abundance, and the high error rates associated with individual reads therefore contribute to over-estimation of diversity since erroneous reads manifest themselves as less abundant but closely related OTUs [15].
A number of attempts have been made to assess and address the impact of 454 single read errors on the estimation of community richness. These efforts have primarily addressed the accuracy of OTU diversity estimates, with special attention paid to enumeration of OTUs within the Rare Biosphere [6], [15]–[17]. One consistent finding has been that standard techniques for processing amplicon pyrosequencing data can result in the detection of several hundred “false” OTUs, mostly at low abundance, even from a single test organism [15]. Those findings have raised concerns that species abundance can be overestimated for amplicon pyrosequencing data. Subsequently, more stringent approaches have been developed that allow the abundances of error-containing reads to be counted toward those of the more abundant, supposedly error-free, reads from which they arose [16]–[21].
Computational strategies employed by these newly developed “de-noising” methods fall into three categories: 1) identity-based clustering, where de-noising is achieved by aligning and clustering nucleotide sequences (e.g., single-linkage pre-clustering, SLP [17]; CD-HIT-OTU, http://weizhong-lab.ucsd.edu/cd-hit-otu; and “otupipe”, http://drive5.com/otupipe/); 2) non-alignment clustering, which utilizes K-mer clustering rather than alignment-based distance calculations to de-noise reads [22] or even directly assign reads to OTUs [20]; 3) flowgram-based clustering, where information obtained by clustering pyrosequencing flowgrams is incorporated into the de-noising pipeline [18], [19], [21], [23]. All these methods also use quality filters perceived to be correlated with low read accuracy, such as abnormal read length, mismatch to barcode and/or PCR primer, and low quality score. To examine and compare the performance of these different approaches in accurately recovering community structures, we chose three published methods, SLP [17], PyroTagger [20], and AmplionNoise [19], to represent the three categories, respectively.
All de-noising pipelines assign the abundance of a “true” amplicon sequence as the sum of its own abundance and those of “noise” reads that arose from it, removing “noise” reads from the dataset in the process. However, different strategies are employed by each de-noising pipeline to determine the sequence identity of the “true” read (i.e., picking the representative sequence of each OTU). Ultimately, the fidelity of representative sequences is important for accurate taxonomic assignment and phylogenetic analysis. Moreover, over-aggressive removal of noise through clustering inevitably leads to incorrect clustering of genuine but closely related sequences that may correspond to highly distinct ecotypes [24].
A wide array of factors affects the determination of microbial community structure from 16S rRNA gene amplicons. PCR amplicon size has been suggested to impact observable diversity [25], ostensibly due to lower amplification/cloning efficiency for longer amplicons; although PCR amplicon size and primer choice are inevitably linked, and their effects are difficult to separate [26], [27]. Additional PCR biases, including primer mismatch [28], [29], differential amplification efficiency [30], [31], and differential annealing efficiency [29], can also affect observed diversity and structure. These issues, when combined with the high error rates discussed above, can distort estimates of community taxonomic richness and abundance.
The de-noising strategies outlined above have not been examined in regards to sensitivity for genuinely rare taxa or accuracy of estimated community structure. For comparative studies in particular, it is essential that the recovered read frequencies can be reliably interpreted as evidence of population abundances. Furthermore, ensuring that rare reads truly indicate rare taxa is important since they constitute the philosophical basis of the modern Rare Biosphere concept [6]. Therefore, the potential influence of the post-PCR pyrosequencing workflow on observed microbial community structure and diversity remains under-examined. A thorough investigation of this topic requires a priori knowledge of community composition and structure.
In this study, we utilized six different in vitro-simulated communities (iv-SCs) of 16S rRNA gene PCR amplicons to characterize biases associated with microbial community structure reconstruction using pyrosequencing data. To achieve this, potential skews in observed community structure, the practical detection limit for rare taxa, and the effects of PCR bias in the initial PCR step were all examined and assessed for their implications on the application of this technique for microbial ecology research.
Results
Community Diversity and Structure from PCR-Neutral Communities
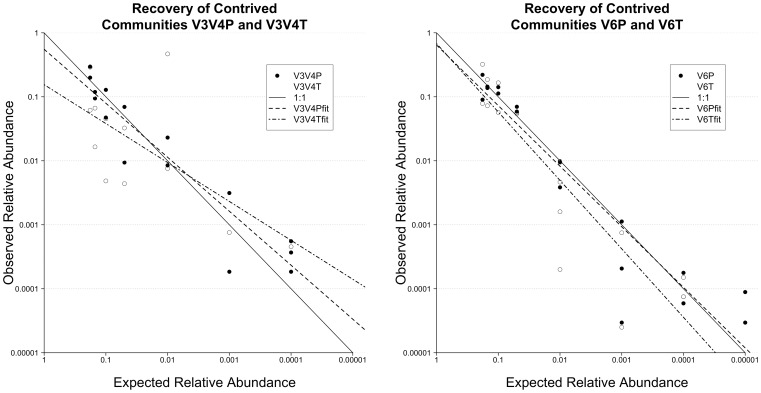
PCR-independent in vitro-simulated communities (iv-SCs) V3V4P and V6P tested the neutrality of 454-Ti pyrosequencing as they were constructed using individually generated amplicons pooled at known abundances (Table 1). Of the 20 original sequences present in each dataset, 19 (95%) were recovered for V6P (36,394 reads), but only 15 (75%) were recovered for V3V4P (9,787 reads, Table S1). The frequency of each known sequence within these iv-SCs was recovered based on the numbers of corresponding error-free reads (i.e., sequences generated by the 454 base-calling software with default parameters that perfectly matched known sequences) (Table S2). The sole sequence missing from V6P was clone LMMI-24 in the lowest frequency tier (0.001%). Clone sequences absent from the V3V4P iv-SCs included all three sequences at 0.001% frequency, one sequence at 0.1%, and one of the three sequences expected at 1%. However, the higher number of sequences recovered from V6P was likely due to its higher accurate read count. Of the sequences recovered from iv-SCs V3V4P and V6P, observed relative abundances were generally in agreement with expected frequencies, although deviations exceeding 10-fold did occur at low expected frequencies (Figure 1). The correlation between observed and expected frequencies was consistent for both the V3V4P and V6P (PCR-controlled) communities (Table 2), with V6P resampled to match the number of error-free reads for V3V4P.
Table 1. Expected relative abundances of each 16S rRNA gene-containing plasmid (E and T) or amplicon (P) in the in vitro-simulated communities (iv-SCs).
| Community | |||
| 16S rRNA gene clone | Equal (E) | Tiered (T) | Tiered PCR Product (P) |
| 4–3Okaro10‡ | 0.05 | 0.18 | 0.18 |
| SC8-3† | 0.05 | 0.18 | 0.18 |
| SC7-1† | 0.05 | 0.15 | 0.15 |
| LMM1-5‡ | 0.05 | 0.15 | 0.15 |
| SC1-5† | 0.05 | 0.1 | 0.1 |
| 3-9‡ | 0.05 | 0.1 | 0.1 |
| 23-7‡ | 0.05 | 0.05 | 0.05 |
| 30-1‡ | 0.05 | 0.05 | 0.05 |
| 19-3† | 0.05 | 0.01 | 0.01 |
| 16-1† | 0.05 | 0.01 | 0.01 |
| 1216C** | 0.05 | 0.01 | 0.01 |
| SC5-2† | 0.05 | 0.001 | 0.001 |
| 29-2† | 0.05 | 0.001 | 0.001 |
| Forsyth-N6† | 0.05 | 0.001 | 0.001 |
| Waahi-22‡ | 0.05 | 0.0001 | 0.0001 |
| SC4-1‡ | 0.05 | 0.0001 | 0.0001 |
| 3-1† | 0.05 | 0.0001 | 0.0001 |
| 6-1† | 0.05 | 0.00001 | 0.00001 |
| EF222209‡ | 0.05 | 0.00001 | 0.00001 |
| LMM1-24‡ | 0.05 | 0.00001 | 0.00001 |
Rueckert et al. 2007.
Rueckert Personal Communication.
Banks et al. 2009.
Figure 1. The relative abundances of recovered reads in V3V4P and V3V4T iv-SCs ( Figure 1A ) and V6P and V6T iv-SCs ( Figure 1B ) are plotted against their respective theoretical relative abundances.
The solid lines represent the ideal 1∶1 scenario (i.e., observed matching expected perfectly).
Table 2. Spearman rank (ρ) and log-log transformed Pearson (r) correlation coefficients of error-free sequences with their respective theoretical frequencies.
| Community | Spearman ρ | Pearson r | |
| V3V4 | V3V4P | 0.941 | 0.943 |
| V3V4T | 0.596 | 0.669 | |
| V6 | V6P | 0.928 {0.899, 0.950} | 0.961 {0.923, 0.986} |
| V6T | 0.923 {0.887, 0.952} | 0.911 {0.855, 0.967} | |
The pools of error-free sequences for V6P and V6T (33,804 and 39,978 reads respectively) were resampled 10,000 times with replacement to match the numbers of V3V4P and V3V4T error-free sequences (5,424 and 6,607 reads respectively). The correlation coefficients for each bootstrap were calculated and presented as means and 95% confidence intervals. The bootstrapping p values (testing the V6x correlation coefficients as higher than the V3V4x equivalents) were 0.814 (ρ) and 0.152 (r) for resampled V6P vs. V3V4P and <0.001 (ρ and r) for resampled V6T vs. V3V4T.
Effects of PCR Biases
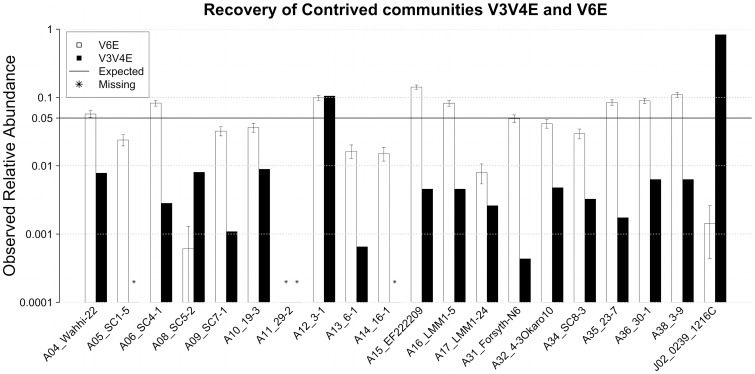
To examine the degree to which PCR biases are sufficient to induce a non-uniform community structure into a uniform community of template DNA, an iv-SC set was constructed with twenty plasmids at equal abundances (Table 1). This set of iv-SC (V3V4E & V6E) was generated using two separate PCR assays, targeting the V3–V4 and V6 regions of 16S rRNA gene, respectively. Analysis of error-free reads from these iv-SCs revealed non-uniform frequency distributions of sequences (Figure 2). The observed bias does not appear to have been caused by quantification error, as the bias observed for sequence 1216C in V3V4E was so extreme that it accounted for 83% of the total dataset. Meanwhile, this clone was significantly under-represented in V6E, accounting for only 0.14% of the reads.
Figure 2. The observed relative abundances of all error-free sequence in the equal-abundance iv-SCs (V3V4E: red; V6E: blue).
The pool of error-free sequences for V6E (14,761 reads) was resampled 10,000 times with replacement to match the number of V3V4E error-free sequences (4,609 reads) and used to calculate 95% confidence intervals for V6E.
The influence of PCR biases on a tiered community structure was also examined using iv-SCs V3V4T and V6T. These amplicons were generated using twenty plasmids, pooled at tiered abundances, as PCR template (Table 1). In general, observed clone frequencies were similar to expected ones (Figure 1 & Table 2). However, as seen in iv-SC V3V4E, the preferential amplification of the V3–V4 region of clone 1216C was severe in V3V4T and resulted in this single sequence comprising nearly half of the total reads obtained (Figure 1A and Table S2). Overall, the observed bias in favor of a single sequence depressed the observed frequencies for other sequences and thus skewed the observed community structure. This resulted in a significantly worse correlation between the observed and expected relative abundances for the longer V3V4T amplicon community than either the V6T community (Table 2) or the PCR-controlled V3V4P community (Table 2). This difference in correlation was ameliorated by removing sequence 1216C from the analysis (Table 3).
Table 3. Spearman rank (ρ) and log-log transformed Pearson (r) correlation coefficients of relative abundances of corresponding sequences in P and T iv-SCs.
| Comparison | N | Spearman ρ | Pearson r |
| V6P vs. V6T | 16 | 0.973 | 0.953 |
| V3V4P vs. V3V4T | 12 | 0.748 | 0.794 |
| V3V4P vs. V3V4T (Excluding 1216C) | 11 | 0.936 | 0.936 |
Impact of PCR Primer Mismatch on Observed Relative Abundances
The original sequences of all 20 clones (obtained using bi-directional Sanger sequencing) were examined for PCR primer mismatches that may have contributed to observed frequency biases (Table S3). Seventeen clones exactly matched both V6 primers (968F & 1046R, Table 4), with single nucleotide mismatches in the remaining three clones (Table S3). Conversely, only one clone (1216C) exactly matched both V3–V4 primers (338F & 806R, Table 4). The remaining 19 clones had mismatches of up to 5 nucleotides (Table S3). For iv-SCs V3V4E and V3V4T, the number of PCR primer mismatches was significantly and negatively correlated with observed/expected ratios (nonparametric Spearman correlation analysis excluding clones 19-3 and 6-1; V3V4E: p = 0.007; V3V4T: p = 0.015;). The same was true for V6E (nonparametric Mann-Whitney test; p = 0.0081), but not V6T (p = 0.0626).
Table 4. Unidirectional hybrid PCR primers; 454 adapter sequence in italic, MID sequence in brackets.
| Primer Name | Primer Sequence |
| V3V4E_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACACGTACAG]ACTCCTACGGGAGGCAGCAG |
| V3V4T_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACACACGTCG]ACTCCTACGGGAGGCAGCAG |
| V3V4P_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACACGTCTCG]ACTCCTACGGGAGGCAGCAG |
| V3V4_Reverse | CCTATCCCCTGTGTGCCTTGGCAGTCTCAG GGACTACCAGGGTATCTAAT |
| V6E_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACAGTACGCG]AACGCGAAGAACCTTACC |
| V6T_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACACTACGAC]AACGCGAAGAACCTTACC |
| V6P_Forward | CCATCTCATCCCTGCGTGTCTCCGACTCAG[ACGACACTAG]AACGCGAAGAACCTTACC |
| V6_Reverse | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGACAGCCATGCANCACCT |
Overview of Pyrosequencing De-noising Strategies
Three recently published algorithms for de-noising 16S rRNA gene PCR amplicon pyrosequencing libraries, SLP [17], PyroTagger [20], and AmpliconNoise [19], were examined for their ability to accurately reconstruct community structure and diversity using the PCR-independent iv-SCs (V3V4P & V6P). Unique reads determined by these de-noising pipelines typically represent multiple error-free and error-containing reads, the latter presumably derived from the former. Each algorithm identifies a set of presumably error-free (“true”) reads, which determine the eventual accuracy of identified OTUs.
Community Structure Estimated in the Presence of Error-Containing Reads
To understand the behavior of each de-noising algorithm and workflow, we devised a classification scheme for OTUs comprised of read predictions. An OTU containing at least one unique read prediction (predicted by de-noising algorithm) that correctly matched one of the twenty reference clone sequences was designated a “true” OTU. An OTU containing raw reads that correctly mapped to one of the 20 reference clone sequences but whose read predictions all contained at least one error was designated as a “miscalled” OTU. An OTU comprised entirely of reads that did not match any of the 20 reference sequences was designated a “false-derived” OTU. Other designations included “near-match” OTUs, which contained sequences matching closely to a reference sequence not found in any “true” or “miscalled” OTUs; “contamination” OTUs, which generally represented E. coli vector contamination; and “chimeric” OTUs, which contained chimeric sequences not identified by the chimera-check algorithm. OTUs classified in this manner for iv-SCs V3V4P and V6P are summarized in Table 5 (details in Table S4). Since recommended clustering procedures differ for each de-noising pipeline, the 20 known sequences were clustered using each procedure in a “clustering control” (Table 5). Based on the number of OTUs obtained from the clustering controls, it was clear that the PyroTagger clustering algorithm was overly aggressive since only 12 OTUs were obtained from the 20 V3–V4 reference sequences, considerably fewer than were found by the SLP (16 OTUs) or AmpliconNoise (17 OTUs) clustering procedures (Table 5). The number of OTUs obtained from the V6 clustering controls was the same for all three pipelines.
Table 5. Summary statistics for the community analysis using several de-noising algorithms on the iv-SCs containing 20 known sequences.
| Total OTU | Chao1 Index | True OTU | Miscalled OTU | Near-Match OTU | Contami-nation OTUs | Chimeric OTU | False-Derived OTU | Missing OTU* | Clustering Control | ||
| V3V4P | SLP | 14 | 17 | 8 | 0 | 0 | 6 | 0 | 0 | 7 | 16 |
| AmpliconNoise | 23 | 51 | 4 | 7 | 3 | 5 | 4 | 0 | 1 | 17 | |
| PyroTagger | 15 | 15 | 10 | 0 | 1 | 2 | 2 | 0 | 3 | 12 | |
| V6P | SLP | 35 | 48.75 | 13 | 3 | 2 | 2 | 6 | 9 | 0 | 18 |
| AmpliconNoise | 33 | 49.5 | 14 | 2 | 1 | 3 | 5 | 8 | 0 | 18 | |
| PyroTagger | 22 | 36 | 13 | 1 | 1 | 1 | 3 | 3 | 3 | 18 | |
For each methodology a “clustering control” was run to determine how many OTUs would be expected in the absence of errant reads.
Missing OTU numbers exclude reference sequences that were unidentifiable in the raw dataset (5 missing in V3V4P dataset and 1 missing in V6P dataset).
The ability of de-noising algorithms to identify true OTUs was better for the shorter V6 region than for the longer V3–V4 regions (Table 5). However, a better estimate of the actual number of OTUs was obtained through analysis of the V3––V4 regions (14–23 observed OTUs vs. 22–35 observed OTUs for iv-SC V6P, Table 5). All three de-noising algorithms appear to function similarly well for analysis of the V6 region. For the V3V4P iv-SC, PyroTagger and SLP appear better at predicting true OTUs (10 and 8 OTUs, respectively) than AmpliconNoise (4 OTUs). However, it should be noted that several OTUs were missing completely from the community reconstructions performed with SLP and PyroTagger (7 and 3 OTUs, respectively), whereas AmpliconNoise produced the highest number of relevant (i.e., true + miscalled + near-match) OTUs (Table 5). A closer examination of OTUs missing from SLP reconstruction revealed that reads that should comprise these missing OTUs were present in the original quality-screened dataset and that the SLP de-noising algorithm itself had over-clustered these reads into a single read prediction represented by a true sequence (Table S4). This behavior was only observed for SLP de-noising of the V3V4P iv-SC, and it performed well for V6P iv-SC.
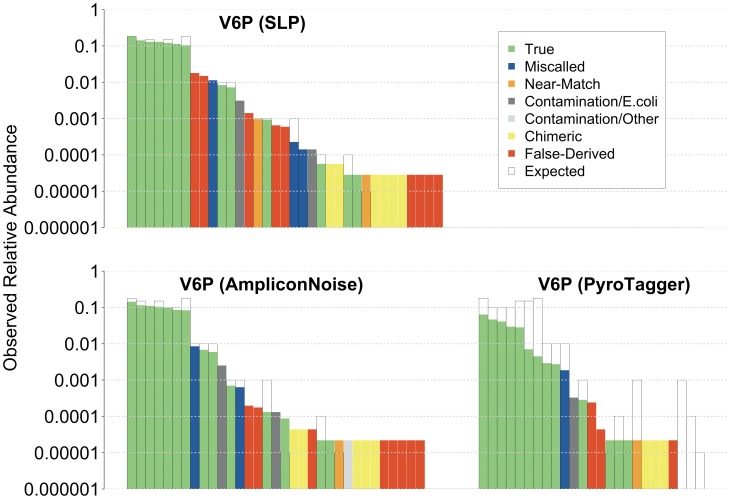
Rank-frequency plots of OTU types generated from the V6P iv-SC (Figure 3) compare observed and expected frequencies for a given clone sequence. Chimeric and false-derived OTUs made up a significant portion of the rare OTUs identified by each de-noising algorithm, and these were indistinguishable from true OTUs at similarly low abundances. False-derived OTUs were observed at >1% relative abundance with SLP, suggesting that even relatively abundant OTUs may be attributable to methodological artifact and that the frequencies of these false-derived OTUs can number as high as 12–13% of the true OTUs from which they are derived (Table S4). The frequencies of false-derived OTUs detected using PyroTagger and AmpliconNoise were notably lower (<0.1%, see Figure 3 and Table S4), but similarly, some of the rare false-derived OTUs had rather high relative abundances to the true OTUs from which they are derived (>27%).
Figure 3. Rank-frequency plots of V6P OTUs generated by SLP, AmpliconNoise, and PyroTagger.
Abundances are shown in log scale. True OTUs (green): OTUs with a reference sequence as its representative; Miscalled OTUs (blue): OTUs containing a reference sequence, but not as its representative; False-Derived OTUs (red): OTUs composed entirely of erroneous reads that are not chimeric, contamination, or closely matching a reference sequence not found in any True or Miscalled OTUs; Near-Match OTUs (orange): OTUs containing sequence(s) that closely match a reference sequence not found in any True or Miscalled OTUs; Contamination/E. coli (dark gray): OTUs composed of sequences affiliated with E. coli (cloning host); Contamination/Other (light gray): OTUs composed of sequences affiliated with potential contaminants; Chimeric OTUs (yellow): OTUs composed of manually identified chimeric sequences; Theoretical (white): expected OTUs.
Correlations between observed and expected OTU frequencies were examined using true, miscalled, and near-match (i.e., relevant) OTUs (Table 6). For the relevant OTUs, community structure estimates based on de-noised reads were not significantly different from those based on error-free reads. It should be noted that the community reconstructed using AmpliconNoise was in marginally better agreement with the expected structure for both the V3V4P and V6P iv-SCs than that from error-free reads. SLP performed similarly well for the V6P iv-SC, but not for V3V4P (Table 6).
Table 6. Spearman rank (ρ) and log-log transformed Pearson (r) correlation coefficients of true and miscalled OTUs identified by de-noising algorithms with their respective expected frequencies.
Pyrosequencing-Specific Chimera Identification
Unique among the pipelines evaluated, AmpliconNoise explicitly integrated a chimera removal algorithm, Perseus, into its analysis pipeline [19]. Perseus was also applied to de-noised reads from SLP and PyroTagger. Examination of datasets inclusive of chimeric reads revealed that although chimeric reads represent a small portion of the overall iv-SC (<1%) (Table 7), they can contribute significantly to overall estimates of OTU richness. Inclusion of chimeric reads increased the number of V3V4P iv-SC OTUs reconstructed using both AmpliconNoise and PyroTagger. A close examination of V3V4P iv-SC OTUs reconstructed with SLP revealed that 128 of the 345 chimeric sequences in the dataset de-noised using AmpliconNoise were also found in the SLP dataset, but these chimeric reads had been “absorbed” into a true OTU by aggressive clustering in the SLP algorithm. Similarly, 20 of these 345 chimeric reads had been “absorbed” into non-chimeric predicted reads by PyroTagger. Perseus did not identify any chimeric reads in the V6P iv-SC, regardless of the de-noising pipeline used. Despite these efforts, several OTUs composed of chimeric reads that had evaded Perseus were manually identified in V3V4P and V6P iv-SCs, and they typically comprised ∼15% of the observed OTUs (Table 5).
Table 7. Summary statistics for chimera-check of iv-SCs.
| Unique De-noised Reads | Replicated De-noised Reads | % of unique reads removed with Perseus (#) | % of replicated reads removed with Perseus (#) | Perseus-Pass OTUs | OTUs Removed by Perseus | ||
| V3V4P | SLP | 15 | 36949 | 0% (0) | 0% (0) | 14 | 0 |
| AmpliconNoise | 70 | 43645 | 34% (24) | 0.8% (345) | 23 | 22 | |
| PyroTagger | 20 | 4475 | 25% (5) | 0.09% (7) | 15 | 5 | |
| V6P | SLP | 35 | 35032 | 0% (0) | 0% (0) | 35 | 0 |
| AmpliconNoise | 36 | 34855 | 0% (0) | 0% (0) | 33 | 0 | |
| PyroTagger | 22 | 10443 | 0% (0) | 0% (0) | 22 | 0 | |
Discussion
The use of Roche 454 GS FLX next generation sequencing has played an instrumental role in introducing the concept of a Rare Biosphere, with this long tail of rare taxa being reported for nearly every community characterized using 454 pyrosequencing [2], [4], [7], [9], [32]–[36]. Although the presence of rare taxa in various environments has been shown using a variety of independent methods [37]–[42], the true frequencies of these taxa, particularly as characterized using pyrosequencing data, remain in question [15], [17]. Moreover, little is known about the accuracy of community structural information derived from the frequency distribution of 16S rRNA gene amplicons within 454 pyrosequencing libraries obtained using the newer Titanium chemistry with longer read lengths.
Overall, our findings show that the 454-Ti sequencing platform provides useful information about microbial community structure since observed and expected frequencies of error-free reads exhibited good correlations (Table 2). Effects of pyrosequencing-specific biases (based on “P” iv-SCs) were exceeded by the impact of PCR biases in mixed template samples (“T” and “E” iv-SCs). For example, nearly half of the error-free reads in V3V4T originated from a single sequence (1216C) at only 1% relative abundance within the template DNA (Table S2), and the positive PCR bias for this sequence in the V3–V4 regions resulted in a significant skew in recovered community structure information (Table 2 and Table 3). Therefore, the presence of one or a few sequences prone to PCR bias can drastically skew observed relative abundances, but the rank frequency distribution of other sequences appears to be preserved (Table 3). Meanwhile, the V6 iv-SCs did not appear to have been subject to significant PCR bias.
Error-containing reads comprised a significant portion of the total reads for all iv-SCs. However, this was not due to quality issues with the sequencing process, as the observed proportions are in fact consistent with a high per-base accuracy (>99.5%, Table S1). Therefore, it would have been impossible to systematically isolate the error-containing reads without a priori knowledge of the community. To resolve this issue, “de-noising” algorithms that employ clustering techniques were used to assign error-containing sequences to the true sequences from which they arose [17]–[23]. Our findings showed that these approaches occasionally infer the wrong “true” sequence from clusters of mixed error-free and error-containing reads, and invariably produced low-abundance false OTUs that are indistinguishable from real ones. These false OTUs can lead to an overestimation of the total number of OTUs in the iv-SCs. In some cases, over-clustering by the de-noising algorithm compensated, albeit incorrectly, for this OTU inflation. Nevertheless, these de-noising algorithms represent a marked improvement over simple, arbitrary quality filters [6], [16] in that they effectively reduce the number of unique error-containing reads that can be mistaken for real sequences.
Although these de-noising pipelines were evaluated in their respective primary publications for the accuracy of recovered richness [17] and relative abundances [19]–[21], this study provides the first independent, explicitly quantitative assessment of their performance using carefully constructed and well quantified in vitro-simulated communities. Given that researchers interpreting results from these pipelines inevitably treat them as quantitatively representative of the biological communities, the results presented here provide a useful assessment of information obtained and disseminated using such methodology. A step-by-step comparison between the three de-noising algorithms was unfeasible due to their integrated pipeline design.
The process of clustering sequencing reads into OTUs traditionally involves three distinct steps: quality filtering, alignment, and clustering. The SLP and AmpliconNoise de-noising step constitutes an independent procedure that occurs after quality filtering but before alignment [17], [19]. PyroTagger instead combines de-noising, alignment and clustering into a single, final step [20]. It should be noted that PyroTagger’s authors pointed out that it may not be suitable for 454-Ti data due to supposedly lower read quality, but given that 454-Ti has become the de facto technology for amplicon sequencing, we felt that an assessment of the unique approach employed by PyroTagger needed to be included. AmpliconNoise was chosen over alternative flowgram-based clustering algorithms for several reasons: 1) it incorporates a number of significant performance improvements over PyroNoise [21]; 2) its implementation allows it to be run on a computer cluster to speed up analysis; 3) it does not incorporate a greedy/heuristic step and thus has better reproducibility (vs. Qiime Denoiser [23], Figure S1). We note that the two central components of AmpliconNoise, PyroNoise and SeqNoise [19], have recently been re-implemented in Mothur as the Shhh.flows command, which was shown to perform comparably to AmpliconNoise under similar circumstances [18].
Correlations between OTU frequencies calculated from de-noised reads and expected OTU relative abundances were similar to those calculated from error-free reads, indicating that these methods can effectively recover error-containing reads while maintaining approximate community structure. All three de-noising approaches identified similar numbers of OTUs that reflected real iv-SC taxa (i.e., true, miscalled and near-known OTUs), but differed in the numbers of false OTUs detected, with PyroTagger outperforming both SLP and AmpliconNoise (Figure 3 and Table 5). However, PyroTagger produced the poorest correlation between observed and expected relative abundances (Table 6) and incorrectly merged reference V3–V4 sequences, indicating a tendency to over-cluster. The stringent quality-based filtering used by PyroTagger also discarded a greater number of raw sequencing reads (data not shown), resulting in the absence of several expected low-abundance taxa from the de-noised dataset (Figure 3 and Table 5).
SLP performed similarly to PyroTagger in predicting species richness within the V3V4P community, but did so by an over-aggressive de-noising procedure that resulted in several real taxa being erroneously grouped into one OTU. This occurred at the de-noising step and was not related to post de-noising clustering procedures (data not shown). Moreover, SLP inferred abundant (>1%) OTUs comprised entirely of error-containing reads in the reconstruction of the V6P iv-SC. Compared to SLP, false-derived OTUs were observed at much lower frequencies (<0.1%) for the V6P iv-SC reconstructed using either PyroTagger or AmpliconNoise. Although more computationally intensive, AmpliconNoise models the distribution of pyrosequencing errors at the flowgram level and is able to robustly assign error-containing reads to their parent error-free reads. AmpliconNoise appears to be free from the over-clustering effect observed with both PyroTagger and SLP, and therefore tends to overestimate OTU richness (Table 5). However, it incorrectly identified the highest number of OTU representative sequences with the V3V4P iv-SC, which may have ramifications for downstream analyses that rely on precise phylogenetic resolution.
Because AmpliconNoise includes a built-in chimera checker, Perseus, it bypasses the need for multiple sequence alignment (MSA) [43] or reference sequences, as recommended for PyroTagger [20]. For typical pyrosequencing amplicon datasets containing thousands of unique sequences, MSA is impractical, as are the use of reference sequences and a priori assumptions about the identity of environmental sequences. The outcome of our analyses shows that AmpliconNoise is the de-noising algorithm least likely to allow chimeric reads to be “absorbed” into read predictions (Table 7), thus affecting abundance estimates. This may partially explain why the correlation between the expected and the observed frequencies of relevant OTUs was highest for the AmpliconNoise pipeline (Table 6).
Rather than using mixtures of genomic DNA preparations, plasmids containing cloned 16S rRNA genes were used for this study. This approach avoided the issues of inter-genomic variations in rrn operon copy numbers, intra-genomic variation in rrn operon sequences, and quantification inaccuracies due to genome size differences [44], thus allowing greater quantitative accuracy. We limited the richness of the iv-SCs to twenty sequences to allow reliable quantification of libraries using both mixed plasmids and PCR products. Given the high proportion of artifactual rare OTUs recovered by all three de-noising pipelines with these relatively simple communities, it is unlikely that a more complex simulated community would have improved their performance. Nineteen of the twenty clones included in the study were from Cyanobacteria isolated from similar environments and are therefore comparatively similar in sequence. This resulted in some of the reference sequences being clustered together, even by the most lenient clustering approach (Table 5), but it also exposed PyroTagger’s tendency to over-cluster and mask genuine diversity (Table 5). The inclusion of one Actinobacteria clone (1216C) allowed us to explore the effects of primer bias on different phylogenetic groups.
Although we had a priori knowledge of the iv-SC sequences, we elected not to customize PCR primers to account for known mismatches and performed the experiment using “universal” primers commonly used for microbial community analyses [25], [27], [28], [45]. Thus, our analyses were subject to the same biases common to any study utilizing these common universal primers against environmental DNA. We also avoided using primers with degenerate bases since primer degeneracy can reduce specificity, lead to exhaustion of effective primers as the reaction progresses [31], [46], and impose biases of its own [47]. Recently, an alternative of using a mixture of non-degenerate primers has been proposed [46], which may significantly increase “universality” while avoiding the pitfalls of degenerate primers.
Numerous mechanisms can contribute to PCR bias, including polymerase error [48], formation of chimeric and heteroduplex molecules [49]–[51], and differential amplification efficiency [30], [31], [52]. Our study incorporated many of the wet bench techniques known to be effective toward reducing these biases [30], [31], [48], [49], [52], including low cycle numbers (30 cycles), pooling multiple reactions (3×30 µl), high template concentration (>4 ng of 16S rRNA gene clones), and the use of a proofreading DNA polymerase. Differential primer annealing efficiency provides another mechanism for PCR bias, and although factors such as annealing temperature and primer GC content can influence the outcome of PCR [29], [46], [47], primer mismatch may have the greatest impact for PCR studies of 16S rRNA gene diversity.
The lack of a truly “universal” pair of 16S rRNA gene PCR primers has long been acknowledged [28], [29], [45], [46], [53]. Although some have suggested that the number of taxa recovered is not necessarily linked to the taxonomic specificity (i.e., universality) of a primer set [25], our findings suggest that mispriming is a major, if not the main, factor leading to errors in the observation frequency of taxa within a community (Table S3). Mispriming near the 5′ end of the priming region is thought to have little effect on PCR since extension occurs from the 3′ end [54]. However, it has been reported that 454 Fusion primers containing the 454 adapter sequence at the 5′ end may be more susceptible to the effects of mispriming, resulting in the over-representation of templates that are not misprimed [20]. The adoption of a two-step PCR for amplicon pyrosequencing may ameliorate this issue [55]. Moreover, our findings highlight the complications associated with comparing community structures obtained using different primer sets.
Certain aspects of our experimental protocol may have exacerbated effects of PCR primer mismatch. For example, preferential amplification of perfectly matching template would be expected since the annealing temperature in our PCR protocol started high and decreased with each cycle (see Information S1) rather than starting at a lower temperature [28], [29]. Our modified PCR protocol was chosen because it resulted in an increased DNA yield and thus enabled accurate quantification of PCR amplicons (a prerequisite of pyrosequencing of PCR amplicons). This limitation can be addressed by new instruments that enable small quantities of DNA to be precisely characterized (e.g., Agilent 2100 Bioanalyzer, Agilent Technologies), fractionated (e.g., LabChip XT, Caliper Life Sciences), and quantified (e.g., Kapa Library Quant Kits, Kapa Biosystems). Although these methods were not available for this study, we recommend that they be adopted for the preparation of 16S rRNA gene amplicon libraries for 454-Ti sequencing in addition to adopting PCR conditions such as very low Tm [29] and low (<25) PCR cycles (in conjunction with higher template quantity where possible) [48], [49].
Our results have shown that while de-noising methods for pyrosequencing data need further development, they are an essential processing step for the recovery of usable community structure information. Overall, the largest hurdle to accurate estimation of microbial community structure appears to be PCR bias, which is independent of sequencing technology. Although a variety of measures may be taken to reduce the impact of PCR bias, it cannot be eliminated outright, and our findings highlight the need to better characterize this phenomenon using simulated communities. Another source of error also arises from PCR in the form of chimeric sequences, which are difficult to eliminate. Even though Perseus was able to effectively remove a large portion of chimeric sequences, a small portion of chimeric sequences contributed disproportionately to the number of OTUs observed, especially the infrequent (i.e., rare) OTUs (Figure 3 and Table S4). Therefore, chimeras can significantly inflate OTU estimates, even with short PCR amplicons generated from presumably “immune” 16S regions such as the V6 hypervariable region [17] (Table S4). These realities, combined with the observed prevalence of artifactual rare OTUs (Figure 3), caution against singular interpretations of community structure, especially those that involve within-sample relative OTU frequencies or estimations of Rare Biosphere diversity. Instead, the strength of the 454-Ti platform more likely lies in comparative studies and identifying the presence of specific rare taxa. Lastly, our findings highlight the dangers in quickly adopting technological advances without statistically robust validation, given that substantial portions of the Rare Biosphere identified using up-to-date de-noising algorithm are still artifacts. The impressively high microbial diversities reported by some past studies [3], [5], [9], [33], [56] based on less developed pyrosequencing quality filters should therefore be re-examined.
Materials and Methods
Preparation of 16S rRNA Gene PCR Clones
Twenty bacterial 16S rRNA gene PCR clones were obtained from two previous studies: 19 taken from fresh water habitats in New Zealand [57], and one from Adelie penguin fecal swab samples taken from Antarctica [58]. The primers used to generate initial PCR products (338F/modified 23S30R and EubB/ITSReub) and PCR cloning procedures were as described previously [57], [58]. Briefly, PCR products were gel-purified and cloned using the TOPO TA Cloning Kit (Invitrogen Corp., Carlsbad, CA) following the manufacturer’s instructions. The resulting clones were screened, isolated, and sequenced bi-directionally on an ABI 3730×l DNA Analyzer (Applied Biosystems, Foster City, CA). All 20 plasmids were verified to contain a unique and known insert of the 16S rRNA gene including the V3–V4 and V6 hypervariable regions. All clones except one (1216C: unclassified Clostridia) affiliate with members of Cyanobacteria.
Generation of in vitro-Simulated Communities and Pyrosequencing
Plasmid preparations were quantified using a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE) and the QuBit dsDNA HS fluorometric kit (Invitrogen); both methods were repeated in triplicate. Purified plasmid preparations were pooled at known abundances to construct two in vitro-simulated communities (iv-SCs): uniformly equal (E) and tiered (T) (Table 1). The pooled plasmid DNA sample was treated with Plasmid-Safe ATP-Dependent DNase (EPICENTRE Biotechnologies, Madison, WI) to remove contaminating genomic DNA from cloning hosts (i.e., E. coli). PCR amplicon libraries of the V3–V4 (iv-SCs: V3V4E & V3V4T) and V6 (iv-SCs: V6E & V6T) hypervariable regions were generated using these mixed plasmid communities as templates (454 Fusion PCR primers listed in Table 4). See Information S1 for PCR components and conditions, and quality control for PCR amplicons. An additional set of PCR-neutral iv-SCs (P) was constructed using PCR products individually amplified from each plasmid and subsequently pooled in tiered compositions (iv-SCs: V3V4P & V6P) after gel extraction and quantification as described in Information S1. The resulting iv-SCs were shipped frozen to the J. Craig Venter Institute, where emPCR was performed separately on pooled V3–V4 and V6 iv-SCs. The iv-SCs were pooled at the following ratios: “T”, 40%; “E”, 20%; and “P”, 40%. The two emPCR libraries were pooled together and sequenced from the A adapter using the Roche GS FLX with Titanium chemistry using one of two regions on a GS FLX Titanium PicoTiterPlate. Original pyrosequencing flowgram files are available from Sequence Read Archive (http://www.ebi.ac.uk/ena/data/view/ERP001633).
Identification of Error-Free Reads
Read sequences and corresponding quality files were generated using standard Roche software. Reads were compared to the expected amplicon products from V3–V4 and V6 regions of known clone sequences to determine the numbers of error-free reads corresponding to each target. Reads were required to match known sequences exactly over the amplified region, excluding primer sequences. Sequences with a perfect match to the known plasmid insert sequence and spanning the entire V3–V4 or V6 region were used in frequency calculations. In the case of the longer V3–V4 amplicons, sequences were also allowed to terminate prematurely if they were at least 216 nt in length (post primer trim), the minimum needed for each known sequence to be unequivocally identified.
Sequence Processing and OTU Determination
Prior to workflow-specific quality filtering and de-noising procedures, read sequences and corresponding quality files were generated using standard Roche software. Reads that did not perfectly match the expected primer and MID sequences were discarded. Among the remaining reads, primer and MID sequences were trimmed after reads were separated into individual files by iv-SCs.
Single-linkage preclustering (SLP) [17]
Reads with one or more ambiguous bases (N, quality score = 0) were removed. Average quality score was then calculated for every remaining read: those with an average quality score of less than 30 were discarded. Reads shorter than a specified length (50 nt) were also discarded. The SLP Perl script was used to assign low-frequency reads to higher frequency reads (http://vamps.mbl.edu/resources/software.php, downloaded in May 2011). Pairwise distances were calculated using Esprit [59]. For pre-clustering, a width of 0.02 was used, and an OTU size of 10 sequences was used for iterative clustering. The resulting datasets were screened for chimeras using Perseus (α = −7.5, β = 0.5) [19]. Esprit was used to calculate pairwise distances for unique sequences, which were then clustered into OTUs using Mothur 1.17.0 [60] at an average neighbor distance of 0.03, as recommended by the SLP authors [17].
PyroTagger [20]
Reads were length-trimmed to a specific length (60 nt for V6 amplicons and 216 nt for V3–V4 amplicons) after removal of primer sequences. All remaining reads with ≥3% bases having Q-scores ≤27 were removed from the dataset. PyroTagger, with the pyroclust option, was used to assign quality-filtered reads directly into OTUs without an alignment-based distance calculation step. To do this, sequences were first sorted by abundance and de-replicated. Chimeras were removed using Perseus (α = −7.5, β = 0.5) [19]. Unique reads were then clustered to form OTUs at 97% sequence identity using pyroclust’s default parameters.
AmpliconNoise [19]
Raw flowgrams (.sff files) were filtered based on primer and MID sequences match, and the occurrence of the first noisy cycle (i.e., 0.5–0.7 or no signal in all four nucleotide flows). For V6 amplicon reads, flowgrams were truncated at the first noisy cycle, whereas V3–V4 amplicon reads were dropped if the first noisy cycle occurred before cycle 360. The flowgrams were then de-noised using PyroNoise (cluster size = 60, initial cutoff = 0.01), and the resulting sequences were truncated at 400 nt for V3–V4 amplicons and 200 nt for V6 (although no V6 actually exceeded this length). In the final de-noising step, SeqNoise (cluster size = 30, initial cutoff = 0.08) was used. MID and primer sequences were trimmed from the resulting sequence predictions. Chimeras were removed using Perseus (α = −7.5, β = 0.5) [19]. The resulting de-noised, unique reads were aligned using mafft [61], [62], and the alignment was imported into Mothur [60] to construct a pairwise distance matrix using the dist.seqs function, ignoring terminal gaps. Sequences were then clustered into OTUs with an average neighbor clustering distance of 0.03.
Supporting Information
Comparison of AmpliconNoise vs. Qiime Denoiser workflows.
(TIFF)
Summary statistics for 16S rRNA gene amplicon sequence libraries of each iv -SC.
(DOCX)
Actual relative abundances of each sequence in each iv -SC based on error-free reads.
(DOCX)
PCR primer mismatches and their impact on the ratio of observed vs. expected frequencies (O:E). F or R indicates forward or reverse primer, respectively; number designates the position of the mismatch numbered from the 5' end. Observed to Expected Ratios (O:E) were calculated from Table S2.
(DOCX)
Identity of OTUs produced by AmpliconNoise (with Perseus) and SLP from V3V4P and V6P.
(DOCX)
Additional Material and Methods.
(DOCX)
Acknowledgments
We would like to acknowledge the support from JCVI sequencing specialists, in particular John Gill for his assistance with preparation and troubleshooting of 454-Ti PCR amplicon libraries.
Funding Statement
This research was supported by grants from the Office of Science (BER), U.S. Department of Energy (Cooperative Agreement No. De-FC02- 02ER63453) and the National Science Foundation to KEW, SJW, and SCC (MCB-0731916) and SCC (ANT-0739648 and ANT-0229836). The New Zealand Marsden Fund provided financial support for CWH, IRM, SCC (UOW0802), and CKL (UOW1003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C (2009) Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869 doi:10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 2. Gilbert JA, Field D, Swift P, Newbold L, Oliver A, et al. (2009) The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139 doi:10.1111/j.1462–2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 3. Huber JA, Welch DBM, Morrison HG, Huse SM, Neal PR, et al. (2007) Microbial population structures in the deep marine biosphere. Science 318: 97–100 doi:10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 4. Kirchman DL, Cottrell MT, Lovejoy C (2010) The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12: 1132–1143 doi:10.1111/j.1462–2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- 5. Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, et al. (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290 doi:10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sogin ML, Morrison HG, Huber JA, Welch DBM, Huse SM, et al. (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere.”. P Natl Acad Sci USA 103: 12115–12120 doi:10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Will C, Thürmer A, Wollherr A, Nacke H, Herold N, et al. (2010) Horizon-Specific Bacterial Community Composition of German Grassland Soils, as Revealed by Pyrosequencing-Based Analysis of 16S rRNA Genes. Appl Environ Microbiol 76: 6751–6759 doi:10.1128/AEM.01063–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, et al. (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12: 2998–3006 doi:10.1111/j.1462–2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 9. Youssef NH, Couger MB, Elshahed MS (2010) Fine-Scale Bacterial Beta Diversity within a Complex Ecosystem (Zodletone Spring, OK, USA): The Role of the Rare Biosphere. PLoS ONE 5: e12414 doi:10.1371/journal.pone.0012414.t004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fierer N, Hamady M, Lauber CL, Knight R (2008) The influence of sex, handedness, and washing on the diversity of hand surface bacteria. P Natl Acad Sci USA 105: 17994–17999 doi:10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Contreras M, Costello EK, Hidalgo G, Magris M, Knight R, et al. (2010) The bacterial microbiota in the oral mucosa of rural Amerindians. Microbiology 156: 3282–3287 doi:10.1099/mic.0.043174-0. [DOI] [PubMed] [Google Scholar]
- 12. Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, et al. (2009) Community-Wide Response of the Gut Microbiota to Enteropathogenic Citrobacter rodentium Infection Revealed by Deep Sequencing. Infection and Immunity 77: 4668–4678 doi:10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, et al. (2010) Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 5: e9085 doi:10.1371/journal.pone.0009085.t003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 378–380 doi:10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123 doi:10.1111/j.1462–2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 16. Huse SM, Huber JA, Morrison HG, Sogin M, Welch DBM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143 doi:10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898 doi:10.1111/j.1462–2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6: e27310. Available:http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0027310. [DOI] [PMC free article] [PubMed]
- 19. Quince C, Lanzén A, Davenport RJ, Turnbaugh PJ (2011) Removing Noise From Pyrosequenced Amplicons. BMC Bioinformatics 12: 38 doi:10.1186/1471–2105–12–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kunin V, Hugenholtz P (2010) PyroTagger: A fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. The Open Journal 1: 1–8. [Google Scholar]
- 21. Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, et al. (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6: 639–641 doi:10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 22.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW (2012) Fast, high specificity error-correction of amplicon pyrosequences for accurate microbial community analyses. In Review: 1–16.
- 23. Reeder J, Knight R (2010) Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7: 668–669 doi:10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, et al. (2010) Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. P Natl Acad Sci USA 107: 2383–2390 doi:10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huber JA, Morrison HG, Huse SM, Neal PR, Sogin M, et al. (2009) Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ Microbiol 11: 1292–1302 doi:10.1111/j.1462–2920.2008.01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, et al. (2010) Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4: 642–647 doi:10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 27. Youssef NH, Sheik CS, Krumholz LR, Najar FZ, Roe BA, et al. (2009) Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75: 5227–5236 doi:10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huws SA, Edwards JE, Kim EJ, Scollan ND (2007) Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J Microbiol Methods 70: 565–569 doi:10.1016/j.mimet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 29. Sipos R, Székely AJ, Palatinszky M, Révész S, Márialigeti K, et al. (2007) Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol Ecol 60: 341–350 doi:10.1111/j.1574–6941.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 30. Suzuki M, Rappé MS, Giovannoni SJ (1998) Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol 64: 4522–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polz MF, Cavanaugh CM (1998) Bias in Template-to-Product Ratios in Multitemplate PCR. Appl Environ Microbiol 64: 3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fonseca VG, Carvalho GR, Sung W, Johnson HF, Power DM, et al. (2010) Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat Comm 1: 98 doi:10.1038/ncomms1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, et al. (2010) Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4: 829–838 doi:10.1038/ismej.2010.3. [DOI] [PubMed] [Google Scholar]
- 34.Bahl J, Lau MCY, Smith GJD, Vijaykrishna D, Cary SC, et al.. (2011) Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat Comm 2: 163–. doi:10.1038/ncomms1167. [DOI] [PMC free article] [PubMed]
- 35. Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, et al. (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5: 908–917 doi:10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, et al. (2011) An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5: 879–895 doi:10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zinger L, Shahnavaz B, Baptist F, Geremia RA, Choler P (2009) Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME J 3: 850–859 doi:10.1038/ismej.2009.20. [DOI] [PubMed] [Google Scholar]
- 38. Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, et al. (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4: e6669 doi:10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, et al. (2008) A latitudinal diversity gradient in planktonic marine bacteria. P Natl Acad Sci USA 105: 7774–7778 doi:10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, et al. (2008) Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol 74: 5422–5428 doi:10.1128/AEM.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D (2007) Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl Environ Microbiol 73: 4532–4542 doi:10.1128/AEM.02956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urich T, Lanzén A, Qi J, Huson DH, Schleper C, et al. (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE 3: e2527 doi:10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319 doi:10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 44. Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF (2004) Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55: 541–555. [DOI] [PubMed] [Google Scholar]
- 46. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, et al. (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74: 2461–2470 doi:10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lueders T, Friedrich MW (2003) Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69: 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol 71: 8966–8969 doi:10.1128/AEM.71.12.8966–8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu X, Wu L, Huang H, McDonel PE, Palumbo AV, et al. (2001) Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl Environ Microbiol 67: 880–887 doi:10.1128/AEM.67.2.880–887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson JR, Marcelino LA, Polz MF (2002) Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by “reconditioning PCR.”. Nucleic Acids Res 30: 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurata S, Kanagawa T, Magariyama Y, Takatsu K, Yamada K, et al. (2004) Reevaluation and Reduction of a PCR Bias Caused by Reannealing of Templates. Appl Environ Microbiol 70: 7545 doi:10.1128/AEM.70.12.7545–7549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62: 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hugenholtz P, Goebel BM (2001) The polymerase chain reaction as a tool to investigate microbial diversity in environmental samples. In: Rochelle PA, editor. Environmental Molecular Microbiology: Protocols and Applications. Norfolk, England: Horizon Scientific Press.
- 54. Bru D, Martin-Laurent F, Philippot L (2008) Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol 74: 1660–1663 doi:10.1128/AEM.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tiao G, Lee CK-W, McDonald IR, Cowan DA, Cary SC (2012) Rapid microbial response to the presence of an ancient relic in the Antarctic Dry Valleys. Nat Comm 3: 660 doi:10.1038/ncomms1645. [DOI] [PubMed] [Google Scholar]
- 56. Acosta-Martinez V, Dowd S, Sun Y, Allen V (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40: 2762–2770. [Google Scholar]
- 57. Rueckert A, Wood SA, Cary SC (2007) Development and field assessment of a quantitative PCR for the detection and enumeration of the noxious bloom-former Anabaena planktonica. Limnol Oceanogr Methods 5: 474–483. [Google Scholar]
- 58. Banks JC, Cary SC, Hogg ID (2009) The phylogeography of Adelie penguin faecal flora. Environ Microbiol 11: 577–588 doi:10.1111/j.1462–2920.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- 59. Sun Y, Cai Y, Liu L, Yu F, Farrell ML, et al. (2009) ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res 37: e76 doi:10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541 doi:10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Katoh K, Misawa K, Kuma K-I, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 doi:10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Katoh K, Asimenos G, Toh H (2009) Multiple Alignment of DNA Sequences with MAFFT. In: Posada D, editor. Methods in Molecular Biology. Totowa, NJ: Humana Press, Vol. 537: 39–64 doi:10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of AmpliconNoise vs. Qiime Denoiser workflows.
(TIFF)
Summary statistics for 16S rRNA gene amplicon sequence libraries of each iv -SC.
(DOCX)
Actual relative abundances of each sequence in each iv -SC based on error-free reads.
(DOCX)
PCR primer mismatches and their impact on the ratio of observed vs. expected frequencies (O:E). F or R indicates forward or reverse primer, respectively; number designates the position of the mismatch numbered from the 5' end. Observed to Expected Ratios (O:E) were calculated from Table S2.
(DOCX)
Identity of OTUs produced by AmpliconNoise (with Perseus) and SLP from V3V4P and V6P.
(DOCX)
Additional Material and Methods.
(DOCX)