Abstract
The development of pre-erythrocytic Plasmodium vivax vaccines is hindered by the lack of in vitro culture systems or experimental rodent models. To help bypass these roadblocks, we exploited the fact that naturally exposed Fy− individuals who lack the Duffy blood antigen (Fy) receptor are less likely to develop blood-stage infections; therefore, they preferentially develop immune responses to pre-erythrocytic–stage parasites, whereas Fy+ individuals experience both liver- and blood-stage infections and develop immune responses to both pre-erythrocytic and erythrocytic parasites. We screened 60 endemic sera from P. vivax-exposed Fy+ or Fy− donors against a protein microarray containing 91 P. vivax proteins with P. falciparum orthologs that were up-regulated in sporozoites. Antibodies against 10 P. vivax antigens were identified in sera from P. vivax-exposed individuals but not unexposed controls. This technology has promising implications in the discovery of potential vaccine candidates against P. vivax malaria.
Introduction
Plasmodium vivax is the second most common human malaria parasite, with an estimated 2.85 billion people at risk of infection.1 Unlike its more deadly counterpart P. falciparum, P. vivax malaria can relapse months to years after the initial blood-stage infection has cleared, and increasing numbers of P. vivax cases involving severe complications or chloroquine resistance are being reported.2 Furthermore, the prevalence of P. falciparum malaria is falling at a faster rate than the prevalence of P. vivax malaria in many endemic areas, potentially because of the targeted efforts at reducing P. falciparum mortality. Currently, 25 of 32 malaria-eliminating countries are fighting solely or mainly against P. vivax.3 Thus, an efficient P. vivax vaccine that acts against the pre-erythrocytic stages of the parasite and prevents infection as well as disease and transmission may be useful to enable the reduction and eventually, the elimination of P. vivax malaria.
The near total absence of P. vivax infection in West Africa led to the discovery that P. vivax uses the Duffy antigen/receptor for chemokines (DARC) expressed on the surface of red blood cells to invade.4–9 Over 95% of Africans in malaria-endemic areas and approximately 70% of African-Americans do not express DARC.1,6 These individuals, referred to as Fy−, have been thought to be completely refractory to blood-stage infection with P. vivax, even when challenged with massive inocula of blood-stage parasites,10 though they are susceptible to infection with P. falciparum.4,8 Recent data suggest that Fy− individuals can, in fact, be infected with blood-stage parasites,11–13 indicating that P. vivax parasites may be able to use receptors other than DARC to invade erythrocytes. Previously, we showed that Fy− individuals exposed to P. vivax in Colombia responded predominantly to pre-erythrocytic antigens rather than erythrocytic antigens.14 The lack of DARC on Fy− erythrocytes and the resulting prevention or reduction of blood-stage parasitemia may consequently reduce the frequency of exposure to erythrocytic parasites in the Fy− population compared with the Fy+ population.14–16 Furthermore, it has been shown that, in both murine (P. yoelii) and human (P. falciparum) malaria, blood-stage infections suppress T-cell responses against liver-stage antigens.17–19 Immune responses to P. vivax pre-erythrocytic–stage antigens may, therefore, be stronger in Fy− than Fy+ individuals, because exposure of Fy− individuals to erythrocytic parasites in the bloodstream would be limited. Altogether, Fy− individuals mount immune responses presumably focusing on pre-erythrocytic antigens. Immune sera or cells from Fy− donors may, thus, be useful for identifying vaccine candidate antigens expressed during the sporozoite invasion and liver parasite development.
The availability of the genome sequence20 and the transcriptome21 of the P. vivax Sal 1 strain provides the means to analyze antigen-specific immune responses in different endemic populations to select optimal P. vivax antigens for vaccine development. Of the ∼5,500 genes encoded by the P. vivax genome,20 few have been studied as potential vaccine candidate antigens, and none of these genes are liver stage-specific genes.22–30 Nevertheless, studies in P. falciparum suggest that the breadth and magnitude of the antibody responses to parasite antigens determine the level of protection,31,32 highlighting the need to fully characterize the natural antibody response after P. vivax exposure to develop an effective anti-infection and anti-disease vaccine. Therefore, in this study, we used an advanced high-throughput screening technology33–37 to identify novel P. vivax pre-erythrocytic–stage antigens in populations living in malaria-endemic areas of Colombia.
Materials and Methods
Recruitment of P. vivax-exposed and -naïve donors.
Donors were enrolled from the malaria-endemic areas of La Delfina (N = 47) and Quibdó (Chocó State; N = 13), two cities on the Pacific coast of Colombia. The sociodemographic characteristics, genotype characteristics, and malaria incidence of the population were described previously.15 All donors were P. falciparum negative as determined by blood smear at the time of donation. Seven donors were recruited from Cali, where there is no malaria transmission, as malaria-naïve controls. All donors were over 18 years of age and gave informed consent using protocols approved by the Institutional Review Board of the Universidad del Valle.15 As previously described, a 5-mL blood sample was collected from each donor, plasma was isolated, and samples were stored at −70°C until use.15
Screening of sera by enzyme-linked immunosorbent assay and immunofluorescent assays to assess reactivity to pre-erythrocytic and erythrocytic antigens.
Sera from all donors were tested for reactivity against both pre-erythrocytic and blood stages by enzyme-linked immunosorbent assay (ELISA) against known antigens or immunofluorescent assay (IFA) against air-dried P. vivax sporozoites and fixed P. vivax blood-stage schizonts as described previously.15 Briefly, recombinant proteins containing region II of the Pv Duffy binding protein (DBP; rPvRII) and a fragment of the amino terminal region of the Pv merozoite surface protein 1 (MSP-1; 200-L fragment), both expressed in P. vivax asexual blood stages, were used to assess antibody responses to the erythrocytic phase. Two long synthetic peptides derived from Pv circumsporozoite protein (CSP) were used to assess the responses to the pre-erythrocytic stages.15 Peptide N includes the nonrepeat region of PvCSP (amino acids 22–125), and the peptide P11 is composed of tandem repeats of a 48-mer peptide (amino acids 96–104). A pool of control sera from healthy volunteers without a history of malaria exposure was used as the negative control. ELISA absorbance < 1:100 and IFA absorbance < 1:80 were detected in control sera and therefore, were considered negative. Antibody absorbance from exposed donors was considered positive when antibody absorbance from the test sera was at least 3 SDs greater than the mean of the antibody absorbance from the average of the control sera.
P. vivax pre-erythrocytic–stage proteins.
The P. vivax genome was explored for genes orthologous (blastp E value < 1 × 10−40) to those genes expressed in the P. falciparum sporozoite transcriptome.38 From this set of likely P. vivax pre-erythrocytic–stage genes, 109 genes were selected based on the following characteristics: (1) gene length < 5 kb, (2) encoded by a single exon, (3) contained a predicted signal peptide or signal anchor or one or more transmembrane domains, and (4) orthologs in the published proteomes of other Plasmodium species39,40 (Supplemental Table 1). Two P. vivax genes encoding CSP and DBP were used as controls for the pre-erythrocytic– and erythrocytic-stage antigens, respectively.
P. vivax protein array fabrication.
We cloned the selected P. vivax genes into the Escherichia coli expression vector pXT7.33 Custom polymerase chain reaction (PCR) primers comprising 20-bp gene-specific sequences with 33-bp adapter sequences were used to amplify target amplicons from P. vivax genomic DNA (P. vivax Sal I strain [MRA-552 and MR4]). The adapter sequences, which flank the target amplicons, are homologous to the adapter sequences at the ends of the linearized T7 expression vector pXT733 into which they were cloned. The homology allows the amplified PCR products to be cloned into the expression vector by in vivo homologous recombination in competent DH5α cells.33,41 The resulting clone mixtures were then verified by PCR using sequence-specific primers and subsequently sequenced. To fabricate the antigen arrays chips, we expressed P. vivax proteins using the E. coli-based cell-free in vitro transcription and translation (IVTT) system (Rapid Translation System 100 High Yield [RTS 100 HY] kits from 5 PRIME, Gaithersburg, MD) according to the manufacturer's instructions. P. vivax protein arrays were printed onto nitrocellulose-coated glass FAST slides (Whatman, Piscataway, NJ) using an OmniGrid Accent microarray printer (DigiLab, Piscataway, NJ); each expressed protein was spotted in triplicate. After being printed, protein expression was verified using monoclonal anti-polyhistidine (clone His-1; Sigma-Aldrich, St. Louis, MO) and anti-hemagglutinin (clone 3F10; Roche, Indianapolis, IN) antibodies. The arrays were rehydrated in 1× Blocking Buffer (Whatman, Piscataway, NJ) for 30 minutes and probed with the monoclonals diluted at 1:1,000 in blocking buffer overnight at 4°C with constant agitation. The next day, slides were washed three times in 1× hydroxymethyl-aminomethane (Tris pH 7.25–7.55) buffer containing 0.05% (vol/vol) Tween 20 (TTBS), and bound antibodies were detected by incubating with Cy3-conjugated goat anti-mouse immunoglobulin G (anti-IgG; Jackson Immuno Research, West Grove, PA) or Cy3-conjugated goat anti-rat IgG (Jackson Immuno Research, West Grove, PA) diluted 1:400 in blocking buffer. After washing the slides three times in TTBS followed by three additional washes in 1× Tris buffer (TBS) and a final rinse in ultrapure water, the slides were briefly centrifuged and air dried. The intensity of the fluorophore was detected and analyzed using the Perkin Elmer, Waltham, MA ScanArray Express HT microarray scanner. All signal intensities were corrected for spot-specific background (the signal from the substrate surrounding the spots). Successful expression of the proteins was determined based on a cutoff of the mean signal of the no DNA control spots plus 1 SD.
Serum screening using P. vivax protein arrays.
Sera were diluted to 1:100 in 1× Blocking Buffer containing 1 mg/mL E. coli lysate, and they were incubated at room temperature for 30 minutes with constant mixing. The arrays were rehydrated in 1× Blocking Buffer for 30 minutes and probed with the pre-treated sera in triplicates overnight at 4°C with constant agitation. The slides were then washed three times in TTBS and incubated in Cy3-labeled goat anti-human Ig (H + L; Jackson Immuno Research, West Grove, PA) diluted 1:400 in 1× Blocking Buffer. The slides were then washed three times in TTBS and three times in TBS, followed by an ultrapure water wash. The slides were then air dried after brief centrifugation and analyzed using a Perkin Elmer ScanArray Express HT microarray scanner(Perkin Elmer, Waltham, MA). Intensities were quantified using QuantArray software.
Protein array data analysis.
Triplicate data points were averaged before normalizing the data. The statistical analysis was performed as described previously.42 Briefly, the data were calibrated and transformed using the variance stabilizing normalization (vsn) package43 in the R statistical environment (www.r-project.org). Differential reactivity analysis was then performed using a one-way regularized analysis of variance (ANOVA).44 All reported P values are not corrected unless otherwise noted. Finally, the data were retransformed into an approximate raw scale for bar plot visualizations prepared in GraphPad Prism.
Bioinformatic analyses of novel P. vivax antigens.
To infer the stage specificity of the novel P. vivax antigens identified in this study, we used datasets including the P. vivax sporozoite21 and blood-stage transcriptomes,45 the P. falciparum sporozoite proteome,40 and the P. yoelii liver-stage transcriptome and proteome,38 The percentiled expression levels for each gene displayed in PlasmoDB (v7.1; www.plasmodb.org) were converted to quintiles. Statistics were performed using GraphPad Prism. Significance was considered at P < 0.05.
Results
Novel P. vivax antigens were identified by antibody profiling with sera from P. vivax-exposed individuals.
Before using the arrays to screen the sera from P. vivax-exposed donors, we first probed the arrays with antibodies to the N- and C-terminal tags to show that the Plasmodium antigens expressed contained both the N and C termini. As described previously,33 the arrays were probed with either anti-His or anti-hemagglutinin (HA) monoclonal antibodies and developed with appropriate secondary antibodies conjugated to Cy3 (data not shown). The arrays were scanned and quantified, and the triplicate spots were averaged. Proteins with signal intensities that were 1 SD above the mean of the no DNA control spots were designated positive for the detection of the tag. Of the 109 P. vivax proteins expressed on the array, 91 (83.5%) proteins were detected by anti-HA or anti-His antibodies, confirming that the majority of the malarial proteins were synthesized (Supplemental Table 1).
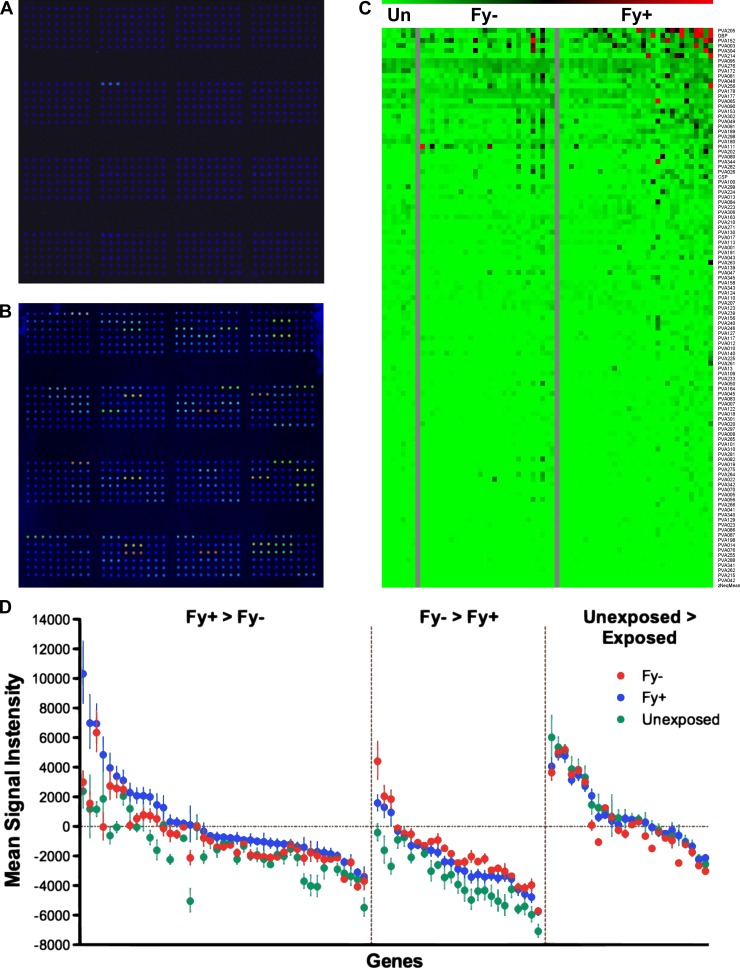
Fy− and Fy+ populations are both susceptible to pre-erythrocytic–stage infections, whereas Fy− individuals are less likely to develop blood-stage infection, because they lack the DARC receptor on their erythrocytes.5,8 These donors have been shown to develop significantly fewer antibodies to blood-stages antigens compared with Fy+ donors.46,47 To investigate the antibody responses induced by natural exposure to P. vivax infection, the protein arrays containing 91 P. vivax proteins were interrogated with 60 sera from P. vivax-exposed individuals and 7 control sera from unexposed individuals (Figure 1). The exposed donors were further subdivided into two groups according to their Fy genotype (Fy+ or Fy−). In this study, both Fy− (N = 28) and Fy+ (N = 32) populations were of similar age (41.8 ± 19.2 and 36.5 ± 14.5, respectively; Mann–Whitney P = 0.69) and were recruited from the same endemic area, and as such, they would be expected to have similar exposures to P. vivax.15 In accordance with this information, Fy− and Fy+ donors in this study exhibited similar antibody responses to both whole sporozoites and two CSP peptides as detected by IFA and ELISA, respectively (Table 1). Fy+ donors had stronger responses to blood-stage parasites and DBP (Table 1).
Figure 1.
P. vivax antigen array. (A and B) Examples of arrays probed with sera from a control donor (A) and an exposed donor (B). (C) Heat map showing the magnitude of responses to 91 genes in unexposed donors, Fy−- and Fy+-exposed donors. (D) Mean signal intensity to 91 genes in unexposed (green), Fy−-exposed (red), and Fy+-exposed (blue) donors. Error bars represent the standard error. Values are corrected for the array background by subtracting the mean of the no-DNA control spots.
Table 1.
Comparison of antibody responses to pre-erythrocytic and erythrocytic parasites or antigens between Fy− and Fy+ donors
| Median response (IQR) | Frequency of response (%) | |||||
|---|---|---|---|---|---|---|
| Fy− (N = 28) | Fy+ (N = 32) | P value* | Fy− (N = 28) | Fy+ (N = 32) | P value† | |
| Sporozoites‡ | 1:320 (80–320) | 1:120 (80–320) | 0.42 | 14 (50) | 22 (71) | 0.7 |
| CSP (P11) | 1:200 (200–250) | 1:200 (200–500) | 0.06 | 14 (50) | 6 (19) | 0.1 |
| CSP (N) | 1:200 (200–200) | 1:200 (200–200) | 0.09 | 15 (54) | 16 (52) | 1.4 |
| iRBC‡ | 1:320 (160–640) | 1:2,560 (1,280–3,200) | < 0.0001 | 15 (54) | 26 (84) | 0.41 |
| DBP | 1:200 (200–300) | 1:600 (400–800) | 0.01 | 5 (18) | 14 (45) | 0.2 |
| MSP1 (Pv200) | 1:200 (200–400) | 1:200 (200–400) | 0.6 | 20 (71) | 24 (77) | 1.3 |
IQR = interquartile range.
Mann–Whitney test was used to compare median responses.
Fisher two-tailed exact test was used to compare frequencies.
IFAs were performed on P. vivax-infected red blood cell (iRBC) and sporozoites.
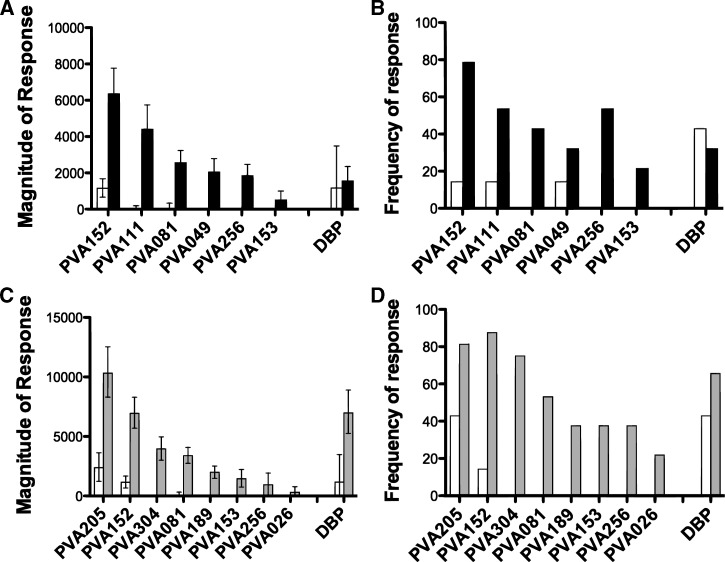
We first compared the magnitude and frequency of the serological reactivity between Fy− and unexposed donors (Figure 2A and B). As a positive control, the known P. vivax blood-stage antigen DBP was among the 91 vivax proteins on the array. As expected, there were no differences between the magnitudes of the responses to DBP in the unexposed group and the Fy− group (Figure 2A). However, the magnitude of the response to six P. vivax proteins (PVA152, PVA111, PVA081, PVA049, PVA256, and PVA153) was significantly increased in the Fy−-exposed group compared with the control group (Figure 2A and Table 2). The magnitude of the response in the unexposed donors was below background in all cases but one (PVA152), where reactivity was detected in the unexposed donors (Figure 2A). However, it was at a fivefold lower magnitude than the magnitude seen in the Fy−-exposed group. The frequency of the response mirrored the magnitude of the response (Figure 2B). Responses to these six proteins were detected in very few unexposed donors; PVA081, PVA256, and PVA153 were not recognized by any unexposed donors, and PVA152, PVA111, and PVA049 were recognized by just 1 of 7 unexposed donors compared with 22, 15, and 9 of 28 Fy− donors, respectively (Figure 2B and Table 2). Although the frequency of response was higher in the Fy− group in all cases, this difference was only significant for PVA152 and PVA256 (Fisher exact test P = 0.003 and P = 0.0125, respectively) (Table 2).
Figure 2.
Novel antigens recognized by P. vivax-exposed donors. Magnitude (A and C) and frequency (B and D) of proteins that are differentially recognized by sera from unexposed donors (N = 7, white bars) and Fy−- (A and B; N = 28, black bars) or Fy+-exposed donors (C and D; N = 32, grey bars). Error bars represent the standard error. Positivity was defined as signal intensities 1 SD above the mean of the no-DNA controls spots. DBP = Duffy binding protein.
Table 2.
Mean fluorescent intensity and frequency of identified genes
| Unexposed (N = 7) | Exposed (Fy−; N = 28) | Exposed (Fy+; N = 32) | Statistical analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MFI (±SEM) | Frequency (%) | MFI (±SEM) | Frequency (%) | MFI (±SEM) | Frequency (%) | P value* (Fy− Un) | P value* (Fy+ Un) | P value* (Fy− Fy+) | |
| Antigens recognized by Fy− donors | |||||||||
| PVA049 | 0.00 (0–0) | 1 (14.3) | 2,048.58 (1,351.8–2,786.7) | 9 (32.1) | 1,298.12 (584.3–2,058.7) | 8 (25.0) | 0.01 | 0.06 | 0.41 |
| PVA081 | 0.00 (0–336.6) | 0 (0) | 2,565.24 (1,926.0–3,237.7) | 12 (42.9) | 3,392.53 (2,740.6–4,077.0) | 17 (53.1) | 0.04 | 0.02 | 0.44 |
| PVA152 | 1,158.22 (622.6–1,675.9) | 1 (14.3) | 6,353.09 (5,046.4–7,770.3) | 22 (78.6) | 6,947.48 (5,687.9–8,306.3) | 28 (87.5) | 0.03 | 0.02 | 0.79 |
| PVA153 | 0.00 (0–0) | 0 (0) | 504.51 (24.1–1,007.0) | 6 (21.4) | 1,459.59 (740.2–2,225.9) | 12 (37.5) | 0.04 | 0.04 | 0.33 |
| PVA256 | 0.00 (0–0) | 0 (0) | 1,843.58 (1,249.5–2,468.0) | 15 (53.6) | 942.90 (29.7–1,936.8) | 12 (37.5) | 0.00 | 0.04 | 0.40 |
| PVA111 | 0.00 (0–0) | 1 (14.3) | 4,408.03 (3,178.5–5,749.0) | 15 (53.6) | 1,589.50 (1,043.9–2,161.3) | 10 (31.3) | 0.03 | 0.12 | 0.02 |
| Antigens recognized by Fy+ donors | |||||||||
| PVA026 | 0.00 (0–0) | 0 (0) | 0.00 (0–117.4) | 5 (17.9) | 309.80 (0–779.2) | 7 (21.9) | 0.11 | 0.01 | 0.33 |
| PVA304 | 0.00 (0–0) | 0 (0) | 2,737.39 (1,804.9–3,741.1) | 13 (46.4) | 3,954.86 (3,005.7–4,971.7) | 24 (75.0) | 0.06 | 0.02 | 0.41 |
| PVA189 | 0.00 (0–0) | 0 (0) | 66.32 (0–393.6) | 6 (21.4) | 2,286.06 (1,663.9–2,940.5) | 12 (37.5) | 0.13 | 0.02 | 0.10 |
| PVA205 | 2,377.63 (1,235.4–3631.4) | 3 (43.8) | 2,995.01 (2,263.5–3,768.8) | 14 (50) | 10,315.13 (8,310.4–12,535.7) | 26 (81.3) | 0.73 | 0.03 | 0.00 |
| DBP | 1,174.87 (0–3487.3) | 3 (43.8) | 1,558.04 (809.2–2,356.9) | 9 (32.1) | 6,985.15 (5,258.7–8,903.1) | 21 (65.6) | 0.85 | 0.09 | 0.00 |
MFI = mean fluorescent intensity; Un = unexposed donors.
P value calculated using a regularized t test between MFIs.
We then compared the magnitude and frequency of the responses between the Fy+-exposed and unexposed groups. The magnitude of the response to the DBP, despite a nearly sixfold increase in mean signal intensity, was not significantly higher in the exposed Fy+ donors compared with the control donors (Figure 2C and Table 2). However, the magnitude of the responses to eight proteins (PVA205, PVA152, PVA304, PVA081, PVA189, PVA153, PVA256, and PVA026) was significantly higher in Fy+-exposed donors than unexposed donors (Figure 2C and Table 2). Although detectable responses were observed in the unexposed donors for PVA205, PVA152, and PVA189, the magnitudes of responses to these proteins in the Fy+ group were 1.9-to 5.9-fold higher than the magnitudes of the unexposed group. Again, the frequency of the response mirrored the magnitude of the response. Six of eight vivax antigens (PVA304, PVA081, PVA189, PVA153, PVA256, and PVA026) were recognized by Fy+-exposed donors but not unexposed donors. The frequency of the responses ranged from 17.9% to 78.6% in the Fy+-exposed donors. Two proteins (PVA205 and PVA152) were recognized by 3 and 1 unexposed donors, respectively, but by 26 and 28 of 32 Fy+-exposed donors, respectively. The frequencies of responses to PVA152, PVA304, and PVA081 were significantly higher in the Fy+ group (Fisher exact test P = 0.0004, P = 0.0004, and P = 0.0124, respectively).
Differential recognition of P. vivax antigens by Fy− and Fy+ individuals.
We exploited the DARC phenotypes of the volunteers in an attempt to divide P. vivax antigens into possible stage-specific groups based on their reactivity to sera from the two different populations. We expected that P. vivax pre-erythrocytic proteins would be recognized similarly by sera from both Fy− and Fy+ individuals in terms of frequency and magnitude of responses. However, if the responses to a novel vivax protein were significantly stronger in Fy+ than Fy− populations, this finding would indicate that the antigen was more likely to be expressed by P. vivax erythrocytic-stage parasites.14,16
Pre-erythrocytic and multistage proteins.
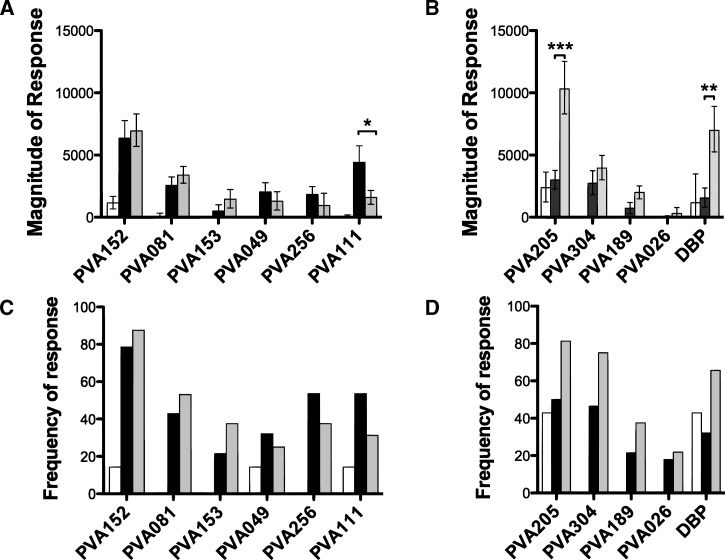
Reactivity to proteins on the array was compared using sera from 28 Fy− and 32 Fy+ individuals (Figure 3). Four proteins (PVA152, PVA081, PVA153, and PVA256) were recognized by sera from both Fy+ and Fy− individuals and in both groups at significantly higher magnitudes than in the unexposed group. These four proteins were recognized at a similar magnitude and frequency in the Fy− and Fy+ groups. Two proteins (PVA049 and PVA111) were recognized by the Fy− at significantly higher magnitudes than the unexposed group but not by the Fy+ donors. Interestingly, PVA111 showed significantly higher magnitudes of responses in the Fy− group than the Fy+ group, with over 2.7-fold higher signal intensities in the Fy− group (P = 0.022) (Figure 3A and Table 2). Although the frequency of the responses to PVA111 was higher in sera from the Fy− group than the Fy+ group (53.6% and 31.1%, respectively), it did not reach statistical significance (Fisher exact test P = 0.11) (Figure 3C and Table 2).
Figure 3.
Differential recognition of proteins by Fy− and Fy+ donors. Reactivity to proteins on the microarray was compared between 28 Fy− (black bars) and 32 Fy+ (grey bars) and 7 unexposed donors (white bars). Magnitude (A and B) and frequency (C and D) of responses to antigens that are recognized by either Fy− and Fy+ donors or Fy− donors only (A and C) or antigens that are preferentially recognized by Fy+ donors (B and D). Error bars represent standard errors. Positivity was defined as signal intensities 1 SD above the mean of the no-DNA controls spots. *P value between 0.05 and 0.01. **P value between 0.01 and 0.001. ***P value between 0.001 and 0.0001.
Overall, a total of six proteins was either recognized by sera from exposed Fy+ and Fy− donors or recognized better in Fy− donors (PVA111) compared with Fy+ donor (Figure 3A and C). We predict that these six proteins are most likely to be expressed by pre-erythrocytic and/or both pre-erythrocytic and erythrocytic parasites.
Erythrocytic-stage proteins.
Five P. vivax proteins, including two known blood-stage antigens DBP and PVA205 (putative MSP-8)48 and newly identified PVA189, PVA304, and PVA026, were recognized by the Fy+ donors at significantly higher magnitudes than the unexposed group but not the Fy− donors (Figure 3B). Responses to DBP were fourfold higher in Fy+ donors than Fy− donors (P = 0.0004) (Figure 3B and Table 2). This finding confirms our previous report with the sera tested from the same endemic area,15 and it is consistent with the notion that Fy+ donors have higher reactivity to erythrocytic-stage proteins than Fy− donors. The frequency of recognition to DBP was twofold higher in the Fy+ group (65.6%) than the Fy− group (32.1%), but there was no statistically significant difference between the two groups (Table 2). Like DBP, PVA205 (MSP-8) was also recognized with significantly higher frequency by sera from Fy+ than Fy− donors (P = 0.0005) (Figure 3B and Table 2). For these two proteins, the mean signal intensity in the Fy+ group was, on average, five times higher than in the control or Fy− groups (Figure 3B). In addition, the magnitude of responses to protein PVA189, PVA304, and PVA026 was significantly higher in Fy+ than control donors (P = 0.02, P = 0.02, and P = 0.01, respectively) (Figure 3B and D and Table 2). However, no significant response to these proteins was detected in Fy−-exposed or control donors.
Bioinformatics confirmation of protein expression profiles.
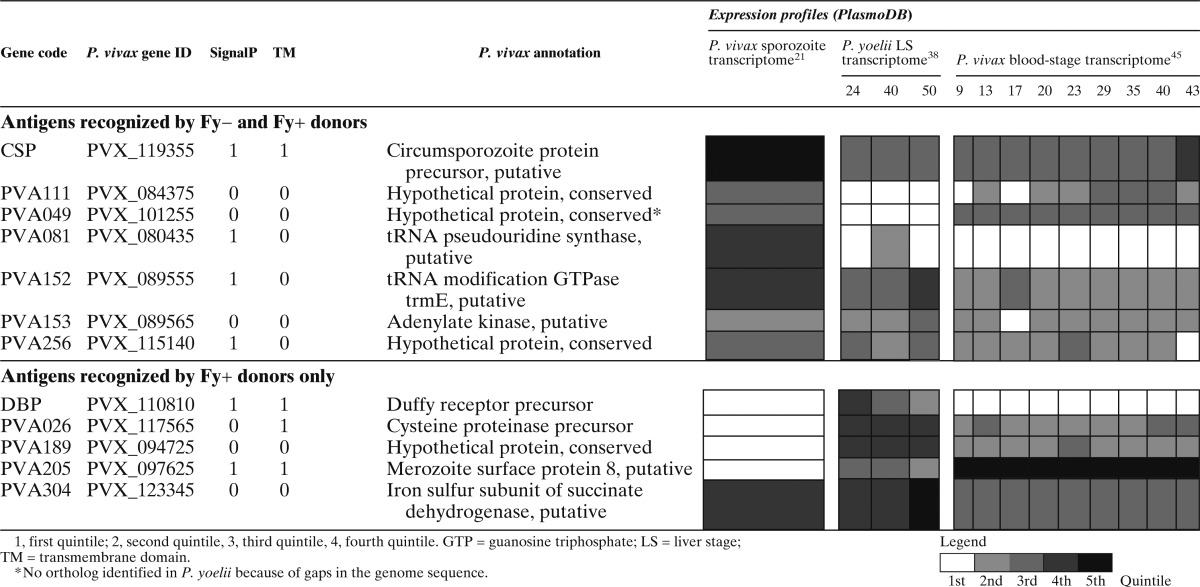
To support our analysis and prediction of stage specificity of the proteins that we identified in this study, we conducted a comparative analysis with proteomic and transcriptomic data from PlasmoDB (www.plasmodb.org) derived from studies in both P. vivax and P. yoelii (Table 3). Although the P. vivax genes were selected based on expression of their P. falciparum orthologs at the sporozoite stage,40 transcriptome data are consistent with all but 1 (PVA081) of 10 proteins identified as being expressed at some level at the blood-stage of infection. As expected, the six proteins recognized by the Fy− group were all highly expressed at the sporozoite stage, whereas the five proteins recognized by Fy+ donors but not Fy− donors were expressed at very low levels in sporozoites (Table 3). The expression profiles of proteins recognized specifically by Fy+ donors were compared with proteins recognized similarly in both Fy− and Fy+ donors (Table 3). Although most proteins were expressed in the blood stages, the proteins recognized specifically by Fy− donors had significantly higher expression levels in sporozoites compared with proteins recognized by Fy+ donors (average quintile = 3.4 and 1.6, respectively, t test P = 0.02). However, these proteins also had significantly lower expression in liver-stage parasites (2.6 versus 3.6, P = 0.05). There were no differences in the expressions of proteins during the blood stage (2.2 versus 2.7, P = 0.34).
Table 3.
Expression profile of identified genes
Interestingly, 4 of 10 proteins identified (PVA152, PVA153, PVA256, and PVA304) are expressed in all stages of the parasites. Three of these four proteins (PVA152, PVA153, and PVA256) were recognized by both Fy−- and Fy+-exposed donors, whereas PVA304 was only recognized by Fy+ donors. The levels of expression profiles are consistent with the frequency and magnitudes of the antibody responses to these proteins (Figure 2A and B and Table 3). This finding may suggest that these proteins might elicit strong antibody responses because of their longer exposure to the immune system.
Discussion
Advances in genomics and proteomics have enabled the study of entire organism proteomes rather than a limited subset of previously characterized antigens without the need for in vitro cultures. Because of this vast increase in the number of potential candidate antigens, cost-effective and time-efficient methods are required to screen all possible antigens and select those antigens most immunogenic for additional development of an anti-P. vivax malaria vaccine. Here, we have developed a P. vivax antigen array containing 91 P. vivax sporozoite proteins. Using this P. vivax protein chip, we have identified 10 antigens that were recognized by semi-immune sera from individuals living in P. vivax-endemic areas in Colombia. None of these proteins elicited a T-cell response, which was measured by enzyme-linked immunospot interferon γ (ELISPOT IFN-γ) using peripheral blood mononuclean cells (PBMC) from the same P. vivax-endemic areas (Finney OC and Wang R, unpublished data).
Because long-term culture systems for P. vivax are still under development, we used differential responsiveness between donors with or without DARC to predict the stage specificity of protein expression. Fy− donors, with erythrocytes that are less susceptible to P. vivax infection, recognize proteins from the pre-erythrocytic stages of the parasite: sporozoite, liver, and hepatic merozoite. Fy+ donors recognize these proteins as well as proteins from erythrocytic stages, including erythrocytic merozoites.46,47 Because hepatic and erythrocytic merozoites seem to be identical, recognition of at least some erythrocytic-stages antigens by Fy− individuals is unsurprising. As expected, DBP was recognized at higher levels in Fy+ than Fy− donors.
Our protein microarray method has been used successfully to identify novel antigens for several different organisms, including Francisella tularensis,36 Burkholderia pseudomallei,49 and P. falciparum.32 In this study, we did not detect robust responses to PvCSP, the immunodominant sporozoite antigen; only 8 of 60 exposed donors displayed detectable levels of antibodies against PvCSP. Using the ELISA assay, 20–31 donors displayed reactivity to CSP depending on the peptide used. Previous protein microarray serology results from our group showed that, although high levels of anti-PfCSP reactivity are seen in irradiated sporozoite-immunized volunteers,34,50 anti-PfCSP reactivity is much lower in naturally exposed individuals.32 Reactivity to IVTT-produced P. falciparum CSP (PfCSP) and purified recombinant PfCSP was compared on the same chip; although good correlation was observed between the proteins, the magnitude of the response to the IVTT-produced PfCSP was five times lower than reactivity to the purified PfCSP.32 A limitation of using the IVTT technology to produce proteins for the arrays is that folding, multimerization, or post-translational modifications, such as phosphorylation or glycosylation, of the arrayed proteins may differ from the native proteins, such that some epitopes present in native proteins are absent in arrays made using IVTT. However, the ability to screen a wide range of novel antigens simultaneously is a substantial advantage for identifying and prioritizing antigens for future research.
The liver-stage parasite, as an intracellular pathogen, primarily induces a cellular rather than humoral response; as such, anti-malarial antibodies target predominantly the sporozoite and blood stages rather than the liver stage.51 Sporozoites travel from the site of infection to the liver in approximately 1–3 hours,52 leaving relatively little time for the host to develop a humoral response to sporozoite proteins. In comparison, the large number of liver merozoites released from late liver schizonts that enters the blood stream and the successive rounds of asexual blood-stage replication and merozoite releases produce an abundant and diverse pool of blood-stage antigens. This finding may explain why only 10 of 91 selected sporozoite proteins showed reactivity in our analysis. Furthermore, the small sample size, particularly of the unexposed group, reduces the power of detection in this study.
Of the 10 proteins identified, five antigens were recognized by Fy+ but not Fy− individuals, suggesting that these antigens are expressed in the blood-stage parasites. Indeed, four of five proteins, including DBP and MSP-8, showed very low expression in the sporozoite (Table 3). MSP-8 (PVA205) was previously characterized as a glycosylphosphatidylinisotol-anchored protein, and it was recognized by P. vivax-infected patients in Columbia53 as well as P. vivax-infected patients from Korea.54 Proteins recognized by both Fy+ and Fy− donors were expressed highly in the sporozoite; however, five of six were also expressed in the blood stages of the parasite. These responses could be caused by antigens expressed by hepatic merozoites. However, the inability of P. vivax to invade red blood cells in Fy− donors has been contested.11 Although Fy− individuals are not usually susceptible to blood-stage P. vivax infection, some may, in certain conditions, carry very low levels of erythrocytic parasites. This finding would indicate that, in addition to the Duffy blood antigen, other receptors may also be involved in P. vivax blood-stage invasion in humans, which may help to explain why the antigens recognized by both Fy+ and Fy− donors were, in some cases, expressed at higher levels during the blood stage. Four antigens identified are expressed in all stages of the parasite life cycle rather than at one particular stage. This finding may provide an advantage for vaccine development. The current interest in designing multistage, multivalent vaccines for malaria may be replaced by rational selection of single antigens that are consecutively expressed in multiple stages of the parasite. These antigens may induce immune responses against parasites at different stages of the life cycle, and therefore, they could be candidates for multifunctional anti-infection and anti-disease malaria vaccines.
Additional characterization of all antigens identified will be necessary to verify their expression in sporozoites and/or blood-stage parasites by immunolocalization as well as their roles in parasite development and growth by gene knockout. Such studies will support the selection of the best candidates for vaccine development for endemic populations, because essential genes will be less likely to evade vaccine-induced immunity through antigenic variation under immune selection.
In conclusion, we have identified 10 P. vivax antigens with potential roles in naturally acquired immunity to P. vivax. Finding antigenic proteins that may derive from pre-erythrocytic–stage parasites may render the liver-stage parasites a reasonable target for vaccine development. These antigens were identified using a high-throughput method and serve as proof of concept for our approach to identify and prioritize antigens for vaccine research. This method can be expanded to quantitatively and comprehensively investigate the humoral response to the entire P. vivax genome and potentially, find new diagnostic, vaccine, or surveillance markers for use in the field with accuracy, efficiency, and speed.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the volunteers without whom this study could not have been possible. We also thank Omaira Vera and the field team at Centro Internacional de Vacunas (Colombia) for collection of samples and Aki Takagi (Seattle BioMed) for running the enzyme-linked immunosorbent assays.
Footnotes
Financial support: This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Disease Grant R01 AI05759206, National Institutes of Health/National Institute of Allergy and Infectious Disease Small Business Innovation Research Grant AI075692, and National Institutes of Health/National Heart, Lung, and Blood Institute Grant R01 RHLO86488-04. This work was also supported by the Columbian Administrative Department of Science, Technology and Innovaton, known as Colciencias, contract 527-2009, code 2304-493-26209.
Authors' addresses: Douglas M. Molina and Xiaowu Liang, Antigen Discovery Inc., Irvine, CA, E-mails: dmolina@antigendiscovery.com and xliang@antigendiscovery.com. Olivia C. Finney, Malcolm J. Gardner, and Ruobing Wang, Seattle Biomedical Research Institute, Seattle, WA, E-mails: Olivia.finney@seattlebiomed.org, Malcolm.gardner@seattlebiomed.org, and ruobing.wang@seattlebiomed.org. Myriam Arevalo-Herrera and Socrates Herrera, Centro Internacional de Vacunas, Instituto de Inmunología, Universidad del Valle, Cali, Colombia, E-mails: marevalo@inmuno.org and sherrera@inmuno.org. Phillip L. Felgner Departments of Infectious Disease and Epidemiology, School of Medicine, University of California, Irvine, CA, E-mail: pfelgner@uci.edu.
References
- 1.Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IRF, Baird JK, Snow RW, Hay SI. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010;4:e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 3.Feachem RGA, Phillips AA, Hwang J, Cotter C, Wielgosz B, Greenwood BM, Sabot O, Rodriguez MH, Abeyasinghe RR, Ghebreyesus TA, Snow RW. Shrinking the malaria map: progress and prospects. Lancet. 2010;376:1566–1578. doi: 10.1016/S0140-6736(10)61270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 5.Gelpi AP, King MC. Duffy blood group and malaria. Science. 1976;191:1284. doi: 10.1126/science.1257752. [DOI] [PubMed] [Google Scholar]
- 6.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for Plasmodium knowlesi malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 7.Mason SJ, Miller LH, Shiroishi T, Dvorak JA, McGinniss MH. The Duffy blood group determinants: their role in the susceptibility of human and animal erythrocytes to Plasmodium knowlesi malaria. Br J Haematol. 1977;36:327–335. doi: 10.1111/j.1365-2141.1977.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 8.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 9.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young MD, Eyles DE, Burgess RW, Jeffrey GM. Experimental testing of the immunity of Negroes to Plasmodium vivax. J Parasitol. 1955;41:315. [PubMed] [Google Scholar]
- 11.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod J-F, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosario VE, Benito A, Berzosa P, Arez AP. Duffy negative antigen is no longer a barrier to Plasmodium vivax: molecular evidences from the African west coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5:e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurtz N, Mint Lekweiry K, Bogreau H, Pradines B, Rogier C, Ould Mohamed Salem Boukhary A, Hafid JE, Ould Ahmedou Salem MS, Trape J-F, Basco L, Briolant S. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. Malar J. 2011;10:336. doi: 10.1186/1475-2875-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Arevalo-Herrera M, Gardner MJ, Bonelo A, Carlton JM, Gomez A, Vera O, Soto L, Vergara J, Bidwell SL, Domingo A, Fraser CM, Herrera S. Immune responses to Plasmodium vivax pre-erythrocytic stage antigens in naturally exposed Duffy-negative humans: a potential model for identification of liver-stage antigens. Eur J Immunol. 2005;35:1859–1868. doi: 10.1002/eji.200425807. [DOI] [PubMed] [Google Scholar]
- 15.Herrera S, Gomez A, Vera O, Vergara J, Valderrama-Aguirre A, Maestre A, Mendez F, Wang R, Chitnis CE, Yazdani SS, Arevalo-Herrera M. Antibody response to Plasmodium vivax antigens in Fy-negative individuals from the Columbian Pacific Coast. Am J Trop Med Hyg. 2005;73((Suppl 5)):44–49. doi: 10.4269/ajtmh.2005.73.44. [DOI] [PubMed] [Google Scholar]
- 16.Maestre A, Muskus C, Duque V, Agudelo O, Liu P, Takagi A, Ntumngia FB, Adams JH, Sim KL, Hoffman SL, Corradin G, Velez ID, Wang R. Acquired antibody responses against Plasmodium vivax infection vary with host genotype for Duffy antigen receptor for chemokines (DARC) PLoS One. 2010;5:e11437. doi: 10.1371/journal.pone.0011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urban BC, Roberts DJ. Inhibition of T cell function during malaria: implications for immunology and vaccinology. J Exp Med. 2003;197:137–141. doi: 10.1084/jem.20022003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ocana-Morgner C, Mota MM, Rodriguez A. Malaria blood stage suppression of liver stage immunity by dendritic cells. J Exp Med. 2003;197:143–151. doi: 10.1084/jem.20021072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban BC, Ferguson DJ, Pain A, Willcox N, Plebanski M, Austyn JM, Roberts DJ. Plasmodium falciparum-infected erythrocytes modulate the maturation of dendritic cells. Nature. 1999;400:73–77. doi: 10.1038/21900. [DOI] [PubMed] [Google Scholar]
- 20.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RMR, Crabb BS, del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TWA, Korsinczky M, Meyer EVS, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, Collins WE, Hoffman SL, Zhou Y, Vinetz JM, Winzeler EA. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Templeton TJ, Kaslow DC. Cloning and cross-species comparison of the thrombospondin-related anonymous protein (TRAP) gene from Plasmodium knowlesi, Plasmodium vivax and Plasmodium gallinaceum. Mol Biochem Parasitol. 1997;84:13–24. doi: 10.1016/s0166-6851(96)02775-2. [DOI] [PubMed] [Google Scholar]
- 23.del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991;88:4030–4034. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas AW, Trape JF, Rogier C, Goncalves A, Rosario VE, Narum DL. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am J Trop Med Hyg. 1994;51:730–740. doi: 10.4269/ajtmh.1994.51.730. [DOI] [PubMed] [Google Scholar]
- 25.Barnwell JW, Galinski MR. Plasmodium vivax: a glimpse into the unique and shared biology of the merozoite. Ann Trop Med Parasitol. 1995;89:113–120. doi: 10.1080/00034983.1995.11812941. [DOI] [PubMed] [Google Scholar]
- 26.Barnwell JW, Galinski MR, DeSimone SG, Perler F, Ingravallo P. Plasmodium vivax, P. cynomolgi, and P. knowlesi: identification of homologue proteins associated with the surface of merozoites. Exp Parasitol. 1999;91:238–249. doi: 10.1006/expr.1998.4372. [DOI] [PubMed] [Google Scholar]
- 27.Galinski MR, Ingravallo P, Corredor-Medina C, Al-Khedery B, Povoa M, Barnwell JW. Plasmodium vivax merozoite surface proteins-3beta and-3gamma share structural similarities with P. vivax merozoite surface protein-3alpha and define a new gene family. Mol Biochem Parasitol. 2001;115:41–53. doi: 10.1016/s0166-6851(01)00267-5. [DOI] [PubMed] [Google Scholar]
- 28.Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW. Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol Biochem Parasitol. 1999;101:131–147. doi: 10.1016/s0166-6851(99)00063-8. [DOI] [PubMed] [Google Scholar]
- 29.Chitnis CE. Molecular insights into receptors used by malaria parasites for erythrocyte invasion. Curr Opin Hematol. 2001;8:85–91. doi: 10.1097/00062752-200103000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Tsuboi T, Kappe SH, al-Yaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osier FHA, Fegan G, Polley SD, Murungi L, Verra F, Tetteh KKA, Lowe B, Mwangi T, Bull PC, Thomas AW, Cavanagh DR, McBride JS, Lanar DE, Mackinnon MJ, Conway DJ, Marsh K. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect Immun. 2008;76:2240–2248. doi: 10.1128/IAI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, Crotty S, Karem KL, Damon IK, Ahmed R, Villarreal L, Felgner PL. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 36.Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, Randall A, Mu Y, Hirst S, Molina DM, Davies DH, Milne T, Griffin KF, Baldi P, Titball RW, Felgner PL. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 37.Vigil A, Davies DH, Felgner PL. Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol. 2010;5:241–251. doi: 10.2217/fmb.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SHI. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci USA. 2008;105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, James K, Rutherford K, Harris B, Harris D, Churcher C, Quail MA, Ormond D, Doggett J, Trueman HE, Mendoza J, Bidwell SL, Rajandream M-A, Carucci DJ, Yates JR, Kafatos FC, Janse CJ, Barrell B, Turner CMR, Waters AP, Sinden RE. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 40.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 41.Liang X, Teng A, Braun DM, Felgner J, Wang Y, Baker SI, Chen S, Zelphati O, Felgner PL. Transcriptionally active polymerase chain reaction (TAP): high throughput gene expression using genome sequence data. J Biol Chem. 2002;277:3593–3598. doi: 10.1074/jbc.M110652200. [DOI] [PubMed] [Google Scholar]
- 42.Sundaresh S, Randall A, Unal B, Petersen JM, Belisle JT, Hartley MG, Duffield M, Titball RW, Davies DH, Felgner PL, Baldi P. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 43.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 44.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 45.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NPJ, White NJ, Carlton JM, Preiser PR. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R, Arevalo-Herrera M, Gardner MJ, Bonelo A, Carlton JM, Gomez A, Vera O, Soto L, Vergara J, Bidwell SL, Domingo A, Fraser CM, Herrera S. Immune responses to Plasmodium vivax pre-erythrocytic stage antigens in naturally exposed Duffy-negative humans: a potential model for identification of liver-stage antigens. Eur J Immunol. 2005;35:1859–1868. doi: 10.1002/eji.200425807. [DOI] [PubMed] [Google Scholar]
- 47.Herrera S, Bonelo A, Perlaza BL, Fernandez OL, Victoria L, Lenis AM, Soto L, Hurtado H, Acuna LM, Velez JD, Palacios R, Chen-Mok M, Corradin G, Arevalo-Herrera M. Safety and elicitation of humoral and cellular responses in Colombian malaria-naive volunteers by a Plasmodium vivax circumsporozoite protein-derived synthetic vaccine. Am J Trop Med Hyg. 2005;73:3–9. doi: 10.4269/ajtmh.2005.73.3. [DOI] [PubMed] [Google Scholar]
- 48.Drew DR, O'Donnell RA, Smith BJ, Crabb BS. A common cross-species function for the double epidermal growth factor-like modules of the highly divergent Plasmodium surface proteins MSP-1 and MSP-8. J Biol Chem. 2004;279:20147–20153. doi: 10.1074/jbc.M401114200. [DOI] [PubMed] [Google Scholar]
- 49.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, Hirst S, Chew JS, Wang D, Tan G, Duffield M, Yang R, Neel J, Chantratita N, Bancroft G, Lertmemongkolchai G, Davies DH, Baldi P, Peacock S, Titball RW. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci USA. 2009;106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trieu A, Kayala MA, Burk C, Molina DM, Freilich DA, Richie TL, Baldi P, Felgner PL, Doolan DL. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007948. M111007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beeson JG, Osier FHA, Engwerda CR. Recent insights into humoral and cellular immune responses against malaria. Trends Parasitol. 2008;24:578–584. doi: 10.1016/j.pt.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Yamauchi LM, Coppi A, Snounou G, Sinnis P. Plasmodium sporozoites trickle out of the injection site. Cell Microbiol. 2007;9:1215–1222. doi: 10.1111/j.1462-5822.2006.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Leal O, Sierra AY, Barrero CA, Moncada C, Martinez P, Cortes J, Lopez Y, Torres E, Salazar LM, Patarroyo MA. Plasmodium vivax merozoite surface protein 8 cloning, expression, and characterisation. Biochem Biophys Res Commun. 2004;324:1393–1399. doi: 10.1016/j.bbrc.2004.09.202. [DOI] [PubMed] [Google Scholar]
- 54.Chen J-H, Jung J-W, Wang Y, Ha K-S, Lu F, Lim CS, Takeo S, Tsuboi T, Han E-T. Immunoproteomics profiling of blood stage Plasmodium vivax infection by high-throughput screening assays. J Proteome Res. 2010;9:6479–6489. doi: 10.1021/pr100705g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.