Abstract
Leishmania species of the Viannia subgenus are responsible for most cases of New World tegumentary leishmaniasis. However, little is known about the vectors involved in disease transmission in the Amazon regions of Peru. We used a novel real-time polymerase chain reaction (PCR) to assess Leishmania infections in phlebotomines collected in rural areas of Madre de Dios, Peru. A total of 1,299 non-blood fed female sand flies from 33 species were captured by using miniature CDC light traps. Lutzomyia auraensis was the most abundant species (63%) in this area. Seven of 164 pools were positive by PCR for Leishmania by kinetoplast DNA. The real-time PCR identified four Lu. auraensis pools as positive for L. (Viannia) lainsoni and L. (V.) braziliensis. The minimum infection prevalence for Lu. auraensis was estimated to be 0.6% (95% confidence interval = 0.20–1.42%). Further studies are needed to assess the importance of Lu. auraensis in the transmission of New World tegumentary leishmaniasis in hyperendemic areas of Peru.
Introduction
Leishmaniasis is a complex of vector-borne diseases caused by protozoan parasites of the genus Leishmania. It occurs in as many as 70 countries worldwide, with more than 2 million new cases a year.1 In South America, New World tegumentary leishmaniasis (NWTL) is mainly caused by species of the Viannia subgenus, including L. (V.) braziliensis, Leishmania (V.) peruviana, Leishmania (V.) guyanensis, and Leishmania (V.) panamensis.2,3 In Peru, NWTL is endemic in 74% of the country,3,4 and there were 6,761 reported cases in 2010.5
The transmission cycle of Leishmania begins with the bite of an infected phlebotomine sand fly. Of the 500 known sand fly species in South America, only approximately 30 are known vectors for NWTL.4,6,7 In Peru, four species have been incriminated as vectors of L. (V.) peruviana, and all are in the Andean Mountain regions.8–11 However, little is known regarding vectors of NWTL in the Peruvian Amazon Basin despite the large number of human cases reported.8,9,11
Vector surveillance studies are often focused on studying the distribution of potential vectors and the prevalence of Leishmania infections in field-captured sand flies. These studies are critical to better understand the dynamics of disease transmission and to identify new parasite-vector associations.12
The detection of Leishmania in sand fly specimens has traditionally been based on the finding of flagellate forms in sand fly midgut dissections. However, this method requires advanced expertise13 and is cumbersome and time-consuming. Consequently, its use is limited under field conditions or in laboratories in remote-endemic areas where technical resources may be scarce. Improved tools to identify Leishmania species in sand fly specimens are greatly needed to enhance entomologic surveillance studies.
Tsukayama and others reported Leishmania species-specific DNA polymorphisms in the genes encoding mannose phosphate isomerase (MPI) and 6-phosphogluconate dehydrogenase (6PGD).14 On the basis of these polymorphisms, we have designed a fluorescence resonance energy transfer (FRET)–based real-time PCR that can identify up to five Leishmania (V.) species (Nuñez JH and others, unpublished data). In the present study, we applied this assay to identify Leishmania parasites in several species of Lutzomyia sand flies collected from a rural community in Madre de Dios, Peru, a region where NWTL is hyperendemic.
Materials and Methods
Study area.
The study was conducted in the community of Flor de Acre in the District of Iberia, Province of Tahuamanu, Department of Madre de Dios, Peru. This community is located along the recently inaugurated Peru–Brazil inter-oceanic highway on the Iberia-Pacahuara route. The site is located at approximately 11°23′09″S, 69°30′55″ (Figure 1) at 300 meters above sea level and has humid sub-tropical climate. Annual temperature in this region ranges from 22°C to 34°C, and annual precipitation ranges from 4,000 mm to 8,000 mm. In 2010, the Department of Madre de Dios reported 415 cases of cutaneous leishmaniasis and 47 cases of mucocutaneous leishmaniasis.5 Previous reports show that L. (V.) braziliensis, L. (V.) guyanensis, L. (V.) lainsoni, and Leishmania (V.) shawi are Leishmania species identified in human samples from the study area.3,15–17
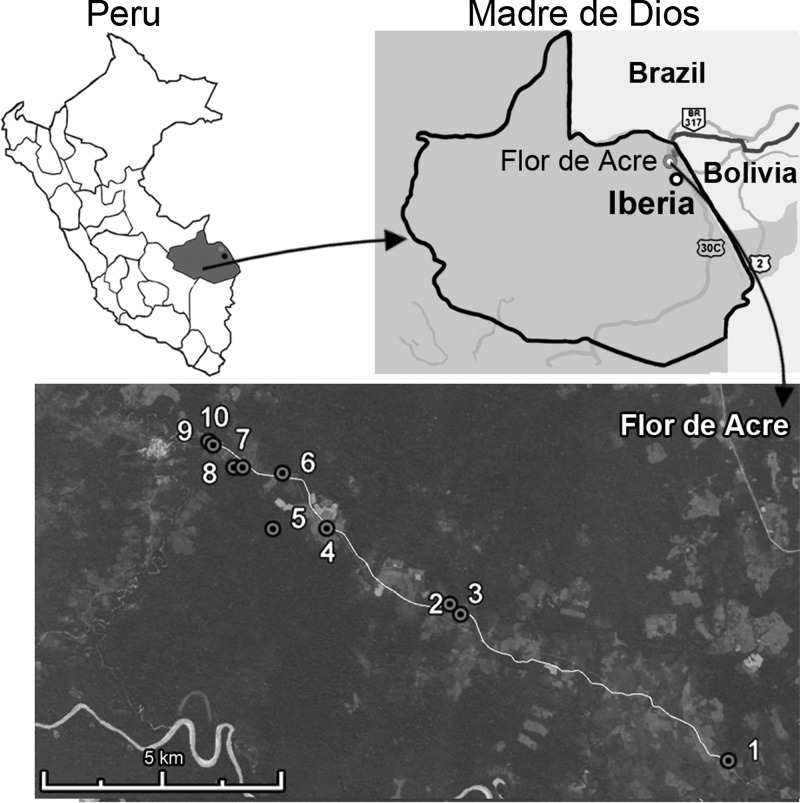
Figure 1.
Flor de Acre, Iberia in Madre de Dios, Peru. Inset shows satellite images of 10 houses around where sand flies were collected. Source: “Flor de Acre” S11° 23′ 09″ and W69° 30′ 55″ Google Earth.
Capture of sand flies and identification.
Peridomiciliary sand fly specimens were captured in September, November, and December of 2009 (before and during the rainy season) in the vicinity of 10 arbitrarily selected houses during 10 consecutive days each month (Figure 1). This period was chosen on the basis of the seasonal increase in sand fly density regularly observed in the region at this time. Miniature CDC light traps equipped with a CO2 generator were placed outside each house for 12 hours per night. No humans or animals were used for collection of specimens, and no collection of information on humans was obtained as part of this study. Phlebotomines were stored in 70% ethanol and transported at room temperature to the central laboratory of the US Naval Medical Research Unit No. 6 in Lima.
Species determination was based on keys developed by Young and Duncan.7 To preserve DNA integrity for PCRs, sand fly species identification was carried out by using a modified procedure that uses lactophenol only on specific parts of the sand flie body, thus preserving the remainder of the specimen for PCR analysis. Sand flies were sexed and non–blood-fed females were pooled in groups of 1–10 specimens according to species and capture location and stored again in 70% ethanol until total DNA was isolated. Male sand flies were used as negative controls because they are not infected by Leishmania.
DNA extraction.
DNA isolation was performed by using the Chelex-100 method according to reported techniques.18 In brief, pooled sand flies were homogenized in 0.85% NaCl. Homogenized pools were treated with 5% (w/v) Chelex-100 for 5 minutes, followed by incubation at 100°C for 10 minutes. Samples were then centrifuged at 16,000 × g for 10 minutes. Supernatants were carefully separated from the pellet and then mixed with 200 μL of 100% ethanol and centrifuged at 16,000 × g for 15 minutes. The supernatants were mixed with 100 μL of 70% ethanol and pellets were dried by using a SpeedVac (Savant, Irvine, CA) and resuspended in 50 μL of nuclease-free distilled water.
Polymerase chain reaction for detection of sand fly DNA.
For the detection of Lutzomyia DNA, a published PCR protocol was used19 with primers T1B 5′-AAA CTA GGA TTA GAT ACC CT-3′ and T2A 5′-AAT GAG AGC GAC GGG CGA TGT-3′ specific for 12S ribosomal DNA. The reactions were carried out in a volume of 25 μL containing 1X Taq polymerase buffer (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 125 μM dNTPs (Invitrogen), 0.5 μM of each primer, 1 unit of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. The PCR was run on a thermocycler (GeneAmp PCR system 9700; Applied Biosystems, Foster City, CA) for 5 minutes at 94°C; followed by 35 cycles for 20 seconds at 94°C, 30 seconds at 56°C, and 25 seconds at 72°C; followed by a final extension step for 5 minutes at 72°C. A PCR product of approximately 400 basepairs confirms the presence of sand fly DNA in the extracted samples and the absence of potential PCR inhibitors in the DNA preparations.
Polymerase chain reaction identification of Leishmania Viannia subgenus.
A modified PCR protocol20 was used to detect Leishmania (Viannia) subgenus from sand fly pools. A conserved region of the kinetoplast DNA minicircle was amplified by using primers MP1-L 5′-TAC TCC CCG ACA TGC CTC TG-3′ and MP3-H 5′-GAA CGG GGT TTC TGT ATG C-3′.20 The reactions were carried out in a volume of 20 μL containing 1X Taq polymerase buffer (Invitrogen), 1.5 mM MgCl2, 125 μM dNTPs (Invitrogen), 0.5 μM of each primer, 1 unit of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. The PCR was run on the aforementioned thermocycler for 5 minutes at 94°C; followed by 35 cycles for 45 seconds at 94°C, 45 seconds at 58°C, and 1 minute at 72°C; followed by a final extension step for 5 minutes at 72°C. This reaction generates a 70-basepair DNA fragment that is specific for the Viannia subgenus.
Fluorescence resonance energy transfer–based real-time PCR for speciation of Leishmania.
This PCR approach is based on known mutations in the 6PGD and MPI genes that yield distinct melting peaks, which are used to distinguish between five Leishmania species.14 The validation of this real-time assay will be published separately. For the 6PGD gene, we used a 50-μL reaction containing 1X Taq polymerase buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTPs (Invitrogen), 0.8 μM of each primer, 1.5 units of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. For the MPI gene, reactions were performed in a volume of 50 μL containing 1X Taq polymerase buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTPs (Invitrogen), 1 μM of each primer, 1.5 units of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. The PCR was run on a thermocycler (GeneAmp PCR system 9700; Applied Biosystems).
After the first round of amplification reactions, a FRET based real-time assay was performed. Both gene reactions were conducted in a volume of 20 μL containing 1X LightCycler® 480 Genotyping Master (Roche, Indianapolis, IN), 1.25 μM of forward primer, 0.25 μM of reverse primer, 0.75 μM of anchor and sensor probes, and 5 μL of PCR product from the first reaction. The real-time PCR was run on a LightCycler 480 (Roche) (Nuñez JH and others, unpublished data). To estimate the analytical sensitivity of the real-time PCR, known amounts of parasite DNA were spiked into parasite-free male sand fly DNA preparations. The limit of detection was the lowest parasite DNA concentration that yielded an amplification product visualized on the FRET real-time PCR.
Results
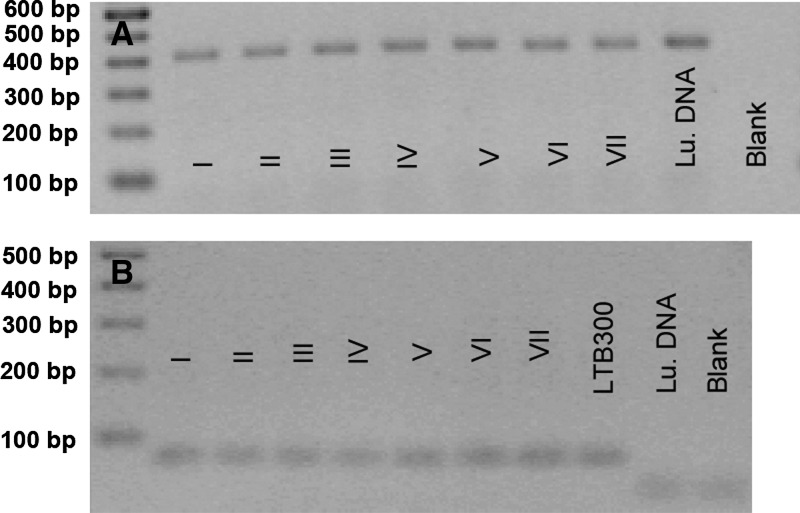
We collected 1,299 female specimens belonging to 33 sand fly species. As shown in Table 1, the most prevalent species were Lutzomyia auraensis (62.9%), followed by Lu. davisi (8.2%), Lu. choti (6.6%), Lu. llanosmartinsi (5.2%), and Lu. hirsuta (5.1%). The PCR analysis of 12S ribosomal DNA confirmed the identity of these sand flies as belonging to the genus Lutzomyia. In addition, the positive results indicated good DNA quality and the absence of PCR inhibitors and served as an internal control to rule out false-negative results (Figure 2A).
Table 1.
Species of Lutzomyia sand flies captured in Flor de Acre, Iberia, Madre de Dios, Peru
| Species | No. sand flies | % |
|---|---|---|
| Lu. auraensis | 821 | 62.9 |
| Lu. davisi | 107 | 8.2 |
| Lu. choti | 86 | 6.6 |
| Lu. llanosmartinsi | 68 | 5.2 |
| Lu. hirsuta | 67 | 5.1 |
| Lu. carrerai | 33 | 2.5 |
| Lutzomyia spp.* | 28 | 2.2 |
| Lu. yucumensis | 20 | 1.5 |
| Lu. aragaoi | 16 | 1.2 |
| Lu. shawi | 8 | 0.6 |
| Lu. richardwardi | 7 | 0.5 |
| Lu. umbratilis | 4 | 0.3 |
| Lu. falcata | 3 | 0.2 |
| Lu. trinidadensis | 3 | 0.2 |
| Other Lutzomyia† | 28 | 2.8 |
| Total | 1,299 | 100 |
The species of these sand flies could not be determined. They were only identified as belonging to the genus Lutzomyia.
Other Lutzomyia species: Lu. barrettoi, Lu. migonei, Lu. nevesi, Lu. quechua, Lu. saulensis, Lu. serrana, Lu. sherlocki, Lu. Nyssomyia whitmanni, Lu. Psychodopygus amazonensis, Lu. antunesi, Lu. claustrei, Lu. cortelezzi, Lu. evangelistai, Lu. furcata, Lu. guyanensis, Lu. nematoducta, Lu. punctigeniculata, Lu. nuneztovari, Lu. preclara, Lu. walkeri.
Figure 2.
Detection of Lutzomyia 12S ribosomal DNA and of Leishmania kinetoplast DNA by polymerase chain reaction (PCR) of pools of field-collected Lutzomyia sand flies. A, Agarose gel electrophoresis of 12S ribosomal DNA PCR products in a representative subset of pools. Lane 1, 100-basepair (bp) ladder; lanes 2–8, DNA from Lutzomyia pools; lane 9, positive control of male Lutzomyia spp.; lane 10, blank. B, Agarose gel electrophoresis of kinetoplast DNA PCR products. Lane 1, 100-bp ladder; lanes 2–8, positive Lutzomyia pools; lane 9, positive control of L. (V.) braziliensis strain MHOM/BR/84/LTB300; lane 10, negative control of male Lutzomyia spp.; lane 11, blank.
Sand fly specimens were grouped into 164 pools on the basis of sand fly species and collection site. Seven of 164 sand fly pools were positive for parasites of the Leishmania (Viannia) subgenus by kinetoplast DNA PCR (pools I–VII; Table 2 and Figure 2B). Four of the seven pools were composed of 10 specimens each of Lu. auraensis, and the remaining three comprised 1, 3, and 10 specimens of Lu. punctigeniculata, Lu. trinidadensis, and Lu. davisi, respectively. DNA samples from male sand flies, which are known not to be infected by Leishmania, did not yield any amplification product.
Table 2.
Identification of Leishmania species by real-time polymerase chain reaction in seven kinetoplast DNA PCR-positive sand fly pools*
| Lutzomyia species | Pool no. | House† | Kinetoplast DNA PCR | Real-time PCR | No. insects in pool | Month of capture |
|---|---|---|---|---|---|---|
| Lu. punctigeniculata | I | 9 | + | – | 1 | September |
| Lu. trinidadensis | II | 3/9 | + | – | 3 | September |
| Lu. davisi | III | 9 | + | L. (Viannia) braziliensis | 10 | September |
| Lu. auraensis | IV | 1 | + | L. (V.) braziliensis | 10 | November |
| Lu. auraensis | V | 9 | + | L. (V.) lainsoni | 10 | December |
| Lu. auraensis | VI | 9 | + | L. (V.) lainsoni | 10 | December |
| Lu. auraensis | VII | 9 | + | L. (V.) lainsoni | 10 | December |
PCR = polymerase chain reaction.
House number from which sand fly pools were collected.
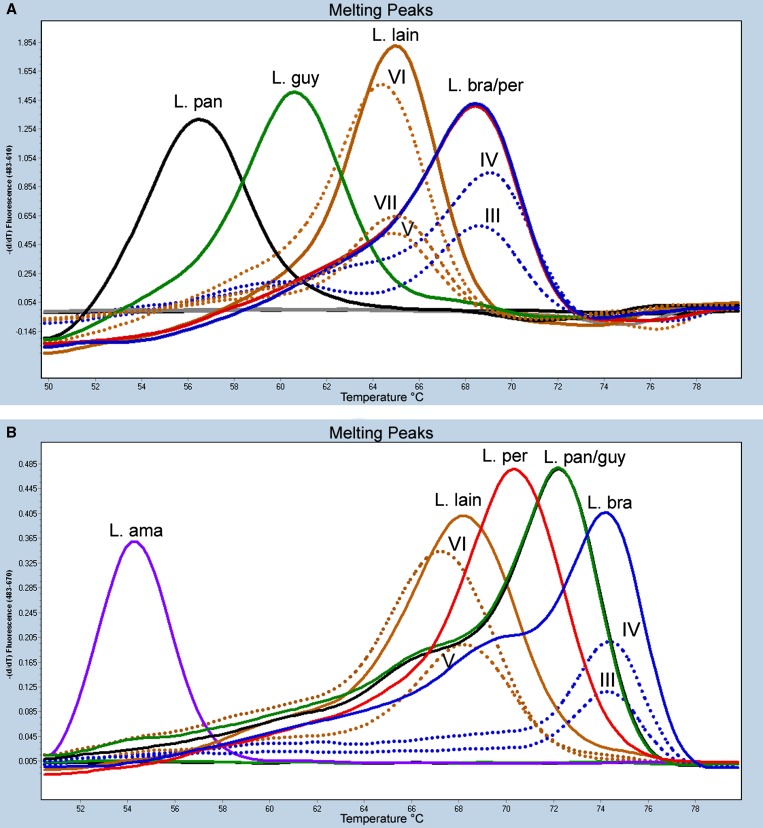
Our FRET-based real-time PCR was used to identify the species of Leishmania in kinetoplast DNA-positive pools. This assay detects down to 60 fg of parasite DNA, which is equivalent to less than five parasites per reaction.21 We identified the species of Leishmania from five of the seven kinetoplast DNA-positive pools (III–VII). The real-time PCR identified three positive pools as L. (V.) lainsoni (V–VII), and two positive pools (III and IV) as L. (V.) braziliensis (Table 2 and Figure 3). All but one of these pools were Lu. auraensis. Pool VII did not yield a positive result for the MPI locus, possibly because of the larger amplicon size for MPI (1,464 basepairs) than for kinetoplast DNA (70 basepairs) and 6PGD (628 basepairs). Two additional kinetoplast DNA-positive, real-time PCR-negative pools did not yield any amplification product for 6PGD or MPI. Therefore, we could not identify the Leishmania species.
Figure 3.
Melting curve analysis of real-time polymerase chain reaction of 6-phosphogluconate dehydrogenase (6PGD) and mannose phosphate isomerase (MPI). A, Melting curves corresponding to 6PGD-based amplification reactions of seven sand fly pools (I–VII) were run and compared with a battery of six standard strains of the New World Leishmania. B, Melting curves of MPI-based real-time amplification reactions of seven sand fly pools (I–VII) were run and compared with a battery of six standard strains of New World Leishmania. Species were identified on the basis of melting curves that had the same or similar peaks. pan = panamensis; guy = guyanensis; lain = lainsoni; bra = braziliensis; per = peruviana; ama = amazonensis.
Five of the seven kinetoplast DNA-positive pools were collected around house no. 9. Another positive pool was collected near house no. 1, which was approximately 12 km from house no. 9 (Figure 1). The minimal infection prevalence for Lu. auraensis and Lu. davisi was 0.60% (5 of 821) (95% confidence interval = 0.20–1.42%) and 0.90% (1 of 107), (95% confidence interval = 0.02–5.10%), respectively. We were unable to estimate the minimal infection prevalence for Lu. punctigeniculata and Lu. trinidadensis because there were only one and three insects captured of each species, respectively.
Discussion
We report infection of Lu. auraensis with New World species of Leishmania. Using a newly developed real-time PCR that can discriminate among five Leishmania species, we identified that Lu. auraensis is a natural carrier of L. (V.) lainsoni and L. (V.) braziliensis in the study area. Positivity among non–blood-fed females supports the likelihood of infection as opposed to only parasite carriage. There are no previous reports of natural Leishmania infection in this sand fly species, despite being highly prevalent in hyperendemic settings for leishmaniasis in Brazil and Peru.22,23
This study also confirmed the large predominance of Lu. auraensis, which comprised 63% of all collected sand flies in the study area. The abundance of this species in the southeastern Peruvian rainforest has been reported,22 and reached approximately 80% of all collected sand flies in Alto Tambopata, Peru. Lu. auraensis has been reported in the State of Acre, Brazil, and comprised 19% of all collected sand flies.23 According to Young and Duncan, there are also reports of Lu. auraensis in Venezuela, Suriname, Bolivia, and Colombia.7 Moreover, Lu. auraensis may have anthropophilic behavior on the basis of studies in Puno, Peru,22 although existing evidence lacks confirmation.24 These findings underscore the urgency to determine the importance of this species in the transmission of NWTL in this region.
To determine the vectorial role of Lu. auraensis, future studies will need to assess if this sand fly species is capable of sustaining the parasite and transmitting it to the vertebrate host.25 The possibility that Lu. auraensis transmits L. (V.) braziliensis is a particularly important consequence of our findings, given that this parasite species is the major etiologic agent of the disfiguring mucosal presentation of the disease. Overall, our results highlight the limited knowledge we have regarding the vectors implicated in the transmission of leishmaniasis in many parts of South America.
The PCR-based techniques that detect low amounts of parasite DNA are highly desirable because of their increased sensitivity and specificity in detecting Leishmania parasites12,13,26,27 compared with midgut dissection techniques.6 In this study, the real-time PCR could detect and discriminate Leishmania species in field-collected sand fly preparations with high reliability. This method is ideal for field conditions because entomologic specimens can be collected and preserved in alcohol at room temperature until transported to the laboratory for processing.13 The real-time assay was evaluated in 192 clinical samples from patients with suspected leishmaniasis throughout Peru and showed a sensitivity of 91% and a specificity of 77%, when compared to kinetoplastid DNA PCR for leishmania diagnosis (Nunez JH, unpublished data). In addition, the real-time PCR assay can identify species within the Leishmania Viannia complex with 100% of concordance compared to multilocus sequencing typing (tested in 72 positive clinical samples). Furthermore, unlike reported PCRs,20,28,29 no post-PCR processing such as electrophoresis, restriction fragment length polymorphism, or sequencing was required.
Most (5 of 7) positive sand fly pools were collected near a single house. Two other houses with positive pools were located 5 and 12 km away from this house and the pools from seven other houses were negative. These findings are consistent with the clustering and heterogeneous spatial distribution observed in a small area of Leishmania-infected sand flies in Rio Branco, Brazil, where the location with the most positive phlebotomines also had the highest number of new cases of cutaneous leishmaniasis.30 Future studies in the Amazon basin need to assess larger number of more dispersed collection points to better describe if transmission risk variability remains over broader geographic ranges.
Lutzomyia davisi represented the second most frequent sand fly species (8.2%), and one pool of this species was positive for L. (V.) braziliensis. Lutzomyia davisi is a vector for NWTL and is commonly found in Brazil, Colombia, and French Guiana.31,32 Although Lu. davisi has been reported in the Peruvian rainforest,22 Leishmania infections have not been reported in this species in Peru. The presence of natural infection by L. (V.) braziliensis confirmed in this study indicates that this sand fly species may also act as vector for leishmaniasis transmission in the study area, although experimental confirmation is lacking.
Less common sand fly species such as Lu. trinidadensis and Lu. punctigeniculata were positive by PCR for kinetoplast DNA, which indicated infection with New World Leishmania species. However, the real-time PCR showed negative results for these samples and consistent results after three repetitions, leaving us unable to identify the infecting Leishmania species. We found that the detection limits of the kinetoplast DNA and real-time PCRs were equivalent when using the same parasite DNA preparation. Therefore, it is unlikely that this discordance could reflect lower sensitivity of the real-time PCR. The kinetoplast DNA PCR may have amplified other Leishmania (Viannia) species that are not within the five species that the real-time PCR detects.
The specificity of the real-time PCR was 100% and the sensitivity was also high but could be improved. Therefore, the two kinetoplast DNA-positive, real-time PCR-negative pools could be infected with other Leishmania species less commonly reported in the study region but not evaluated in our assays. For example, L. (V.) shawi was reported in the same region of Peru where our study was conducted.17 Also, a study in Venezuela detected Leishmania (L.) venezuelensis in Lu. trinidadensis, the same sand fly species found to be Leishmania positive in our study but without an identifiable species.33 However, it is unlikely that the Leishmania species found in Lu. auraensis could be a misclassification of other species. For example, L. (V.) braziliensis and L. (V.) shawi have different 6PGD multilocus enzyme electrophoresis patterns.34 Also, the MPI real-time PCR can identify Leishmania (L.) amazonensis/mexicana (Nuñez JH, unpublished data) and probably other Leishmania subgenus species as a complex, differentiating them from L. (V.) braziliensis and L. (V.) lainsoni. Therefore, none of these potential limitations affect our main conclusion, which is that Lu. auraensis is a common and important carrier of multiple Leishmania species in the southern Amazon basin. Expanding the list of species detected by our assay and more extensive vector studies will further enhance our knowledge of the distribution of Leishmania vectors in this little-described disease-endemic region.
In summary, we identified natural Leishmania infection in Lu. auraensis, a widespread sand fly species in Peru and Brazil. Its role as a new vector for leishmaniasis remains to be determined to better understand the risk of leishmaniasis transmission in the southern Amazon Basin. Our real-time PCR proved to be a viable and efficient tool for identification of Leishmania species in field-collected sand fly specimens, compared with traditional dissection and microscopic examination techniques. The use of this assay in field conditions could be useful in identifying Leishmania-carrying sand fly species and enhancing research and prevention efforts.
ACKNOWLEDGMENTS
We thank Arturo Marino Sanchez for his support with sand fly captures, Dr. Juan Francisco Sanchez for helping with geographic positioning analysis, Noah Nattell for critically reading the manuscript, and Dr. Rosa Pacheco for advice and support as thesis advisor to Hugo Valdivia.
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Footnotes
Financial support: This study was supported by training grant NIH/FIC 2D43 TW007393 awarded to the US Naval Medical Research Unit No. 6 by the Fogarty International Center of the U.S. National Institutes of Health and by grants CO497_11_L1 and CO466_11_L1 of the Global Emerging Infections Surveillance and Response System of the U.S. Department of Defense.
Disclosure: Several authors of this manuscript are military service members or employees of the U.S. Government. This work was prepared as part of their duties. Title 17 U.S.C. § 105 provides that Copyright protection under this title is not available for any work of the United States Government. Title 17 U.S.C. § 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Authors' addresses: Hugo O. Valdivia, Maxy B. De Los Santos, Roberto Fernandez, G. Christian Baldeviano, Victor O. Zorrilla, Carmen M. Lucas, Kimberly A. Edgel, Andrés G. Lescano, and Kirk D. Mundal, U.S. Naval Medical Research Unit No. 6, Av. Venezuela Cuadra 36, Callao 2, Peru, E-mails: hugo.valdivia@med.navy.mil, maxy.delossantos@med.navy.mil, roberto.fernandez@med.navy.mil, christian.baldeviano@med.navy.mil, victor.zorrilla@med.navy.mil, carmen.lucas@med.navy.mil, kimberly.edgel@med.navy.mil, willy.lescano@med.navy.mil, and kirk.mundal@hotmail.com. Hubert Vera, Direccion Regional de Salud de Madre de Dios, Ernesto Rivero 475, Puerto Maldonado, Madre de Dios, Peru, E-mail: hubvera@hotmail.com. Paul C. F. Graf, E-mail: paul.graf@med.navy.mil.
References
- 1.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 2.Banuls AL, Hide M, Prugnolle F. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 3.Lucas CM, Franke ED, Cachay MI, Tejada A, Cruz ME, Kreutzer RD, Barker DC, McCann SH, Watts DM. Geographic distribution and clinical description of leishmaniasis cases in Peru. Am J Trop Med Hyg. 1998;59:312–317. doi: 10.4269/ajtmh.1998.59.312. [DOI] [PubMed] [Google Scholar]
- 4.Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990;4:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 5.Peruvian Ministry of Health General Epidemiology Directorate Report. 2010. http://www.dge.gob.pe/salasit.php. Available at.
- 6.Kato H, Gomez EA, Caceres AG, Uezato H, Mimori T, Hashiguchi Y. Molecular epidemiology for vector research on leishmaniasis. Int J Environ Res Public Health. 2010;7:814–826. doi: 10.3390/ijerph7030814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young DG, Duncan MA. Guide to the Identification and Geographic Distribution of Lutzomyia Sand Flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae) Gainesville, FL: Associated Publishers; 1994. [Google Scholar]
- 8.Caceres AG, Villaseca P, Dujardin JC, Banuls AL, Inga R, Lopez M, Arana M, Le Ray D, Arevalo J. Epidemiology of Andean cutaneous leishmaniasis: incrimination of Lutzomyia ayacuchensis (Diptera: Psychodidae) as a vector of Leishmania in geographically isolated, upland valleys of Peru. Am J Trop Med Hyg. 2004;70:607–612. [PubMed] [Google Scholar]
- 9.Kato H, Caceres AG, Gomez EA, Mimori T, Uezato H, Marco JD, Barroso PA, Iwata H, Hashiguchi Y. Molecular mass screening to incriminate sand fly vectors of Andean-type cutaneous leishmaniasis in Ecuador and Peru. Am J Trop Med Hyg. 2008;79:719–721. [PubMed] [Google Scholar]
- 10.Davies CR, Fernandez M, Paz L, Roncal N, Llanos-Cuentas A. Lutzomyia verrucarum can transmit Leishmania peruviana, the aetiological agent of Andean cutaneous leishmaniasis. Trans R Soc Trop Med Hyg. 1993;87:603–606. doi: 10.1016/0035-9203(93)90103-w. [DOI] [PubMed] [Google Scholar]
- 11.Perez JE, Veland N, Espinosa D, Torres K, Ogusuku E, Llanos-Cuentas A, Gamboa D, Arevalo J. Isolation and molecular identification of Leishmania (Viannia) peruviana from naturally infected Lutzomyia peruensis (Diptera: Psychodidae) in the Peruvian Andes. Mem Inst Oswaldo Cruz. 2007;102:655–658. doi: 10.1590/s0074-02762007005000077. [DOI] [PubMed] [Google Scholar]
- 12.Paiva BR, Secundino NF, Nascimento JC, Pimenta PF, Galati EA, Junior HF, Malafronte RS. Detection and identification of Leishmania species in field-captured phlebotomine sandflies based on mini-exon gene PCR. Acta Trop. 2006;99:252–259. doi: 10.1016/j.actatropica.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Schonian G, Kuhls K, Mauricio IL. Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology. 2011;138:405–425. doi: 10.1017/S0031182010001538. [DOI] [PubMed] [Google Scholar]
- 14.Tsukayama P, Lucas C, Bacon DJ. Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol. 2009;39:355–362. doi: 10.1016/j.ijpara.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Verastegui C, Lazo M, Loayza-Muro R, De Doncker S, Maurer A, Chappuis F, Dujardin JC, Llanos-Cuentas A. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis. 2007;195:1846–1851. doi: 10.1086/518041. [DOI] [PubMed] [Google Scholar]
- 16.Yardley V, Croft SL, De Doncker S, Dujardin JC, Koirala S, Rijal S, Miranda C, Llanos-Cuentas A, Chappuis F. The sensitivity of clinical isolates of Leishmania from Peru and Nepal to miltefosine. Am J Trop Med Hyg. 2005;73:272–275. [PubMed] [Google Scholar]
- 17.Kato H, Caceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, Iwata H, Uezato H, Velez LN, Gomez EA, Hashiguchi Y. Use of FTA cards for direct sampling of patients' lesions in the ecological study of cutaneous leishmaniasis. J Clin Microbiol. 2010;48:3661–3665. doi: 10.1128/JCM.00498-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lardeux F, Chavez T, Rodriguez R, Torrez L. Anopheles of Bolivia: new records with an updated and annotated checklist. C R Biol. 2009;332:489–499. doi: 10.1016/j.crvi.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Beati L, Caceres AG, Lee JA, Munstermann LE. Systematic relationships among Lutzomyia sand flies (Diptera: Psychodidae) of Peru and Colombia based on the analysis of 12S and 28S ribosomal DNA sequences. Int J Parasitol. 2004;34:225–234. doi: 10.1016/j.ijpara.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Lopez M, Inga R, Cangalaya M, Echevarria J, Llanos-Cuentas A, Orrego C, Arevalo J. Diagnosis of Leishmania using the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg. 1993;49:348–356. doi: 10.4269/ajtmh.1993.49.348. [DOI] [PubMed] [Google Scholar]
- 21.Salotra P, Sreenivas G, Pogue GP, Lee N, Nakhasi HL, Ramesh V, Negi NS. Development of a species-specific PCR assay for detection of Leishmania donovani in clinical samples from patients with kala-azar and post-kala-azar dermal leishmaniasis. J Clin Microbiol. 2001;39:849–854. doi: 10.1128/JCM.39.3.849-854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arístides H. La leishmaniasis tegumentaria en el Alto Tambopata, Departamento de Puno, Perú. Rev Med Exp. 1999;15:15–24. [Google Scholar]
- 23.Azevedo AC, Costa SM, Pinto MC, Souza JL, Cruz HC, Vidal J, Rangel EF. Studies on the sandfly fauna (Diptera: Psychodidae: Phlebotominae) from transmission areas of American cutaneous leishmaniasis in state of Acre, Brazil. Mem Inst Oswaldo Cruz. 2008;103:760–767. doi: 10.1590/s0074-02762008000800003. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez R, Galati EB, Carbajal F, Wooster MT, Watts DM. Notes on the phlebotomine sand flies from the Peruvian southeast–I. Description of Lutzomyia (Helcocyrtomyia) adamsi n. sp. (Diptera: Psychodidae) Mem Inst Oswaldo Cruz. 1998;93:41–44. doi: 10.1590/s0074-02761998000100009. [DOI] [PubMed] [Google Scholar]
- 25.Farrell J. Leishmania. Boston, MA: Kluwer Academic Publishers; 2002. [Google Scholar]
- 26.Talmi-Frank D, Nasereddin A, Schnur LF, Schonian G, Toz SO, Jaffe CL, Baneth G. Detection and identification of old world Leishmania by high resolution melt analysis. PLoS Negl Trop Dis. 2010;4:e581. doi: 10.1371/journal.pntd.0000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antinori S, Calattini S, Piolini R, Longhi E, Bestetti G, Cascio A, Parravicini C, Corbellino M. Is real-time polymerase chain reaction (PCR) more useful than a conventional PCR for the clinical management of leishmaniasis? Am J Trop Med Hyg. 2009;81:46–51. [PubMed] [Google Scholar]
- 28.Hajjaran H, Vasigheh F, Mohebali M, Rezaei S, Mamishi S, Charedar S. Direct diagnosis of Leishmania species on serosity materials punctured from cutaneous leishmaniasis patients using PCR-RFLP. J Clin Lab Anal. 2011;25:20–24. doi: 10.1002/jcla.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacha HA, Tuon FF, Zampieri RA, Floeter-Winter LM, Oliveira J, Nicodemo AC, Quiroga MM, Mascheretti M, Boulos M, Amato VS. Leishmania (Viannia) braziliensis identification by PCR in the state of Para, Brazil. Trans R Soc Trop Med Hyg. 2010;105:173–178. doi: 10.1016/j.trstmh.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Miranda JC, Reis E, Schriefer A, Goncalves M, Reis MG, Carvalho L, Fernandes O, Barral-Netto M, Barral A. Frequency of infection of Lutzomyia phlebotomines with Leishmania braziliensis in a Brazilian endemic area as assessed by pinpoint capture and polymerase chain reaction. Mem Inst Oswaldo Cruz. 2002;97:185–188. doi: 10.1590/s0074-02762002000200006. [DOI] [PubMed] [Google Scholar]
- 31.Gil LH, Basano SA, Souza AA, Silva MG, Barata I, Ishikawa EA, Camargo LM, Shaw JJ. Recent observations on the sand fly (Diptera: Psychodidae) fauna of the State of Rondonia, Western Amazonia, Brazil: the importance of Psychdopygus davisi as a vector of zoonotic cutaneous leishmaniasis. Mem Inst Oswaldo Cruz. 2003;98:751–755. doi: 10.1590/s0074-02762003000600007. [DOI] [PubMed] [Google Scholar]
- 32.Grimaldi G, Jr, Momen H, Naiff RD, McMahon-Pratt D, Barrett TV. Characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the Amazon region of Brazil. Am J Trop Med Hyg. 1991;44:645–661. doi: 10.4269/ajtmh.1991.44.645. [DOI] [PubMed] [Google Scholar]
- 33.Bonfante-Garrido R, Urdaneta R, Urdaneta I, Alvarado J. Natural infection of Lutzomyia trinidadensis (Diptera: Psychodidae) with Leishmania in Barquisimeto, Venezuela. Mem Inst Oswaldo Cruz. 1990;85:477. doi: 10.1590/s0074-02761990000400015. [DOI] [PubMed] [Google Scholar]
- 34.Brito ME, Andrade MS, Mendonca MG, Silva CJ, Almeida EL, Lima BS, Felix SM, Abath FG, da Graca GC, Porrozzi R, Ishikawa EA, Shaw JJ, Cupolillo E, Brandao-Filho SP. Species diversity of Leishmania (Viannia) parasites circulating in an endemic area for cutaneous leishmaniasis located in the Atlantic rainforest region of northeastern Brazil. Trop Med Int Health. 2009;14:1278–1286. doi: 10.1111/j.1365-3156.2009.02361.x. [DOI] [PubMed] [Google Scholar]