Abstract
Bartonella infections were investigated in bats in the Amazon part of Peru. A total of 112 bats belonging to 19 species were surveyed. Bartonella bacteria were cultured from 24.1% of the bats (27/112). Infection rates ranged from 0% to 100% per bat species. Phylogenetic analyses of gltA of the Bartonella isolates revealed 21 genetic variants clustering into 13 divergent phylogroups. Some Bartonella strains were shared by bats of multiple species, and bats of some species were infected with multiple Bartonella strains, showing no evident specific Bartonella sp.–bat relationships. Rarely found in other bat species, the Bartonella strains of phylogroups I and III discovered from the common vampire bats (Desmodus rotundus) were more specific to the host bat species, suggesting some level of host specificity.
Introduction
Bats are some of the most abundant, diverse, and geographically dispersed mammals.1 The high mobility, broad geographic distribution, social behavior (communal roosting and fission–fusion social structure), and longevity of bats make them ideal reservoir hosts and sources of infection for various etiologic agents. Multiple studies have highlighted that bats may play an important role in serving as natural reservoirs to numerous pathogens.2–4 Although the knowledge of bats was mostly associated with viruses, several studies also reported the detection of bacterial agents in bats and their ectoparasites5–13 and have suggested potential for human infection.14
The genus of Bartonella includes a group of gram-negative bacteria that are highly adapted to intracellular persistence in a wide variety of vertebrates, including rodents, insectivores, carnivores, ungulates, and other mammals. A number of Bartonella species have been associated with human illnesses and are responsible for a growing spectrum of emerging diseases.15–23 Knowledge of the transmission of Bartonella bacteria between mammalian hosts is incomplete. However, hematophagous arthropods, such as fleas, flies, lice, mites, and ticks, have been found naturally infected and are frequently implicated in transmitting Bartonella species.24–28 Other modes, such as direct animal contact, may also facilitate transmission of bartonellae. For example, detection of Bartonella DNA in the saliva of dogs suggests that dog biting might be a potential route for Bartonella transmission.29 Data on host specificity of Bartonella species are controversial. A study from the United Kingdom reported no specificity between Bartonella spp. and rodents,30 whereas investigations from North America and Asia suggested definite host specificity of Bartonella strains among rodent species.31–34
Several recent studies have reported Bartonella infection in bats from different parts of the world, including England, Kenya, and Guatemala.5,7,8 Studies from Kenya and Guatemala reported that Bartonella infections are highly prevalent in bats, and the Bartonella strains exhibit high genetic diversity. Nevertheless, Bartonella strains recovered from Kenyan bats exhibited specificity to their bat hosts, whereas those strains obtained from Guatemalan bats showed very little specificity to their bat hosts. To better understand the ecology of Bartonella infections in bats, the role of bats as reservoir hosts of Bartonella, and therefore, their potential significance to human and veterinary medicine, we investigated Bartonella infection in bats from Peru. The specific goals of this study were to (1) culture Bartonella bacteria from bat blood and estimate the prevalence of Bartonella infections in bat populations in Peru, (2) characterize Bartonella strains obtained from the Peruvian bats using citrate synthase gene (gltA) and compare them with strains obtained from bats in other studies and those strains available in the public domain, and (3) evaluate the genetic diversity of the Bartonella strains circulating among the bat populations.
Materials And Methods
Study sites and bat blood sample collections.
Bats were captured from two villages (Santa Marta and Truenococha) in Loreto Department, the Republic of Peru, between May 1 and May 6, 2010. Located in the northern part of the country, Loreto is situated in the Amazon rainforest, and it is sparsely populated. Bats were collected using mist nets placed near village homes, livestock, and fruit trees as authorized by the Peru Ministry of Agriculture permit RD-0389-2010-DGFFS-DGEFFS. Captured bats were anesthetized by an intramuscular injection of ketamine hydrochloride (25–55 mg/kg) in compliance with the field protocol approved by the Animal Institute Care and Use Committee of the Centers for Disease Control and Prevention (CDC; Atlanta, GA). The bats were weighed, sexed, morphologically identified to species by a field expert, and exsanguinated by cardiac puncture. Serum and red blood cell pellets were separated for each blood sample. Both were immediately stored in liquid nitrogen in the field and transferred to a −80°C freezer in the laboratory, and the red blood cell pellets were then sent to the Bartonella Laboratory, CDC, Fort Collins, Colorado.
Culturing.
Red blood cell pellets were resuspended 1:4 in brain heart infusion broth supplemented with 5% amphotericin B to reduce likelihood that bacterial and fungal contaminants would overgrow the slow-growing Bartonella bacteria; then, they were plated on heart infusion agar containing 10% rabbit blood and incubated in an aerobic atmosphere with 5% carbon dioxide at 35°C up to 4 weeks. Bacterial growth was monitored at the end of each week. Bacterial colonies were presumptively identified as Bartonella based on their morphology. Morphologically distinct colony types from each sample were identified individually. Subcultures of Bartonella colonies from the original agar plate were streaked onto secondary agar plates that were also supplemented with 10% rabbit blood, and they were incubated at the same conditions until sufficient growth was observed, usually between 5 and 7 days. Pure cultures were harvested and stored in 10% glycerol.
Verification of Bartonella by polymerase chain reaction amplification of gltA.
Bartonella isolates were verified by polymerase chain reaction (PCR) amplification of a specific region in the gltA of the genus of Bartonella using primers BhCS781.p (5′-GGGGACCAGCTCATGGTGG-3′) and BhCS1137.n (5′-AATGCAAAAAGAACAGTAAACA-3′) to generate a 379-bp amplicon of the Bartonella gltA. Genomic DNA was prepared from heating a heavy suspension of microorganisms for 10 minutes at 95°C followed by centrifugation of the lysed cells for 1 minute at 8,000 rpm. The supernatant was then transferred to a clean centrifuge tube to be used as the template DNA. PCR amplifications were performed in a PTC 200 Peltier Thermal Cycler (MJ Research, Inc., Cambridge, MA). Positive and negative controls were included within each PCR assay to evaluate the presence of appropriately sized amplicon and contamination, respectively. PCR products were separated and visualized by 1.5% agarose gel electrophoresis with ethidium bromide staining.
Sequencing and phylogenetic analysis.
PCR products of appropriate size were purified with the QIAquick PCR Purification Kit (Qiagen, Germantown, MD) according to the manufacturer's instructions and sequenced in both directions using an Applied Biosystems Model 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequencing reactions were carried out in a PTC 200 Peltier Thermal cycler using the same primers as the initial PCR assay at a concentration of 1.6 μM.
Sequences were analyzed using Lasergene (DNASTAR, Madison, WI) sequence analysis software to determine consensus of sequences for the amplified region of the gltA. The Clustal V program within MegAlign of Lasergene was used to align and compare homology of bartonella gltA sequences obtained from bat samples in the present study with other bat-associated Bartonella genotypes from a Guatemala study, other genotypes available in the public domain, and known Bartonella species. The neighbor-joining method by Kimura's two-parameter distance method and bootstrap calculation was carried out with 1,000 replicates. Novel sequences from the current study were submitted to GenBank and assigned with unique accession numbers. We follow up the study carried out in Guatemala5 and continue using the Roman numbers (I–XIII and so on) used in the Guatemala study for the same definition of phylogroup identification.
Results
Bat species composition and distribution.
A total of 112 bats were tested for Bartonella infection. These bats included 67 bats from Santa Marta and 45 bats from Truenococha. The bats represented 19 species of 14 genera. Carollia perspicillata (Seba's short-tailed bat), Artibeus planirostris (flat-faced fruit-eating bat), and Desmodus rotundus (common vampire bat) were the three most prevalent species among the captured bats, comprising 25.9% (29/112), 16.1% (18/112), and 14.3% (16/112), respectively. The majority of Seba's short-tailed bats (89.7%; 26/29) were captured in Santa Marta, whereas the majority of the flat-faced fruit-eating bats (87.5%; 14/16) were captured in Truenococha. The common vampire bats were distributed in both sites (66.7% [12/18] in Santa Marta and 33.3% [6/18] in Truenococha). For each of the other 16 species, the number of captures was from 1 to 10 and accounted for smaller proportions of total bats (0.9–8.9%); the bats were distributed in either one or both study sites (Table 1).
Table 1.
Prevalence of Bartonella spp. in bats from two villages in the Peruvian Amazon in 2010
| Bat species | Number positive/number cultured (%) | ||
|---|---|---|---|
| Santa Marta | Truenococha | Overall | |
| Dermanura glaucus | 0/1 | 0/1 | |
| Artibeus lituratus | 0/3 | 0/3 | |
| Artibeus obscurus | 0/1 | 1/9 (11.1) | 1/10 (10) |
| Artibeus planirostris | 0/2 | 2/14 (14.3) | 2/16 (12.5) |
| Carollia brevicauda | 2/2 (100) | 2/2 (100) | |
| Carollia castanea | 0/2 | 0/2 | |
| Carollia perspicillata | 4/26 (15.4) | 0/3 | 4/29 (13.8) |
| Desmodus rotundus | 6/12 (50) | 4/6 (66.7) | 10/18 (55.6) |
| Diphylla ecaudata | 0/1 | 0/1 | |
| Glossophaga soricina | 1/1 (100) | 0/1 | 1/2 (50) |
| Molossus molossus | 0/10 | 0/10 | |
| Myotis sp. | 1/6 (16.7) | 1/6 (16.7) | |
| Phyllostomus discolor | 1/1 (100) | 1/1 (100) | 2/2 (100) |
| Phyllostomus hastatus | 1/2 (50) | 1/2 (50) | |
| Platyrrhinus recifinus | 0/1 | 0/1 | |
| Rhinophylla pumilio | 0/2 | 0/2 | |
| Sturnira lilium | 1/1 (100) | 1/1 (100) | |
| Uroderma bilobatum | 0/1 | 0/1 | |
| Vampyricus bidens | 2/3 (66.7) | 2/3 (66.7) | |
| Total | 16/67 (23.9) | 11/45 (24.4) | 27/112 (24.1) |
Bartonella prevalence.
Bartonella cultures were obtained from 27 of 112 tested bats, resulting in 24.1% prevalence of infection. The infected bats included 16 individuals from Santa Marta and 11 individuals from Truenococha. Although the bat community of the two sites varied in species composition, the prevalence of Bartonella infection in bats was similar across the two sites, with 23.9% (16/67) in Santa Marta and 24.4% (11/45) in Truenococha. During the primary plating, colonies with different morphological characteristics were observed from blood of two Phyllostomus discolor (pale spear-nosed bat) and one common vampire bat, resulting in a total of 30 isolates. Of the 19 tested bat species, Bartonella cultures were obtained in 11 species with prevalence as follows: A. obscurus, 1/10 (10%); A. planirostris, 2/16 (12.5%); C. brevicauda, 2/2 (100%); C. perspicillata, 4/29 (13.8%); D. rotundus, 10/18 (55.6%); Glossophaga soricina, 1/2 (50%); Myotis sp., 1/6 (16.7%); P. discolor, 2/2 (100%); P. hastatus, 1/2 (50%); Sturnira lilium, 1/1 (100%); and Vampyriscus bidens, 2/3 (66.7%). No bartonella was discovered in bats of Molossus molossus (0/10) and seven other species, with one to three individuals tested from each species (Table 1). The prevalence of bartonella infection in common vampire bats was significantly higher compared with all other bats (χ2 = 4.4, P < 0.05).
Heterogeneity of bat-associated Bartonella strains.
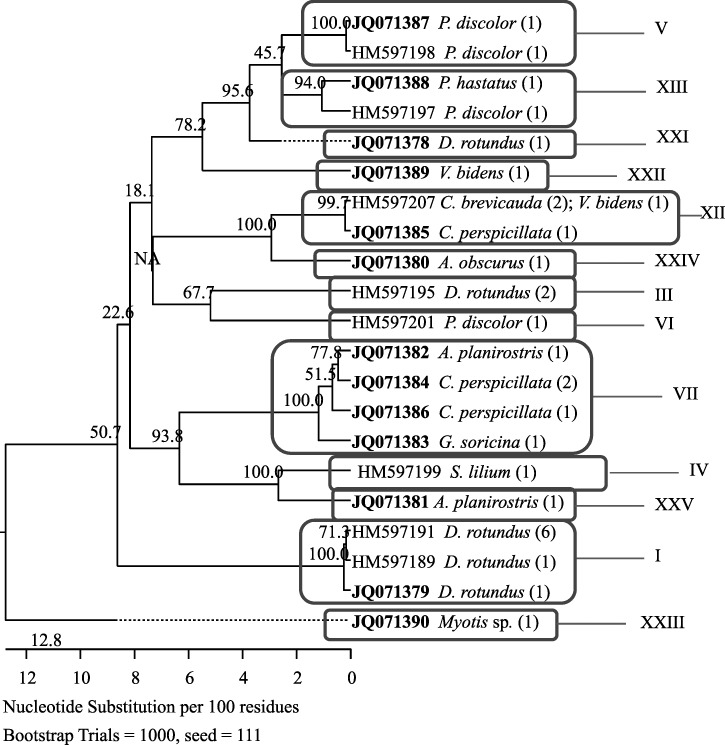
The gltA sequences from the Peruvian Bartonella cultures exhibited considerable sequence diversity. Sequencing analyses of the 30 Bartonella isolates revealed 21 genetic variants with 0.3–20.1% divergence. Of the variants, 13 variants were newly identified in the present study with GenBank accession numbers JQ071378–JQ071390 (Table 2), whereas the other 8 variants were identical to some previously described variants from bats in Guatemala.5 Phylogenetic analyses grouped these variants into 13 phylogroups (Figure 1) with genetic distance of 4.2–20.1% among the groups. Five phylogroups (XXI–XXV) were distinct from all described Bartonella species and previously reported Bartonella genogroups; the other eight groups contained variants that are identical or close to other known variants in bats from Guatemala.5
Table 2.
GenBank accession number (AC#) of the 13 novel Bartonella genetic variants identified in bats from Peru and the distribution of each variant in bats with indication of the number of isolates cultured from a certain bat species
| AC# | Type strain | Host species | Sequences | Distribution | Phylogroup |
|---|---|---|---|---|---|
| JQ071379 | B32945 | Desmodus rotundus | 1 | D. rotundus (1) | I |
| JQ071387 | B32947 | Phyllostomus discolor | 1 | P. discolor (1) | V |
| JQ071382 | B32954 | Artibeus planirostris | 1 | A. planirostris (1) | VII |
| JQ071383 | B32946 | Glossophaga soricina | 1 | G. soricina (1) | VII |
| JQ071384 | B32943 | Carollia perspicillata | 2 | C. perspicillata (2) | VII |
| JQ071386 | B32960 | Carollia perspicillata | 1 | C. perspicillata (1) | VII |
| JQ071385 | B32955 | Carollia perspicillata | 1 | C. perspicillata (1) | XII |
| JQ071388 | B32854 | Phyllostomus hastatus | 1 | P. hastatus (1) | XIII |
| JQ071378 | B32855 | Desmodus rotundus | 1 | D. rotundus (1) | XXI |
| JQ071389 | B32856 | Vampyriscus bidens | 1 | V. bidens (1) | XXII |
| JQ071390 | B32942 | Myotis sp. | 1 | Myotis sp. (1) | XXIII |
| JQ071380 | B32851 | Artibeus obscurus | 1 | A. obscurus (1) | XXIV |
| JQ071381 | B32953 | Artibeus planirostris | 1 | A. planirostris (1) | XXV |
Figure 1.
Phylogenetic relationships of the Bartonella genotypes based on partial sequences of gltA detected in bats from Peru compared with previously described Bartonella genotypes detected in Guatemalan bats. The phylogenetic tree was constructed by the neighboring-joining method, and bootstrap values were calculated with 1,000 replicates. A total of 21 Bartonella genotypes were identified among the 30 isolates obtained from bats in Peru, of which 13 genotypes were novel. The other eight genotypes were identical to Bartonella genotypes previously described from bats in Guatemala. Each genotype is indicated by its GenBank accession number, with novel genotypes in boldface. After each accession number is the bat species from which bartonella isolates of the genotype were obtained. Numbers in parentheses are the number of isolates obtained from bats of the indicated species. The Bartonella genotypes formed 13 phylogroups. Each square-circled clade represents a phylogroup and is marked by a unique Roman number using the description in the work by Bai and others.5
Association of Bartonella phylogroups and bat species.
A total of 11 isolates were obtained from 10 individuals of D. rotundus, the common vampire bat, with 2 isolates cultured from a single individual. The gltA sequences indicated that they were of five unique variants, among which two variants were newly identified in the present study (JQ071378–JQ071379) and the other three variants were identical to those variants previously described in the common vampire bats from Guatemala (HM597189, HM597191, and HM597195). The variants clustered into three phylogroups, named I, III, and XXI (Figure 1), with genetic distance > 13% among groups. Group I contained eight isolates of three variants, group III contained one variant with two isolates, and group XXI contained a novel variant that is distinct from all other known Bartonella species or genotypes. The two isolates derived from the same individual common vampire bat were distinct from each other and fell into groups I and III, respectively (Figure 1).
Six isolates were obtained from Carollia bats, including two from C. brevicauda and the other four from C. perspicillata, and four variants were identified. The two isolates from C. brevicauda were identical to a previously described variant (HM597207) found in a Micronycteris microtis bat from Guatemala and fell into phylogroup XII; the other four isolates from C. perspicillata belonged to three novel variants (JQ071384–JQ071386) and clustered into two phylogroups. Variant JQ071385 fell into the group XII with isolates from C. brevicauda; the other two variants of three isolates clustered into group VII with a previously identified variant (HM597202) in bats (G. soricina and Pteronotus davyi) from Guatemala (Figure 1).
All five isolates obtained from three Phyllostomus bats (two P. discolor bats each yielded two isolates based on colony morphology differences) differed from each other, and each represented a unique gltA variant. Two of them were first identified in the present study (JQ071387–JQ071388), whereas the other three variants were each identical to a previously described variant from P. discolor and Dermanura tolteca (formerly Artibeus toltecus) bats from Guatemala (HM597197, HM597198, and HM597201). These variants clustered into groups V (HM597198 and JQ071387), VI (HM597201), and XIII (HM597197 and JQ071388). The two isolates from each P. discolor bat belonged to groups V/VI and V/XIII, respectively (Figure 1).
Three isolates were obtained from Artibeus bats, including one from A. obscurus and two from A. planirostris. Sequences of each isolate represent a unique novel variant with distance > 12% among the variants. One variant from A. planirostris (JQ071382) fell into group VII with variants from Carollia bats; the other two variants were distinct from all other described Bartonella species, falling into two novel defined phylogroups XXIV (JQ071380) and XXV (JQ071381), respectively (Figure 1).
Two variants were identified in Vampyriscus bidens bats, with one isolate for each individual bat. One variant was identical to the variant found in a M. microtis bat from Guatemala (HM597207) and clustered into group XII with variants from Carollia bats. The other variant was distant from any other variants and formed phylogroup XXII (JQ071389) (Figure 1).
One isolate was obtained from G. soricina, S. lilium, and Myotis sp. bats, and each isolate represented a unique variant. The variant from G. soricina (JQ0713/83) was novel and fell into group VII with some isolates from C. perspicillata and others; the isolate from S. lilium was identical to those isolates previously described in S. lilium bat from Guatemala (HM597199), which formed group IV. Finally, the variant from Myotis sp. (JQ071390) differed from all other variants, falling into phylogroup XXIII (Figure 1).
Discussion
After reports of Bartonella infections in bats from Kenya in eastern Africa and Guatemala in Central America,5,8 the present investigation provided additional evidence that Bartonella infections are also prevalent in bat populations from Peru in South America. Overall prevalence (24.1%) of the infections was similar to the other two reports but varied by bat species. The common vampire bat, a locally abundant species across its geographic range, exhibited high prevalence of Bartonella infection, with more than one-half of the population infected in the present study. Such high prevalence is more likely a result of persistent infection considering the long life span (average 10–20 years) of bats, which is the case with some bat viruses.8,35 Some bat species, such as Artibeus bats, showed much lower prevalence of infection. Interestingly, in Guatemala, there was no Bartonella found in Artibeus bats as well. Similarly, Bartonella was not detected in any of the 10 tested M. molossus bats. Although the small sample size could have contributed to such results, it is possible that less frequent interactions occur between Bartonella species and these bats.
Several Bartonella strains representing a variety of distinct phylogroups were identified in bats from Peru, indicating a diverse Bartonella community associated with Peruvian bats. These phylogroups, with gltA sequence identity < 96%, each likely indicate a novel Bartonella species according to the proposal in the work by La Scola and others.36 Some bat species are infected with multiple Bartonella strains and seem to be infected with species-specific strains. For example, Bartonella strains of phylogroups I and III were rarely found in bats other than common vampire bats. However, some Bartonella strains, such as those strains from phylogroups VII and XII, infected bats of different species, suggesting no specific Bartonella sp.–bat relationships. These observations were similar to the observations reported from Guatemala5 but contrast the findings of Bartonella spp. in bats from Kenya, where there seemed to be a specific relationship between bat species and Bartonella strains.8 This contrasting host specificity is very similar to the interaction observed in Bartonella sp.–rodent relationships from different regions of the world.30–34 Dramatic differences in observations of host specificity between Bartonella and bats from different geographic areas presented an interesting model for understanding evolutionary trends in bacterium–bat coadaptation considering the high level of natural Bartonella infection among diverse bat species and wide genetic diversity of the strains. Ancient origins for some zoonotic viruses, such as henipaviruses37 and lyssaviruses,38 and their association with bat reservoirs suggest a long history of coevolution. Apparent congruence between the phylogeny of Bartonella and their hosts in Africa and absence of similar congruence in America suggest that bartonellae have long adapted to African bat species, whereas American bartonellae have not established balanced relations with their bat hosts. The lack of host specificity observed in the present study may be partially explained by differences in the community of arthropod vectors that parasitize bats. Future studies including the ectoparasite fauna would allow us to test hypotheses about whether any of the arthropods may be important vectors in the Bartonella transmission cycle and whether ectoparasite specificity contributes to the lack of Bartonella–bat host specificity. Because some bat species (such as Carollia and Glossophaga bats) share roosts with other species, may reach large population densities, and exhibit crowded roosting behavior, there is a potential for frequent intra- and interspecies transmission of infections, which was shown in the study of bat rabies viruses.39 In line with this hypothesis, our results indicate that active interspecies transmission of Bartonella species likely occurs among bats in Peru, although the specificity of ectoparasite arthropod vectors among the bat fauna remains unclear and may also contribute to transmission of Bartonella among bats.
Evidence of apparent disease in bats caused by Bartonella species has not been reported to date; however, bats have very long life spans compared with other mammals of similar body size,40 such as rodents, and may play very important roles as reservoirs contributing to the maintenance and transmission of Bartonella to other animals and/or humans. Some bat species have been known to directly transmit infections to humans. For example, throughout South and Central America, the common vampire bat has been long recognized in the transmission of rabies virus to humans by bite.41 The common vampire bat is one of three hematophagous bats in the New World, and it typically feeds on the blood of mammals, including domestic animals, such as cattle, horses, pigs, and dogs; it may also feed on the blood of humans.42 If Bartonella, indeed, can be transmitted to humans through the bite of bats, it is imperative that additional studies be conducted with vampire bats, because more than one-half of the population of bats included in our study was infected with Bartonella spp.; additionally, the population is at significant risk for being infected by Bartonella spp. Predation of vampire bats on humans seems to be a problem in Latin America.41 Presumably, if Bartonella species are transmitted by ectoparasites, some, if not all, bat-associated Bartonella species could be transmitted to humans, because bats are frequent hosts to a wide range of ectoparasites, including bat flies, fleas, soft ticks, and mites.43 Studies of ectoparasites collected from bats from Egypt and the United States have reported the detection of Bartonella-specific DNA,6,9,11–13 which suggests a role of arthropods as vectors of Bartonella to other wildlife. Humans, as opportunistic hosts, are at risk of being bitten by ectoparasites that feed on bats, especially vampire bats. However, transmission potential might vary with the degree of synanthropic roosting or foraging behavior among the bat community.
In summary, our study provides information on the distribution and genetic diversity of Bartonella strains in the bat populations in Peru. Additional information is needed to understand the interaction of Bartonella species with their bat hosts under natural conditions. Because more and more Bartonella species are being found to be associated with human illness, there is an increasing need to identify the animal reservoirs of these novel Bartonella species and understand their disease ecology. Additional study is required to determine whether bat-associated Bartonella spp. are responsible for the etiology of undiagnosed illnesses in humans in Peru.
ACKNOWLEDGMENTS
Funding for the study was provided by a collaborative Centers for Disease Control and Prevention–University of Georgia Seed Award. The authors thank Brett Petersen, Victor A. Laguna-Torres, Ivan Vargas, Jose Peña, Juan Ramon Meza, and the San Lorenzo Ministry of Health post for technical assistance in the field. We also thank Carolina Guevara from the Virology Department of Naval Medical Research Unit-6 (NAMRU-6) for technical assistance.
Footnotes
Authors' addresses: Ying Bai, Centers for Disease Control and Prevention (CDC), Division of Vector-Borne Disease (DVBD), Bacterial Diseases Branch (BDB), Fort Collins, CO, E-mail: YBai1@cdc.gov. Sergio Recuenco, CDC, Division of High-Consequence Pathogens and Pathology (DHCPP), Rabies Program (RP), Atlanta, GA, E-mail: fni9@cdc.gov. Amy Turmelle Gilbert, CDC/DHCPP/RP, Atlanta, GA, E-mail: fcj6@cdc.gov. Lynn M. Osikowicz, CDC/DVBD/BDB, Fort Collins, CO, E-mail: vir5@cdc.gov. Jorge Gómez, Ministerio de Salud, Dirección General de Epidemiología, Lima, Peru, E-mail: jgomez@dge.gob.pe. Charles Rupprecht, CDC/DHCPP/RP, Atlanta, GA, E-mail: cyr5@cdc.gov. Michael Y. Kosoy, CDC/DVBD/BDB, Fort Collins, CO, E-mail: mck3@cdc.gov.
References
- 1.Simmons NB. Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd Ed. Baltimore, MD: Johns Hopkins University Press; 2005. pp. 312–529. [Google Scholar]
- 2.Messenger SL, Smith JS, Orciari LA, Yager PA, Rupprecht CE. Emerging pattern of rabies deaths and increased viral infectivity. Emerg Infect Dis. 2003;9:151–154. doi: 10.3201/eid0902.020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turmelle AS, Olival KJ. Correlates of viral richness in bats (order Chiroptera) EcoHealth. 2009;6:522–539. doi: 10.1007/s10393-009-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, Turmelle A, Ellison J, Garcia DL, Estevez A, Lindblade K, Rupprecht C. Bartonella spp. in bats, Guatemala. Emerg Infect Dis. 2011;17:1269–1271. doi: 10.3201/eid1707.101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billeter SA, Hayman DTS, Peel AJ, Baker K, Wood JLN, Cunningham A, Suu-Ire R, Dittmar K, Kosoy MY. Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology. 2012;139:324–329. doi: 10.1017/S0031182011002113. [DOI] [PubMed] [Google Scholar]
- 7.Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ. Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology. 2005;131:489–496. doi: 10.1017/S0031182005008097. [DOI] [PubMed] [Google Scholar]
- 8.Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, Agwanda B, Breiman RF, Rupprecht CE. Discovery of Bartonella bacteria in bats from Kenya. Emerg Infect Dis. 2010;16:1875–1881. doi: 10.3201/eid1612.100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loftis AD, Gill JS, Schriefer ME, Levin ML, Eremeeva ME, Gilchrist MJ, Dasch GA. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Aragasidae) J Med Entomol. 2005;42:473–480. doi: 10.1093/jmedent/42.3.473. [DOI] [PubMed] [Google Scholar]
- 10.Matthias MA, Díaz MM, Campos KJ, Calderon M, Willig MR, Pacheco V, Gotuzzo E, Gilman RH, Vinetz Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by bayesian phylogenetic analysis of 16S ribosomal DNA sequences. Am J Trop Med Hyg. 2005;73:964–974. [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves WK, Loftis AD, Gore JA, Dasch GA. Molecular evidence for novel Bartonella species in Trichobius major (Diptera: Streblidae) and Cimex adjunctus (Hemiptera: Cimicidae) from two southeastern bat caves, USA. J Vector Ecol. 2005;30:339–341. [PubMed] [Google Scholar]
- 12.Reeves WK, Streicker DG, Loftis AD, Dasch GA. Serologic survey of Eptesicus fuscus from Georgia, USA for Rickettsia and Borrelia and laboratory transmission of a Rickettsia by bat ticks. J Vector Ecol. 2006;31:386–389. doi: 10.3376/1081-1710(2006)31[386:ssoeff]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Reeves WK, Rogers TE, Durden LA, Dasch GA. Association of Bartonella with the fleas (Siphonaptera) of rodents and bats using molecular techniques. J Vector Ecol. 2007;32:118–122. doi: 10.3376/1081-1710(2007)32[118:aobwtf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Gill JS, Rowley WA, Bush PJ, Viner JP, Gilchrist MJ. Detection of human blood in the bat tick Carios (Ornithodoros) kelleyi (Acari: Argasidae) in Iowa. J Med Entomol. 2004;41:1179–1181. doi: 10.1603/0022-2585-41.6.1179. [DOI] [PubMed] [Google Scholar]
- 15.Bass JW, Vincent JM, Person DA. The expanding spectrum of Bartonella infections. II. Cat scratch disease. Pediatr Infect Dis J. 1997;16:163–179. doi: 10.1097/00006454-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O'Connor SP. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, Ferraro MJ, Holden JM, Nicholson WL, Dasch GA, Koehler JE. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;256:2381–2387. doi: 10.1056/NEJMoa065987. [DOI] [PubMed] [Google Scholar]
- 18.Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosoy MY, Murray M, Gilmore RD, Bai Y, Gage K. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–650. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosoy M, Morway C, Sheff K, Bai Y, Colborn J, Chalcraft L, Dowell SF, Peruski LF, Maloney SA, Baggett H, Sutthirattana S, Sidhirat A, Maruyama S, Kabeya H, Chomel BB, Kasten R, Popov V, Robinson J, Kruglov A, Petersen LR. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from human patients from Thailand. J Clin Microbiol. 2008;46:772–775. doi: 10.1128/JCM.02120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of a febrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–1700. doi: 10.1128/jcm.38.4.1698-1700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch DF, Pickett DA, Slater LN, Steigerwalt AG, Brenner DJ. Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol. 1992;30:275–280. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch DF, Carroll KC, Hofmeister EK, Persing DH, Robison DA, Steigerwalt AG, Brenner DJ. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Caceres U, Garcia FU. Bartonellosis: an immunodepressive disease and the life of Daniel Alcides Carrion. J Clin Pathol. 1991;95((Suppl)):58–66. [PubMed] [Google Scholar]
- 25.Chomel BB, Kasten RW, Floyd-Hawkins KA, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield AN, Abbott RC, Pedersen NC, Koehler JE. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins JA, Radulovic S, Jaworski DC, Azad AF. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae) J Med Entomol. 1996;33:490–495. doi: 10.1093/jmedent/33.3.490. [DOI] [PubMed] [Google Scholar]
- 27.Pappalardo BL, Correa MT, York CC, Peat CY, Breitschwerdt EB. Epidemiologic evaluation of the risk factors associated with exposure and seroreactivity to Bartonella vinsonii in dogs. Am J Vet Res. 1997;58:467–471. [PubMed] [Google Scholar]
- 28.Roux V, Raoult D. Body lice as tools for diagnosis and surveillance of reemerging diseases. J Clin Microbiol. 1999;37:596–599. doi: 10.1128/jcm.37.3.596-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duncan AW, Maggi RG, Breitschwerdt EB. Bartonella DNA in dog saliva. Emerg Infect Dis. 2007;13:1948–1950. doi: 10.3201/eid1312.070653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birtles RJ, Harrison TG, Molyneux DH. Grahamella in small woodland mammals in the UK—isolation, prevalence and host-specificity. Ann Trop Med Parasitol. 1994;88:317–327. doi: 10.1080/00034983.1994.11812872. [DOI] [PubMed] [Google Scholar]
- 31.Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Leepitakrat W, Monkanna T, Khlaimanee N, Jones JW, Coleman RE. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–433. [PubMed] [Google Scholar]
- 32.Jardine C, McColl D, Wobeser G, Leighton FA. Diversity of Bartonella genotypes in Richardson's ground squirrel populations. Vector Borne Zoonotic Dis. 2006;6:395–403. doi: 10.1089/vbz.2006.6.395. [DOI] [PubMed] [Google Scholar]
- 33.Kosoy MY, Regery RL, Tzianabos T, Marston EL, Jones DC, Green D, Maupin GO, Olson JG, Childs JE. Distribution, diversity, and host specificity of Barotnella in rodents from the Southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 34.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 35.Sulkin SE, Allen R. Virus infections in bats. Monogr Virol. 1974;8:1–103. [PubMed] [Google Scholar]
- 36.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–321. doi: 10.1016/s0966-842x(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 37.Gould AR. Comparison of the deduced matrix and fusion protein sequences of equine morbillivirus with cognate genes of the Paramyxoviridae. Virus Res. 1996;43:17–31. doi: 10.1016/0168-1702(96)01308-1. [DOI] [PubMed] [Google Scholar]
- 38.Badrane H, Tordo N. Host switching in lyssavirus history from the Chiroptera to the Carnivora orders. J Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streichker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 40.Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- 41.Schneider MC, Romijn PC, Uieda W, Tamayo H, da Silva DF, Belotto A, da Silva JB, Leanes LF. Rabies transmitted by vampire bats to humans: an emerging zoonotic disease in Latin America? Rev Panam Salud Publica. 2009;25:260–269. doi: 10.1590/s1020-49892009000300010. [DOI] [PubMed] [Google Scholar]
- 42.Turner DC, Bateson P. The Vampire Bat: A Field Study in Behavior and Ecology. Baltimore, MD: Johns Hopkins University Press; 1975. pp. 1–145. [Google Scholar]
- 43.Autino AG, Claps GL, Barquez RM, Díaz MM. Ectoparasitic insects (Diptera: Streblidae and Siphonaptera: Ischnopsyllidae) of bats from Iquitos and surrounding areas (Loreto, Peru) Mem Inst Oswaldo Cruz. 2011;106:917–925. doi: 10.1590/s0074-02762011000800004. [DOI] [PubMed] [Google Scholar]