Abstract
We describe sequences of six strains of dengue virus (DENV): three DENV-1 isolates and two DENV-4 isolates from Puerto Rico, and a DENV-1 strain from Key West, Florida, obtained from blood donors during 2010 epidemics. Phylogenetic analysis revealed that the Puerto Rico DENV-1 strains constitute a new lineage within genotype V different from those that circulated in Puerto Rico during the past two decades. The newer Puerto Rico DENV-1 strains associated with strains from the Caribbean and South America. The DENV-1 strain from Key West, Florida clustered with a strain isolated from mosquito pools collected in that area and with a number of strains from Nicaragua and Mexico circulating during 2006–2009. The Puerto Rico DENV-4 isolates of genotype II associated with strains that have circulated on the island throughout the 1980s and 1990s and with strains from the Caribbean region and Central America. Introduction and circulation of novel DENV lineages in dengue-endemic regions have the potential to increase the severity of dengue cases.
Dengue is caused by any of the four dengue virus types (DENV-1 to DENV-4), family Flaviviridae, genus Flavivirus. Dengue virus is primarily transmitted by the urban mosquito Aedes aegypti and most dengue infections are asymptomatic or sub-clinical. Dengue disease spectrum ranges from a mild, influenza-like disease (dengue fever) to a potentially life-threatening condition known as severe dengue/dengue hemorrhagic fever.1
The four DENV types are genetically distant from each other, and phylogenetic analysis shows epidemic genotypes that vary in number and geographic distribution depending on the DENV type. Additionally, sylvatic cycles exist in Africa and Asia where sylvatic strains of DENV-2 and DENV-4 have been isolated from arboreal mosquitoes, humans, and non-human primates.2 Four DENV-1 epidemic genotypes (I, II, IV, and V) and a sylvatic genotype have been described. However, for the sylvatic genotype recent evidence suggest a human origin, therefore being included as genotype III in newer classifications. Conversely, for DENV-4, three epidemic genotypes (I–III) and a sylvatic genotype have been described.3 Some DENV genotypes have shown to be more virulent, with better fitness, and have been associated with increased clinical severity of dengue cases (e.g., American/Asian genotype of DENV-2).4
In the United States, dengue is endemic to the Commonwealth of Puerto Rico, which in 2010 experienced the largest epidemic in its history with more than 21,000 dengue suspected cases reported, of which approximately 75% were laboratory confirmed.5 Sporadic dengue outbreaks have occurred in the Territories of Guam and American Samoa, the States of Hawaii and Texas,6–8 and more recently in Key West, Florida during 2009–2011.9,10 We report sequences and phylogenetic analyses of three DENV-1 and two DENV-4 strains isolated from Puerto Rico and a DENV-1 strain from Key West, Florida circulating during the 2010 epidemics.
Six plasma samples obtained from blood donors infected with DENV that were asymptomatic at the time of collection were used for further testing by using a TaqMan quantitative reverse transcription polymerase chain reaction (qRT-PCR). All specimens were subjected to RNA extraction by using the QIAamp Viral RNA mini kit (QIAGEN, Valencia, CA), and the presence of DENV RNA was confirmed at the Food and Drug Administration by using a modified type-specific qRT-PCR in a singleplex format based on a published protocol.11 Plasma specimens were also evaluated for infectious virus by culture in mosquito cells. In brief, 200–250 μL of plasma were added to a semi-confluent monolayer of C6/36 cells (American Type Culture Collection, Manassas, VA) propagated at 32°C in an atmosphere of 5% CO2. Supernatants from the first and second passages in C6/36 cells were tested for viral production by qRT-PCR and infectivity was tested by focus-forming assay.
Cell culture supernatants from the second passages in C6/36 cells were used for sequencing. Extracted viral RNA was subjected to RT-PCR by using DENV-specific primers to amplify the DENV structural genes region (capsid-premembrane-envelope). In brief, fragments of approximately 3,700 and 3,500 nucleotides for DENV-1 and DENV-4, respectively, were generated by using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), LA Taq polymerase (Takara, Otsu, Japan), and specific DENV-1 and DENV-4 primers (Supplemental Table 1). Amplified products were electrophoresed in agarose gels, purified from the gels and subjected to bidirectional Sanger sequencing with a minimum coverage of four-fold by using appropriate sequencing primers (Supplemental Table 1). Sequences were assembled and analyzed by using Sequencher version 5 (GeneCodes Corp., Ann Arbor, MI) and deposited in the GenBank database under accession numbers JQ045561–JQ045566.
Phylogenetic analyses were conducted with envelope protein (E) gene sequences (1,485 nucleotides) in datasets containing a total of 36 DENV-1 and 30 DENV-4 strains. Each dataset was comprised of randomly selected sequences from the Caribbean, Central America, and South America and representative sequences from diverse geographic origins encompassing all DENV genotypes retrieved from the GenBank by using the Virus Variation Tool.12 BLAST (National Center for Biotechnology Information, Bethesda, MD) searches were conducted for each newly sequenced DENV-1 and DENV-4 strain and the closest hits for each were included in the datasets. The E gene sequences were selected for conducting the phylogenetic analyses because a considerable number of DENV-1 and DENV-4 strains from Puerto Rico and the Caribbean region available in the database were limited to only sequences of that gene. A sylvatic sequence (for the DENV-4 analysis) and each of the DENV prototype strain sequences; i.e., DENV-2 NGC, DENV-3 H87, and DENV-4 H241 or DENV-1 Hawaii (for both DENV-1 and DENV-4 datasets, respectively) were used as outgroup to root the trees. Genotypes were classified according to Chen and Vasilakis.3 The list of analyzed sequences is shown in Table 1.
Table 1.
DENV-1 and DENV-4 strains used in this study*
| Strain | Place of collection | Year of collection | Genotype | GenBank accession no. |
|---|---|---|---|---|
| DENV-1 | ||||
| US-HI/Hawaii/1944 | Hawaii, USA | 1944 | I | AB609588 |
| MY/36000/2005 | Malaysia | 2005 | I | FR666924 |
| TH/TH-Sman/1954 | Thailand | 1954 | II | D10513 |
| TH/2543-63/1963 | Thailand | 1963 | II | AF425629 |
| MY/P72-1244/1972 | Malaysia | 1972 | III | AF231721 |
| MY/36046/2005 | Malaysia | 2005 | III | FN825674 |
| US-HI/HawM2540/2001 | Hawaii, USA | 2001 | IV | DQ672562 |
| US-HI/HawO3663/2001 | Hawaii, USA | 2001 | IV | DQ672564 |
| VG/BID-V2937/1985 | British Virgin Islands | 1985 | V | GQ868601 |
| HT/DB067/2010 | Haiti | 2010 | V | JF969282 |
| US-FL/KW10AG/2010 | Key West, FL, USA | 2010 | V | JF519855 |
| US-FL/FDA-ARC-39-10/2010† | Key West, FL, USA | 2010 | V | JQ045564 |
| MQ/481013035/2008 | Martinique | 2008 | V | JN022599 |
| MX/BID-V3739/2007 | Mexico | 2007 | V | GQ868527 |
| MX/BID-V3679/2007 | Mexico | 2007 | V | GU131966 |
| MX/BID-V3758/2008 | Mexico | 2008 | V | GQ868537 |
| NI/BID-V653/2004 | Nicaragua | 2004 | V | EU596501 |
| NI/BID-V2343/2006 | Nicaragua | 2006 | V | FJ562104 |
| NI/BID-V746/2006 | Nicaragua | 2006 | V | JN819403 |
| NI/BID-V2646/2008 | Nicaragua | 2008 | V | GQ199858 |
| NI/BID-V2652/2008 | Nicaragua | 2008 | V | GQ199859 |
| NI/BID-V5504/2009 | Nicaragua | 2009 | V | JF937635 |
| NI/BID-V5067/2009 | Nicaragua | 2009 | V | JF937644 |
| US-PR/BID-V2136/1992 | Puerto Rico | 1992 | V | FJ410186 |
| US-PR/BID-V2135/1992 | Puerto Rico | 1992 | V | FJ547087 |
| US-PR/BID-V2133/1993 | Puerto Rico | 1993 | V | FJ410184 |
| US-PR/BID-V2134/1993 | Puerto Rico | 1993 | V | FJ410185 |
| US-PR/BID-V1743/1995 | Puerto Rico | 1995 | V | FJ205874 |
| US-PR/BID-V1744/1995 | Puerto Rico | 1995 | V | FJ205875 |
| US-PR/BID-V2139/1996 | Puerto Rico | 1996 | V | FJ410188 |
| US-PR/BID-V1739/1998 | Puerto Rico | 1998 | V | FJ205872 |
| US-PR/BID-V852/2006 | Puerto Rico | 2006 | V | EU482591 |
| US-PR/FDA-ARC-12-10/2010† | Puerto Rico | 2010 | V | JQ045561 |
| US-PR/FDA-ARC-30-10/2010† | Puerto Rico | 2010 | V | JQ045562 |
| US-PR/FDA-ARC-31-10/2010† | Puerto Rico | 2010 | V | JQ045563 |
| VE/BID-V2423/2004 | Venezuela | 2004 | V | JN819425 |
| DENV-4 | ||||
| PH/H241-P/1956 | Philippines | 1956 | I | FJ439174 |
| JP/D4-61NIID/1961 | Japan | 1961 | I | AB111090 |
| SL/No17/1978 | Sri Lanka | 1978 | I | AY550909 |
| CR/D4.108/1996 | Costa Rica | 1996 | II | AY152104 |
| DM/814669/1983 | Dominica | 1981 | II | AF326573 |
| DR/DB040/1997 | Dominican Republic | 1997 | II | JF804053 |
| SV/D4.110/1993 | El Salvador | 1993 | II | AY152300 |
| MQ/D4.112/1995 | Martinique | 1995 | II | AY152100 |
| MX/D4.111/1995 | Mexico | 1995 | II | AY152304 |
| US-PR/D4M.25/1982 | Puerto Rico | 1982 | II | AY152296 |
| US-PR/D4M.9/1982 | Puerto Rico | 1982 | II | AY152324 |
| US-PR/D4.115/1986 | Puerto Rico | 1986 | II | AY152224 |
| US-PR/D4.116/1986 | Puerto Rico | 1986 | II | AY152272 |
| US-PR/D4.3/1987 | Puerto Rico | 1987 | II | AY152108 |
| US-PR/D4.67/1987 | Puerto Rico | 1987 | II | AY152236 |
| US-PR/D4.85/1994 | Puerto Rico | 1994 | II | AY152096 |
| US-PR/D4.86/1994 | Puerto Rico | 1994 | II | AY152116 |
| US-PR/D4.88/1994 | Puerto Rico | 1994 | II | AY152288 |
| US-PR/BID-V2435/1996 | Puerto Rico | 1996 | II | GQ199881 |
| US-PR/D4.19/1998 | Puerto Rico | 1998 | II | AY152040 |
| US-PR/D4.44/1998 | Puerto Rico | 1998 | II | AY152088 |
| US-PR/BID-V1093/1998 | Puerto Rico | 1998 | II | EU854296 |
| US-PR/BID-V1082/1998 | Puerto Rico | 1998 | II | FJ024424 |
| US-PR/BID-V2443/1998 | Puerto Rico | 1998 | II | FJ850059 |
| US-PR/FDA-ARC-19-10/2010† | Puerto Rico | 2010 | II | JQ045565 |
| US-PR/FDA-ARC-42-10/2010† | Puerto Rico | 2010 | II | JQ045566 |
| US-VG/SC/DB045/1994 | U.S. Virgin Islands | 1994 | II | JF804058 |
| TH/ThD4 0439 01/2001 | Thailand | 2001 | III | AY618940 |
| TH/ThD4 0164 99/1999 | Thailand | 1999 | III | AY618986 |
| MY/P75-514/1975 | Malaysia | 1975 | Sylvatic | AF231723 |
Phylogenetic reconstruction was conducted by the maximum-likelihood method using MEGA5.13 For the DENV-1 dataset the Tamura-Nei + Γ4 + I model was determined to be the best fitted model for the data. For the DENV-4 dataset, the general time reversible (GTR) + Γ4 was the best model as determined according to the Bayesian information criterion (BIC) determined by using MEGA5. A bootstrap test of 1,000 replicates was used for both datasets.
In addition, Bayesian phylogenetic (BA) and time-scale analyses were conducted by using Mr. Bayes version 3.1.214 and BEAST version 1.6.2,15 respectively. For BA analysis, the GTR + Γ4 + I model with successive branch swapping was used as the substitution model. Four Markov Chain Monte Carlo chains were run for 10,000,000 generations, sampling every 100 generations, and the first 10,000 sampled trees were discarded as burn in. A 50% majority rule consensus tree was constructed from the posterior distribution of trees. To calculate the age for the nodes in which the newly sequenced DENV-1 and DENV-4 were located, a time-scale analysis was performed for the same datasets used for the phylogenetic analysis (excluding other DENV type sequences) by using the GTR + Γ4 + I nucleotide substitution model, the relaxed uncorrelated lognormal clock, and constant population size tree prior as implemented in BEAST. Three independent Markov Chain Monte Carlo analyses, each run for 10,000,000 steps, were performed and combined with a burn in value set to 10% generations. A maximum clade credibility tree was generated for each DENV type, and the 95% highest posterior density intervals were obtained to determine the uncertainty in the parameters estimated.
We detected DENV RNA in all six plasma specimens using our qRT-PCR. Four of them were positive for DENV-1 and two were positive for DENV-4. All samples were infectious in cell culture as determined by positive focus-forming assay results in supernatants from infected C6/36 cells. DENV-1 and DENV-4 accounted for most of the confirmed dengue cases during the 2010 epidemic in Puerto Rico, and DENV-2 cases were also detected but at a significantly lower incidence,5 which can explain why no DENV-2 cases were detected in the studied blood donor cohort.
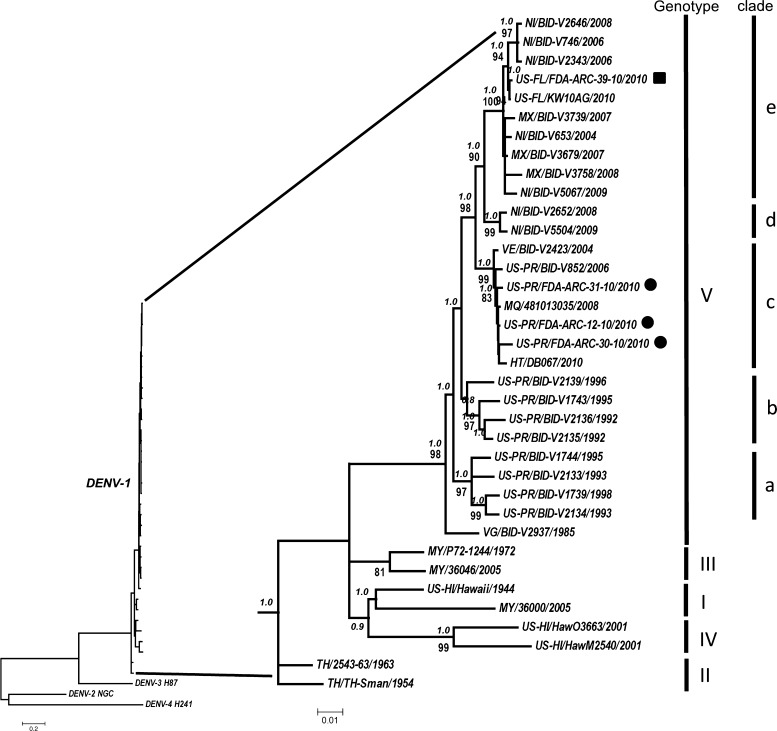
Maximum-likelihood and BA analyses showed phylogenetic trees with similar topology for each DENV type and our findings are supported by high bootstrapping and/or Bayesian posterior probability values (Figures 1 and 2). Analysis of the phylogenetic trees enabled us to identify the three PR DENV-1 strains from 2010 as belonging to a different lineage within genotype V, which is distant from those that had circulated in the island during 1980s and 1990s (Figure 1). The 2010 strains represent a lineage related to those that have been detected in Central America, Mexico, and the Caribbean during the past decade,16 which are closely associated with strains from Venezuela, Haiti, Martinique, and strain DENV-1/US/BID-V852/2006, which is the most recent strain from Puerto Rico deposited in the database that contains the entire E gene sequence (Figure 1).
Figure 1.
Consensus maximum-likelihood phylogenetic tree constructed with sequences of the dengue virus type 1 (DENV-1) envelope protein gene. Genotypes are identified by roman numerals. Clades of genotype V are identified by the letters a–e. Strains collected from blood donors from Puerto Rico are represented by black circles and the strain from Key West, Florida, are represented by a black square. Bayesian posterior probability (in italics) and bootstrapping values ≥ 80 are shown above and under the branch nodes, respectively. Branch lengths are proportional to the scale bar and the scale bar represents number of nucleotide substitutions between the analyzed viruses. The tree is mid-point rooted.
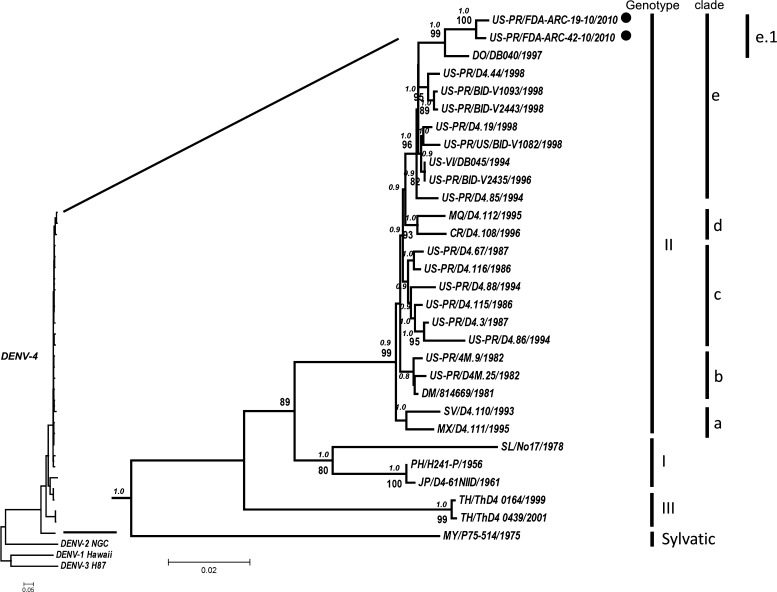
Figure 2.
Consensus maximum-likelihood phylogenetic tree constructed with sequences of the dengue virus type 4 (DENV-4) envelope protein gene. Genotypes are identified by roman numerals. Clades of genotype II are identified by the letters a–e. Sub-clade e.1 identifies the cluster where the 2010 Puerto Rican DENV-4 strains are located, and these strains are represented by black circles. Bayesian posterior probability (in italics) and bootstrapping values ≥ 80 are shown above and under the branch nodes, respectively. Branch lengths are proportional to the scale bar and the scale bar represent the number of nucleotide substitutions between the analyzed viruses. The tree is mid-point rooted.
As expected, the DENV-1 strain obtained from a blood donor in Key West, Florida clustered with strain KW10AG obtained from mosquito pools collected in Key West, Florida.17 Comparison of E protein gene sequences between these two strains showed 99.9% identity among them (1,484/1,485 nucleotides). The Key West, Florida DENV-1 strains are closely related to strains from Nicaragua sampled in 2006 and 2008, and constitute a different lineage of DENV-1 within a cluster that includes a number of strains from Mexico collected during 2007–2008 (Figure 1). The age of the DENV-1 node containing the 2010 Puerto Rico strains (clade c) is 7–15 years (mean = 11 years). For clades d and e (clade e contains the 2010 DENV-1 strain from Key West, Florida), the age is 16–34 years (mean = 25 years). The age of the parent node for these three clades is 19–40 years (mean = 29 years).
Two DENV-4 strains isolated from Puerto Rico in 2010 are closely related to a strain from the Dominican Republic, which together form a separate cluster within genotype II. This cluster includes a number of strains from Puerto Rico, Central America, and the Caribbean region that have circulated throughout 1980s and 1990s (Figure 2). Although no additional Puerto Rico DENV-4 strains collected after 2000 were available for inclusion in the study to enable a better temporal resolution of the tree, our phylogenetic analysis of the DENV-4 strains circulating during the 2010 epidemic in Puerto Rico is consistent with previous studies, which indicate that the more recently sampled strains available for analysis (from the late 1990s) belong to a lineage that has been recurrently isolated in the island for a period ≥ 10 years. Our findings and those of previous reports18,19 suggest that this lineage has survived numerous epidemic and inter-epidemic periods. DENV-4 has not been the dominant type for most of the past decade in Puerto Rico, and this low transmission frequency may have contributed to the lower genetic diversity observed for DENV-4 in Puerto Rico.19,20 The calculated age for the node defining clade e is 19–24 years (mean = 21 years). For sub-clade e.1 (containing the 2010 strains), the calculated age is 14–18 years (mean = 16 years).
Introduction of new lineages of DENV in dengue-endemic regions have the potential to cause increased severity for dengue cases. This suggestion can be illustrated by a recent report from Nicaragua, which showed that introduction and replacement of the circulating lineage of DENV-2 with a new lineage displaying increased fitness correlated with an augment in disease severity.21 A shortcoming of our study is the scarcity of Puerto Rico DENV-1 and DENV-4 sequences collected after 2000 available for analysis. Nevertheless, our results enabled us to identify circulation of a new DENV-1 lineage on the island and to speculate that its circulation can potentially lead to increased severity for cases in future epidemics. Long-term epidemiologic and in vitro studies are needed to determine if strains from the new DENV lineage circulating in Puerto Rico have increased fitness in comparison with older strains, and how this fitness may contribute to the already complex epidemiology of DENV on the island. In summary, our results on the molecular epidemiology of DENV-1 and DENV-4 in Puerto Rico and Florida during 2010 support the notion that an exchange of DENV strains occurrs within the Caribbean islands and between the Caribbean region and other dengue-endemic countries in Central and South America,18–22 and that these strains and lineages circulating may have an impact on the clinical outcome of dengue cases in the region.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Community Blood Centers of Florida for providing samples and the Dengue Branch of the Centers for Disease Control and Prevention for performing additional confirmatory testing of specimens (data not reported in this manuscript).
Footnotes
Financial support: This study was supported in part by the Office of Blood Research and Review/Center for Biologics Evaluation and Research/Food and Drug Administration (OBRR/CBER/FDA). Financial support for blood donation screening in Puerto Rico provided by Federal Earmark 1H75CK000159-01.
Authors' addresses: Germán Añez, Daniel A. R. Heisey, Luz M. Espina, and Maria Rios, Center for Biologics Evaluation and Research, U.S. Food and Drug Administration, HFM-315, Bethesda, MD, E-mails: german.anez@fda.hhs.gov, daniel.heisey@fda.hhs.gov, luz.espinagomez@fda.hhs.gov, and maria.rios@fda.hhs.gov. Susan L. Stramer, American Red Cross, Gaithersburg, MD, E-mail: stramers@usa.redcross.org.
References
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. 2011;9:532–541. doi: 10.1038/nrmicro2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen R, Vasilakis N. Dengue - Quo tu et quo vadis? Viruses. 2011;3:1562–1608. doi: 10.3390/v3091562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rico-Hesse R. Dengue virus evolution and virulence models. Clin Infect Dis. 2007;44:1462–1466. doi: 10.1086/517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention Largest Dengue Outbreak in Puerto Rico History. 2011;Vol. 3 http://www.cdc.gov/dengue/dengue_upd/resources/DengueUpdateVo3No1.pdf The Dengue Update. No. 1. February 16, 2011. Available at. Accessed November 28, 2011. [Google Scholar]
- 6.Centers for Disease Control and Prevention Dengue hemorrhagic fever - U.S.–Mexico border, 2005. MMWR Morb Mortal Wkly Rep. 2007;56:785–789. [PubMed] [Google Scholar]
- 7.Effler PV, Pang L, Kitsutani P, Vorndam V, Nakata M, Ayers T, Elm J, Tom T, Reiter P, Rigau-Perez JG, Hayes JM, Mills K, Napier M, Clark GG, Gubler DJ, Hawaii Dengue Outbreak Investigation Team Dengue fever, Hawaii, 2001–2002. Emerg Infect Dis. 2005;11:742–749. doi: 10.3201/eid1105.041063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubler D. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler D, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. New York, NY: CAB International; 1997. pp. 175–198. [Google Scholar]
- 9.Centers for Disease Control and Prevention Locally acquired dengue - Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep. 2010;59:577–581. [PubMed] [Google Scholar]
- 10.Adalja AA, Sell TK, Bouri N, Franco C. Lessons learned during dengue outbreaks in the United States, 2001–2011. Emerg Infect Dis. 2012;18:608–614. doi: 10.3201/eid1804.110968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BW, Russell BJ, Lanciotti RS. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resch W, Zaslavsky L, Kiryutin B, Rozanov M, Bao Y, Tatusova TA. The virus variation resources at the National Center for Biotechnology Information: dengue virus. BMC Microbiol. 2009;9:65. doi: 10.1186/1471-2180-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2719. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303:110–119. doi: 10.1006/viro.2002.1686. [DOI] [PubMed] [Google Scholar]
- 17.Graham AS, Pruszynski CA, Hribar LJ, DeMay DJ, Tambasco AN, Hartley AE, Fussell EM, Michael SF, Isern S. Mosquito-associated dengue virus, Key West, Florida, USA, 2010. Emerg Infect Dis. 2011;17:2074–2075. doi: 10.3201/eid1711.110419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20:1650–1658. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- 19.Bennett SN, Drummond AJ, Kapan DD, Suchard MA, Muñoz-Jordán JL, Pybus OG, Holmes EC, Gubler DJ. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27:811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McElroy KL, Santiago GA, Lennon NJ, Birren BW, Henn MR, Muñoz-Jordán JL. Endurance, refuge, and reemergence of dengue virus type 2, Puerto Rico, 1986–2007. Emerg Infect Dis. 2011;17:64–71. doi: 10.3201/eid1701.100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborío S, Nuñez A, Lennon NJ, Birren BW, Gordon A, Henn MR, Harris E. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 2011;3:114–128. doi: 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Añez G, Morales-Betoulle ME, Rios M. Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS ONE. 2011;6:e27459. doi: 10.1371/journal.pone.0027459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.