Abstract
The study deals with the survey of different bat populations (Pteropus giganteus, Cynopterus sphinx, and Megaderma lyra) in India for highly pathogenic Nipah virus (NiV), Reston Ebola virus, and Marburg virus. Bats (n = 140) from two states in India (Maharashtra and West Bengal) were tested for IgG (serum samples) against these viruses and for virus RNAs. Only NiV RNA was detected in a liver homogenate of P. giganteus captured in Myanaguri, West Bengal. Partial sequence analysis of nucleocapsid, glycoprotein, fusion, and phosphoprotein genes showed similarity with the NiV sequences from earlier outbreaks in India. A serum sample of this bat was also positive by enzyme-linked immunosorbent assay for NiV-specific IgG. This is the first report on confirmation of Nipah viral RNA in Pteropus bat from India and suggests the possible role of this species in transmission of NiV in India.
Bats are known reservoirs for many highly infectious viruses such as Ebola virus (EBOV), Marburg virus (MARV), severe acute respiratory syndrome corona virus, and Nipah virus (NiV).1 Nipah virus was first recognized in India during an encephalitis outbreak in Siliguri, West Bengal, 20012 and in Nadia District, West Bengal, in 2007.3 The adjacent country, Bangladesh has had multiple outbreaks of NiV during 2001–2011.4–7
Nipah virus is a member of the family Paramyxoviridae and the genus Henipavirus. The henipaviruses are naturally harbored by Pteropoid fruit bats (flying foxes) and are characterized by a large genome, a wide host range, and their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans.8 Pteropus bats are the known vectors of this disease, and their involvement has been reported in many countries (Malaysia, Bangladesh, Australia, Thailand, Cambodia, Indonesia, and Madagascar).1,9–11
The route of infection of NiV from bats to humans is by ingestion of NiV-contaminated fresh date palm sap, consumption of partially eaten fruits, or contact with infected domestic animals (cattle, pigs, and goats). Large numbers of persons in Bangladesh and India showed development of disease after person-to-person transmission of the virus.12 Pteropus giganteus bats are widely distributed across Bangladesh and India.13 A recent study reported the seropositivity for NiV in P. giganteus bats in northern India.14 Similar finding have been reported from Bangladesh.4 We have surveyed two states (Maharashtra and West Bengal) in India for viral RNA and IgG against NiV, MARV, and REBOV viruses in different bat populations.
With the permission of Chief Conservator of Forests, West Bengal and Maharashtra State, India, bats were caught at roosts either by using mist nets or harp traps. Bats were anesthetized and bled from either the brachial or cephalic arteries; in some cases, blood samples were collected by cardiac puncture. One hundred forty bats of three species, P. giganteus (31), Cynopterus sphinx (30), and Megaderma lyra (79), were captured in Maharashtra and West Bengal States during May 2009–October 2010.
Some bats were euthanized with isoflurane, dissected, and organs were collected. Organs were stored in liquid nitrogen and blood was stored at 4°C during transport. Sex, age, body condition score, pregnancy status, lactation status, weight, and morphologic details were recorded in field laboratory. During capturing the bats and postmortem surgery, proper protecting equipments were used to avoid any infection to the workers. Bat specimens were transported to Biosafety Level 3 Laboratory of National Institute of Virology, Pune, India for further testing.
Liver and spleen were homogenized in phosphate-buffered saline by using a homogenizer (GenoGrinder 2000; BT&C Inc., Lebanon, NJ). Tissue homogenates were used to extract RNAs by using an RNA extraction machine (ABI 6100; Applied Biosystems, Foster City, CA). Quantitative reverse transcription–polymerase chain reactions (RT-PCRs) specific for REBOV, MARV, and NiV were performed.15,16 A diagnostic nested RT-PCR for the NiV nucleocapsid gene was performed as described.2 Partial glycoprotein, phosphoprotein, and fusion gene sequences were amplified. Mitochondrial DNA of bat was amplified by using PCR and sequenced for further species confirmation.17
Serum was separated from blood and tested by indirect enzyme-linked immunosorbent assay for IgG against MARV, NiV, and REBOV by using kits provided by the Centers for Disease Control and Prevention, Atlanta, GA and described protocols.18 Each serum sample was tested at 4 dilutions (1:100, 1:400, 1:1,600, and 1:6,400). Isolation of virus was attempted from liver and kidney tissue suspensions in Vero E6 cells that were blindly passaged three times.
Of 140 bat specimens, only one liver homogenate was positive for NiV viral RNA in a P. giganteus bat from Myanaguri, West Bengal, India. The NiV-positive bat was a healthy male adult (weight = 650 grams). The quantitative RT-PCR result for the liver homogenate was positive for NiV (cycle threshold value = 31.0), and the kidney homogenate was negative. However, results of a diagnostic NiV-specific RT-PCR were positive for liver and kidney homogenates. This bat serum sample was also found positive for NiV-specific IgG at dilutions of 1:100 and 1:400. Serum and tissues from this Pteropus bat were negative for REBOV and MARV by ELISA and quantitative RT-PCR. The remaining 139 bat specimens were negative for IgG against all three viruses and for viral RNA.
During virus isolation attempts in Vero E6 cells, NiV-positive liver and kidney homogenates from P. giganteus did not show a cytopathic effect. The sample was blind passaged in Vero E6 cells for three passages without evidence of a cytopathic effect.
A 2,021-basepair fragment of the viral genome was sequenced for NiV, which includes partial gene fragments for the nucleocapsid gene (205 basepairs), the glycoprotein gene (954 basepairs), the phosphoprotein gene (338 basepairs,), and the fusion protein gene (524 basepairs) (Genbank accession nos. JF899339–JF8993420). Partial mitochondrial gene of a Pteropus bat was sequenced, and the identity of the bat was confirmed as P. giganteus.
The nucleotide and amino acid sequences of partial nucleocapsid gene of Pteropus-derived NiV showed 100.0% homology with NiV sequences reported during 2001 (Siliguri) and 2007 (Nadia) outbreaks in India and with Bangladesh sequences. Partial nucleocapsid gene sequences of NiV showed 96.0% identity with Cambodia and Malaysia strains and 81.0% with Hendra virus from Australia at the nucleotide level. Pteropus NiV amino acid sequences from India showed 100.0% identity with all NiV sequences except with AF017149 (Australia) and NiV from Pteropus bats from Malaysia (AY858110) (98.0% identity).
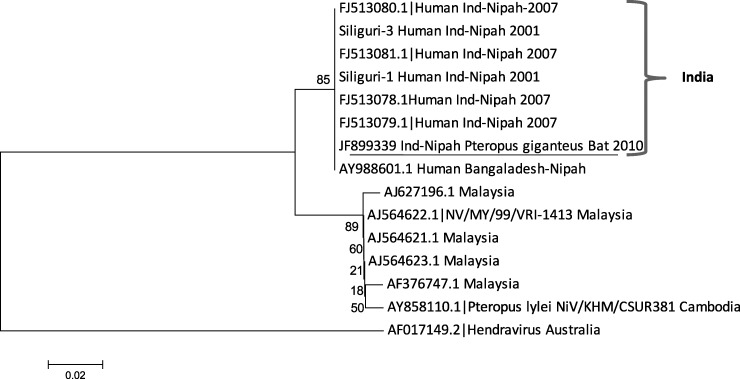
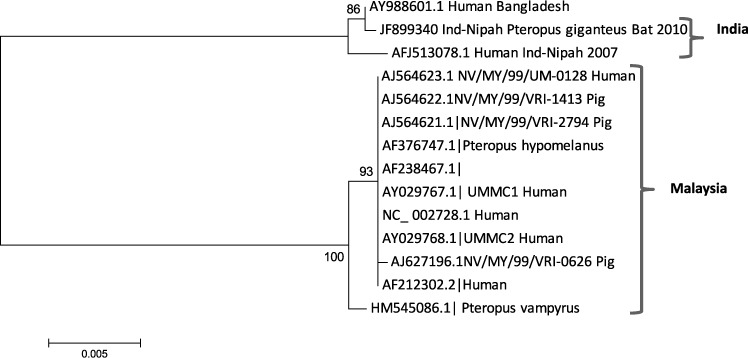
Bat NiV glycoprotein gene sequence showed 99.0% and 100.0% identity with virus sequences from Nadia outbreak (AFJ513078), and with Bangladesh sequence (Ay988601) and 100% identity at the amino acid level. Bat NiV sequence had identities of 92.0% and 96.0% with Malaysia strains at the nucleotide and amino acids levels, respectively, except for HM545086.1 sequence (derived from P. vampyrus), which showed 97.0% identity at the amino acid level. Two phylogenetic lineages were formed for NiV nucleocapsid and glycoprotein gene sequences: Bangladesh and India sequences and Malaysia and Cambodia sequences (Figures 1 and 2).
Figure 1.
Phylogenetic tree of partial nucleocapsid gene (159 basepairs) sequence of Nipah virus from Pteropus giganteus from India and other reported Nipah viruses from GenBank constructed by using the neighbor-joining method with 1,000 bootstrap replicates. Scale bar indicates nucleotide substitutions per site.
Figure 2.
Phylogenetic tree of partial glycoprotein gene (954 basepairs) sequence of Nipah virus from Pteropus giganteus from India and other reported Nipah viruses from GenBank constructed by using the neighbor-joining method with 1,000 bootstrap replicates. Scale bar indicates nucleotide substitutions per site.
Partial sequences of glycoprotein and nucleocapsid genes of NiV from India detected in Pteropus bats showed that NiV was present in the captured bat. Viral sequences from India were similar to those from Bangladesh in the same phylogenetic lineage. The same NiV strain might circulate in India and Bangladesh. Thus, we believe that when the bat was captured, the viremia phase had ended but there was still some virus in the liver. The healthy body condition of the bat, the positive quantitative RT-PCR result for liver, and the presence of IgG suggest that the virus infection likely had no adverse effect on the bat, which overcome the infection. Our results suggest that Pteropus bats may be a reservoir for NiV in India. A systematic survey is required in different states of India to understand the distribution of this virus.
ACKNOWLEDGMENTS
We thank Rajen Lakra, Uttam K Shende, Dinesh K Singh, and Sanjay V. Gopale for technical assistance and Dr. Vidya Arrankalle (Head, Hepatitis Department) for sharing unpublished primer sequences of Nipah virus.
Footnotes
Authors' addresses: Pragya D. Yadav, Chandrashekhar G. Raut, Anita M. Shete, Akhilesh C. Mishra, and Devendra T. Mourya, Microbial Containment Complex, National Institute of Virology, Pashan, Pune, India, E-mails: hellopragya22@gmail.com, yadavpd@icmr.org.in, raut.cg@niv.co.in, anitamicro@yahoo.com, acm1750@rediffmail.com, mourya.dt@niv.co.in, and dtmourya@gmail.com. Jonathan S. Towner and Stuart T. Nichol, Viral Special Pathogens Branch, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: jit8@cdc.gov and stn1@cdc.gov.
References
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chadha MS, Comer JA, Lowe L, Rota PA, Rollin P, Bellini WJ, Ksiazek TG, Mishra AC. Nipah virus–associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arankalle VA, Bandyopadhyay BT, Ramdasi AY, Jadi R, Patil DR, Rahman M, Majumdar M, Banerjee PS, Hati AK, Goswami RP, Neogi DK, Mishra AC. Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis. 2011;17:907–909. doi: 10.3201/eid1705.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu VP, Hossain MJ, Paeashar UD, Ali MM, Ksiazek TG, Kuzmin I, Niezgoda M, Rupprecht C, Bresee J, Breiman RF. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;12:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein JH, Field HE, Luby S, Pulliam JR, Daszak P. Nipah virus: impact, origins, and causes of emergence. Curr Infect Dis Rep. 2006;8:59–65. doi: 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luby SP, Hossain MJ, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. Recurrent Nipah virus outbreaks in Bangladesh, 2001–2007. Emerg Infect Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh WJ, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 8.Sawatsky B, Ranadheera C, Weingartl HM, Czubet M. Hendra and Nipah Virus. Animal Viruses: Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- 9.Yob JM, Field H, Rashdi AM, Morrissy C, van der Heide B, Rota P, bin Adzhar A, White J, Daniels P, Jamaluddin A, Ksiazek T. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis. 2001;7:439–441. doi: 10.3201/eid0703.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynes JM, Counor D, Ong S, Faure C, Seng V, Molia S, Walston J, Georges-Courbot MC, Deubel V, Sarthou JL. Nipah virus in Lyle's flying foxes, Cambodia. Emerg Infect Dis. 2005;11:1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson JG, Rupprecht C, Rollin PE, An US, Niezgoda M, Clemins T, Walston J, Ksiazek TG. Antibody to Nipah-like virus in bats (Pteropus lylei), Cambodia. Emerg Infect Dis. 2002;2:987–988. doi: 10.3201/eid0809.010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis. 2009;49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowak R. Walker's Bats of the World. Baltimore, MD: Johns Hopkins University Press; 1994. [Google Scholar]
- 14.Epstein JH, Prakash VB, Smith CS, Daszak P, McLaughlin AB, Meehan G, Field HE, Cunningham AA. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis. 2008;14:1309–1311. doi: 10.3201/eid1408.071492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh W, Batten B, Sealy TK, Carrillo C, Moran KE, Bracht AJ, Mayr GA, Cruz MS, Catbagan DB, Lautner EA, Ksiazek TG, White WR, McIntosh MT. Discovery of swine as a host for the Reston Ebolavirus. Science. 2009;325:204. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 16.Guillaume V, Lefeuvre A, Faure C, Marianneau P, Buckland R, Lam S, Wild TF, Deubel V. Specific detection of Nipah virus using real-time RT-PCR (Taq Man) J Virol Methods. 2004;120:229–237. doi: 10.1016/j.jviromet.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Frieder M, Otto VH. Cryptic diversity in European bats. Proc Biol Sci. 2001;268:1825–1832. doi: 10.1098/rspb.2001.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Towner JS, Amman BR, Tara KS, Carroll SA, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, Formenty PB, Albarino CG, Miller DM, Reed ZD, Kayiwa ZT, Mills JN, Cannon DL, Greer PW, Byaruhanga E, Farnon EC, Atimnedi P, Okware S, Katongole-Mbidde E, Downing R, Tappero JW, Zaki SR, Ksiazek TG, Nichol ST, Rollin PE. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]