Abstract
Background
Mice that are deficient for glutathione peroxidases 1 and 2 (GPX) show large variations in the penetrance and severity of colitis in C57BL/6J and 129S1/SvImJ backgrounds. We mapped a locus contributing to this difference to distal chromosome 2 (∼119–133 mbp) and named it glutathione peroxidase-deficiency-associated colitis 1 (Gdac1). The aim of this study was to identify the best gene candidates within the Gdac1 locus contributing to the murine colitis phenotype.
Method/Principal Findings
We refined the boundaries of Gdac1 to 118–125 mbp (95% confidence interval) by increasing sample size and marker density across the interval. The narrowed region contains 128 well-annotated protein coding genes but it excludes Fermt1, a human inflammatory bowel disease candidate that was within the original boundaries of Gdac1. The locus we identified may be the Cdcs3 locus mapped by others studying IL10-knockout mice. Using in silico analysis of the 128 genes, based on published colon expression data, the relevance of pathways to colitis, gene mutations, presence of non-synonymous-single-nucleotide polymorphisms (nsSNPs) and whether the nsSNPs are predicted to have an impact on protein function or expression, we excluded 42 genes. Based on a similar analysis, twenty-five genes from the remaining 86 genes were analyzed for expression-quantitative-trait loci, and another 15 genes were excluded.
Conclusion/Significance
Among the remaining 10 genes, we identified Pla2g4f and Duox2 as the most likely colitis gene candidates, because GPX metabolizes PLA2G4F and DUOX2 products. Pla2g4f is a phospholipase A2 that has three potentially significant nsSNP variants and showed expression differences across mouse strains. PLA2G4F produces arachidonic acid, which is a substrate for lipoxygenases and, in turn, for GPXs. DUOX2 produces H2O2 and may control microbial populations. DUOX-1 and -2 control microbial populations in mammalian lung and in the gut of several insects and zebrafish. Dysbiosis is a phenotype that differentiates 129S1/SvImJ from C57BL/6J and may be due to strain differences in DUOX2 activity.
Introduction
Mice deficient in the glutathione peroxidase isoenzymes, GPX1 and GPX2 (Gpx1/2-Double Knock Out [DKO]) have spontaneous ileocolitis that is driven by gut microbiota [1], [2]. Both proteins are members of the selenium-dependent GPX family and are classic antioxidant enzymes that reduce potentially noxious H2O2 and fatty acid hydroperoxides to water and alcohols. In humans, fourteen genes affecting oxidative stress, including GPX1 and GPX4, are candidate genes for inflammatory bowel disease (IBD) and oxidative stress has been associated with IBD [3], [4]. Although the GPX2 gene is hypomorphic [5], it is regulated by nuclear factor erythroid-derived 2-like 2 (NFE2L2/NRF2) [6]. An NFE2L2/NRF2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population, implying that GPX2 may modify IBD [7]. Thus, Gpx1/2-DKO mice may represent an extreme case of oxidative stress-associated intestinal inflammation useful for understanding how oxidative stress affects IBD.
The impact of the Gpx1/2-DKO construct is dependent on mouse strain background. Colitis in C57BL/6 (B6) mice is rare, and it is mild when it occurs. In contrast, colitis occurs with 90% penetrance in the 129S1/SvlmJ (129) strain and often leads to morbidity before weaning. We reported that the GPX-deficiency-associated-colitis 1 locus (Gdac1), containing the B6 allele, confers resistance to colitis in the 129 background. Gdac1 maps to chromosome (Chr) 2: 119–133 mbp [8]. The large size of the reported interval accounts for differences in the 95% confidence intervals (CI), calculated by R/QTL, among the four phenotypes used to characterize strain difference in colitis susceptibility. The Gdac1 locus overlaps cytokine-deficiency-induced-colitis susceptibility 3 locus (Cdcs3), which was localized based on mapping in resistant B6 IL10-KO vs. sensitive C3H/HeJBir (C3H) IL10-KO mice [9]. Here we compared nsSNPs in the genes at Gdac1 and Cdcs3 locus to determine the likelihood that Gdac1 replicates Cdcs3.
The original Gdac1 locus covers a region containing ∼300 genes, which makes thorough candidate analysis a daunting task. It contains two gene clusters separated by a gene-sparse region (122.6–124.4 mbp). The distal 125–133 mbp region includes a candidate human IBD gene, FERMT1, which encodes kindlin-1 localized at focal adhesions [10], [11], a spermine oxidase gene (Smox), an antioxidant transporter for ascorbate uptake (Slc23a2), several immunity genes (IL1a, IL1b and Sirpa), a major cell-cycle check-point gene (Bub1; marker SNP at 127.65 mbp) and a proliferative-cell-nuclear antigen gene (Pcna). Any of these genes could be envisioned to modify disease in Gpx1/2-DKO mice based on the colon pathology (crypt apoptosis, hyper-proliferation, acute inflammation, chronic inflammation, dysbiosis and/or antioxidant deficiency). The proximal region, ∼119–124 mbp, contains an equally compelling list of candidates, whose functions mirror those of the distal candidates. For example, BUB1B and CASC5 produced from the proximal region physically associate with BUB1 produced from the distal region to regulate mitosis [12]. The proximal region has two dual oxidase (Duox) genes, whereas the distal region has the Smox gene, all three oxidases generate H2O2 [13]. The proximal region has two potential autophagy genes, encoding vacuolar protein sorting (VPS)-18 and -39, whereas the distal region encodes VPS16 [14].
Here we report our analysis of 155 additional mice with an increased number of single-nucleotide polymorphism (SNP) markers throughout the Gdac1 region. The analysis eliminated the distal 126–133 mbp of the original locus from consideration, essentially halving the number of potential genes. The common ancestry of the proximal region in C3H and 129 better supports the notion that Cdcs3 replicates the proximal region of Gdac1 rather than the distal region. Using in silico analysis, we evaluated 128 well-annotated protein-encoding genes, largely from the proximal region as well as 1 putative and 2 validated microRNA (miRNA) genes. Based on gene function in the colon and/or pathology observed in DKO mice, we selected the top 25 protein-encoding colitis gene candidates for e-QTL analysis. In this report, we summarize the process used for gene selection and elimination. We then explain the rationale for why we chose Pla2g4f and Duox2 as the top colitis gene candidates rather than eight other genes that included Bub1b, Plcb2, Casc5, Chac1, Oip5, Pla2g4e, Trp53bp1 and Slc28a2.
Results
Refined Mapping of the Gdac1 Locus
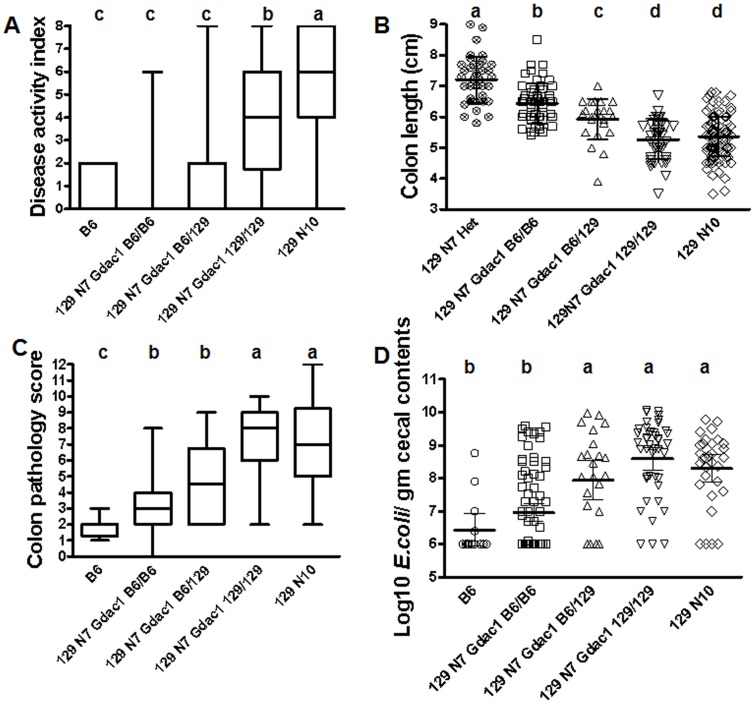
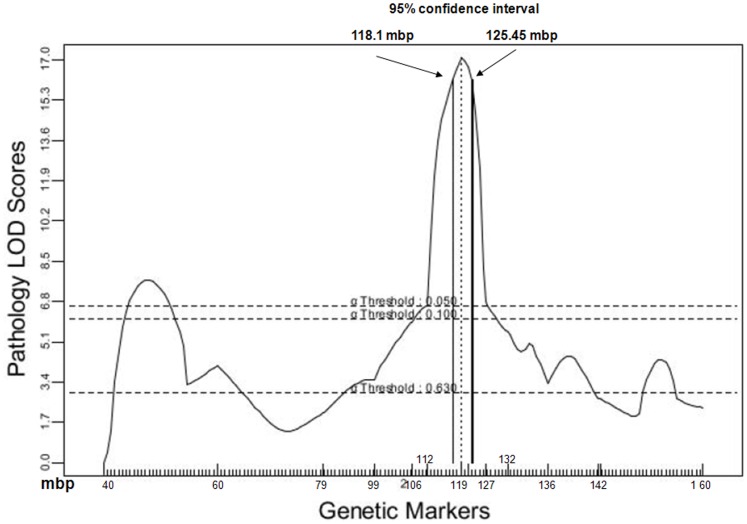
The increased marker SNP density enabled us to detect multiple recombination events between the original markers at 118.8 mbp and 142 mbp and estimate the location of recombination within the Gdac1 interval (Table 1). The increased numbers of mice provided more recombinants in the interval for analysis. The impact was that the 95% confidence interval (CI) was limited to the region of 118 mbp to 125 mbp with good agreement among the four phenotypes. The phenotypes measured were disease activity index (DAI; the criteria are defined in Table 2), colon length, colon pathology score (H&E histology) and E. coli overgrowth (Fig. 1). R/QTL calculated LOD ranged from ∼11–21 within the 95% CI (Fig. 2 and Fig. S1, S2, S3).
Table 1. Additional SNP markers for this study.
| Location | Reference SNP | Gene | Comment |
| Chr1: 87687800 | rs32549111 | –* | polymorphism eliminated** |
| Chr1: 87838162 | rs30671689 | Gpr55 | polymorphism eliminated |
| Chr1:1422002036 | rs13459053 | Cfh | polymorphism eliminated |
| Chr2: 40553058 | rs27175338 | Lrp1b | no polymorphism expected |
| Chr2: 79162839 | rs28305948 | Itga4 | |
| Chr2: 106189893 | rs3148954 | Dcdc5 | |
| Chr2: 112319953 | rs27491511 | Chrm5 | |
| Chr2: 122117242 | rs27453362 | Duox2 | |
| Chr2: 122509506 | rs2650268 | Slc30a4 | backup for Duox2 |
| Chr2: 127649702 | rs2826904 | Bub1 | |
| Chr2: 132737018 | rs2724570 | Fermt1 | |
| Chr2: 136711224 | rs2726691 | Mkks | |
| Chr3: 11824234 | rs29713670 | – | no polymorphism# |
| Chr8: 64173319 | rs33548981 | Palld | no polymorphism |
“-” means there is no gene with the SNP. Analysis in R/QTL showed no association of chromosome 1 alleles with disease phenotypes.
Selective incrosses were set up to eliminate B6 alleles at these loci.
B6 alleles indicated by genome-wide analysis are not detected by in-house markers and methodology.
Table 2. Disease activity index.
| Score* | Growth or weight loss | Diarrhea index |
| 0 | On par with non-DKO littermates | None |
| 1 | Growth arrest** | |
| 2 | 10% loss | Wet tail# |
| 3 | 10% to 20% loss | |
| 4 | ≥20% loss¶ | Diarrhea§ |
The final score is based on adding the results from the Growth and Diarrhea columns.
Cessation of growth commencing on day 8 to 13.
Matted and discolored fur at tail base. No accumulation of feces.
Lethargy was not scored. It was taken into consideration to euthanize mice and occurred independent of scores.
Large accumulation of partially or completely dried stool at tail base.
Figure 1. Phenotypes used to refine the Gdac1 locus.
Panel A. A box-and-whisker plot of disease activity index of DKO mice grouped by strain background and Gdac1 genotype. Gdac1 alleles were based on typing at Rpusd2 (118.8 mbp) and Duox2 (122.1 mbp) and/or Slc30a4 (122.5 mbp). Statistics was evaluated with 1-way ANOVA (Kruskal-Wallis); Dunn’s post test; α = 0.05. Different letters above the boxes indicate means are different, where a>b>c. The number of mice in each group is 16, 74, 35, 38 and 233, from left to right. Panel B. A scatter plot of colon lengths. B6 DKO mice were not included as a reference because they and the B6 WT colon lengths fell in the middle of the collective129 data. This might be contributed by the smaller stature of B6 mice at weaning. Instead, Gpx1−/−Gpx2+/− littermates of the Gdac1 N7 DKO mice were used as the reference (129 N7 Het). One-way ANOVA; Tukey’s post test; N = 39, 56, 23, 43, 119. Different letters above the boxes indicate means are different, where a>b>c>d. Panel C. A box-and-whisker plot of colon pathology scores. One way ANOVA (Kruskal-Wallis), Dunn’s post test; N = 21, 52, 24, 38, 54. Different letters above the boxes indicate means are different, where a>b>c. Panel D. A scatter plot of Log10 CFU E. coli per gm cecal contents. One-way ANOVA (Kruskal-Wallis), Dunn’s post test; N = 14, 79, 24, 42, 31. Different letters above the boxes indicate means are different, where a>b.
Figure 2. LOD plot for the distal colon pathology score phenotype across Chr 2 from 40.5 to 160 mbp.
Marker locations (mbp from centromere) are indicated on the X axis. Arrows demark the 95% CI. LOD plots for colon length, DAI and Log10 E. coli CFU/gm cecal contents are in Figure S1, S2, S3.
The Gdac1 gene count was cut by nearly one-half compared with our previous report [8]. Because many strong candidates were eliminated, we manually analyzed 4 genotypes of Gpx1/2-DKO mice to assess the results of the R/QTL analysis (Fig. S4A). We identified a group of mice that were 129/129 for markers at 118.8 and 122.1–122.5 mbp and B6/B6 at 127.6 and 132.7 mbp (the reciprocal was not found). This group shared the phenotypic properties of 129N10 mice (129/129 throughout the interval) and distinguished them both from N7 mice B6/B6 throughout the 118.8–132.7 mbp interval and a congenic line (B6/B6; 118.8–137 mbp) (P≤0.05; 1-way ANOVA; Fig. S4B–S4E). This was consistent with the R/QTL output, which indicated there was little impact on disease severity caused by any variation in the distal region. We have a congenic line in which the proximal end of the differential segment is delineated by a SNP marker at 118.8 mbp. We observed the congenic DKO mice were significantly healthier than the reference 129 N10 population based on all 4 phenotypes, which was consistent with the R/QTL analysis indicating that the proximal boundary of Gdac1 is near 118 mbp.
Influence of Gdac1 on Dysbiosis
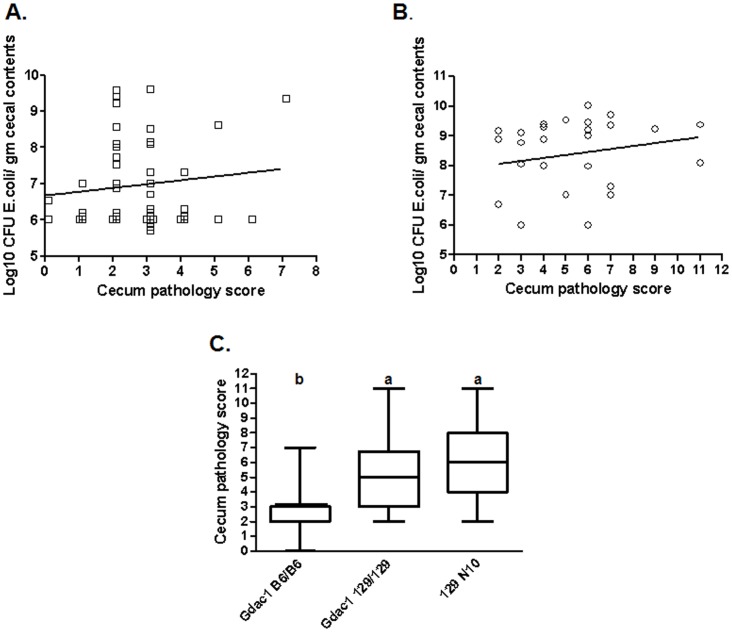
In a previous study, we examined the composition of the microflora in the ceca of WT and Gpx1/2-DKO mice of the B6 and 129 strains [15]. This was determined by non-culture-based, automated ribosomal-intragenic-spacer analysis on DNA isolated from total cecal contents. The consistent feature was the overgrowth of E. coli and Enterococcus sp. in the 129 Gpx1/2-DKO mice that suggested their association with pathology. E. coli overgrowth had high enough penetrance in 129 DKO mice to be a good marker for dysbiosis, whereas Enterococcus overgrowth had too low penetrance to be useful (Fig. 1D).
We found that Gdac1 could influence E. coli overgrowth in the cecum (Fig. 1D, 3A and 3B). The cecum was a disease site, although it had milder disease than the distal colon (Fig. 1C and 3C). When we plotted bacterial CFU against pathology scores, we did not find any correlation between them within Gdac1 genotype groups (Gdac1B6/B6, R2 = 0.013; Gdac1129/129, R2 = 0.04) (Fig. 3A and 3B). Therefore, dysbiosis was not a result of gross inflammation (pathology scores of 7 and above) but was associated with the underlying conditions that promoted apoptosis, hyper-proliferation and mucin depletion (all of which contributed to scores of 1 to 6). This was the first indication that Gdac1 affected processes that precede or promote development of inflammation rather than affecting the intensity of inflammation.
Figure 3. Correlation between E. coli overgrowth and cecum pathology in 129 N7 Gpx1/2-DKO Gdac1 B6/B6 mice (Panel A; N = 53) and Gdac1 129/129 mice (Panel B; N = 25).
The slopes of the linear regression lines are not significantly different from 0. Data clustered at 6 for Log10 CFU have been offset on both the X and Y axes to show the points. Panel C shows the cecum pathology scores from Panel A and B in box-and-whisker format along with reference 129 N10 (N = 29). Different letters above the groups indicate significant differences (a>b; P<0.05; Kruskal-Wallis with Dunn’s multiple comparison test).
The Genetic Landscape of Gdac1 and Relationship to Cdcs3 and Dssc2
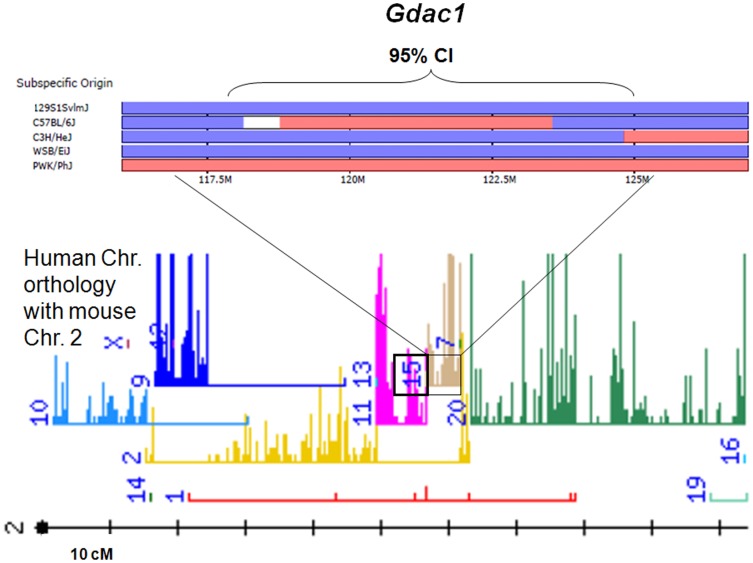
Gdac1 lies adjacent to Cdcs3 and Dssc2, which are colitis loci that were identified and analyzed in B6 vs. C3H IL10-KO mice and dextran sodium sulfate (DSS)-treated wild-type (WT) mice, respectively [9], [16]. Our refined Gdac1 was close to Cdcs3 (peak LOD at 117.95 mbp; Thbs1; Table 3) and could be confidently distinguished from Dssc2 (∼80 mbp). We found Gdac1 was conserved in humans, who had a nearly identical gene list for Chr.15q: 38–49 mbp (Fig. 4).
Table 3. Gdac1 QTL candidate genes (shortlist in proximal to distal order).
| Gene | Location | Primers∧ | nsSNPs** | PolyPhen-2** | eQTL#evidence | KO/mutant¶ |
| Spred1 | 116947186 | GAGATGACTCAAGTGGTGGATGTCTGAAAGGTAAGGCCAAACTTC | No | No | KO-airway eosinophilia | |
| Thbs1 | 117937658 | GCCGGCGGTAGACTAGGCCT GCGAGGAAAGGCGATCCCGG | Possible | All benign | No | KO-bad DSS response |
| Bmf | 118354493 | GGAGCGGGCGTATTTTGGAAACACTCGATTGGGAAGAAGGG | No | No | KO-urogenital defect | |
| Bub1b * | 118423992 | GAGGCGAGTGAAGCCATGTTCCAGAGTAAAAGCGGATTTCAG | Yes | No | KO-hemopoetic defects | |
| Plcb2 * | 118533253 | AGGATAGCTGTGATGGAAGAAGGGCCCAGGTGTCAGGTATGTAG | Yes | No | KO-enhanced chemotaxis | |
| Bahd1 | 118727351 | GGGCAGCTTCTACCTGTACTGGCTGTAGCCATTCGCTCCA | No | No | KO- listeriosis decreased | |
| Rpusd2 | 118860526 | AGGACGCCTGCATCTCAACCGGCTTGTCTATAACAACCACAT | No | No | None | |
| Casc5 * | 118872855 | TCCGACTCCAAGGAGCGCGATGCAAAGCTGACTCGACGAGAGC | Yes | No | None | |
| Rad51 | 118938553 | GTCCACAGCCTATTTCACGGTACAGCCTCCACTGTATGGTAAC | No | No | KO-embryonic lethal | |
| Spint1 | 119063096 | CCAGTGTGCTCCGGCAGCTGGTGGCCTCGTCGGAGCCATC | No | No | KO-peri-natal lethal | |
| Vps18 | 119124189 | ATACACTGCTCCGCATTGACTGGTTCATGTAAAGGACCTCGGT | No | No | (Zebra Fish) | |
| Dll4 | 119151565 | CAGTTGCCCTTCAATTTCACCTAGCCTTGGATGATGATTTGGC | Yes | All benign | No | KO-no impact (complements Dll1) |
| Chac1 * | 119176992 | CTGTGGATTTTCGGGTACGGCTCGGCCAGGCATCTTGTC | Possible | No | None | |
| Ino80 | 119198778 | CCACACAACAAGCACAGAACAGAGACCACGTTTCTTGCCG | No | No | None | |
| Oip5 * | 119435268 | CTCGTCTAAACTCACTGACGGGCAGGATCTTTGGTGATGCTGTT | Yes | No | None | |
| Mapkbp1 | 119798435 | AGAAGGGTCAACCATTACTAGCCGCTGGGTAGGCAACTAAGCC | Yes | All benign | No | None |
| Pla2g4b | 119860328 | CGGCCAGCCTCCAAGACAGCAGAGCTCGGCGATCCTGCCA | Yes | All benign | No | None |
| Pla2g4e * | 119992148 | AACGGCGTGCTGGTGTCTCGAGGCGCCTCTGCGTAAAGCTGAAACCCATGTGAAGTTCCCACCCTCCAGGCAAGAGACTTGG | Yes | Yes | None | |
| Pla2g4f * | 120125702 | CCCAGTGCTGAGCCCCAAGCTGCTGGCCCTCCTCCAGGTC | Yes | No | None | |
| Trp53bp1 * | 121023987 | ATTGAACGGTTACCTCAGCCACCCAACTGTGATGAAGCAGAAT | Possible | No | KO-immune deficient | |
| Duox2 * | 122106173 | TCACAACGGACGGCTTGCCCCCCGGCCACTCCATTGCTGG | Yes | No | Mutant-hypothyroid | |
| Duoxa2 | 122124636 | GCACTCGCGCTGGTTCTGGTATGTTAACGCCCGCCAGCCC | No | No | Mutant-hypothyroid | |
| Slc28a2 * | 122252213 | AGTGGAGAATTGCATGGAGAACGACCAAGCAGGATCTTTCTGAA | Possible | No | None | |
| Slc30a4 | 122506975 | GCTGACCATCGCTGCCGTCCTGGCCGACAAAACCTCTAGGCG | Yes | All benign | No | Mutant-lethal milk |
| Cops2 | 125656040 | ATGAGGAGGACTACGACCTGGTGAATCCCCATTCTCCCTTCTC | No | No | KO-embryonic lethal |
The primers are listed at 5′ to 3′ direction. Each set of primers are listed with forward primer on the top and reverse primer in the bottom. Two primer sets were tested for Pla2g4e.
means those genes are viable candidates.
nsSNPs- “No” means no nsSNPs reported in databases for B6 vs. 129 and often among all strains. “Possible” refers to incomplete annotations for B6 and/or 129; an nsSNP was noted at this location involving at least one strain among all inbred strains in databases. For Trp53bp1, PolyPhen-2 rejected several amino acid calls, which prohibited SNP evaluation. The latter also applied to several other genes shown in Table S1. “All benign” means that all nsSNPs were predicted as “benign” by PolyPhen-2.
Gene expression levels in colon examined by qPCR are normalized against β-actin or 36B4. P-values between 0.05 and 0.1 were rated as “Possible.” However the pattern of expression in the sets of mice often suggests that this was a reaction to pathology rather than true cis-e-QTL status.
Knockout or mutants available for reference. “None”-no KO or mutant studies in mice, except Vps18-KO study was performed in Zebra fish. However, Online Mendelian Inheritance in Man (OMIM) reports at least 20 mutations/deletions in humans for this region (Chr: 15q 38–49 mbp) with associated syndromes.
Figure 4. The subspecific origin of Gdac1 and the relationship to a human locus.
The upper portion of the figure shows chromosome ancestry analysis. The Gdac1 95% CI in B6 is derived from M.m. musculus (small section of indeterminate ancestry at proximal end; reference wild-derived M.m. musculus strain is PWK/PhJ). This contrasts with both 129 and C3H, where M.m. domesticus is the source of the chromosome (wild-derived WSB/EiJ is the reference M.m. domesticus strain). The boundaries of Gdac1 coincide with a portion of human chromosome 15q (brown), shown in the lower portion of the figure (data retrieved from CGD- http://msub.csbio.unc.edu/ −and MGD at the MGI website; 04/2012-mouse phylogeny viewer and mouse; human orthology map).
The mouse phylogeny viewer (http://msub.csbio.unc.edu/) showed that the refined B6 allele of Gdac1 coincided with a chromosome block that originated from M.m. musculus (PWK/PhJ), flanked by blocks that originated from M.m. domesticus (WSB/EiJ). For C3H and 129, which were used to define Cdcs3 and Gdac1 vs. B6, respectively, the entire stretch originated from domesticus. This suggested that Gdac1 and Cdcs3 could be replicates (Fig. 4). In fact, C3H and 129 shared haplotypes in the gene-dense 118.8–119.3 mbp region, which indicated sequence identity (Fig. S5). A similar circumstance arose at 123–124 mbp; this region had only 2 poorly annotated genes. Distal to 124 mbp, B6 shared ancestry with the129 and C3H strains. At ∼125–129 mbp, C3H did not share the common ancestry of B6 and 129. Thus, it appeared unlikely that Cdcs3 and Gdac1 had the same polymorphism in the region distal to 124 mbp.
Preliminary Analysis of Gdac1 to Identify Unlikely Candidate Genes
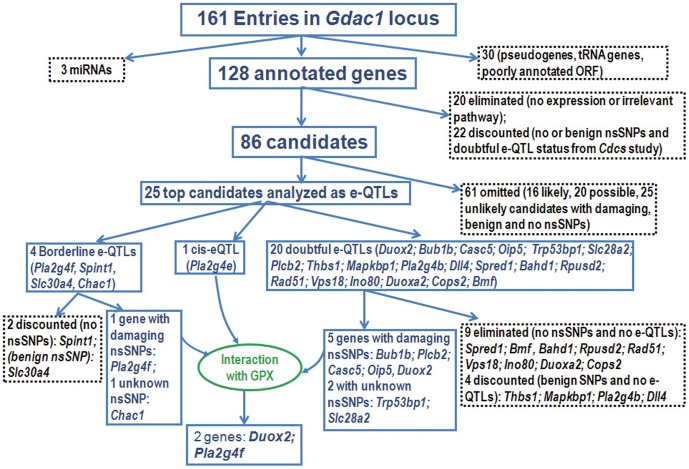
There were 128 well-annotated, protein-encoding gene entries in the Gdac1 region (161 total entries before we excluded pseudogenes, tRNA genes, miRNA genes and poorly annotated, presumptive open-reading frames) (Fig. 5 and Table S1). Non-coding RNAs are discussed in a separate section, below. Sixteen genes were eliminated by in silico analysis based on lack of expression in the colon or involvement in pathways that fall outside of those implicated in IBD (flagged as N in column 14 and “pathway” or “no expression” in column 15 and highlighted in grey in Table S1) [3]. Below are examples of what we considered to be irrelevant pathways. A Tyro3 47 knockout affected spermatogenesis by disruption of Sertoli cell-specific signaling pathways (the subscript 47 refers to the gene number in Table S1) [17]. Cdan1 66 defects produced anemia without other discernable effects [18]. The codanin-1 protein chaperones the heterochromatin protein 1 homolog α from the Golgi to the nucleus in erythroblasts [19], and Myef2 118 codes for a factor that regulates the myelin basic protein gene [20]. An additional 4 genes were eliminated based on inference from gene deletions or mutations in human studies. Fbn1 123 codes for fibrillin-1; deletions in this gene are implicated in aneurysm [21], [22]. Tmg5 72, Spg11 101 and Cep152 124 defects likewise produced human syndromes (skin peeling, spastic paraplegia and Seckel syndrome, respectively) without impacting the gastrointestinal (GI) tract [23], [24], [25]. Seven of the 20 genes (Tyro3 47, Cdan1 66, Strc 82, Catsper2 83, Spg11 101, Slc24a5 117 and Slc12a1 120) were eliminated using multiple criteria that included analysis of human gene mutations, results obtained from mouse or zebrafish knockouts, involvement in irrelevant pathways or lack of expression in the colon [17], [18], [24], [26], [27], [28], [29], [30], [31].
Figure 5. Outline of the candidate categorization process.
The eliminated and discounted genes as well as the genes for 3 microRNAs and omitted genes are shown in dotted-black boxes. The viable candidates at each step are shown in solid blue boxes.
Thirteen genes (Rasgrp1 3, Srp14 9, Ivd 19, Snap23 62, Lrrc57 63, Ccndbp1 70, Tubgcp4 77, Ckmt1 81, Pdia3 84, Serf2 86, Shf 107, Dut 121 and Eid 126) were discounted based on lack of nsSNPs and doubtful status as e-QTLs (Cdcs3 analysis by de Buhr et al.) [32]. Another 9 (Eif2ak4 8, Dnajc17 28, Zfyve19 30, Ndufaf1 42, Stard9 65, Ttbk2 67, Ubr1 68, B2m 98 and Sqrdl 114) were provisionally discounted because PolyPhen-2 analysis indicated the nsSNPs were benign (i.e. did not affect protein function) and they had doubtful e-QTL status. PolyPhen-2 is a second generation web-based tool that evaluates the impact of amino acid substitutions caused by nsSNPs on protein function [33]. Because PolyPhen-2 analysis is not regarded as definitive (72% accuracy) and because we did not perform our own e-QTL analysis, these 22 genes cannot be eliminated from candidacy [34]. In order to eliminate a discounted gene, we would need to demonstrate that it 1) lacks nsSNPs and has no e-QTL status; or 2) has benign nsSNPs and has no e-QTL status. In summary, we eliminated 20 genes and discounted 22 from candidacy.
Selection Criteria for e-QTL Analysis of Gdac1 Candidates
Based on gene function in the colon and/or pathology observed in DKO mice, we selected 25 genes from the remaining 86 candidates for e-QTL analysis (Table 3). Among the 86 genes, there were at least 11 genes in the DNA repair and mitosis spindle formation/regulation/check-point group (Bub1b 11, Casc5/Blinkin24, Rad51 25, Ino80 37, Oip5/Lint-2540, Nusap1 41, Trp53bp1 78, Cep152 124, Haus2/Cep2764, Ccndbp1 70 and Tubgcp4 77 ) [35]. We selected six; Bub1b 11, Casc5 24, Rad51 25, Trp53bp1 78, Ino80 37 and Oip5 40. Cep152 124 was eliminated because in humans a mutation causes Seckel syndrome without specific GI symptoms [25]. Nusap1 41, Tubgcp4/D2Ertd435e 77 and Ccndbp1 70 were analyzed by de Buhr et al. as e-QTLs and did not reach the 1.5x threshold of biological significance [32]. Haus2/Cep2764 was not analyzed in order to allow analysis of genes in other pathways.
The H2O2-producing oxidases DUOX1 and DUOX2 are encoded by the Duox1 106 and Duox2 103 genes. They are clustered in Gdac1 with their respective maturation factors, Duoxa1 105 and Duoxa2 104 [36]. Duox1 is expressed at a very low level in the intestine, while Duox2 is readily detectable ([37] and data not shown). There is a cluster of three Pla2g4 genes producing intracellular PLA2s, and a PLA1, which is encoded by Pla2g4e 54 [38]. Pla2g4d 55 is not expressed in the intestine, and is involved in psoriasis [39], [40], [41]. Therefore, we included Duox2 103, Duoxa2 104, Pla2g4b 51, Pla2g4e 54 and Pla2g4f 56 in further analysis (Table 3).
Seven genes, Spred1 1, Thbs1 4, Bmf 10, Plcb2 14, Bahd1 20, Mapkbp1 50 and Slc28a2 108, were selected from 12 genes potentially linked to colitis through either immune signaling or suggestive evidence from DSS and/or gene knock out studies. The remaining five, Rasgrp1 3, Ndufaf1 42, Ltk 45, Pdia3 84 and B2m 98, were previously analyzed as Cdcs3 candidate e-QTLs but showed no differences in levels at the significance threshold of 1.5x [32], and so were not analyzed in this study.
Seven additional genes were selected based on either possible overlap with human IBD candidates (Rpusd2 23), pathway defects implicated in IBD (Vps18 34; Chac1 36; Cops2 128), involvement with epithelial differentiation and development (Dll4 35) or involvement in mouse development (Spint1 32; Slc30a4 112). Rpusd2 is a possible analog of a human IBD candidate, PUS10; Spint1 regulates development; Vps18 is in the autophagy pathway; Dll4 encodes a Notch1 ligand; the Chac1 product links ER stress to apoptosis; a Slc30a4 112 mutant causes lethal milk (zinc deficiency); the Cops2 protein is involved in the ubiquitin-proteasome COP9 signalosome that functions during general embryonic proliferation, T-cell development and T-cell antigen-stimulated proliferation [14], [42], [43], [44], [45], [46], [47].
Gdac1 Candidates Evaluated as e-QTLs
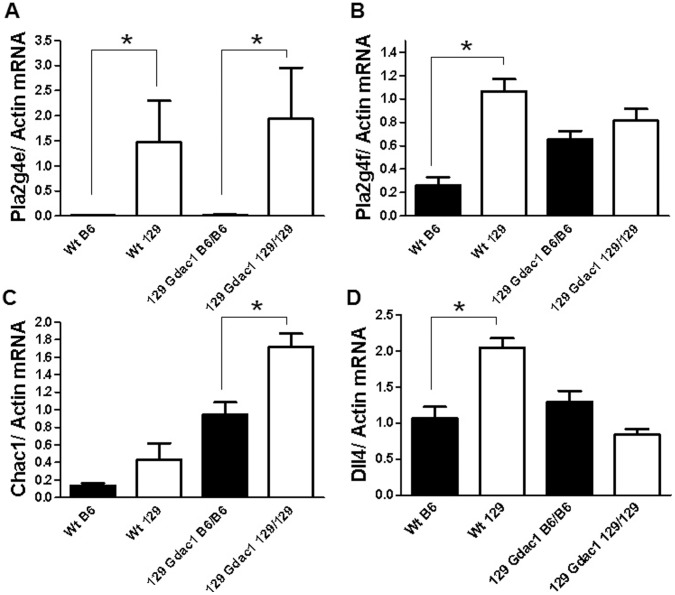
Among the 25 genes analyzed, Pla2g4e 54 was the lone definitive cis-e-QTL found in this survey; RT-qPCR with two primer sets confirmed it had different expression levels between 129-Gdac1B6/B6 and 129-Gdac1129/129 DKO mice and between the parental strains (Fig. 6A). We and others found that B6 colon barely expressed Pla2g4e mRNA [41]. In contrast, the current results showed that the 129 colon does express the Pla2g4e mRNA. Pla2g4f 56 was a borderline e-QTL (Fig. 6B); between B6 and 129 the difference in expression levels was significant, but between 129-Gdac1B6/B6 and 129-Gdac1129/129 mice the difference was not significant. However, the 129 allele had higher expression levels in replicate RT-PCR analyses using separate standards. Pla2g4f mRNA in B6 colon was detectable on Northern blots, a result consistent with the RT-PCR results [41]. The e-QTL status of Spint1 32 and Slc30a4 112 was identical to Pla2g4f. Because Pla2g4e and Pla2g4f also had predicted damaging nsSNPs, they were considered to be good candidates. Spint1 had no nsSNPs and Slc30a4 nsSNPs were rated as benign, so their candidacy was discounted. Chac1 36 (Fig. 6C) was a fourth borderline e-QTL. The 3-fold difference in gene expression levels between the parental strains was not significant. The difference between the Gdac1 DKO mice sets was significant and consistent with the trend observed in the parental strains. The borderline e-QTL status and uncertainty about the Chac1 nsSNPs resulted in the gene being classified as undecided. However, the fact that Chac1 responds to oxidative stress indicated that it may warrant further analysis. Another 7 genes (Bub1b 11, Plcb2 14, Casc5 24, Oip5 40, Trp53bp1 78, Duox2 103 and Slc28a2 108) were not e-QTLs, but they remained candidates because they had potentially damaging nsSNPs.
Figure 6. e-QTL analysis by RT-PCR. Relative mRNA levels of Pla2g4e (A), Pla2g4f (B), Chac1 (C) and Dll4 (D) in the distal colons of 129 strain Gpx1/2-DKO Gdac1 mice by genotype and in WT B6 and WT 129.
Dll4 represents a common pattern among candidates, where the pattern suggests expression was affected due to pathology in the Gdac1129/129 mice. For all panels, N = 6, 6, 6, 6. An * indicates significant difference, α = 0.05, for pair-wise t-tests between parental strains or between the Gdac1 sets (B–D). Pla2g4e uses the non-parametric Mann-Whitney test, because the distributions were not Gaussian.
Due to negative or conflicting outcomes from our e-QTL analysis, we considered the potential candidacy of another 13 genes to be poor. In the case of the Dll4 35 gene (Fig. 6D), the significant difference in levels between 129 WT and 129-Gdac1129/129 mice (P<0.05) can be attributed to the loss of goblet cells, where it is expressed [48]. Thbs1 4, Dll4 35, Mapkbp1 50 and Pla2g4b 51 were discounted due to nsSNPs being rated as benign, and/or no expression differences, or inconsistent expression differences. Nine genes (Spred1 1, Bmf10, Bahd1 20, Rpusd2 23, Rad51 25, Vps18 34, Ino80 37, Duoxa2 104 and Cops2 128 ) were eliminated because they had no nsSNPs and showed either no expression differences or inconsistent expression differences.
Of the 25 genes selected for evaluation as e-QTLs, 10 remained as candidates (Bub1b 11, Plcb2 14, Casc5 24, Chac1 36, Oip5 40, Pla2g4e 54, Pla2g4f 56, Trp53bp1 78, Duox2 103 and Slc28a2 108). However, this was generally due to the nsSNPs found in these genes rated as damaging by PolyPhen-2. The clear exception was Pla2g4e; Pla2g4f and Chac1 represented borderline e-QTLs. Among the 10, Pla2g4f and Duox2 were prioritized as the strongest candidates because they were associated with oxidative stress through possible interactions with GPXs.
Non-coding RNAs
There were 2 validated miRNAs in the MGI databases that mapped within Gdac1; Mir6741a (117 mbp) and Mir147111a (122.4 mbp). There appears to be one SNP within Mir674 and 13 more were found within 2 kb of Mir674 using available mouse strains. However, the B6 sequence is not yet determined and a function for Mir674 has not been reported. There were no SNPs in Mir147 but 4 were within 2 kb of its location and it is possible that 129 and B6 could have different variants at all 4 sites. Mir147 is expressed in the spleen but not in normal colon and is involved with TLR4-stimulated macrophage production of TNFα and IL6 [49]. This possible anti-inflammatory role made Mir147 a candidate colitis gene in Gdac1 [50]. However the paucity of SNPs around the miRNA and within the AA467197/NMES1 gene (7 SNPs within 2 kb), where Mir147 resided, suggested that there may be no expression difference between 129 and B6.
In the corresponding human 15q region we identified 5 miRNAs (miR-626, miR-4310, miR-627, miR-1282, miR-147B) and one antisense RNA, OIP5-AS1. miR-147B is the human ortholog of Mir147. The regulation of miR-147B is distinct from Mir147 suggesting a different function in humans [50]. We found a match for miR-128287a in the Gdac1 locus using BLAST (100% match; 100% coverage; ∼121.28 mbp). No matches were found for miR-626, miR-4310, miR-627 and OIP5-AS1 even with a lower stringency search. Collectively, there were two validated and one potential miRNA in Gdac1.
Discussion
Based on the low LOD score in the distal portion of our previously defined Gdac1 locus on mouse Chr. 2, we have objectively eliminated close to 50% of genes [8]. The major constraints for screening through the remaining 128 annotated protein-encoding and 3 miRNA genes were the high number of SNPs between the B6 and 129 strains and the multiple pathways involved in colitis [3]. Consequently, in silico methods were only moderately useful. An immediate rejection of 20 genes was based on instances where there was no expression in the colon and/or the pathways the genes acted in were unlikely to impact IBD. This was sometimes inferred from human mutations. We were able to use microarray data from de Buhr et al. to directly exclude a few genes due to lack of expression in mouse colon (de Buhr’s Supplemental Gene Expression Omnibus (GEO) data) [32]. Relying on web tools to cull more genes involved some uncertainty because the mouse SNP databases are not yet complete and in silico tools, such as PolyPhen, are still being refined. However, altogether we were able to discount 22 more genes due to an absence of nsSNPs and doubtful e-QTL status, or the nsSNPs present rated as benign and no differences in expression levels between strains.
After reviewing the literature on the remaining 86 genes, we selected 25 as the best colitis gene candidates based on gene function in the colon and/or pathology observed in DKO mice (i.e. apoptosis, proliferation, mucin depletion, colitis and dysbiosis). Using individual e-QTL analysis, we eliminated 9 genes because they had no nsSNPs and no evidence of cis-e-QTL status. We also downgraded the importance of 6 more genes because they had no disease-associated nsSNPs and no expression differences.
Four of the 10 remaining candidates were involved in cell cycle checkpoint/DNA repair pathways. Bub1b 11 and Casc5 24 could affect the pathology of Gpx1/2-DKO mice via DNA damage check-point regulation and apoptosis. Because their products interact, it is possible that the nsSNPs of either gene could have a significant impact on this pathway [51]. Oip5 40, which functions in the cell-cycle regulation pathway, has damaging nsSNPs [52]. Trp53bp1 78, which affects DNA damage responses and contributes to immune regulation, might be eliminated upon the completion of the database for B6 and 129 nsSNPs [53], [54]. Because these four genes did not have a direct interaction with GPX they were not prioritized as top candidates, but they remain of interest for future investigation as are the pathways they represent.
Three other candidates may be linked to IBD for various reasons and all have significant nsSNPs. The solute carrier, Slc28a2 108, is involved in the control of extracellular adenosine pools, which, in turn, are involved in inflammation pathways. The lower expression level of Slc28a2 108 we observed in the colons of 129-Gdac1 129/129 DKO mice was consistent with an expected anti-inflammatory reaction to maintain high extracellular levels of adenosine; however the levels in the 129-Gdac1129/129 mice were not statistically less than the other groups of mice [55]. Plcb2 14 produces a phospholipase C, which has definitive roles in inflammation [56]. Pla2g4e 54 is a cis-e-QTL with one disease-associated nsSNP. However, because the PLA2G4E protein does not have PLA2 activity to produce arachidonic acid, we did not choose Pla2g4e as a top candidate [38]. Although the Slc28a2, Plcb2 and Pla2g4e genes were good candidates, they were not prioritized as top candidates because their protein products do not interact with GPX directly.
Chac1 36 has been linked to ER stress downstream of Chop and to an apoptosis-promoting pathway [42]. Chac1 responds to oxidized 1-palmitoyl-2-arachidonyl-sn-3-glycerophosphorylcholine treatment of aortic endothelial cells [57]. This links Chac1 to oxidative stress, although not directly to GPX1 or GPX2 [58]. The up-regulation of Chac1 in 129-DKO mice was consistent with the observed elevated apoptosis but no ER stress was detected in 129 Gpx1/2-DKO mice [8]. Therefore, the cause of the significant 4–7 fold up-regulation in the colon may be linked to other pathways down-stream of oxidative stress. Also, it is unclear in which cell types Chac1 is expressed and induced. Lusis et al. showed that Chac1 induction by oxidized phospholipid occurred in HAEC (human aortic endothelial cells) but not HEK (human embryonic kidney) or HeLa (human cervical cancer) cells [42]. There was a non-significant, but noticeable difference in Chac1 expression levels between the parental strains but the difference was significant between the Gdac1B6/B6 DKO set and the Gdac1129/129 DKO set. Although Chac1 may not be a true cis-e-QTL, its synergy with the pathology may be relevant in candidate assessment. Currently, the nsSNPs status of Chac1 is undetermined due to discrepancies in the databases. The Chac1 and Casc5 genes are located in two haplotype blocks shared by 129 and C3H. Their candidacy does support the assumption that Gdac1 and Cdcs3 are based on the same polymorphism. The uncertainties in Chac1 nsSNP and e-QTL status prevented us from rating it as a top candidate.
The best colitis gene candidates were Pla2g4f 56 and Duox2 103, because their products can interact with GPXs directly and have been implicated in immune responses and colitis [37], [38], [59]. The Pla2g4 cluster at the distal Chr. 2 produces an obscure set of intracellular enzyme activities in contrast to the well characterized Pla2g4a and Pla2g4c genes on Chr. 1 and 7, respectively [60], [61], [62]. Pla2g4f is the only gene in the Pla2g4 cluster that is a candidate equal to Duox2. By virtue of its colon expression, possible e-QTL status, 3 significant nsSNPs, PLA2 activity and an interaction with GPX [38], [41], we considered it to be a top candidate. The arachidonic acid generated by PLA2 is essential for activation of phagocyte NADPH oxidase that is required for microbicidal activity [63]. Pla2gIVF (encoded by Pla2g4f) mobilizes to membrane ruffles, and possibly contributes to intestinal epithelial restitution [40]. The nsSNPs rated as damaging/disruptive by PolyPhen-2 for Pla2g4f and Duox2 were shared between 129 and C3H with B6 as the outlier. Although the genes were not on shared haplotype blocks, candidacy based on nsSNPs supported the assumption of replication of Gdac1 and Cdcs3.
Of the disease phenotypes used in this study, E. coli overgrowth was not linked to intense inflammation. E. coli overgrowth occurred with roughly 85% penetrance in the cecum of the 129 Gpx1/2-DKO mice, whereas Gdac1B6/B6 reduced the penetrance to 37%. The tendency for overgrowth was shared by 129 IL10-KO mice, and others have shown the overgrowth phenotype to be a property of WT 129 rather than B6 [64], [65]. Gulati et al. suggest that variation in Paneth cell numbers and antimicrobial products may mediate this tendency [65]. Gdac1 does not have genes encoding antimicrobial peptides. However, the DUOXs have antimicrobial activities in several eukaryotic systems including Caenorhabditis elegans, Drosophila, Anopheles, zebra fish intestine and mammalian lung [66], [67], [68], [69], [70]. NOD2 is encoded by the IBD1 gene and interacts with DUOX2, which functions as a NOD2 effecter [71]. Furthermore, we have recently characterized lactoperoxidase (LPO) expression in colon epithelium [72]. LPO and DUOX may form a potent antimicrobial defense team in the colon to fend off microbial invasion as they do in other tissues [37], [70]. Therefore, we have selected Duox2 and Pla2g4f as prime candidates for further analysis of their roles in murine colitis models and human IBD.
Materials and Methods
Mice
The original Gpx1/2-DKO colony was a mixed line of the B6 and 129 strains [8]. This line was backcrossed to B6 for 7 generations, and to 129 to produce N5, N7 and N10 cohorts [8]. All data for Gdac1 genetics were obtained from mice fed semi-purified diets (Harland Teklad; casein, sucrose, corn oil; TD 06306 and TD 06307), designed to mimic LabDiets 5020 (10% corn oil, Purina) and 5001 (5% corn oil) for calories and macronutrients and using AIN76A vitamin and micronutrient specifications [15]. These diets reduced disease severity relative to LabDiet formulations [8], [15]. To produce homozygous DKO offspring efficiently, it was necessary to use semi-purified diets to prevent high mortality of homozygous DKO male breeders before reaching 35 days of age when on LabDiets. Studies reported here were approved by the City of Hope Institutional Animal & Use Committee.
Refined Gdac1 Marker Panel
A denser SNP marker panel was established concentrating on the Gdac1 region of Chr. 2 and some of the flanking area (Table 1) [8]. Additional SNPs were examined on Chr 1: 88 and 142 mbp as well as Chr 3: 64 and 118 mbp because B6 alleles were detected by a genome-wide scan of ten 129 N7 mice performed by the Jackson Laboratory Genome Scanning Service (Bar Harbor, ME) using 141 markers to cover the 19 autosomes at ∼20 mbp interval (Table 1 and [8]). SNPs were obtained from the Jackson Laboratory MGI SNP database (www.Jax.org) and the flanking sequences were screened for repeats in RepeatMasker (www.repeatmasker.org/cgi/bin/WEBRepeatMasker). Primers were made for the MassARRAY iPLEX Gold system by Sequenom, San Diego, CA and primer sequences are available on request. All DNA samples from the 129 N7 cohort described previously were rescreened [8].
Phenotypes Used in Mapping
Disease activity index (DAI): One hundred ninety-nine mice were analyzed for DAI, a post-hoc semi-quantitative appraisal of mouse health from 8 to 22 days of age or presentation of morbidity. DAI criteria (listed in Table 2) were modified from traditional criteria. Our criteria accounted for growth arrest and did not account for blood in the stool [73]. The final score was based on adding the findings from the growth/wasting column and the diarrhea column, so that the scores ranged from 0 to 8.
Colon length and pathology: Colon length was measured on 117 mice, and colon pathology scores (based on H&E stained sections) were appraised on 92 mice [8]. Pathology scores of 0–6 generally reflect presence of crypt apoptosis/hyperproliferation and mucin depletion without overt signs of inflammation. When the score is above 6, acute inflammation is generally evident with neutrophil infiltration, gland abscesses or erosion of the epithelium [8], [15].
E. coli overgrowth: The cecum contents were analyzed for E. coli colony forming units (CFU)/gm on LB plates grown aerobically at 37°C for 18–22 hours. The cecum is a disease site in these mice [8], [15]. Large colonies were scored as E. coli, and less frequently detected small colonies were identified as Enterococcus sp. (hirae, gallinarum or faecalis). The colony identity was established from the sequence of rDNA amplified from single colonies for both E. coli and Enterococcus sp. and E. coli colonies also were verified by the Clinical Microbiology Laboratory at COH [15]. Spot checks were performed on randomly selected large colonies throughout the project to confirm their identities. Single dilutions of cecal contents were plated with an approximate sensitivity of 2×106–1×107 CFU/gm [8], [15]. Zero colonies were entered as a default of 1x106 CFU/gm for statistical analysis in R/QTL, which was empirically determined to be the upper limit for healthy mice at this age [15], [74]. Log10 transformed CFU/gm was used as the phenotype parameter. One hundred seventeen samples were used for R/QTL analysis.
R/QTL Interval Mapping Analysis
Statistical associations of markers and phenotypes were performed to identify the loci underlying the traits. Interval mapping was performed with the R/QTL interface, J/QTL (version 1.3.1; cgd.jax.org). The LOD thresholds were calculated using 2000 permutations. The marker physical locations were converted to genetic locations using the Mouse Map Converter (cgd.jax.org/tools/tools.shtml). The genetic length was increased to adjust for the multiple generations. Log of the odds (LOD) scores and the 95% confidence interval were established by the program (Bayesian credible interval) [75]. Our original marker spacing across Chr. 2 was at 10 cM intervals [8]. Here, we decreased the marker intervals to 2.5–3.5 cM in the core of the Gdac1 region to detect recombination internal to the original markers and obtain an estimate of the location (Table 1). The QTLs were positioned by the interval mapping program that calculates maximum likelihood estimates (LOD scores) at and between markers using quantitative phenotype data. The scores are a measure of the strength of association of a trait and genotype stated as the log10 of the likelihood of the odds ratio (LOD). LOD scores of 3.3 and 4.3 or greater are generally considered statistically significant evidence of association in backcrosses and intercrosses involving one generation, respectively [76]. In this case, the thresholds for the colon pathology, DAI and CFU phenotypes (6.8, 15.5 and 8.4; α Threshold = 0.05) were higher as a consequence of the adjustment for multiple generations.
RT-PCR for Evaluation of Genes as e-QTLs
Mice were euthanized by CO2 asphyxiation. Distal colon tissues were dissected out and stored in RNAlater (Qiagen). For the synthesis of cDNA, colon tissues were homogenized with a Polytron homogenizer (PT 1200E: Brinkmann Kinematica, Fisher Scientific) and sonicated. Total RNA was isolated using the RNeasy Mini kit (Qiagen). cDNA was synthesized from 2 µg of total RNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) in the presence of 1 µg of random hexamers (Invitrogen). Real-time quantitative PCR (qPCR) was performed with the Eva qPCR SuperMix kit containing SYBR green dye (Biochain Institute, Hayward, CA, USA) using the iQ5 Detection system (Bio-Rad Laboratories, Hercules, CA, USA).
Data was analyzed with Bio-Rad iQ5 Optical System Standard Edition, version 2.0 software. The primer sequences for 25 genes are listed in Table 3. Briefly, standard curves for each primer set were generated from a serial dilution of pooled test samples plotted with x-axis of log starting quantity (SQ) and y-axis of threshold cycle (Ct). Only those results obtained with PCR efficiency between 80–120% and correlation coefficient (R2) between 0.95–1.00 (obtained from the standard curve) were used. The cDNA quantity of each sample was determined from the Ct value based on the standard curve, and then normalized to β-actin or 36B4 for acidic ribosomal phosphoprotein P0 [77], both showed similar results. Each assay was performed in duplicate. For screening purposes, P≤0.1 was considered of interest. The relevant comparisons in this evaluation were between the parental strains and between 129-Gdac1B6/B6 and 129-Gdac1129/129 mice. Finding a cis-e-QTL requires a significant difference in both sets. 129-Gdac1 mouse designation as B6/B6 and 129/129 was based on genotyping with marker SNPs at 118.8 and 122.1 mbp.
In Silico Analysis
The distal Chr. 2 gene list was obtained from the National Center for Biotechnology Information (NCBI). It was updated for revisions in gene nomenclature, addition of new open-reading frames and annotation of genes in PubMed, MGD (Mouse genome database), NCBI and Ensembl (www.ensembl.org) [78]. Annotated genes were evaluated for involvement in pathways that were relevant to the pathology of Gpx1/2-DKO mice by literature searches and reports of KOs and/or use of agents such as dextran sodium sulfate (DSS), when available, in addition to mutations in human genes. PolyPhen-2 was used to evaluate nsSNPs for potential significant impact on protein function [33]. One GEO dataset, available as supplemental data for de Buhr et al., had colon microarray expression data for approximately one-third of the genes in the Gdac1/Cdcs3 loci from the B6 and C3H strains (NCBI: GEO; GSM39288–GSM39297) [32]. This analysis set the threshold for biological effects at a 1.5x expression level difference between strains. A manual survey of the results suggested that statistical significance was unlikely at less than 1.5x difference. These results were combined with available literature to determine if gene expression was significant in the colon, if the mouse strain background could have a significant impact on levels, and to evaluate selected RT-PCR results performed for this study.
Supporting Information
LOD plot for disease activity index.
(TIF)
LOD plot for log10 E. coli/gm cecal contents.
(TIF)
Verification of R-QTL result. Panel A shows genotypes of 4 groups of mice manually analyzed for R-QTL verification. The number in the arrow is the mbp of SNP markers used for genotyping. B6/B6 and 129/129 genotypes are shown in solid black and white boxes, respectively. The shaded gray box indicates either B6/B6, 129/129 or B6/129 genotypes present in those regions in individual mice. A Gdac1 congenic line established in the 129 strain mice. The differential segment of the mice is anchored at 118.8 mbp at the proximal end and the distal end is at 137 mbp. The group of N7 B6/B6-122 B6/B6 distal consists of mice typed as B6/B6 across the 118.8–132 mbp interval. The N7 129/129-122 B6/B6 distal group consists of mice that typed 129/129 at 118.8 and 122.1 mbp and B6/B6 at 127 and 132 mbp. The 129 N10 mice are 129/129 throughout. Panels B–E show the same phenotypes as in Figure 1. Letters indicate significant differences in means for panels C and E or medians for panels B and D; where a>b; P≤0.05; 1-way ANOVA.
(PPT)
Detailed haplotype block analysis on the B6, 129 and C3H strains across Gdac1 . The comparison of C3H and 129 is shown as diamonds, where a value of zero represents identical haplotypes and y-axis shows the values representing a metric of dissimilarity of haplotypes for each pair of strains. In the region from 117.7 to 124 mbp, the C3H and 129 haplotypes had greater resemblance to each other than to B6 (129 vs. B6: black squares; C3H vs. B6: open triangles). The location of the Pla2g4f (proximal) and Duox2 (distal) genes are indicated by the small horizontal red bars. Data were retrieved from CGD (http://msub.csbio.unc.edu/) and MGD at the MGI website (04/2012).
(PPT)
Gdac1 gene list and analysis of candidacy. The 128 gene list excludes 30 entries that include pseudogenes, tRNA genes and predicted but un-annotated open reading frames. Two validated and one putative miRNA are listed as #a where # is the nearest proximal gene. The shaded entries are genes that were eliminated in the preliminary analysis. Each column is defined below: 1. Number of genes. The subscript numbers following the gene symbols in the text refer to the entry number in the Table. 2. Gene symbols are in the order of proximal to distal (116.9–125.6 mbp) in the physical map. 3. Gene names were obtained from NCBI. 4. nsSNPs were mined from MGD and CGD. nsSNPs are either present (Y), absent (N; means no for B6 vs. 129), maybe (means data is missing for B6 and/or 129, but at least one inbred strain from the full list has nsSNPs at a given position) or problem with IDs (nsSNPs listed at multiple locations or other problems with database listing). 5. Evaluation of nsSNPs (SNP eval) with PolyPhen-2 program. “Benign” means that the amino acid variations are unlikely to alter protein functionality or structure. “Damaging SNP” means that variant amino acids are substantially different and therefore, are likely to have an impact on the protein function. “Uneval” means unevaluated; the nsSNP and corresponding amino acids conflict with amino acid sequence downloaded from MGD, so cannot be analyzed. PolyPhen-2 nsSNP evaluations are not definitive. Therefore, genes with benign nsSNPs and no expression differences cannot be positively eliminated as candidates. 6. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 WT B6 colons after normalization with β-actin. 7. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 WT 129 colons after normalization with β-actin. 8. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 DKO 129-Gdac1B6/B6 colons after normalization with β-actin. 9. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 DKO 129-Gdac1129/129 colons after normalization with β-actin. 10. P shows the p-value for the Gdac1 comparison (columns 8 and 9). 11. e-QTL. N means no variation in levels found between parental strains or Gdac1 variants. Maybe means difference found between Gdac1 variants, however, in all but the Pla2g4e gene, the differences are likely the result of pathology or inflammation. Y means cis-e-QTL based on both parental strain and Gdac1 variants. A cis-e-QTL is the case where the difference observed between the parental strains is preserved between 129-Gdac1129/129 DKO and 129-Gdac1 B6/B6. 12. e-QTL. Clarification of the e-QTL difference based on groupings; for example, WT vs. DKO. A major finding was an outlier 129-Gdac1129/129 DKO expression level, suggesting a reaction to pathology or inflammation. 13. Pathway analysis or expression levels obtained through in silico analysis. “Unlikely” means that either the gene was not expressed in the colon and/or the pathways involved are unlikely to impact colitis. 14. Determination of candidate genes. D is discounted for the present. Undecided is no expression data, and/or SNPs can’t be evaluated, or expression passed 1.5x threshold in de Buhr et al. analysis. Y is a candidate gene. N is not a candidate. The order of candidacy is Y>D = Undecided>N. 15. Column 14. Summary of the reason for candidacy. A candidate gene has either potentially disruptive amino acid variants or e-QTL status, or both. No entry means that there was insufficient information for evaluation at this time or that the gene was among the 25 selected for further analysis and is discussed in the text.
(XLS)
Acknowledgments
We thank Sofia Loera and Tina Montgomery at Anatomic Pathology Core for tissue processing and Drs. Margaret Morgan and Keely Walker of the Office of Faculty and Institutional Support for editing the paper.
Funding Statement
This work was partially supported by National Institutes of Health R01CA114569 (FFC)http://projectreporter.nih.gov/reporter_searchresults.cfm. No additional external funding sources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, et al. (2004) Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res 64: 962–968. [DOI] [PubMed] [Google Scholar]
- 2. Esworthy RS, Binder SW, Doroshow JH, Chu FF (2003) Microflora trigger colitis in mice deficient in selenium-dependent glutathione peroxidase and induce Gpx2 gene expression. Biological Chemistry 384: 597–607. [DOI] [PubMed] [Google Scholar]
- 3. Khor B, Gardet A, Xavier RJ (2011) Genetics and pathogenesis of inflammatory bowel disease. Nature 474: 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rezaie A, Parker RD, Abdollahi M (2007) Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig Dis Sci 52: 2015–2021. [DOI] [PubMed] [Google Scholar]
- 5. Hesketh J (2008) Nutrigenomics and selenium: gene expression patterns, physiological targets, and genetics. Annu Rev Nutr 28: 157–177. [DOI] [PubMed] [Google Scholar]
- 6. Banning A, Deubel S, Kluth D, Zhou Z, Brigelius-Flohe R (2005) The GI-GPx gene is a target for Nrf2. Mol Cell Biol 25: 4914–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, et al. (2008) Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population. Hepatogastroenterology 55: 394–397. [PubMed] [Google Scholar]
- 8. Esworthy RS, Kim BW, Larson GP, Yip ML, Smith DD, et al. (2011) Colitis locus on chromosome 2 impacting the severity of early-onset disease in mice deficient in GPX1 and GPX2. Inflamm Bowel Dis 17: 1373–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farmer MA, Sundberg JP, Bristol IJ, Churchill GA, Li R, et al. (2001) A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc Natl Acad Sci U S A 98: 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, et al. (2011) A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet 7: e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ussar S, Moser M, Widmaier M, Rognoni E, Harrer C, et al. (2008) Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet 4: e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kiyomitsu T, Murakami H, Yanagida M (2011) Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol 31: 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casero RA, Pegg AE (2009) Polyamine catabolism and disease. Biochem J 421: 323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao H, Chen D, Fang Z, Xu J, Sun X, et al. (2009) Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell 20: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esworthy RS, Smith DD, Chu FF (2010) A Strong Impact of Genetic Background on Gut Microflora in Mice. Int J Inflam 2010: 986046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahler M, Bristol IJ, Sundberg JP, Churchill GA, Birkenmeier EH, et al. (1999) Genetic analysis of susceptibility to dextran sulfate sodium-induced colitis in mice. Genomics 55: 147–156. [DOI] [PubMed] [Google Scholar]
- 17. Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, et al. (1999) Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398: 723–728. [DOI] [PubMed] [Google Scholar]
- 18. Dgany O, Avidan N, Delaunay J, Krasnov T, Shalmon L, et al. (2002) Congenital dyserythropoietic anemia type I is caused by mutations in codanin-1. Am J Hum Genet 71: 1467–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Renella R, Roberts NA, Brown JM, De Gobbi M, Bird LE, et al. (2011) Codanin-1 mutations in congenital dyserythropoietic anemia type 1 affect HP1{alpha} localization in erythroblasts. Blood 117: 6928–6938. [DOI] [PubMed] [Google Scholar]
- 20. Haas S, Steplewski A, Siracusa LD, Amini S, Khalili K (1995) Identification of a sequence-specific single-stranded DNA binding protein that suppresses transcription of the mouse myelin basic protein gene. J Biol Chem 270: 12503–12510. [DOI] [PubMed] [Google Scholar]
- 21. Pereira L, Lee SY, Gayraud B, Andrikopoulos K, Shapiro SD, et al. (1999) Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci U S A 96: 3819–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Furtado LV, Wooderchak-Donahue W, Rope AF, Yetman AT, Lewis T, et al. (2011) Characterization of large genomic deletions in the FBN1 gene using multiplex ligation-dependent probe amplification. BMC Med Genet 12: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cassidy AJ, van Steensel MA, Steijlen PM, van Geel M, van der Velden J, et al. (2005) A homozygous missense mutation in TGM5 abolishes epidermal transglutaminase 5 activity and causes acral peeling skin syndrome. Am J Hum Genet 77: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Southgate L, Dafou D, Hoyle J, Li N, Kinning E, et al. (2010) Novel SPG11 mutations in Asian kindreds and disruption of spatacsin function in the zebrafish. Neurogenetics 11: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, et al. (2011) CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet 43: 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verpy E, Weil D, Leibovici M, Goodyear RJ, Hamard G, et al. (2008) Stereocilin-deficient mice reveal the origin of cochlear waveform distortions. Nature 456: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Malekpour M, Al-Madani N, Kahrizi K, Zanganeh M, et al. (2007) Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet 44: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quill TA, Ren D, Clapham DE, Garbers DL (2001) A voltage-gated ion channel expressed specifically in spermatozoa. Proc Natl Acad Sci U S A 98: 12527–12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, et al. (2005) SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310: 1782–1786. [DOI] [PubMed] [Google Scholar]
- 30. Ginger RS, Askew SE, Ogborne RM, Wilson S, Ferdinando D, et al. (2008) SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem 283: 5486–5495. [DOI] [PubMed] [Google Scholar]
- 31. Castrop H, Schnermann J (2008) Isoforms of renal Na-K-2Cl cotransporter NKCC2: expression and functional significance. Am J Physiol Renal Physiol 295: F859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Buhr MF, Mahler M, Geffers R, Hansen W, Westendorf AM, et al. (2006) Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol Genomics 25: 426–434. [DOI] [PubMed] [Google Scholar]
- 33. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopes MC, Joyce C, Ritchie GR, John SL, Cunningham F, et al. (2012) A combined functional annotation score for non-synonymous variants. Hum Hered 73: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, et al. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morand S, Ueyama T, Tsujibe S, Saito N, Korzeniowska A, et al. (2009) Duox maturation factors form cell surface complexes with Duox affecting the specificity of reactive oxygen species generation. FASEB J 23: 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL (2003) Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J 17: 1502–1504. [DOI] [PubMed] [Google Scholar]
- 38. Ghomashchi F, Naika GS, Bollinger JG, Aloulou A, Lehr M, et al. (2010) Interfacial kinetic and binding properties of mammalian group IVB phospholipase A2 (cPLA2beta) and comparison with the other cPLA2 isoforms. J Biol Chem 285: 36100–36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiba H, Michibata H, Wakimoto K, Seishima M, Kawasaki S, et al. (2004) Cloning of a gene for a novel epithelium-specific cytosolic phospholipase A2, cPLA2delta, induced in psoriatic skin. J Biol Chem 279: 12890–12897. [DOI] [PubMed] [Google Scholar]
- 40. Ghosh M, Tucker DE, Burchett SA, Leslie CC (2006) Properties of the Group IV phospholipase A2 family. Prog Lipid Res 45: 487–510. [DOI] [PubMed] [Google Scholar]
- 41. Ohto T, Uozumi N, Hirabayashi T, Shimizu T (2005) Identification of novel cytosolic phospholipase A(2)s, murine cPLA(2){delta}, {epsilon}, and {zeta}, which form a gene cluster with cPLA(2){beta}. J Biol Chem 280: 24576–24583. [DOI] [PubMed] [Google Scholar]
- 42. Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, et al. (2011) Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140: 1230–1240 e1231–1237. [DOI] [PMC free article] [PubMed]
- 44. Szabo R, Molinolo A, List K, Bugge TH (2007) Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene 26: 1546–1556. [DOI] [PubMed] [Google Scholar]
- 45. Huang L, Gitschier J (1997) A novel gene involved in zinc transport is deficient in the lethal milk mouse. Nat Genet 17: 292–297. [DOI] [PubMed] [Google Scholar]
- 46. Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, et al. (2003) Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Menon S, Chi H, Zhang H, Deng XW, Flavell RA, et al. (2007) COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8: 1236–1245. [DOI] [PubMed] [Google Scholar]
- 48. Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, et al. (2011) Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mineno J, Okamoto S, Ando T, Sato M, Chono H, et al. (2006) The expression profile of microRNAs in mouse embryos. Nucleic Acids Res 34: 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, et al. (2009) miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A 106: 15819–15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kiyomitsu T, Obuse C, Yanagida M (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676. [DOI] [PubMed] [Google Scholar]
- 52. Naetar N, Hutter S, Dorner D, Dechat T, Korbei B, et al. (2007) LAP2alpha-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci 120: 737–747. [DOI] [PubMed] [Google Scholar]
- 53. Ward IM, Minn K, van Deursen J, Chen J (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. FitzGerald JE, Grenon M, Lowndes NF (2009) 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904. [DOI] [PubMed] [Google Scholar]
- 55. Ye JH, Rajendran VM (2009) Adenosine: an immune modulator of inflammatory bowel diseases. World J Gastroenterol 15: 4491–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang H, Kuang Y, Wu Y, Xie W, Simon MI, et al. (1997) Roles of phospholipase C beta2 in chemoattractant-elicited responses. Proc Natl Acad Sci U S A 94: 7971–7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Romanoski CE, Che N, Yin F, Mai N, Pouldar D, et al. (2011) Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ Res 109: e27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chu FF, Doroshow JH, Esworthy RS (1993) Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI. J Biol Chem 268: 2571–2576. [PubMed] [Google Scholar]
- 59. Chu FF, Esworthy RS, Ho YS, Bermeister M, Swiderek K, et al. (1997) Expression and chromosomal mapping of mouse Gpx2 gene encoding the gastrointestinal form of glutathione peroxidase, GPX-GI. Biomed Environ Sci 10: 156–162. [PubMed] [Google Scholar]
- 60. Bonventre JV (1999) The 85-kD cytosolic phospholipase A2 knockout mouse: a new tool for physiology and cell biology. J Am Soc Nephrol 10: 404–412. [DOI] [PubMed] [Google Scholar]
- 61. Adler DH, Cogan JD, Phillips JA, 3rd, Schnetz-Boutaud N, Milne GL, et al (2008) Inherited human cPLA(2alpha) deficiency is associated with impaired eicosanoid biosynthesis, small intestinal ulceration, and platelet dysfunction. J Clin Invest 118: 2121–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brown JK, Knight PA, Thornton EM, Pate JA, Coonrod S, et al. (2008) Trichinella spiralis induces de novo expression of group IVC phospholipase A2 in the intestinal epithelium. Int J Parasitol 38: 143–147. [DOI] [PubMed] [Google Scholar]
- 63. Dana R, Leto TL, Malech HL, Levy R (1998) Essential requirement of cytosolic phospholipase A2 for activation of the phagocyte NADPH oxidase. J Biol Chem 273: 441–445. [DOI] [PubMed] [Google Scholar]
- 64. Wohlgemuth S, Haller D, Blaut M, Loh G (2009) Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ Microbiol 11: 1562–1571. [DOI] [PubMed] [Google Scholar]
- 65. Gulati AS, Shanahan MT, Arthur JC, Grossniklaus E, von Furstenberg RJ, et al. (2012) Mouse background strain profoundly influences Paneth cell function and intestinal microbial composition. PLoS One 7: e32403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chavez V, Mohri-Shiomi A, Garsin DA (2009) Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun 77: 4983–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ha EM, Oh CT, Bae YS, Lee WJ (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310: 847–850. [DOI] [PubMed] [Google Scholar]
- 68. Flores MV, Crawford KC, Pullin LM, Hall CJ, Crosier KE, et al. (2010) Dual oxidase in the intestinal epithelium of zebrafish larvae has anti-bacterial properties. Biochem Biophys Res Commun 400: 164–168. [DOI] [PubMed] [Google Scholar]
- 69. Oliveira JH, Goncalves RL, Lara FA, Dias FA, Gandara AC, et al. (2011) Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog 7: e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fischer H (2009) Mechanisms and function of DUOX in epithelia of the lung. Antioxid Redox Signal 11: 2453–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lipinski S, Till A, Sina C, Arlt A, Grasberger H, et al. (2009) DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci 122: 3522–3530. [DOI] [PubMed] [Google Scholar]
- 72. Kim BW, Esworthy RS, Hahn MA, Pfeifer GP, Chu FF (2012) Expression of lactoperoxidase in differentiated mouse colon epithelial cells. Free Radic Biol Med 52: 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Murano M, Maemura K, Hirata I, Toshina K, Nishikawa T, et al. (2000) Therapeutic effect of intracolonically administered nuclear factor kappa B (p65) antisense oligonucleotide on mouse dextran sulphate sodium (DSS)-induced colitis. Clin Exp Immunol 120: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Savage DC, Dubos R, Schaedler RW (1968) The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med 127: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- 76. Fisler JS, Warden CH (1997) Mapping of mouse obesity genes: A generic approach to a complex trait. J Nutr 127: 1909S–1916S. [DOI] [PubMed] [Google Scholar]
- 77. Akamine R, Yamamoto T, Watanabe M, Yamazaki N, Kataoka M, et al. (2007) Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J Biochem Biophys Methods 70: 481–486. [DOI] [PubMed] [Google Scholar]
- 78. Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT (2011) The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res 39: D842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LOD plot for disease activity index.
(TIF)
LOD plot for log10 E. coli/gm cecal contents.
(TIF)
Verification of R-QTL result. Panel A shows genotypes of 4 groups of mice manually analyzed for R-QTL verification. The number in the arrow is the mbp of SNP markers used for genotyping. B6/B6 and 129/129 genotypes are shown in solid black and white boxes, respectively. The shaded gray box indicates either B6/B6, 129/129 or B6/129 genotypes present in those regions in individual mice. A Gdac1 congenic line established in the 129 strain mice. The differential segment of the mice is anchored at 118.8 mbp at the proximal end and the distal end is at 137 mbp. The group of N7 B6/B6-122 B6/B6 distal consists of mice typed as B6/B6 across the 118.8–132 mbp interval. The N7 129/129-122 B6/B6 distal group consists of mice that typed 129/129 at 118.8 and 122.1 mbp and B6/B6 at 127 and 132 mbp. The 129 N10 mice are 129/129 throughout. Panels B–E show the same phenotypes as in Figure 1. Letters indicate significant differences in means for panels C and E or medians for panels B and D; where a>b; P≤0.05; 1-way ANOVA.
(PPT)
Detailed haplotype block analysis on the B6, 129 and C3H strains across Gdac1 . The comparison of C3H and 129 is shown as diamonds, where a value of zero represents identical haplotypes and y-axis shows the values representing a metric of dissimilarity of haplotypes for each pair of strains. In the region from 117.7 to 124 mbp, the C3H and 129 haplotypes had greater resemblance to each other than to B6 (129 vs. B6: black squares; C3H vs. B6: open triangles). The location of the Pla2g4f (proximal) and Duox2 (distal) genes are indicated by the small horizontal red bars. Data were retrieved from CGD (http://msub.csbio.unc.edu/) and MGD at the MGI website (04/2012).
(PPT)
Gdac1 gene list and analysis of candidacy. The 128 gene list excludes 30 entries that include pseudogenes, tRNA genes and predicted but un-annotated open reading frames. Two validated and one putative miRNA are listed as #a where # is the nearest proximal gene. The shaded entries are genes that were eliminated in the preliminary analysis. Each column is defined below: 1. Number of genes. The subscript numbers following the gene symbols in the text refer to the entry number in the Table. 2. Gene symbols are in the order of proximal to distal (116.9–125.6 mbp) in the physical map. 3. Gene names were obtained from NCBI. 4. nsSNPs were mined from MGD and CGD. nsSNPs are either present (Y), absent (N; means no for B6 vs. 129), maybe (means data is missing for B6 and/or 129, but at least one inbred strain from the full list has nsSNPs at a given position) or problem with IDs (nsSNPs listed at multiple locations or other problems with database listing). 5. Evaluation of nsSNPs (SNP eval) with PolyPhen-2 program. “Benign” means that the amino acid variations are unlikely to alter protein functionality or structure. “Damaging SNP” means that variant amino acids are substantially different and therefore, are likely to have an impact on the protein function. “Uneval” means unevaluated; the nsSNP and corresponding amino acids conflict with amino acid sequence downloaded from MGD, so cannot be analyzed. PolyPhen-2 nsSNP evaluations are not definitive. Therefore, genes with benign nsSNPs and no expression differences cannot be positively eliminated as candidates. 6. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 WT B6 colons after normalization with β-actin. 7. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 WT 129 colons after normalization with β-actin. 8. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 DKO 129-Gdac1B6/B6 colons after normalization with β-actin. 9. qRT-PCR evaluation of relative mRNA levels of 25 selected genes in 6 DKO 129-Gdac1129/129 colons after normalization with β-actin. 10. P shows the p-value for the Gdac1 comparison (columns 8 and 9). 11. e-QTL. N means no variation in levels found between parental strains or Gdac1 variants. Maybe means difference found between Gdac1 variants, however, in all but the Pla2g4e gene, the differences are likely the result of pathology or inflammation. Y means cis-e-QTL based on both parental strain and Gdac1 variants. A cis-e-QTL is the case where the difference observed between the parental strains is preserved between 129-Gdac1129/129 DKO and 129-Gdac1 B6/B6. 12. e-QTL. Clarification of the e-QTL difference based on groupings; for example, WT vs. DKO. A major finding was an outlier 129-Gdac1129/129 DKO expression level, suggesting a reaction to pathology or inflammation. 13. Pathway analysis or expression levels obtained through in silico analysis. “Unlikely” means that either the gene was not expressed in the colon and/or the pathways involved are unlikely to impact colitis. 14. Determination of candidate genes. D is discounted for the present. Undecided is no expression data, and/or SNPs can’t be evaluated, or expression passed 1.5x threshold in de Buhr et al. analysis. Y is a candidate gene. N is not a candidate. The order of candidacy is Y>D = Undecided>N. 15. Column 14. Summary of the reason for candidacy. A candidate gene has either potentially disruptive amino acid variants or e-QTL status, or both. No entry means that there was insufficient information for evaluation at this time or that the gene was among the 25 selected for further analysis and is discussed in the text.
(XLS)