Abstract
Regulator of G-protein signaling-10 (RGS10) is a GTPase activating protein (GAP) for Gαi/q/z subunits that is highly expressed in the immune system and in a broad range of brain regions including the hippocampus, striatum, dorsal raphe, and ventral midbrain. Previously, we reported that RGS10-null mice display increased vulnerability to chronic systemic inflammation-induced degeneration of nigral dopaminergic (DA) neurons. Given that RGS10 is expressed in DA neurons, we investigated the extent to which RGS10 regulates cell survival under conditions of inflammatory stress. Because of the inherent limitations associated with use of primary DA neurons for biochemical analyses, we employed a well-characterized ventral mesencephalon DA neuroblastoma cell line (MN9D) for our studies. We found that stable over-expression of RGS10 rendered them resistant to TNF-induced cytotoxicity; whereas MN9D cells expressing mutant RGS10-S168A (which is resistant to phosphorylation by protein kinase A (PKA) at a serine residue that promotes its nuclear translocation) showed similar sensitivity to TNF as the parental MN9D cells. Using biochemical and pharmacological approaches, we identified protein kinase A (PKA) and the downstream phospho-cAMP response element-binding (CREB) signaling pathway (and ruled out ERK 1/2, JNK, and NFkB) as key mediators of the neuroprotective effect of RGS10 against inflammatory stress.
Keywords: RGS10, MN9D, dopaminergic, CREB, neuroinflammation, TNF
Introduction
Progressive loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc) gives rise to the motor deficits of Parkinson’s disease (PD), an age-associated neurodegenerative disorder that affects approximately 1% of the population between the ages of 65–70 years of age and 4 – 5% of 85-year old individuals (Tanner and Aston 2000) (Forno 1996). Although the exact mechanisms that lead to degeneration of nigral DA neurons have not been identified, oxidative stress, neuroinflammation, accumulation of abnormal or mis-folded proteins, enhanced activation of pro-apoptotic pathways, and mitochondrial dysfunction are all hypothesized to lead to nigral DA neuron dysfunction and ultimately death (Hirsch et al. 1997; Hirsch et al. 1999; Jenner and Olanow 1998). Therefore, identifying important molecular regulators of DA neuron responses to insults is pre-requisite for development of neuroprotective strategies that could exert disease-modifying effects if advanced to the clinic.
Regulator of G-protein signaling (RGS) proteins belong to a large family of proteins that function as GTPase activating proteins (GAPs) at Gα subunits. RGS proteins are negative regulators of G-protein signaling by accelerating GTP hydrolysis, a step that promotes deactivation of the G-protein and its G-protein coupled receptor (GPCR) (Berman et al. 1996; Ross and Wilkie 2000; Siderovski et al. 1999). There are more than thirty functional RGS genes sub-divided into six families expressed in mice and humans (Ross and Wilkie 2000; Zheng et al. 1999). RGS proteins differ widely in their size and contain a variety of structural domains and motifs that regulate their activity and determine regulatory binding partners (Ross and Wilkie 2000; Zheng et al. 1999). Recent studies have shown that RGS proteins are involved in CNS disorders. Abnormal RGS4 function has been implicated in schizophrenia (Ding and Hegde 2009; Mirnics et al. 2001; Morris et al. 2004; Prasad et al. 2005; Talkowski et al. 2006; Williams et al. 2004), anxiety (Leygraf et al. 2006), and Alzheimer’s disease (Emilsson et al. 2006; Muma et al. 2003), and the striatal-enriched RGS9-2 has been implicated in PD-related motor abnormalities (Gold et al. 2007) and in regulation of opiate analgesia in the dorsal horn (Papachatzaki et al. 2011) and striatum (Psifogeorgou et al. 2011). Polymorphisms in the RGS10 gene have also been reported in a cohort of Japanese schizophrenia patients (Hishimoto et al. 2004) and the modulation of both RGS4 and RGS10 by acute and chronic electroconvulsive seizures has been demonstrated in rat brain (Gold et al. 2002). In humans, genetic susceptibility loci for age-related maculopathy (ARM), a photoreceptor degenerative disease associated with microgliosis, map to the same region as the RGS10 gene on the human chromosome 10q26 (Jakobsdottir et al. 2005; Schmidt et al. 2006), suggesting that loss of RGS10 may predispose an organism to neurodegenerative disease. At 20-kDa, RGS10 is one of the smallest RGS proteins and a member of the R12 subfamily (Ross and Wilkie 2000; Sierra et al. 2002); it is abundantly expressed in the immune system and in a broad range of brain regions including the hippocampus, striatum, and dorsal Raphe (Gold et al. 1997). Although RGS10 protein expression is detectable in a number of subcellular compartments in mouse neurons and microglia (Waugh et al. 2005), the physiological function of RGS10 in DA neurons is unknown. Although phosphorylation of RGS10 by the cAMP-dependent protein kinase (PKA) at Ser-168 induces translocation of RGS10 from the plasma membrane and cytosol into the nucleus (Burgon et al. 2001), it is not known whether RGS10 regulates gene transcription or other nuclear processes. Previously, we reported that RGS10-null mice display increased microglial burden in the CNS and exposure to chronic systemic inflammation induced degeneration of nigral DA neurons (Lee et al. 2008), a parkinsonian phenotype. In addition to demonstrating the presence of RGS10-positive microglia in the ventral midbrain, those studies revealed the presence of RGS10-immunoreactivity in the soma and nuclei of tyrosine hydroxylase (TH)-positive nigral DA neurons. Our most recent study identified RGS10 as a negative regulator of NF-κB-dependent inflammatory factor production in activated microglia, suggesting a novel role for RGS10 in regulation of inflammatory gene transcription (Lee et al. 2011). Given that RGS10 is also expressed in various neuronal populations in the mammalian brain, the purpose of these studies was to determine the role of RGS10 as a direct regulator of DA neurons during periods of inflammatory stress by investigating the extent to which modulation of neuronal RGS10 activity could afford neuroprotective effects against neurotoxin-induced degeneration.
Methods
Materials
Constructs encoding wild type human RGS10 and SA mutant RGS10 were generously provided by Dr. Patrick Burgon at The University of Ottawa. Both constructs were subcloned into a pcDNA3.1 plasmid containing FLAG sequences at the 5’ end using the XbaI and BamH1 restriction site. Inhibitors including H89 and SB203580 were purchased from Cell Signaling Technology. Recombinant mouse (rm) Tumor Necrosis Factor (TNF) was purchased from R&D Systems (Minneapolis, MN).
Cell Culture
The murine clonal hybrid cell line MN9D was developed by A. Heller and colleagues by somatic cell fusion of rostral mesencephalic tegmentum from E14 mice and the murine neuroblastoma cell line N18TG2 (Choi et al. 1991). MN9D cells were grown in DMEM (Sigma-Aldrich) supplemented with 10 % Fetal Clone III (Hyclone) and 1 % penicillin/streptomycin. Cells were serially passaged when they reached 70 % confluence. To induce terminal neuron-like differentiation of MN9D cells, cells were incubated with 5 mM valproic acid (Sigma-Aldrich) in N2 (Invitrogen)-supplemented serum-free DMEM (Sigma-Aldrich) for 3 days.
Quantitative Real-Time Polymerase Chain Reaction (QPCR)
QPCR was performed as previously described (Kurrasch et al. 2004). Briefly, MN9D cells were plated at a density of 300,000 cells per plate on a 6-well plate. Cells were neurally differentiated as previously described (Lee et al. 2011) for 48–72 hrs and then treated with different concentrations of TNF as indicated times. Total RNA was isolated from cells in culture using the RNeasy isolation kit (Qiagen), treated with DNaseI, and reverse transcribed using Superscript II RNase H- reverse transcriptase (Invitrogen). QPCR was performed using SYBR Green in 384-well format using an ABI Prism 7900HT Fast Detection System (Applied Biosystems, Foster City). Oligonucleotide primers for QPCR were obtained from Integrated DNA Technologies (Coralville). RGS10 primer sequences (available upon request) were validated and used for gene amplification. Levels of mRNA expression were normalized to those of the mouse house-keeping genes cyclophilin B and GAPDH. Values represent the mean value of triplicate samples +/− SEM. Data are representative of at least two independent experiments.
Immunocytochemistry
Stably transfected MN9D cell lines expressing WT RGS10 or SA mutant RGS10 were plated at a density of 50,000 cells per well in a 4-well chamber. Cells were fixed and immunostained as described previously (Lee et al. 2008). Anti- FLAG epitope antibody (M2) (Sigma Aldrich) was used at 1:250 dilution and the nuclear counterstain bisbenzamide or Hoechst 33258 (Invitrogen) was used at 1:20,000. Anti-mouse Alexa fluor 594-conjugated secondary antibody (Invitrogen) was used at a dilution of 1:1000. All digital images were acquired on an Olympus BX61 fluorescence microscope with a CoolSnap CCD ES monochromatic camera and analyzed with Metamorph software.
SDS-PAGE and Western blot Analysis
Cells were lysed in a buffer containing 1 % NP-40, 10 mM Tris pH 7.4, 150 mM NaCl, 100 µg/mL PMSF, and protease inhibitor mix (Sigma) for 30 min on ice. Lysates were resuspended in 2X Laemmli sample buffer and loaded on precast 12 % SDS-PAGE gels (Bio-Rad), transferred onto PDVF membranes (Millipore), and probed with anti-cleaved-PARP1, cleaved-caspase 3, phospho-CREB, CREB, Bcl-2, phopho-p38MAPK, p38MAPK , phospho-JNK, JNK, phospho-ERK, ERK, phospho-p65 and p65 antibodies (Cell Signaling Technology) or anti-α-tubulin (1:1000) antibody (Santa Cruz Biotechnology) plus the appropriate HRP-conjugated secondary antibody (1:5000, Jackson ImmunoResearch Lab, West Grove, PA). Immunoreactive bands were visualized with SuperSignal West Femto HRP substrate (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s instructions and imaged on a Syngene G:Box Chemi gel documentation station (Frederick, MD). Membranes were stripped with 0.2 M glycine, 1 % SDS and 0.1 % Tween-20, pH 2.2 and re-probed as necessary.
Cell viability assays
Cell viability was measured by using the CellTiter 96 AQueous Assay reagent (Promega). This reagent utilizes the novel tetrazolium compound (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS) and the electron coupling reagent, phenazine methosulfate (PMS). MTS is chemically reduced by cells into formazan, which is soluble in tissue culture medium. The measurement of the absorbance of the formazan was performed in multi-titer 96-well plates at 492 nm during the last 2 to 4 hrs of a 2-day culture. Each experimental condition was performed in triplicate (or quadruplicate in the case of experiments involving differentiated MN9D cells) and three to four independent experiments were conducted to confirm the results.
Statistical Analyses
Differences treatments among the different groups were analyzed by two-way ANOVA followed by the Bonferroni post hoc test for p values significance. Differences treatments within the group were analyzed by one-way ANOVA followed by the Tukey’s post hoc test for p values significance. Values expressed are the group mean +/− SEM.
Results
TNF attenuates RGS10 expression in neurally differentiated dopaminergic (DA) cells
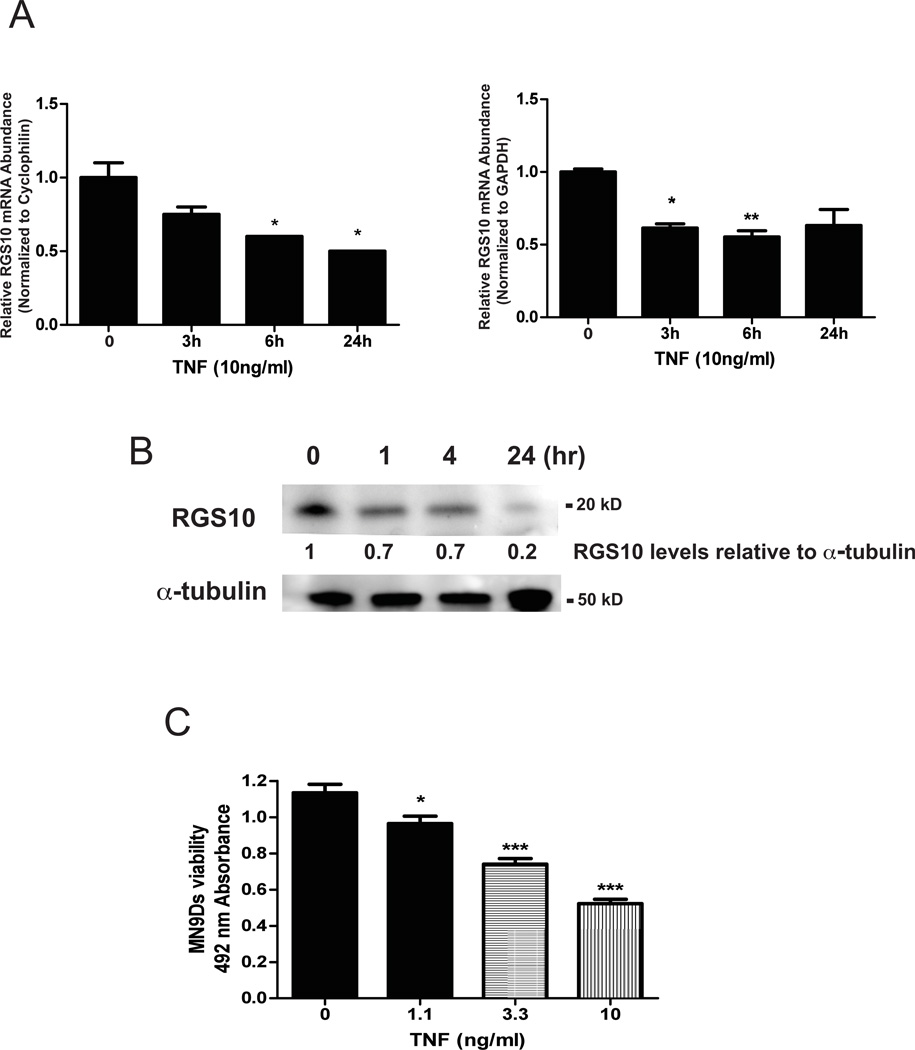
As our previous studies demonstrated that siRNA-mediated knockdown of RGS10 in MN9D cells increased their sensitivity to microglial-derived inflammatory mediators such as TNF (Lee et al. 2008), we hypothesized that inflammation leads to reduced neuronal RGS10 expression. Because of the inherent limitations associated with use of primary DA neurons for biochemical analyses, we employed a well-characterized ventral mesencephalon DA neuroblastoma cell line (MN9D) to test this hypothesis. We treated neurally differentiated ventral mesencephalon MN9D cells with TNF and measured RGS10 mRNA abundance by real-time PCR and RGS10 protein amounts by immunoblotting. (Fig. 1). TNF induced a time-dependent decrease in RGS10 mRNA H (Figure 1A) and decreased RGS10 protein amounts (Figure 1B). To determine whether TNF-induced RGS10 downregulation affected neuronal survival, we assayed the viability of MN9D cells treated with a range of TNF concentrations. Consistent with published studies, we observed dose-dependent cytotoxicity to TNF (Harms et al. 2012). Together, these data suggest that TNF-dependent down-regulation of RGS10 levels contributes to the vulnerability of neuronal DA cells to TNF-induced death and possibly other toxic insults.
Figure 1. TNF-induced down-regulation of RGS10 mRNA and protein in dopaminergic (DA) cells correlates with TNF-induced cytotoxicity.
MN9D cells were plated and neurally differentiated as described under Methods. Cells were treated with 10 ng/mL of TNF for the times indicated. A, Levels of RGS10 mRNA were measured by real-time quantitative PCR and normalized to one of two housekeeping genes GAPDH or cyclophilin. Values represent mean ± SEM. QPCR analysis was performed by one-way ANOVA followed by Tukey’s post hoc test, * denotes p < 0.05 and ** denotes p < 0.01 relative to the untreated sample. B, Levels of RGS10 protein were measured by western blot analysis. Bands immunoreactive for RGS10 were quantified by densitometry and normalized to α-tubulin as a protein loading control. Blots are representative of three independent experiments. C, MN9D cell viability was assessed using the MTS assay as described under Methods. Values represent mean ± SEM. One way ANOVA followed by Tukey’s post hoc test, * denotes p < 0.05 and *** denote p < 0.001 relative to the untreated sample.
Stable overexpression of RGS10 in differentiated dopaminergic (DA) MN9D cells reduces their vulnerability to TNF-induced cytotoxicity
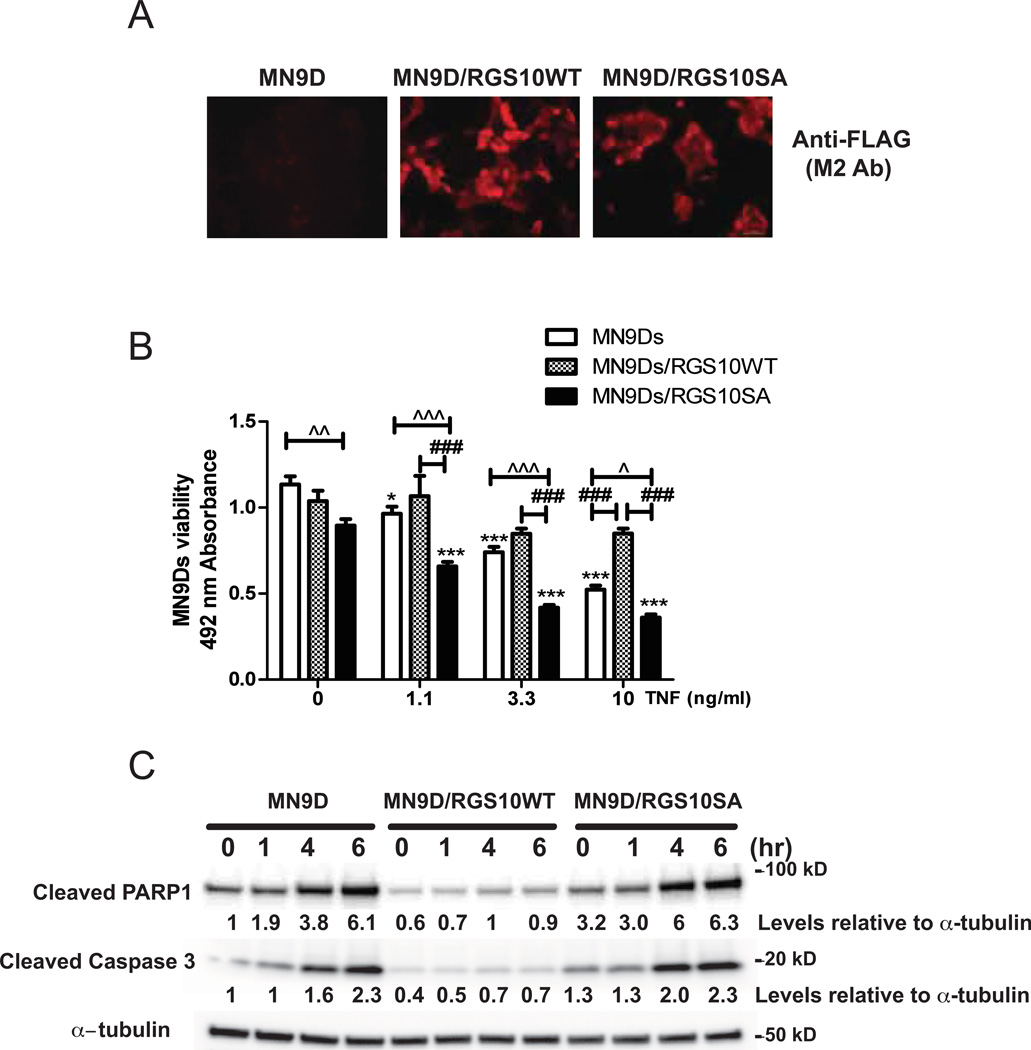
Based on the observations that down-regulation of RGS10 during TNF treatment correlated with TNF-induced cytotoxicity, we predicted that over-expression of RGS10 would protect MN9D cells against TNF-induced cytotoxicity. We generated MN9D cells lines stably expressing wild type (WT) RGS10 protein or the PKA phosphorylation-deficient RGS10 mutant (SA) (Burgon et al. 2001). Immunocytochemical analyses with an anti-FLAG (M2) antibody confirmed that MN9D cells were successfully transfected and expressed FLAG- RGS10 WT or FLAG- RGS10 SA (Figure 2A). In contrast to neurally differentiated parental MN9D cells, RGS10WT-overexpressing MN9D cells displayed significant resistance to TNF-induced toxicity. Interestingly, RGS10 SA MN9D cells showed similar or higher sensitivity to TNF than the parental MN9D cell line (Figure 2B). These data indicated that overexpression of WT RGS10 shields dopaminergic MN9D cells from TNF toxicity and that phosphorylation of RGS10 by PKA is required for such protection. Attempts to extend and confirm these findings in neuron enriched ventral mesencephalon cultures harvested from RGS10-null mice in which restoration of RGS10 expression via transduction of HSV-RGS10 (versus HSV-GFP as negative control) could be assessed for protective effects were technically unsuccessful due to the fact that HSV infection proved cytotoxic at even the lowest titers tested (data not shown).
Figure 2. Overexpression of RGS10 in DA cells attenuates TNF-induced caspase 3-dependent apoptotic signaling and protects DA cells against TNF-induced cytotoxicity.
A, Immunocytochemical detection of FLAG-tagged RGS10 protein in parental MN9D cells, MN9D cell lines stably expressing WT RGS10, or RGS10 SA mutant, B, TNF-induced cytotoxicity was measured in neurally differentiated parental MN9D cells or cell lines stably expressing WT RGS10 or RGS10 SA mutant using an MTS assay. Values represent mean ± SEM. One way ANOVA followed by Tukey’s post hoc test, * and *** denote significant differences between vehicle and TNF within the group at p < 0.05 or p < 0.001 respectively. #, ## or ### (Λ, ΛΛ or ΛΛΛ) denote significant differences between the groups (parental versus RGS10 WT group, RGS10 WT vs. RGS10 SA mutant group, or parental vs. RGS10 SA group) at p < 0.05, p < 0.01 or p < 0.001 respectively. Values shown are group means (n=6) ± S.E.M from one experiment representative of four independent experiments. C, Western blot analyses of cleaved PARP and caspase 3. Neurally differentiated MN9D cell lines stably expressing WT RGS10, or RGS10 SA mutant were treated with 10 ng/mL of TNF for 0, 1, 4 or 6 hrs as indicated. Densitometry and normalization to α-tubulin as a protein loading control was performed. Blots are representative of three independent experiments.
Since it is has been well-established that TNF can activate cell death cascades in various cell types by activating caspase signaling, we next examined whether RGS10 over-expressing MN9D cells displayed attenuated caspase activation upon TNF treatment. Immunoblot analyses revealed increased levels of cleaved PARP-1 and cleaved caspase 3 in parental MN9D cells or RGS10 SA overexpressing MN9D cells treated with TNF. In contrast, RGS10 WT overexpressing MN9D cells displayed significantly lower baseline and TNF-induced PARP-1 or caspase 3 cleavage (Figure 2C). These data strongly suggest that RGS10 enhances neuronal survival by inhibiting caspase-dependent apoptosis. As the mutant RGS10 SA, which has intact GAP activity but is resistant to phosphorylation by PKA, (Burgon et al. 2001) did not protect MN9D cells against TNF toxicity, we hypothesized that RGS10 mitigates neuronal apoptosis pathways through PKA-dependent mechanism(s).
Neuroprotective effect of RGS10 is mediated by phospho-cAMP response element-binding (pCREB) pathway
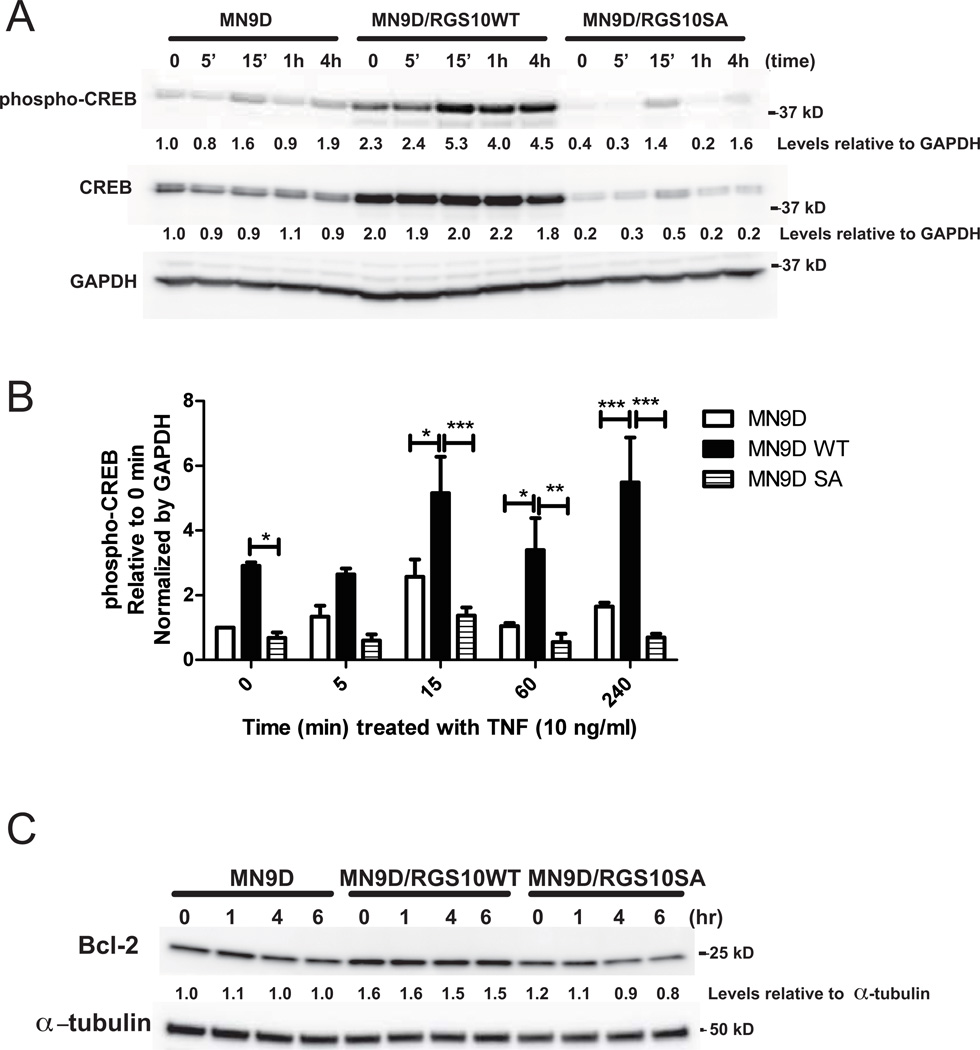
To evaluate the extent to which RGS10 abundance influences signaling downstream of PKA, we measured CREB levels and CREB phosphorylation (at Ser 133) in parental and RGS10-expressing MN9D cells. While treatment of parental MN9D cells with TNF induced very modest phosphorylation of CREB by 15 min, levels of CREB phosphorylation as well as total CREB were much higher in RGS10WT overexpressing MN9D cells relative to parental MN9D cells and RGS10SA mutant overexpressing MN9D cells (Figures 3A and B). Consistent with enhancement of CREB-dependent gene transcription by RGS10, bcl-2 abundance was significantly higher in RGS10WT over-expressing MN9Ds relative to the parent or RGS10SA MN9D cell lines in the presence or absence of TNF (Figure 3C). Importantly, these findings indicate that RGS10 WT clearly affects both CREB levels and CREB activation (phospho-CREB) independent of the changes in total CREB levels, as the difference in phospho-CREB activation observed after RGS10WT overexpression compared to the parental or RGS10SA persist when phospho-CREB is normalized to total CREB (data not shown).
Figure 3. Neuroprotective effect of RGS10 in DA cells is associated with enhanced phospho-CREB signaling and increased levels of the pro-survival protein Bcl-2.
Neurally differentiated parental MN9D cells or cell lines stably expressing WT or SA mutant RGS10 were treated with 10 ng/mL of TNF for 0, 5 min, 15 min, 1, or 4 hrs. A, Levels of phospho-CREB and total CREB by western blot analysis. Densitometry and normalization to GAPDH as a protein loading control was performed. Blots are representative of 3 independent experiments. B, Relative levels of phospho-CREB in parental MN9D cells or cell lines stably expressing WT or SA mutant RGS10 under control or TNF (10 ng/mL) -stimulated conditions. Values represent mean ± SEM. One way ANOVA followed by Tukey’s post hoc test, *, **, and *** denote significant differences between the groups indicated at p < 0.05, p < 0.01, or p < 0.001 respectively. C, Levels of Bcl-2 protein by western blot on parental or stably expressing WT RGS10 or SA mutant RGS10 MN9D cells treated with TNF (10 ng/mL) for 0, 1, 4 or 6 hrs. Densitometry and normalization to α-tubulin as a protein loading control was performed. Blots are representative of three independent experiments.
Neuroprotective effects of RGS10 in dopaminergic (DA) cells is abolished by inhibiting PKA activation
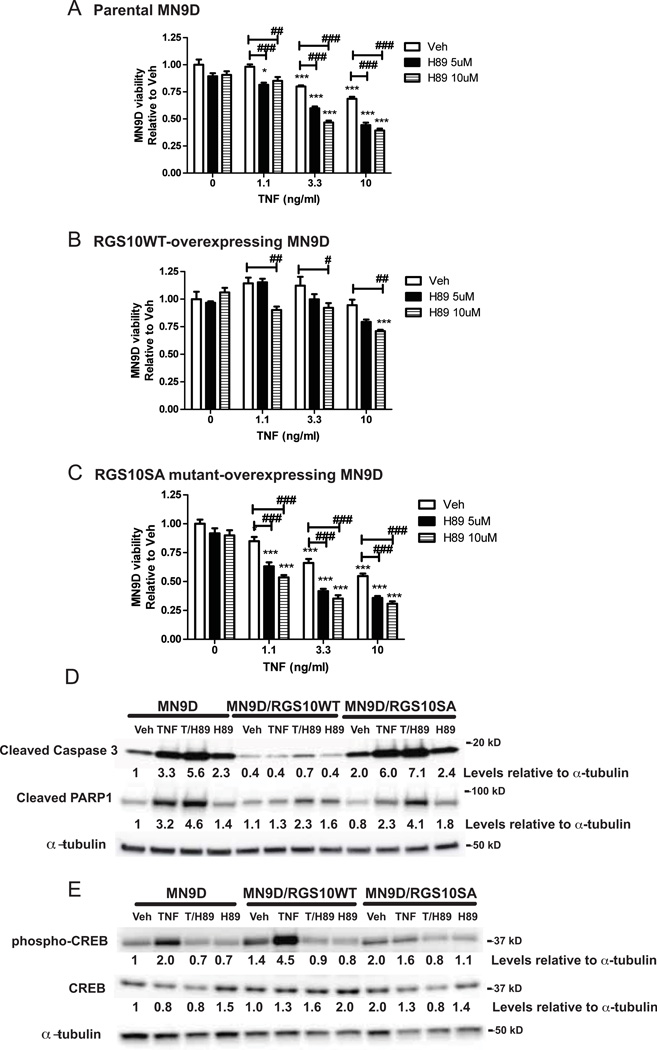
PKA-induced phosphorylation of cAMP response element-binding (CREB) protein is known to promote neuronal survival and counteract the pro-apoptotic effects of TNF by eliciting expression of several genes including brain derived neurotrophic factor and bcl-2 (Bieganska et al. 2011; Krakstad et al. 2004; Mueller et al. 2005). We investigated whether pharmacological inhibition of PKA-CREB signaling could ablate the protective effects of RGS10 WT over-expression in MN9D cells by pre-treating cells with H89, a specific PKA inhibitor, prior to TNF exposure. Inhibition of PKA signaling with H89 increased the sensitivity of parental MN9D (Figure 4A) and RGS10 SA overexpressing MN9D cells (Figure 4C) to TNF-induced cytotoxicity. Similarly, H89 treatment dose-dependently increased the sensitivity to TNF-induced loss of viability in RGS10 WT over-expressing MN9D cells (Figure 4B). Accordingly, H89 pre-treatment of MN9D cells also led to increased TNF-dependent caspase 3 and PARP-1 cleavage (Figure 4D) as well as decreased TNF-dependent CREB phosphorylation (Figure 4E). These data strongly implicate PKA signaling in mediating the anti-apoptotic effect of RGS10 in DA cells via activation of CREB phosphorylation.
Figure 4. Neuroprotective effect of RGS10 in DA cells requires activation of cAMP-dependent protein kinase (PKA).
MN9D cells were neurally differentiated then treated with TNF (1.1, 3.3 or 10 ng/mL) for 48 hrs in the presence or absence of pre-treatment of H89 (5 or 10 µM) for 45 min. Cell viability was assessed using the MTS assay on A, parental MN9D viability, B, MN9D cells stably over-expressing WT RGS10 viability, and C, MN9D cells stably over-expressing RGS10 SA mutant. Values represent mean ± SEM. One way ANOVA followed by Tukey’s post hoc test, *, ** or *** denote significant differences between vehicle and TNF treatments within the group at p < 0.05, p < 0.01 or p < 0.001 respectively. #, ## or ### denote significant differences between the H89 treatments groups at p < 0.05, p < 0.01 or p < 0.001 respectively. Values shown are group means (n=6) ± S.E.M from one experiment representative of three independent experiments. D, Effect of PKA inhibition on TNF-induced caspase 3 and PARP1 cleavage by western blot analysis. Parental MN9D cells or MN9D cell lines stably over-expressing WT RGS10 or RGS10SA mutant were neurally differentiated then treated with TNF (10 ng/mL) in the presence or absence of H89 (10 µM). E, Effect of PKA inhibition on TNF-induced phospho-CREB and CREB levels by western blot analysis. Cells were prepared as described above. Densitometry and normalization to α-tubulin as a protein loading control was performed. All the western blots are representative of three independent experiments.
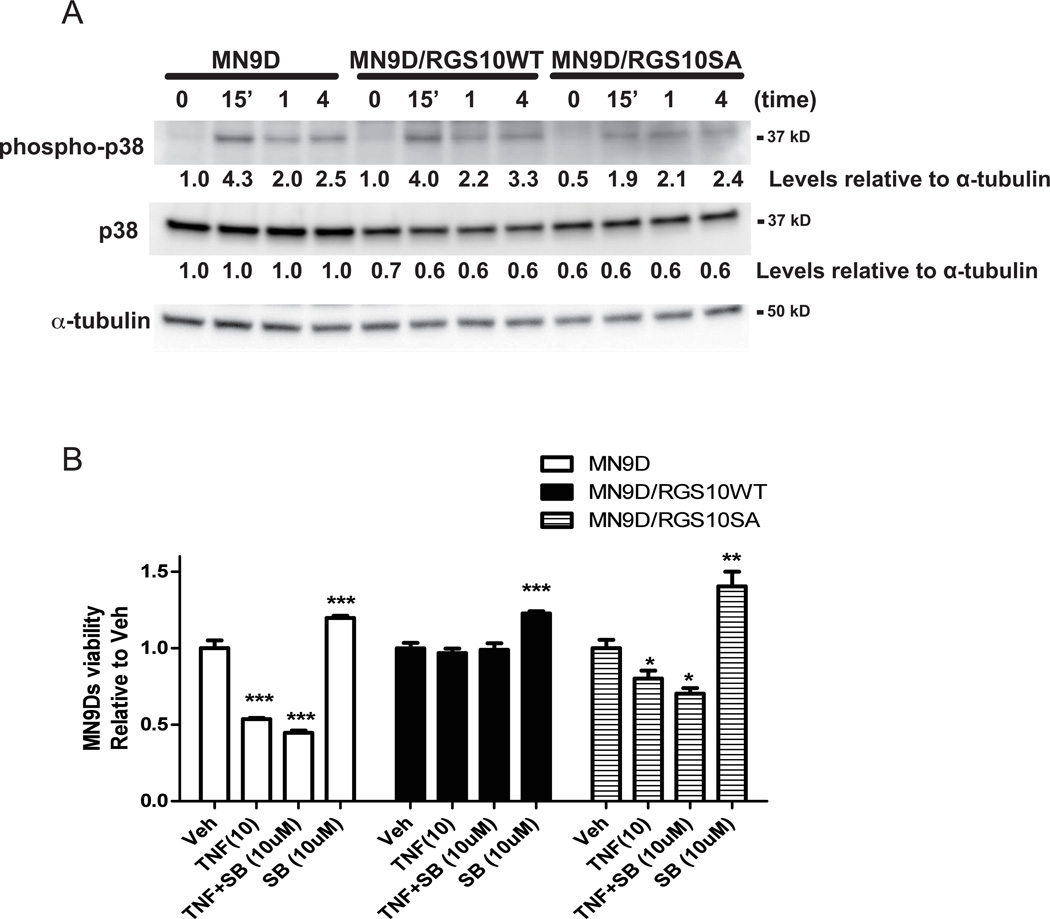
Neuroprotective effects of RGS10 in dopaminergic (DA) cells does not involve modulation of p38MAPK pathway
Our results so far indicated that RGS10 confers protection from neurotoxic stress by modulating PKA-dependent CREB activity and expression. TNF also elicits CREB activation through stimulation of p38 MAP kinase, which in turn leads to direct phosphorylation of CREB by the kinase MSK-1 (Deak et al. 1998; Gustin et al. 2004). We investigated whether RGS10 also affected TNF-dependent activation of p38 in MN9D cells. TNF treatment induced rapid and robust phosphorylation of p38 in both parental MN9D cell lines and the stable cell lines over-expressing WT or SA mutant equivalently (Figure 5A). Next, we pre-incubated MN9D cells with the pharmacological inhibitor SB203580 to determine whether inhibition of the p38MAPK pathway affected cell survival during TNF exposure. While inhibition of p38 MAPK activity by pretreatment of cells with SB203580 enhanced cell viability in the absence of TNF, it did not robustly enhance TNF-induced cytotoxicity in parental MN9D or RGS10 SA over-expressing MN9D cells nor did it affect the ability of RGS10 WT to confer resistance to TNF-induced cytotoxicity (Figure 5B). These results indicate that RGS10 does not regulate TNF-induced p38 MAPK activation, nor is p38 MAPK activation critical for TNF-mediated neurotoxicity.
Figure 5. Neuroprotective effect of RGS10 in DA cells does not require modulation of p38 MAPK signaling.
A, Levels of phospho-p38MAPK and total p38 MAPK upon TNF-treatments. Neurally differentiated parental MN9D cells or cell lines stably expressing WT or SA mutant RGS10 were treated with 10 ng/mL of TNF for 0, 15 min, 1 or 4 hrs as indicated. Densitometry and normalization to α-tubulin as a protein loading control was performed. Blots are representative of three independent experiments. B, Effect of p38 inhibition on TNF-induced cytotoxicity. Neurally differentiated MN9D cells were treated with TNF (10 ng/mL) for 48 hrs in the presence or absence of pretreatment of SB203580 (10 µM) for 45 min. Cell viability was assessed using the MTS assay. Values represent mean ± SEM. One way ANOVA followed by Tukey’s post hoc test, *, ** or *** denote significant differences between vehicle and TNF treatments within the group at p < 0.05, p < 0.01 or p < 0.001 respectively.
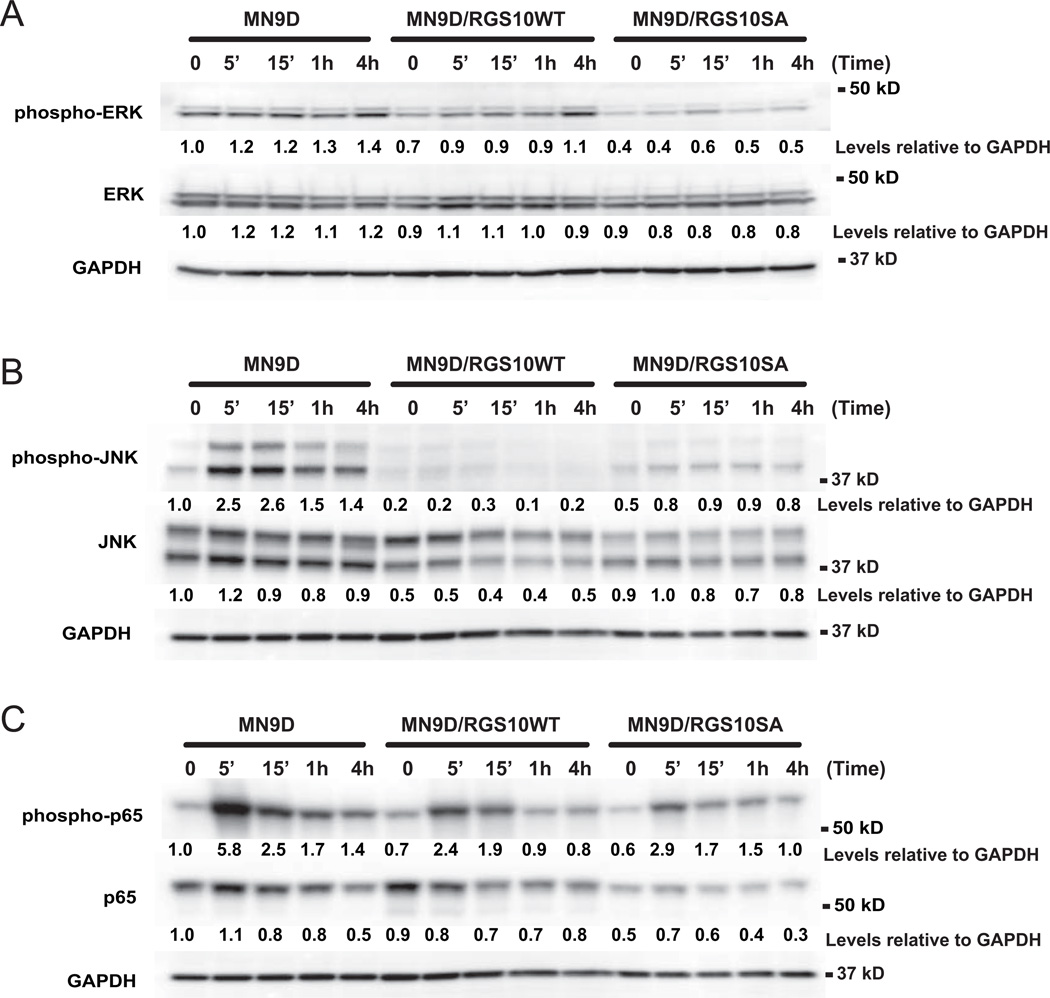
To extend these observations, we also investigated the extent to which overexpression of WT versus SA mutant RGS10 differentially modulated ERK 1/2 and JNK pathway activation. TNF treatment did not induce significant changes in phosphorylation of ERK 1/2 in neurally differentiated parental MN9D or WT RGS10 overexpressing MN9D cells while RGS10 SA mutant cells did have slightly attenuated phospho-ERK 1/2 levels (Figure 6A); no differences were observed in total ERK 1/2 levels. Therefore, these data do not support a role for modulation of ERK 1/2 phosphorylation by RGS10 in its neuroprotective effects against TNF-induced neurotoxicity. With regards to JNK pathway activation, as expected we found that TNF treatment induced robust phosphorylation of JNK and this may be an important mechanism by which TNF compromises viability in dopaminergic neurons. However, because phospho-JNK was similarly attenuated by overexpression of WT and SA mutant RGS10, these responses do not correlate with selective protection from TNF-induced cytotoxicity by WT RGS10 overexpression (Figure 6B). In addition, since mutant RGS10 overexpressing cells have decreased viability despite reduced activation of JNK signaling, attenuation of phospho-JNK is not sufficient to promote survival of dopaminergic cells treated with TNF. Lastly, given that TNF signaling can activate the NFkB pathway in various cell types and previous work from our group demonstrated strong negative regulation of this pathway by RGS10 in microglial cells, we measured phospho-p65 and total p65 in parental, WT RGS10 and mutant RGS10 cell lines. We found that phosphorylation patterns of p65 induced by TNF in MN9D cells were similar between parental MN9D cells and WT RGS10 overexpressing MN9D cells (Figure 6C). Therefore, we conclude that modulation of NFkB signaling by RGS10 does not mediate the neuroprotective role of RGS10 against TNF-induced neurotoxicity in dopaminergic cells. Taken together, biochemical, pharmacologic and metabolic assay data lead us to conclude that RGS10 enhanced DA cell survival by strengthening PKA-CREB-Bcl2 signaling cascades and attenuating caspase-dependent apoptotic signaling induced by TNF.
Figure 6. Neuroprotective effects of WT RGS10 overexpression do not correlate with modulation of NFkB, ERK 1/2, or JNK pathway activation.
Neurally differentiated parental MN9D cells or cell lines stably expressing WT or SA mutant RGS10 were treated with 10 ng/mL of TNF for 0, 5 min, 15 min, 1 or 4 hrs as indicated to quantify levels of A, phospho-ERK 1/2 and total ERK 1/2 B, phospho-JNK and total JNK C, phospho-p65RelA and total p65. Densitometry and normalization to GAPDH as a protein loading control was performed. Blots are representative of three independent experiments.
Discussion
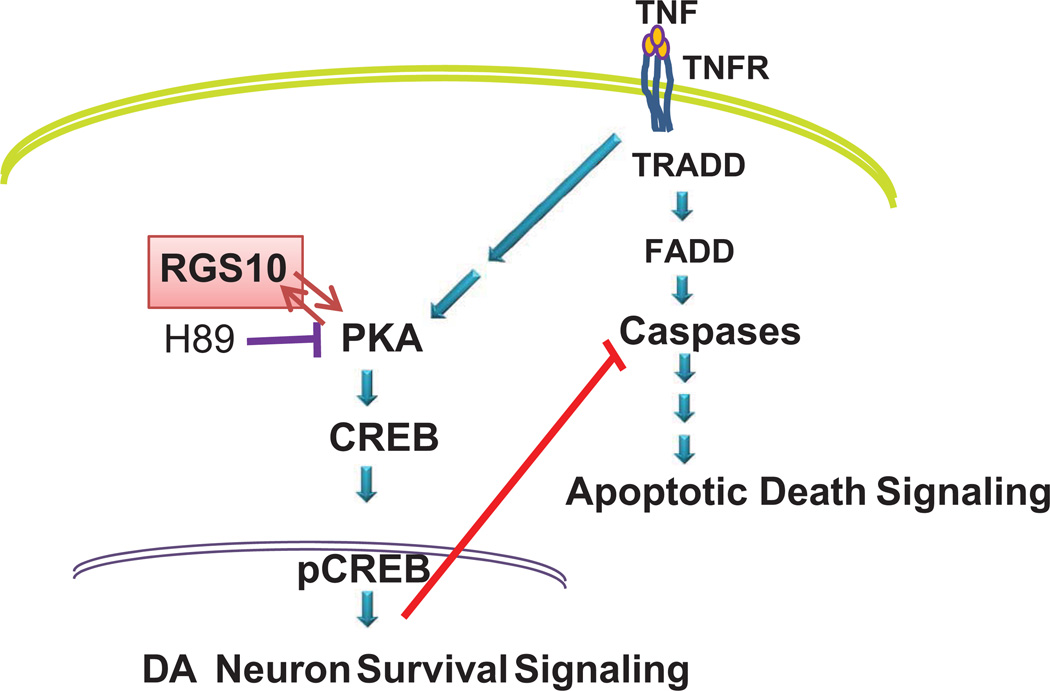
Nigral DA neurons are exquisitely sensitive to inflammatory stimuli and to soluble TNF in particular because of their high levels of expression of TNF receptor 1 (Aloe and Fiore, 1997; McGuire et al., 2001; Gayle et al., 2002; Carvey et al., 2005), the canonical death receptor (Tartaglia et al., 1993). Direct evidence that TNF-dependent inflammatory stress contributes to the progressive loss of nigral DA neurons in rodent models of parkinsonism derives from studies demonstrating that inhibition of soluble TNF signaling can significantly attenuate endotoxin and oxidative neurotoxin-induced degeneration (Harms et al. 2011; McCoy et al. 2006; McCoy et al. 2008). The studies presented here support a molecular model in which reciprocal interactions between RGS10 and PKA defend cells with a dopaminergic fate against inflammatory stress by augmenting CREB signaling and attenuating caspase-dependent apoptotic cascades downstream of TNF receptor activation. Moreover, activation of TNF signaling in neurally differentiated DA cells result in decreased cellular levels of RGS10 and correlated with TNF-induced cytotoxicity, raising the interesting possibility that loss of neuronal RGS10 may contribute to vulnerability to soluble TNF. In support of this idea, previous work from our group demonstrated that siRNA-mediated knockdown of RGS10 in DA cells rendered them more susceptible to TNF-induced cytotoxicity (Lee et al. 2008). We now directly demonstrate that stable overexpression of RGS10 in neurally differentiated DA cells significantly attenuated TNF-dependent cytotoxicity by inhibiting PARP-1 and caspase 3 cleavage while enhancing PKA/CREB pro-survival signaling, including levels of bcl-2. Surprisingly, changes in the activation state of p38MAPK had modest effects on TNF-induced cytotoxicity and no effect on the protective activity of RGS10 overexpression, suggesting p38MAPK may play a secondary role in mediating TNF-induced cytotoxicity in DA cells and neurons. Mechanistically, our biochemical data indicate that a specific serine residue previously shown to be phosphorylated by PKA (Burgon et al. 2001) is critical for mediating the enhanced CREB signaling downstream as RGS10 SA mutant protein cannot mimic the effects of WT RGS10. Consistent with this finding, selective pharmacological inhibition of PKA abolished the neuroprotective effects of RGS10 WT altogether in DA cells treated with TNF. It is important to note that these biochemical and functional studies could not have been performed in primary DA neurons because DA neurons only make up 4% of all neurons in a ventral mesencephalon primary culture and this amount is insufficient for population studies. More importantly, techniques needed to manipulate the levels of RGS10 in these neurons via transfection or electroporation of cDNA or viral transduction is highly inefficient and/or cytotoxic. In summary, our data suggest a molecular model by which reciprocal interactions between RGS10 and PKA promote DA neuron survival by potentiating CREB signaling and attenuating caspase-dependent apoptotic cascades downstream of TNF receptor activation (Figure 7). Given that RGS10 can directly interact with PKA as a substrate for phosphorylation at Ser 168, it will be of interest to determine how this interaction contributes to functional modulation of downstream PKA-dependent CREB signaling by RGS10.
Figure 7. Model for mechanism by which RGS10 exerts anti-apoptotic and pro-survival effects in DA cells.
Reciprocal interactions between RGS10 and PKA promote DA neuron survival by potentiating CREB signaling and attenuating caspase-dependent apoptotic cascades downstream of TNF receptor activation.
The primary mechanism by which RGS proteins regulate cellular responses is through their GTPase activating protein (GAP) activity at Gα subunits, which results in reduced G-protein coupled receptor (GPCR) signaling (Burchett 2003; Chatterjee and Fisher 2000; Lee et al. 2011; Ross and Wilkie 2000; Sierra et al. 2002). However, several RGS proteins, including RGS10, have been shown to traffic to the nucleus (Burchett 2003; Chatterjee and Fisher 2000; Lee et al. 2011). Their extra-cytoplasmic roles, however, remain unclear and underexplored primarily because most GPCRs are localized at the plasma membrane (Huang and Fisher 2009). Recent work from our group revealed that nuclear enrichment of RGS10 in microglia in response to inflammatory stimuli enables it to negatively regulate NFκB, thereby limiting production of microglial-derived inflammatory mediators including soluble Tumor Necrosis Factor (TNF) and lessening the toxic effects on dopaminergic (DA) neurons. Here, we provide compelling evidence to support an important direct role for RGS10 in survival responses of DA cells during inflammatory stress. The significance or our findings are three-fold. First, the results of our studies implicate endogenous RGS10 as an important protective factor in DA neurons against toxic insults by virtue of its anti-apoptotic and pro-survival activities. Second, our studies raise the possibility that selective modulation of RGS10 levels or bioactivity in neurons may permit neuron-specific potentiation of PKA/CREB-dependent gene transcription to promote neuronal survival without non-selectively activating CREB-dependent proliferation in other cell types. Lastly, together with previous work in which we demonstrated an important anti-inflammatory role for RGS10 in microglia, these novel findings raise the interesting possibility that boosting levels or RGS10 activity in both ventral midbrain microglia and DA neurons may be a novel and doubly effective therapeutic modality to protect DA neurons from chronic inflammatory stress and provide strong rationale for investigating this possibility in vivo in pre-clinical models of PD. The growing number of elderly individuals in the U.S. and worldwide at risk for development of chronic neurodegenerative diseases like PD makes it imperative that biomedical science identify feasible and efficacious ways to lessen the detrimental effects of chronic inflammatory stress in the brain to prevent, delay onset, or ameliorate age-related neurodegenerative diseases. Enhancement of RGS10 levels in ventral midbrain DA neurons via gene transfer may represent a novel approach to achieve neuroprotection in vivo and will be investigated using rodent models of progressive PD-like neurodegeneration.
Acknowledgements
This work was supported by a Target Validation grant from the Michael J. Fox Foundation for Parkinson’s Research (M.G.T.), a pilot grant from Emory University Parkinson’s Disease Collaborative Environmental Research Center Development Program (J.-K.L.), and Grant R01NS072467-02 (M.G.T.) from the NINDS at the National Institutes of Health. We would like to thank Patrick Burgon at The University of Ottawa for kindly providing the RGS10-SA mutant cDNA, Lynn Malone for technical help with cell culture, and members of the Tansey Lab for useful discussions.
Abbreviations
- RGS10
Regulator of G-protein Signaling-10
- PKA
cyclic AMP-dependent protein kinase A
- CREB
cyclic AMP response element binding
- GAP
GTPase activating protein
- TNF
tumor necrosis factor
Footnotes
Conflict of Interest Statement
The authors state that they have no conflict of interests to declare.
Contributor Information
Jae-Kyung Lee, Email: jaekyung.lee@emory.edu.
Jaegwon Chung, Email: jaegwon.chung@emory.edu.
Kirk M. Druey, Email: kdruey@niaid.nih.gov.
Malú G. Tansey, Email: malu.tansey@emory.edu.
References Cited
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86(3):445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Bieganska K, Figiel I, Gierej D, Kaczmarek L, Klejman A. Silencing of ICERs (Inducible cAMP Early Repressors) results in partial protection of neurons from programmed cell death. Neurobiol Dis. 2011 doi: 10.1016/j.nbd.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Burchett SA. In through the out door: nuclear localization of the regulators of G protein signaling. J Neurochem. 2003;87(3):551–559. doi: 10.1046/j.1471-4159.2003.02047.x. [DOI] [PubMed] [Google Scholar]
- Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J Biol Chem. 2001;276(35):32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- Chatterjee TK, Fisher RA. Cytoplasmic, nuclear, and golgi localization of RGS proteins. Evidence for N-terminal and RGS domain sequences as intracellular targeting motifs. J Biol Chem. 2000;275(31):24013–24021. doi: 10.1074/jbc.M002082200. [DOI] [PubMed] [Google Scholar]
- Choi HK, Won LA, Kontur PJ, Hammond DN, Fox AP, Wainer BH, Hoffmann PC, Heller A. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552(1):67–76. doi: 10.1016/0006-8993(91)90661-e. [DOI] [PubMed] [Google Scholar]
- Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17(15):4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Hegde AN. Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biol Psychiatry. 2009;65(6):541–545. doi: 10.1016/j.biopsych.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Emilsson L, Saetre P, Jazin E. Low mRNA levels of RGS4 splice variants in Alzheimer's disease: association between a rare haplotype and decreased mRNA expression. Synapse. 2006;59(3):173–176. doi: 10.1002/syn.20226. [DOI] [PubMed] [Google Scholar]
- Forno LS. Neuropathology of Parkinson's disease. J Neuropathol Exp Neurol. 1996;55(3):259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Heifets BD, Pudiak CM, Potts BW, Nestler EJ. Regulation of regulators of G protein signaling mRNA expression in rat brain by acute and chronic electroconvulsive seizures. J Neurochem. 2002;82(4):828–838. doi: 10.1046/j.1471-4159.2002.01002.x. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Hoang CV, Potts BW, Porras G, Pioli E, Kim KW, Nadjar A, Qin C, LaHoste GJ, Li Q, et al. RGS9-2 negatively modulates L-3,4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson's disease. J Neurosci. 2007;27(52):14338–14348. doi: 10.1523/JNEUROSCI.4223-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17(20):8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin JA, Pincheira R, Mayo LD, Ozes ON, Kessler KM, Baerwald MR, Korgaonkar CK, Donner DB. Tumor necrosis factor activates CRE-binding protein through a p38 MAPK/MSK1 signaling pathway in endothelial cells. Am J Physiol Cell Physiol. 2004;286(3):C547–C555. doi: 10.1152/ajpcell.00332.2002. [DOI] [PubMed] [Google Scholar]
- Harms AS, Barnum CJ, Ruhn KA, Varghese S, Trevino I, Blesch A, Tansey MG. Delayed dominant-negative TNF gene therapy halts progressive loss of nigral dopaminergic neurons in a rat model of Parkinson's disease. Mol Ther. 2011;19(1):46–52. doi: 10.1038/mt.2010.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms AS, Lee JK, Nguyen TA, Chang J, Ruhn KM, Trevino I, Tansey MG. Regulation of microglia effector functions by tumor necrosis factor signaling. Glia. 2012;60(2):189–202. doi: 10.1002/glia.21254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch EC, Faucheux B, Damier P, Mouatt-Prigent A, Agid Y. Neuronal vulnerability in Parkinson's disease. J Neural Transm Suppl. 1997;50:79–88. doi: 10.1007/978-3-7091-6842-4_9. [DOI] [PubMed] [Google Scholar]
- Hirsch EC, Hunot S, Faucheux B, Agid Y, Mizuno Y, Mochizuki H, Tatton WG, Tatton N, Olanow WC. Dopaminergic neurons degenerate by apoptosis in Parkinson's disease. Mov Disord. 1999;14(2):383–385. doi: 10.1002/1531-8257(199903)14:2<383::aid-mds1037>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Hishimoto A, Shirakawa O, Nishiguchi N, Aoyama S, Ono H, Hashimoto T, Maeda K. Novel missense polymorphism in the regulator of G-protein signaling 10 gene: analysis of association with schizophrenia. Psychiatry Clin Neurosci. 2004;58(5):579–581. doi: 10.1111/j.1440-1819.2004.01303.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Fisher RA. Chapter 5 nuclear trafficking of regulator of g protein signaling proteins and their roles in the nucleus. Prog Mol Biol Transl Sci. 2009;86:115–156. doi: 10.1016/S1877-1173(09)86005-5. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner P, Olanow CW. Understanding cell death in Parkinson's disease. Ann Neurol. 1998;44(3 Suppl 1):S72–S84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- Krakstad C, Christensen AE, Doskeland SO. cAMP protects neutrophils against TNF-alpha-induced apoptosis by activation of cAMP-dependent protein kinase, independently of exchange protein directly activated by cAMP (Epac) J Leukoc Biol. 2004;76(3):641–647. doi: 10.1189/jlb.0104005. [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time PCR measurement of regulators of G-protein signaling (RGS) mRNA levels in mouse tissues. Methods in Enzymology. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- Lee JK, Chung J, McAlpine FE, Tansey MG. Regulator of G-Protein Signaling-10 Negatively Regulates NF-{kappa}B in Microglia and Neuroprotects Dopaminergic Neurons in Hemiparkinsonian Rats. J Neurosci. 2011;31(33):11879–11888. doi: 10.1523/JNEUROSCI.1002-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, McCoy MK, Harms AS, Ruhn KA, Gold SJ, Tansey MG. Regulator of G-protein signaling 10 promotes dopaminergic neuron survival via regulation of the microglial inflammatory response. J Neurosci. 2008;28(34):8517–8528. doi: 10.1523/JNEUROSCI.1806-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leygraf A, Hohoff C, Freitag C, Willis-Owen SA, Krakowitzky P, Fritze J, Franke P, Bandelow B, Fimmers R, Flint J, et al. Rgs 2 gene polymorphisms as modulators of anxiety in humans? J Neural Transm. 2006;113(12):1921–1925. doi: 10.1007/s00702-006-0484-8. [DOI] [PubMed] [Google Scholar]
- McCoy MK, Martinez TN, Ruhn KA, Szymkowski DE, Smith CG, Botterman BR, Tansey KE, Tansey MG. Blocking soluble tumor necrosis factor signaling with dominant-negative tumor necrosis factor inhibitor attenuates loss of dopaminergic neurons in models of Parkinson's disease. J Neurosci. 2006;26(37):9365–9375. doi: 10.1523/JNEUROSCI.1504-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Ruhn KA, Martinez TN, McAlpine FE, Blesch A, Tansey MG. Intranigral lentiviral delivery of dominant-negative TNF attenuates neurodegeneration and behavioral deficits in hemiparkinsonian rats. Mol Ther. 2008;16(9):1572–1579. doi: 10.1038/mt.2008.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6(3):293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Morris DW, Rodgers A, McGhee KA, Schwaiger S, Scully P, Quinn J, Meagher D, Waddington JL, Gill M, Corvin AP. Confirming RGS4 as a susceptibility gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2004;125B(1):50–53. doi: 10.1002/ajmg.b.20109. [DOI] [PubMed] [Google Scholar]
- Mueller JC, Fuchs J, Hofer A, Zimprich A, Lichtner P, Illig T, Berg D, Wullner U, Meitinger T, Gasser T. Multiple regions of alpha-synuclein are associated with Parkinson's disease. Ann Neurol. 2005;57(4):535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- Muma NA, Mariyappa R, Williams K, Lee JM. Differences in regional and subcellular localization of G(q/11) and RGS4 protein levels in Alzheimer's disease: correlation with muscarinic M1 receptor binding parameters. Synapse. 2003;47(1):58–65. doi: 10.1002/syn.10153. [DOI] [PubMed] [Google Scholar]
- Papachatzaki MM, Antal Z, Terzi D, Szucs P, Zachariou V, Antal M. RGS9-2 modulates nociceptive behaviour and opioid-mediated synaptic transmission in the spinal dorsal horn. Neurosci Lett. 2011;501(1):31–34. doi: 10.1016/j.neulet.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KM, Chowdari KV, Nimgaonkar VL, Talkowski ME, Lewis DA, Keshavan MS. Genetic polymorphisms of the RGS4 and dorsolateral prefrontal cortex morphometry among first episode schizophrenia patients. Mol Psychiatry. 2005;10(2):213–219. doi: 10.1038/sj.mp.4001562. [DOI] [PubMed] [Google Scholar]
- Psifogeorgou K, Terzi D, Papachatzaki MM, Varidaki A, Ferguson D, Gold SJ, Zachariou V. A unique role of RGS9-2 in the striatum as a positive or negative regulator of opiate analgesia. J Neurosci. 2011;31(15):5617–5624. doi: 10.1523/JNEUROSCI.4146-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Hauser MA, Scott WK, Postel EA, Agarwal A, Gallins P, Wong F, Chen YS, Spencer K, Schnetz-Boutaud N, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78(5):852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderovski DP, Diverse-Pierluissi M, De Vries L. The GoLoco motif: a Galphai/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem Sci. 1999;24(9):340–341. doi: 10.1016/s0968-0004(99)01441-3. [DOI] [PubMed] [Google Scholar]
- Sierra DA, Gilbert DJ, Householder D, Grishin NV, Yu K, Ukidwe P, Barker SA, He W, Wensel TG, Otero G, et al. Evolution of the regulators of G-protein signaling multigene family in mouse and human. Genomics. 2002;79(2):177–185. doi: 10.1006/geno.2002.6693. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Seltman H, Bassett AS, Brzustowicz LM, Chen X, Chowdari KV, Collier DA, Cordeiro Q, Corvin AP, Deshpande SN, et al. Evaluation of a susceptibility gene for schizophrenia: genotype based meta-analysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry. 2006;60(2):152–162. doi: 10.1016/j.biopsych.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner CM, Aston DA. Epidemiology of Parkinson's disease and akinetic syndromes. Curr Opin Neurol. 2000;13(4):427–430. doi: 10.1097/00019052-200008000-00010. [DOI] [PubMed] [Google Scholar]
- Waugh JL, Lou AC, Eisch AJ, Monteggia LM, Muly EC, Gold SJ. Regional, cellular, and subcellular localization of RGS10 in rodent brain. J Comp Neurol. 2005;481(3):299–313. doi: 10.1002/cne.20372. [DOI] [PubMed] [Google Scholar]
- Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, McCreadie RG, Buckland P, Sharkey V, Chowdari KV, Zammit S, et al. Support for RGS4 as a susceptibility gene for schizophrenia. Biol Psychiatry. 2004;55(2):192–195. doi: 10.1016/j.biopsych.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Zheng B, De Vries L, Gist Farquhar M. Divergence of RGS proteins: evidence for the existence of six mammalian RGS subfamilies. Trends Biochem Sci. 1999;24(11):411–414. doi: 10.1016/s0968-0004(99)01474-7. [DOI] [PubMed] [Google Scholar]