Abstract
NOTCH1 pathway activation contributes to the pathogenesis of over 60% of T-cell acute lymphoblastic leukemia (T-ALL). While Notch is thought to exert the majority of its effects through transcriptional activation of Myc, it also likely has independent roles in T-ALL malignancy. Here, we utilized a zebrafish transgenic model of T-ALL, where Notch does not induce Myc transcription, to identify a novel Notch gene expression signature that is also found in human T-ALL and is regulated independently of Myc. Cross-species microarray comparisons between zebrafish and mammalian disease identified a common T-ALL gene signature, suggesting that conserved genetic pathways underlie T-ALL development. Functionally, Notch expression induced a significant expansion of pre-leukemic clones; however, a majority of these clones were not fully transformed and could not induce leukemia when transplanted into recipient animals. Limiting-dilution cell transplantation revealed that Notch signaling does not increase the overall frequency of leukemia-propagating cells (LPCs), either alone or in collaboration with Myc. Taken together, these data indicate that a primary role of Notch signaling in T-ALL is to expand a population of pre-malignant thymocytes, of which a subset acquire the necessary mutations to become fully transformed LPCs.
Keywords: thymocyte, relapse, Myc, zebrafish, self-renewal
Introduction
T-cell acute lymphoblastic leukemia is an aggressive malignancy of thymocytes. Despite improved therapies, patients with relapsed disease have a 5-year survival rate of <30% (1). T-ALL is thought to arise from a subset of cells known as leukemia-propagating cells (LPCs), which are rare cells that have the ability to reform the leukemia from a single cell (2,3), and are thus believed to be critical for inducing relapse. Recent studies have demonstrated that clonal evolution frequently occurs in relapsed T-ALL, resulting in new genomic changes within transformed cells. These genetic lesions were selected for at relapse and likely result in a more aggressive malignancy, due in part to their ability to increase the relative frequency of LPCs within the leukemia and thus enhance LPC activity (4,5). Genes frequently mutated at relapse include PTEN, FBXW7, MYC, and NOTCH1; however, it is currently unknown whether these genes enhance LPC frequency and/or promote LPC activity to drive relapse formation.
NOTCH1 is the most commonly mutated oncogene in T-ALL, with >60% of T-ALL patients harboring activating NOTCH1 mutations that confer ligand independent activation or increased stability of the Notch intra-cellular domain (NotchICD) (6,7). Transgenic mouse experiments confirmed that NotchICD initiates a highly penetrant T-ALL when introduced into bone marrow-derived cells and elicits transformation by acting as a transcription factor (8,9). Activating Notch mutations spontaneously occur in several mouse models of T-ALL and exhibit similar mutation spectra as human disease (10–12), suggesting a central role for Notch in T-ALL malignancy. Currently, there are conflicting reports regarding the role of Notch signaling in LPC development and function. Recent studies have suggested that Notch signaling plays a role in LPC maintenance, in that Notch activation in primary human T-ALL cells is critical for their ability to serially xenograft in mice (2) and inhibition of Notch signaling in a mouse model of T-ALL also significantly reduced leukemia development when transplanted into syngeneic recipients (13). However, others have indicated that Notch signaling is not sufficient to induce LPC formation (14) or long-term repopulating potential in normal thymocytes (15). To date, a role for NOTCH in regulating leukemia-propagating cell frequency and function remains controversial.
Understanding the precise role of Notch in LPC biology is confounded by its transcriptional activation of the Myc oncogene in mammalian T-ALL (16–18). Notch was found to promote T-ALL proliferation in human cell lines exclusively through Myc activation (18) and Emerging evidence suggests that Myc can play a critical role in the maintenance of normal tissue stem cells (19). Whether the effects of Notch on LPCs are driven by the Notch/Myc axis is currently unknown. Importantly, Notch also collaborates with Myc to enhance T-ALL development in transgenic mouse models, and synergy between Myc and Notch was identified in retroviral based screens in T-cell malignancy (17,20,21). These latter results would not be expected if the Notch/Myc axis was strictly linear. In normal tissues, where Notch does not necessarily induce Myc expression, Notch signaling can promote stem cell survival and self-renewal (22). Taken together, it is likely that NOTCH has other important roles in T-ALL malignancy, apart from Myc induction.
We have utilized the Myc- (23) and NotchICD-induced (24) zebrafish T-ALL models to better understand the function of each of these transcription factors in T-ALL development and LPC function. These models are uniquely well-suited to this type of study, in that Notch does not transcriptionally activate myc in zebrafish T-ALL (24), and notch1a and notch1b are not mutationally activated in Myc-induced T-ALL, suggesting that Notch and Myc exert independent and non-overlapping roles in regulating diverse oncogenic T-ALL pathways. Our microarray comparisons indicate that zebrafish T-ALL shares molecular similarities to both human and mouse disease, and revealed a novel Notch gene signature that is regulated independently of Myc. Functionally, we found that Notch collaborates with Myc to significantly decrease time to leukemia onset without affecting overall proliferation rates or apoptosis. Clonal analysis of these primary T-ALL cells demonstrates that Notch signaling results in a significant expansion of a pool of T cell clones. However, a majority of these clones are not fully transformed and could not engraft disease into syngeneic recipient animals. Additionally, by utilizing limiting-dilution cell transplantation of leukemic cells into over 1,200 recipient animals, we found that Notch signaling did not increase the overall frequency of LPCs in T-ALL. Together, these data suggest that mutational activation of Notch likely acts as a primary initiating event in T-ALL through expansion of a pre-malignant pool of T cell clones, a subset of which acquire additional mutations to become fully transformed LPCs.
Methods
Stable transgenic zebrafish that develop T-ALL
rag2-GFP (25) and rag2-NOTCH(ICD)-EGFP (human Notch expressing) (24) zebrafish lines have been described. Stable transgenic rag2-EGFP-Myc (murine Myc) zebrafish were re-created using meganuclease transgenic technology. Transgenic fish were monitored for disease onset beginning at 21 days post-fertilization (dpf) and every 7 days thereafter. Thymic hyperplasia was defined by a three-fold increase in thymus size, and lymphoma by the local expansion of GFP-positive lymphoblasts outside the thymus. Leukemia was defined as >50% of the animal being overtaken by GFP-positive lymphoblasts, established previously as a robust surrogate for infiltration of lymphocytes into the marrow (26). Representative animals at various stages of disease progression are shown in Supplemental Figure 1. Kaplan-Meier analyses were completed using the SAS program and results are presented using the Log-rank (Mantel-Cox) test.
Generation of mosaic transgenic animals that develop T-ALL
Transgenes were prepared as previously described (27). 40 ng/µL rag2-GFP was mixed with 20 ng/µL of either rag2-Myc (murine Myc) or rag2-notch1aICD (zebrafish Notch), and for injections with all 3 constructs, 20 ng/µL of each plasmid were used. DNA was microinjected into one-cell stage CG1 embryos. Animals with GFP-positive thymi at day 21 dpf were monitored for disease onset every 7 days. GFP-negative fish were re-screened every 30 days for 6 months to ensure that diseased animals were not missed in our analysis and to verify that T-ALL did not develop in animals that failed to have GFP-labeled thymocytes at day 21 dpf.
Leukemia cell morphology, cell cycle, and apoptosis analysis
Cytospins and FACS analysis of kidney marrow and spleen were completed as described (23), with propidium iodide was used to exclude dead cells. For cell cycle and apoptosis analysis, unsorted cells were isolated from leukemic fish and analyzed for DNA synthesis using Click IT EDU Alexa Fluor 647 (Invitrogen) according to the manufacturer’s protocol. Of note, cells were incubated with EDU for 30’, then fixed in 4% paraformaldehyde before Alexa Fluor conjugation. Propidium iodide was used as a counter-stain. Unsorted cells were also stained with Annexin V Alexa Fluor 647 (Invitrogen) in the presence of propidium iodide, according to manufacturer’s protocol to quantify apoptotic cells. For all experiments, unsorted normal thymocytes were collected by dissecting GFP-positive thymus from 45d rag2-GFP animals, and whole blood was collected from 45d CG1-strain zebrafish for use as controls.
TCRβ clonality assays
T-cell receptor (TCR) recombination has been used extensively to follow minimal residual disease in human T-ALL and is a robust way to assess clonality (28,29). We created an analogous assay to detect each of the 102 individual TCRβ rearrangements in zebrafish (Supplemental Figure 2). RNA was extracted from FACS sorted T-ALL cells, made into cDNA, and PCR was performed utilizing each Vβ and Cβ primer (Supplemental Table 1). A semi-nested PCR was completed using 1 µL of the PCR product and resolved on a 2% agarose gel.
Cell transplantation
GFP-positive cells from T-ALL, lymphoma and hyperplasia were isolated and transplanted as previously described (30,31). Briefly, tumor-bearing fish were macerated in 5%FBS+0.9XPBS, cells were strained through 40 µm nylon mesh (BD Falcon) and stained with PI before FACS. GFP+/PI− cells (≥95% purity, >98% viability) were sorted at the appropriate number into wells of a 96-well plate containing 50 µL 5%FBS+0.9XPBS plus 3×104 whole blood cells isolated from CG1-strain zebrafish. Cells were centrifuged, cell pellets re-suspended in 5 µL 5%FBS+0.9XPBS and transplanted by intra-peritoneal injection into >60 day old CG1-strain recipients. Fish were monitored for T-ALL growth for up to 120 days. Leukemia-propagating cell frequency was calculated using the Extreme Limiting Dilution Analysis software (http://bioinf.wehi.edu.au/software/elda/) (32).
Microarray Analyses
RNA was extracted from 3×105 GFP-positive leukemia cells (FACS isolated as above) and normal thymocytes from 45d rag2-GFP animals, (RNeasy Plus Micro kit, Qiagen), amplified, and labeled with Cy3. Reference control RNA from total CG1-strain fish (2 males and 2 females) was labeled with Cy5. Samples were hybridized together onto the Zebrafish 44K Agilent microarray, scanned, and probes identified at various log-fold change differences with p<0.01 using a standard two-tailed T-test. Zebrafish Agilent probe IDs were mapped to their human homologues using the Beagle Genetic Analysis Software Package (http://chgr.mgh.harvard.edu/genomesrus/index.php).
Gene set enrichment analysis (GSEA) was completed essentially described (33) using microarray data sets of interest including: GSE19499, a study comparing TLX-1 induced murine T-ALL to several other genetic T-ALL models in mouse (34) and GSE7050, a study comparing a β-catenin driven mouse T-ALL model (CD4Cre-Ctnnbex3) to thymocytes from LCKCre control mice (35), as well as GSE13425 and GSE13351 where 190 human diagnosis ALL samples were used to develop a classifier between T-ALL and B-ALL, then confirmed in an independent cohort of 107 patient samples (36). The murine data sets were combined using a trimmed mean followed by quantile normalization to create a merged GCT file. GSEA significance was defined by a p<0.05 and false discovery rate (FDR)≤ 0.25.
Real-time RT-PCR
RNA was extracted from 3×105 FACS sorted zebrafish T-ALL cells and from human T-ALL cells using an RNeasy Mini kit with on-column DNAse treatment (Qiagen). Total RNA was reverse transcribed (Reverse Transciption kit, Applied Biosystems), and real-time PCR was performed using Power Sybr 2X Master Mix (Invitrogen, primers available in Supplemental Table 2). Data were normalized to ef1α expression; fold-change was calculated using the 2−ΔΔCt method.
Results
Notch collaborates with Myc to enhance zebrafish T-ALL progression
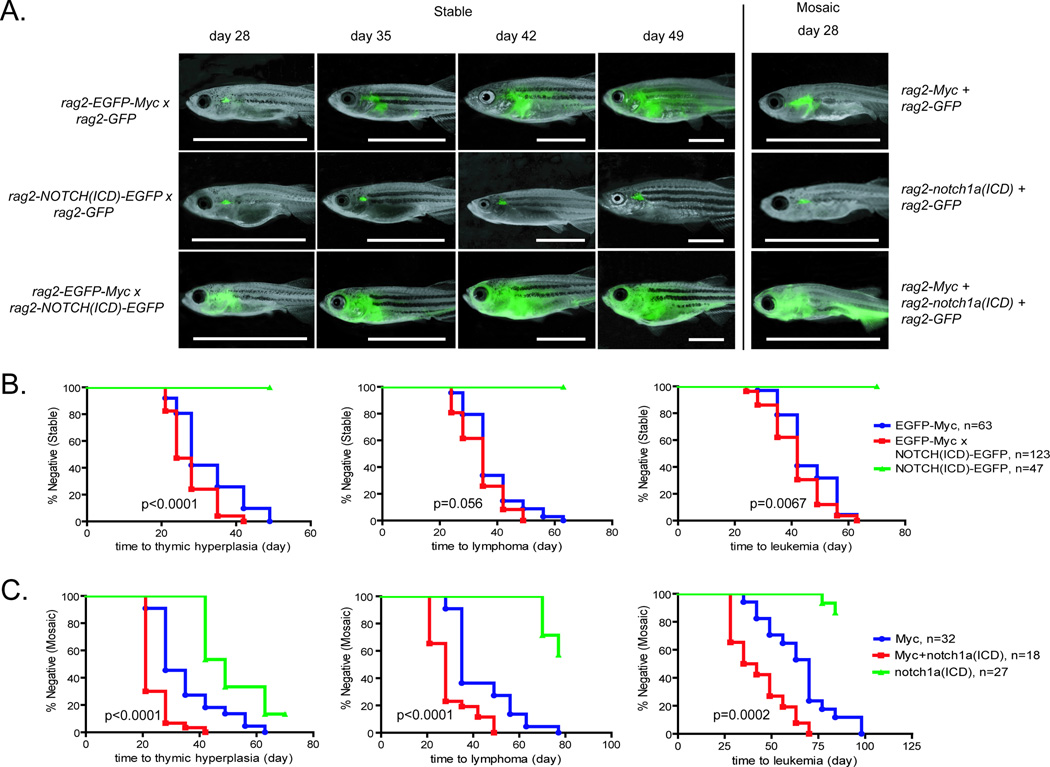
Notch collaborates with Myc to enhance leukemia onset in mouse models of T-ALL. To confirm that Notch also synergizes with Myc in zebrafish T-ALL, heterozygous stable transgenic rag2-EGFP-Myc fish were crossed with rag2-NOTCH(ICD)-EGFP animals. Each of these lines were also crossed with rag2-GFP animals as a control (AB* strain, n=123 double transgenic fish, n=63 Myc and n=47 NOTCH(ICD). Double transgenic animals developed thymic hyperplasia 10 days faster than those that expressed Myc alone (p<0.0001, Log-rank test), which progressed rapidly to lymphoma (Figure 1A–B). The Myc+NOTCH(ICD) double transgenic animals had an accelerated time to T-ALL onset (40.9±9.2 days) compared to Myc expressing zebrafish (46.2±9.3 days, p=0.0067). As previously reported, the stable transgenic rag2-NOTCH(ICD)-GFP transgenic animals did not develop disease within 6 months of life (24).
Figure 1. Notch collaborates with Myc to promote T-ALL progression in both stable and mosaic transgenic zebrafish.
(A) Stable transgenic rag2-EGFP-Myc, rag2-NOTCH(ICD)-EGFP, and double transgenic AB-strain zebrafish were followed for disease onset and photographed at 28, 35, 42 and 49 days of life using epi-fluorescence stereomicroscopy. Alternatively, CG1-strain zebrafish were injected at the one-cell stage of life with rag2-Myc+rag2-GFP, rag2-notch1a(ICD)+rag2-GFP, or rag2-Myc+rag2-notch1a(ICD)+rag2-GFP transgenic constructs. Animals with GFP-labeled thymi were photographed at 28 days of life. Panels are merged fluorescent and brightfield images. Scale bar, 10 mm. (B) Kaplan Meier analysis of disease progression in stable transgenic zebrafish. Fish were scored for thymic hyperplasia (left), lymphoma (middle), or leukemia (right). (C) Kaplan Meier analysis of disease progression in mosaic transgenic zebrafish. The total number of animals used in each experiments are shown, and p-values comparing Myc transgenic fish to Myc+Notch(ICD) expressing fish are denoted in each panel (Log-rank/Mantel-Cox statistic).
While stable transgenic approaches demonstrated a synergy of Myc and NOTCH(ICD) at inducing T-ALL in zebrafish, these experiments required the generation of stable transgenic lines that are difficult to maintain due to their early T-ALL onset (25). Our laboratory has successfully used mosaic transgenic approaches to create zebrafish models of T-ALL, where up to three transgenic constructs can be co-expressed within developing thymocytes that eventually give rise leukemia (33,37). Specifically, linearized DNA constructs are co-injected into one-cell stage embryos; a subset of the resulting mosaic animals incorporate transgenes into thymocytes, leading to transgene co-expression within T cells and subsequently the leukemia. Importantly, unlike stable transgenic approaches that drive high transgene expression within all thymocytes, only a small subset of thymocyte precursor cells integrate the transgene and express Myc in mosaic transgenic zebrafish. Given that a majority of genetic mutations are acquired somatically and sporadically in human T-ALL, we believe that use of mosaic transgenic approaches likely more accurately recapitulates what is observed in human cancer.
To assess if mosaic transgenesis could be used to identify collaborating events in T-ALL, one-cell stage CG1-strain, syngeneic embryos were microinjected with 1) rag2-mouseMyc+rag2-GFP, 2) rag2-zebrafish notch1a(ICD)+rag2-GFP or 3) rag2-Myc+rag2-notch1a(ICD)+rag2-GFP. Mosaic animals were assessed for the development of GFP-positive thymi at 21 dpf and subsequently followed for time to T-ALL onset. Mosaic transgenic animals that co-expressed Myc and notch1a(ICD) (n=27) exhibited significantly shortened time to thymic hyperplasia compared to those expressing Myc (n=32, p<0.0001) or notch1a(ICD) alone (n=18, p<0.0001). Lymphoma arose within 29.3±9.2 days in transgenic Myc+notch1a(ICD) animals, compared to 42.9±13.2 days in Myc expressing animals (p<0.0001). Myc+notch1a(ICD) zebrafish also developed leukemia 20 days earlier than animals that expressed Myc alone (45.5±14.4 days, compared to 65.6±15.1 days, p=0.0002, Figure 1A, 1C). Only 50% of animals that expressed notch1a(ICD) developed leukemia, with an average latency of >6 months. By contrast, all rag2-Myc+rag2-GFP mosaic animals that had GFP-labeled thymi developed leukemia (n=32 of 32). Zebrafish that did not have GFP-positive thymi by 21 dpf never developed GFP-labeled thymocytes or T-ALL later in life (n=362).Strikingly, mosaic transgenic zebrafish (CG1 strain) displayed a longer latency to Myc-induced T-ALL than the stable transgenic lines (AB strain), which may be attributed to differences in the genetic backgrounds of the animals used in each study. Despite this, our experiments demonstrate that Myc and Notch(ICD) collaborate to induce T-ALL and highlight that our mosaic transgenic approach is a robust methodology to identify modulators of T-ALL progression. Subsequent analyses focused exclusively on T-ALL that developed in mosaic transgenic animals.
Characterization of the mosaic transgenic T-ALL models
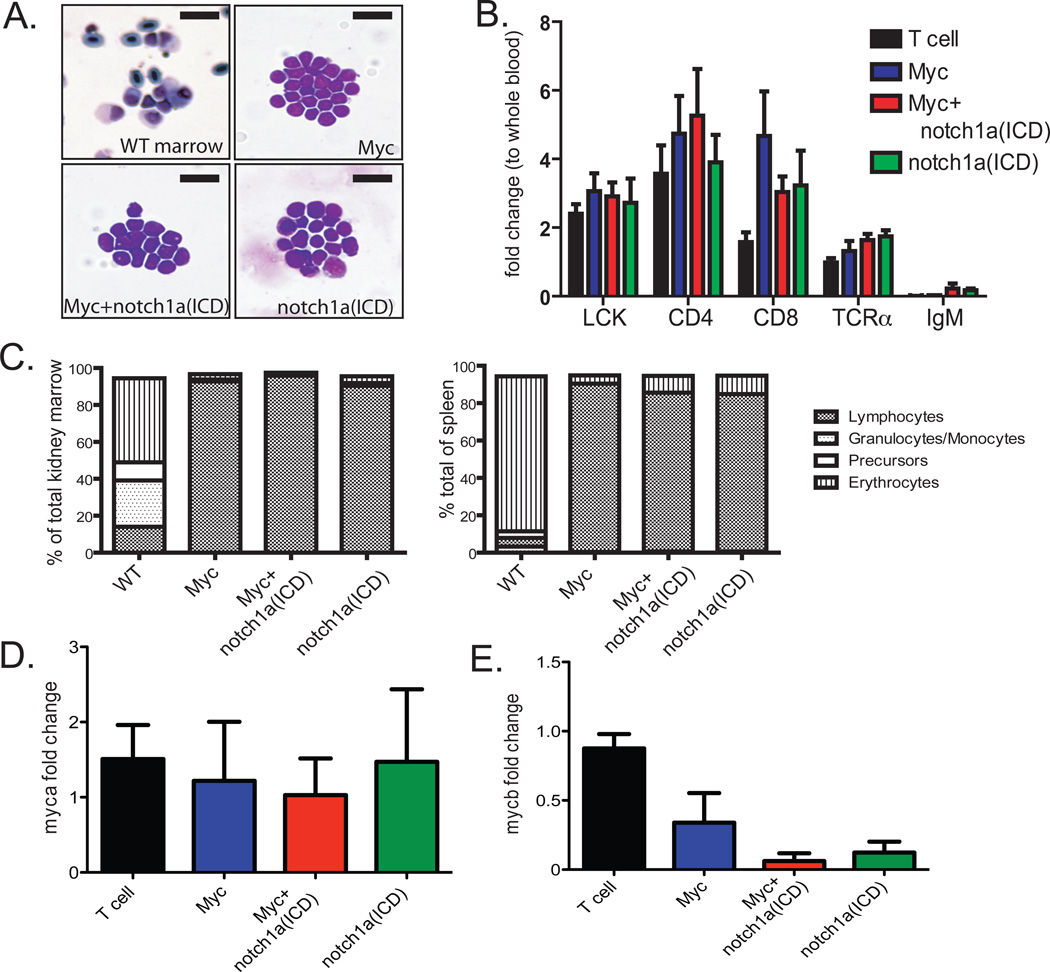
GFP-labeled T-ALL arose from the thymus of mosaic transgenic animals and shared typical lymphoblast morphology without shape or size differences across transgenic strains (Figure 2A). Leukemias expressed high levels of each transgene (Supplemental Figure 3) and T cell specific genes, but not the B cell specific gene IgM (Figure 2B). The expression of cd4, cd8, rag2 and the presence of Tcr-beta rearrangements suggested that the T-ALL blast cells were arrested at the cortical thymocyte stage, irrespective of genotype. FACS demonstrated >75% lymphocyte infiltration into the kidney and spleen of animals with GFP-positive T-ALL spreading over >50% of the body (n=3, Figure 2C, Supplemental Figure 4), validating whole body imaging as a robust marker for leukemia onset and growth, as previously reported (25). Taken together, our results are consistent with previous reports that Myc and Notch-induced T-ALL arises from T-cells confined to the thymus of zebrafish (24,25).
Figure 2. Characterization of mosaic transgenic zebrafish with Myc-, Notch- and Myc+Notch-induced T-ALL.
(A) May-Grünwald and Wright-Giemsa stained cytospins of kidney marrow from wild-type and mosaic transgenic zebrafish from each genotype. Scale bar, 20 µm. (B) Real-time RT-PCR analysis of the T cell-specific genes lck, CD4, CD8 and TCRα, and the B-cell gene IgM in normal rag2-GFP thymocytes, and GFP-positive T-ALL cells from mosaic transgenic animals. Data are the average expression from 4 primary T-ALL per group. Samples were normalized to ef1α expression and gene expression is shown relative to whole blood cells isolated from normal CG1-strain fish. Error bars are the standard deviation (SD). (C) Representative FACS analysis of kidney marrow (left) and spleen (right) from wild-type and mosaic transgenic zebrafish that developed T-ALL. The percentage of lymphocytes, granulocytes/monocytes, hematopoietic precursors and erythrocytes were quantified by analysis of cell size and granularity. (D) Real-time RT-PCR analysis of zebrafish myca and (E) mycb expression in normal thymocytes and T-ALL cells. Data are the average of 6 individual primary T-ALL, normalized to ef1α expression and relative to rag2-GFP thymocytes isolated from 45d animals. Errors bars are +/− SD.
Previous work utilizing the stable rag2-NOTCHICD-EGFP line demonstrated that NOTCHICD does not induce myc expression in zebrafish22. Similarly, notch1a(ICD) did not induce the expression of myc or its related family members in mosaic transgenic zebrafish (Figure 2D–E, Supplemental Figure 5). In human T-ALL, common “hot-spot” mutations found in NOTCH exons 26, 27 and 34 result in ligand-independent NOTCH activation (7). Analysis of these same sites in genomic DNA isolated from 16 Myc-induced T-ALL revealed that notch1a and notch1b were not mutationally activated in zebrafish T-ALL, suggesting that notch mutations are not selected in Myc-induced T-ALL. Thus, the zebrafish Myc and notch1a(ICD)-induced T-ALL models therefore provide a unique opportunity to directly assess the function of Notch signaling in T-ALL apart from its role in Myc induction.
Zebrafish T-ALL is molecularly similar to mouse and human disease
To validate the relevance of the zebrafish T-ALL model and to assess if common gene programs are found in T-ALL across species, microarray gene expression profiling and cross-species comparisons were completed between zebrafish, mouse, and human T-ALL. Up- and down-regulated gene signatures were identified by comparing FACS sorted GFP-positive zebrafish T-ALL cells (n=15) to rag2-GFP thymocytes. Gene sets were identified at various fold change levels and compared to mouse T-ALL normalized to thymocytes using Gene Set Enrichment Analysis (GSEA). Analysis at multiple fold change cut-offs ensured that GSEA significance was not due to the arbitrary creation of the gene list. The zebrafish up- and down-regulated gene sets were significantly associated with murine T-ALL at each of the fold changes examined (p=0.004, Table 1, Supplemental Figure 6, GSEA results at the 3-fold change cut-off listed in Supplemental Table 3 and 4) but were not specifically associated with any transgenic mouse T-ALL model (38), indicating that zebrafish and mouse T-ALL share common molecular pathway regulation, irrespective of initiating oncogene. The up- and down-regulated gene sets found in zebrafish T-ALL were also significantly associated with human T-ALL (p<0.001), but not precursor B-ALL (Table 1, GSEA results at the 3-fold change cut off are shown in Supplemental Table 5 and 6), confirming that zebrafish, mouse, and human T-ALL share common molecular pathway activation. Taken together, our results suggest common molecular pathways are utilized in T-ALL development that are highly conserved across species.
Table 1. Zebrafish T-ALL is molecularly similar to mouse and human disease.
Microarray gene expression signatures were identified by comparing zebrafish T-ALL to normal thymocytes. The up- and down-regulated gene sets were assessed for concordant gene regulation in human and mouse disease. The mouse T-ALL to thymocyte comparisons were made using GSE19499 and GSE7050 data sets, while human T-ALL to B-ALL comparisons were made using GSE13425. Similar results were observed using the human GSE13351 data set (data not shown). GSEA significance was defined by p-value <0.05 and a false-discovery rate of ≤0.25 (FDR) at three different fold cut-offs. Number of common probes found at each fold-change cut off (# of probes), Enrichment Score (ES), Normalized Enrichment Score (NES). Negative ES and NES scores indicate that the gene set is down regulated within the samples being compared.
| GSEA Comparison | Gene Set | # of probes | ES | NES | FDR | p-value |
|---|---|---|---|---|---|---|
| Mouse T-ALL to thymocytes | 2.5 fold Up | 585 | 0.486 | 2.031 | 0.000 | <0.001 |
| Mouse T-ALL to thymocytes | 3.0 fold Up | 361 | 0.428 | 1.841 | 0.000 | <0.001 |
| Mouse T-ALL to thymocytes | 3.5 fold Up | 192 | 0.404 | 1.699 | 0.002 | 0.001 |
| Mouse T-ALL to thymocytes | 2.5 fold Down | 532 | −0.367 | −1.770 | 0.000 | <0.001 |
| Mouse T-ALL to thymocytes | 3.0 fold Down | 348 | −0.361 | −1.584 | 0.009 | 0.004 |
| Mouse T-ALL to thymocytes | 3.5 fold Down | 199 | −0.385 | −1.579 | 0.007 | 0.003 |
| Human T-ALL to B-ALL | 2.5 fold Up | 534 | 0.465 | 2.077 | 0.000 | <0.001 |
| Human T-ALL to B-ALL | 3.0 fold Up | 312 | 0.468 | 2.001 | 0.000 | <0.001 |
| Human T-ALL to B-ALL | 3.5 fold Up | 156 | 0.442 | 1.773 | 0.000 | <0.001 |
| Human T-ALL to B-ALL | 2.5 fold Down | 517 | −0.355 | −1.637 | 0.000 | <0.001 |
| Human T-ALL to B-ALL | 3.0 fold Down | 328 | −0.395 | −1.752 | 0.000 | <0.001 |
| Human T-ALL to B-ALL | 3.5 fold Down | 209 | −0.426 | −1.799 | 0.000 | <0.001 |
Identification of novel Notch target genes in T-ALL
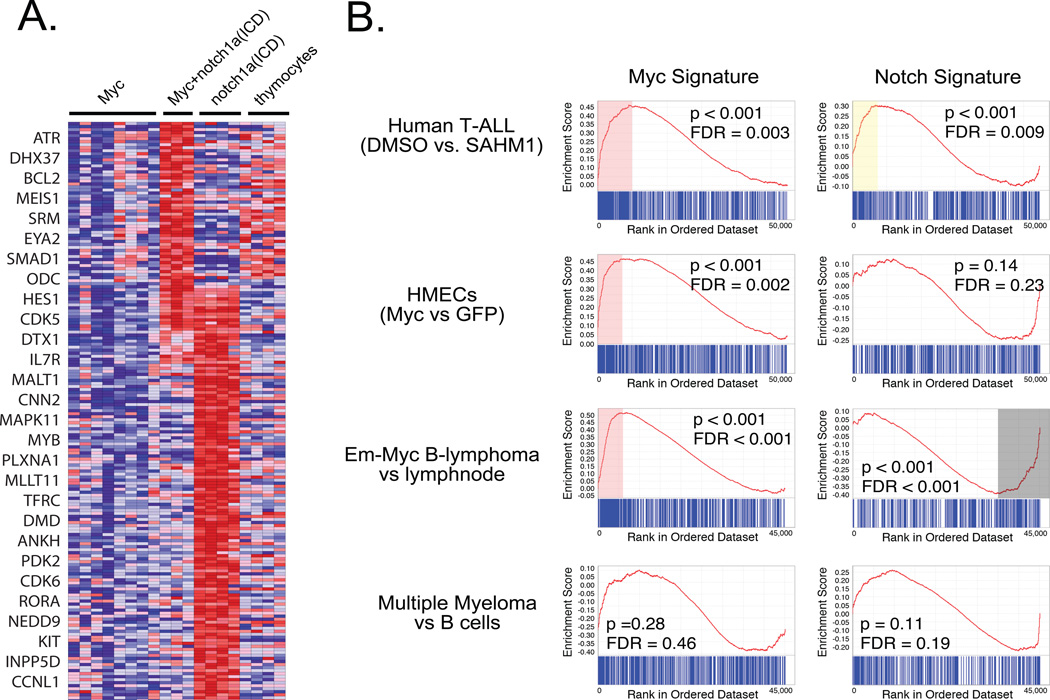
The Notch/Myc axis is critical for T-ALL progression in mammals, and while Notch and Myc may co-regulate gene expression in some instances (16), Notch likely transcriptionally activates many Myc-independent target genes. To identify novel Notch-associated and Myc-associated gene signatures, the following comparisons were completed on GFP-labeled FACS sorted cells: 1) Single mosaic transgenic Myc-expressing T-ALL cells to normal thymocytes isolated from 45-day-old rag2-GFP transgenic animals (Myc signature), 2) Myc+ notch1a(ICD)-expressing T-ALL to Myc-expressing T-ALL, and 3) single mosaic transgenic notch1a(ICD)-induced T-ALL to Myc-expressing T-ALL. The latter two comparisons were combined to identify up and down-regulated genes that were specifically associated with Notch status (the Notch signature) and were independent of Myc expression (Figure 3A, Supplemental Table 7). At the three-fold change cut off, the Notch signature had 171 up-regulated genes (387 probes, Supplemental Table 8) and the Myc signature comprised 280 up-regulated genes (523 probes, Supplemental Table 9). Importantly, only one common gene was shared between these signatures (ankh), suggesting that our gene expression analysis identified unique gene lists that were independently regulated by Myc or notch1a(ICD). Realtime RT-PCR of the primary Myc, Myc+ notch1a(ICD) and notch1a(ICD) expressing T-ALL confirmed that microarray target genes were regulated exclusively by notch1a(ICD) (Supplemental Figure 7), and subsequent analysis of human T-ALL cell lines treated with NOTCH or MYC inhibitors showed that a subset of genes identified in our analysis were bone fide NOTCH targets, regulated independently of MYC, including IL7R, BCL2, MEIS1, HES1, and MYB (Supplemental Figure 8). Finally, the Myc and Notch specific gene signatures showed little overlap with previously identified NOTCH target genes in human T-ALL (Supplemental Figure 9), suggesting that cross-species microarray comparisons provide a powerful model to uncover novel genes associated with oncogene status.
Figure 3. Cross-species microarray comparisons identify Myc and Notch signatures in both human and zebrafish T-ALL.
A) Differentially regulated genes found by comparing zebrafish notch1a(ICD)- or Myc+ notch1a(ICD)-induced T-ALL to both Myc-induced T-ALL and normal thymocytes. A subset of differentially regulated genes is denoted on the left. B) GSEA analysis demonstrate that Myc and notch1a(ICD) signatures are co-regulated in human T-ALL cells following Notch inhibition with the SAHM1 stapled peptide. The Myc signature, but not the notch1a(ICD) gene signature, is coordinately regulated in human mammary epithelial cells (HMECs) transduced with Myc and in mouse models of Myc-induced B-lymphoma. XBP1-induced multiple myeloma does not express either the Myc or Notch signature, confirming that our zebrafish gene signatures are not specific to lymphocytes. The Notch signature is also negatively associated with B-lymphomas, suggesting a role for Notch in B-cell maturation or stroma in normal lymph-nodes.
Given that NOTCH regulates transcriptional expression of MYC in human T-ALL, we expected that both the zebrafish Myc and Notch signature genes should be coordinately regulated in human disease. GSEA analysis was performed using our up-regulated Notch and Myc genes sets and each was assessed for concordant regulation in NOTCH-dependent human T-ALL cell lines that were treated with DMSO (control) or the stapled peptide inhibitor of Notch, SAMH1 (39). DMSO treated cells would presumably have higher Notch activity than cells treated with SAMHI. The zebrafish Myc and Notch gene signatures were significantly up-regulated in control DMSO treated cells, but not cells treated with the Notch inhibitor (Figure 3B, p<0.001). To verify that the zebrafish Notch signature is regulated independent of Myc, each was assessed for concordant regulation in comparing 1) human mammary epithelial cells transduced with either Myc or one of several other oncogenes to GFP-transduced control cells (40), 2) Eμ-Myc induced murine B-cell lymphoma compared to normal lymphnode (41), and 3) XBP1-induced murine multiple myeloma to B-cells (42). The zebrafish Notch signature was not detected in the Myc or other oncogene transduced HMECs or Eμ-Myc induced B-cell lymphoma (Figure 3B) while the zebrafish Myc signature was significantly associated with Myc status in these cells (Figure 3B, p<0.001), but not HMECs transduced with other oncogenes (Supplemental Table 10). Neither gene signature was concordantly regulated in XBP1-induced multiple myeloma, indicating that these signatures were not simply identifying a lymphocyte-specific gene signature. GSEA analysis was also completed at various fold change cut-offs to demonstrate that the results were not biased by arbitrary gene list selection (Supplemental Table 10). In total, the non-overlapping Myc and Notch gene expression signatures identified in zebrafish T-ALL validate our choice of the this model to better define the role of Notch signaling in T-ALL development and LPC function.
Effects of Notch activity on T-ALL proliferation or apoptosis
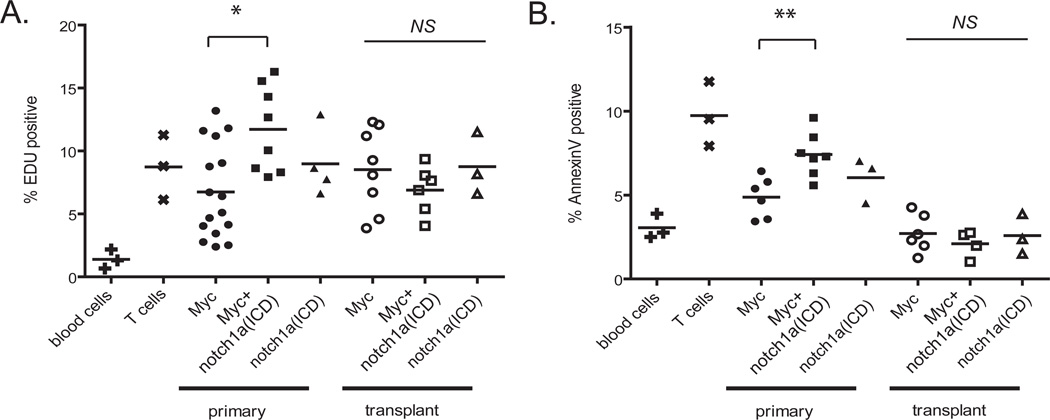
To understand the effects of Notch signaling in T-ALL progression, zebrafish T-ALL were analyzed for effects on cell cycle and proliferation. EDU cell cycle analysis indicated that primary Myc+notch1a(ICD) T-ALL contained a slightly higher percentage of cells in S-phase than Myc primary leukemias (p=0.004, Figure 4A). However, AnnexinV staining revealed that Myc+notch1a(ICD) primary T-ALL cells were more apoptotic than Myc-induced leukemias (p=0.002, Figure 4B). The higher level of apoptosis found in Myc+notch1a(ICD) primary T-ALL was unexpected, but was not significantly different than the apoptosis occurring in normal T cells (p=0.438). Normal thymocytes undergo extensive proliferation and apoptosis during their development and differentiation; Myc+notch1a(ICD) primary T-ALL cells appear to undergo similar cellular turnover, suggesting that these leukemias share characteristics of normal, immature T cells. Interestingly, following tumor re-initiation in recipient animals, there was no significant difference in proliferation or apoptosis between any of the T-ALL groups (p=0.30), suggesting that notch1a(ICD) expression does not augment proliferation rates or alter apoptosis in fully transformed T-ALL cells.
Figure 4. Myc+Notch-induced T-ALL have increased cellular turnover in primary, but not transplanted leukemias.
A) EDU staining to quantify the percentage of cells in S-phase of whole blood cells, rag2-GFP thymocytes, and primary and transplanted T-ALL. Each point is representative of one sample. (B) AnnexinV staining of apoptotic cells of whole blood cells, rag2-GFP thymocytes and primary and transplanted T-ALL. Each point represents one sample. *p=0.0043, Myc compared to Myc+ notch1a(ICD) expressing primary T-ALL. **p=0.0016, Myc compared Myc+notch1a(ICD)-induced primary T-ALL. NS, not significant comparing transplanted T-ALL.
Notch signaling expands a pre-malignant pool of clones within the primary T-ALL
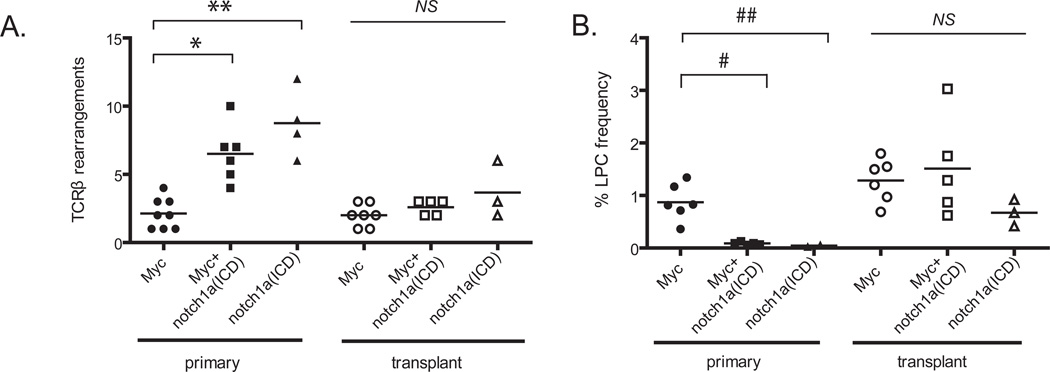
In T-ALL development, normal T cells undergo clonal expansion, and a subset of these cells acquire the necessary mutations to become LPCs over time (43). We wanted to determine whether Notch and/or Myc plays a role in this clonal expansion. Primary GFP-labeled T-ALL cells were isolated by FACS and transplanted at 2.5×105 cells into CG1-strain, syngeneic recipient animals. T-ALL cells were re-isolated from the transplant animals after leukemia development (>45 days), and purified by FACS. The primary and transplanted leukemias were then assessed for expression of TCRβ transcript variants to identify the number of expanded T cell subclones contained within each T-ALL. Primary and transplanted Myc-induced T-ALL contained 2.1±1.1 subclones, with each clone from the primary T-ALL being detected in transplant animals (Figure 5A). Myc-induced hyperplasia and lymphoma contained a similar number of clones as Myc T-ALL (4.0±2.08 and 2.3±0.57 clones, respectively), but were not able to reliably engraft disease in recipient animals (Table 2). These data suggest that Myc expression, alone, is not sufficient to transform T cells, and that additional genetic/epigenetic events are required for LPC formation. Interestingly, the notch1a(ICD) and Myc+ notch1a(ICD) primary T-ALL were comprised of significantly more subclones than the Myc-induced T-ALL (8.8±2.5 and 6.5±2.1 clones, respectively, p=0.0001). However, following engraftment into recipient animals, notch1a(ICD) and Myc+notch1a(ICD) secondary T-ALL contained an average of 3.0±1.3 clones, similar to transplanted Myc T-ALL (Figure 5A). Given that 2.5×105 cells were transplanted into recipient animals and that the same clones were present in multiple recipients (n>3 per tumor) of the same primary T-ALL, the reduced number of subclones following cell transplantation cannot be accounted for by lack of adequate numbers of transferred LPC clones but rather suggests that not all Notch-expressing subclones within the primary T-ALL could reinitiate leukemia growth. Moreover, only 4 of 9 Notch alone-induced leukemias engrafted disease in recipient animals, even when transplanted with 2.5×106 cells (Table 2), but exhibited typical lymphoblast morphology (Supplemental Figure 9). These data suggest that despite GFP-positive cells spreading throughout the primary animals and overtaking >50% of the body, Notch signaling did not confer a fully transformed phenotype to thymocytes.
Figure 5. Notch expands a pool of pre-malignant clones but does not enhance the frequency of LPCs within the leukemia.
(A) A PCR based assay to detect TCRβ rearrangements was used to determine the number of T-cell clones that comprise the primary and transplanted T-ALL. *p<0.0001, comparing primary Myc- and Myc+notch1a(ICD)-induced T-ALL,**p<0.0001, comparing Myc and notch1a(ICD)-expressing T-ALL, NS not significant, comparing transplanted T-ALL. (B) Extreme Limiting Dilution Analysis (ELDA) statistical software was used to calculate the LPC frequency within Myc-, notch1a(ICD)- and Myc+notch1a(ICD)-induced primary and transplanted T-ALL. Each point represents the average LPC frequency of one T-ALL. The raw data are presented in Table 1 (n=1,059 transplant animals). #p<0.0001, comparing primary Myc and Myc+notch1a(ICD)-induced T-ALL, ##p<0.0001, comparing Myc and Myc+notch1a(ICD)-induced T-ALL, NS not significant, comparing transplanted T-ALL.
Table 2. Primary Notch-induced T-ALL have reduced engraftment potential.
Total number of primary T-ALL that were capable of engraftment at various cell doses.
| Cell Number | Myc hyperplasia |
Myc lymphoma |
Myc T-ALL |
Myc+notch1a(ICD) T-ALL |
notch1a(ICD) T-ALL |
|---|---|---|---|---|---|
| 2.5×106 | ND | ND | ND | ND | 4/9 |
| 106 | ND | ND | ND | ND | 4/9 |
| 2.5×105 | 0/2 | ND | ND | ND | 4/9 |
| 105 | ND | 2/3 | ND | ND | 4/9 |
| 104 | ND | ND | 6/6 | 7/7 | 4/9 |
| 1000 | 0/2 | 0/3 | 6/6 | 7/7 | 3/9 |
| 100 | 0/2 | 0/3 | 6/6 | 2/7 | 1/9 |
| 10 | 0/2 | 0/3 | 6/6 | 0/7 | 0/9 |
ND, no data
Notch signaling does not increase the frequency of LPCs in T-ALL
Recent data suggests that Notch signaling is critical for LPC maintenance in that Notch inhibition prevents xenograft growth and can significantly increase T-ALL latency (2,13). However, it is unclear whether Notch signaling acts to increase the overall frequency of LPCs in the leukemia; this latter attribute could function in collaboration with Myc to promote T-ALL onset and enhance the propensity of cells for relapse. Large-scale limiting-dilution cell transplantation analyses were performed on primary T-ALL to determine the frequency of LPCs within each leukemia subtype (22 primary and 16 transplanted T-ALL, n=1,059 transplanted animals). Primary Myc-induced T-ALL had a mean LPC frequency of 1 in 111 cells (95% CI: 1:89–139), while the mean leukemia-propagating cell frequency of notch1a(ICD) and Myc+notch1a(ICD) leukemias was significantly less, with only 1 in 1,639 cells (95% CI: 1:727–3313) and 1 in 1,591 cells (95% CI 1:1088–2477), respectively (Table 3 and Figure 5B, p<0.0001). These data are expected, given that the clonal analysis of primary leukemias and transplant recipients revealed that a majority of subclones in the Myc+notch1a(ICD) and notch1a(ICD)-induced T-ALL were not fully transformed LPCs. We therefore questioned if leukemia-propagating ability was enhanced in LPCs from the Myc+notch1a(ICD) and notch1a(ICD)-expressing T-ALL following leukemia engraftment, which would select for the fully transformed clones. GFP-positive T-ALL cells were re-isolated from transplant recipient animals that received 2.5×105 unsorted primary cells, and then transplanted at limiting dilution into CG1-strain recipient animals. The mean LPC frequency of the Myc-induced leukemias did not change following serial transplantation (1 in 74 cells, 95% CI: 1:55–102). However, serially passaged notch1a(ICD) and Myc+notch1a(ICD) T-ALL exhibited an overall increase in leukemia-propagating ability when compared to the primary leukemias, with 1 in 205 cells (95% CI: 1:119–354) and 1 in 101 cells (95% CI: 1:75–137) now capable of initiating leukemia growth, respectively (Table 3 and Figure 5B, p<0.0001). Despite this overall increase in leukemia-propagating ability, there was no difference in the LPC frequency between secondary leukemias induced from any combination of transgenes (p=0.137), indicating that neither Myc nor Notch signaling augments the frequency of LPCs contained within the leukemia. Myc and Myc+notch1a(ICD)-induced T-ALL expressed both tal1/lmo2 (Supplemental Figure 11), suggesting that T-ALL molecular subtype differences are not responsible for modulating LPC activity in our comparsions. Further, tertiary transplants did not lead to an additional increase in LPC frequency (Supplemental Table 11) or additional loss of T-ALL subclones.
Table 3. Limiting Dilution Analysis of primary and transplanted T-ALL.
Number engrafted/total number of animals transplanted at each cell dose
| Primary | Transplant | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| T-ALL Type | 10,000 cell | 1,000 cell | 100 cell | 10 cell | LPC frequency | 1,000 cell | 100 cell | 10 cell | LPC frequency |
| Myc#1 | 4/4 | 6/6 | 10/20 | 8/40 | 1:93 (152,57) | 5/5 | 6/10 | 5/15 | 1:69 (134,35) |
| Myc#2 | 3/3 | 5/5 | 7/15 | 4/30 | 1:145 (293,71) | 4/4 | 4/8 | 3/15 | 1:105 (220,50) |
| Myc#3 | 3/3 | 7/7 | 15/21 | 7/42 | 1:71 (112,45) | 3/3 | 5/5 | 5/8 | 1:14 (35,7) |
| Myc#4 | 5/5 | 5/5 | 10/21 | 5/42 | 1:120 (200,71) | 5/5 | 5/10 | 3/12 | 1:93(189,45) |
| Myc#5 | 3/3 | 4/4 | 3/9 | 1/12 | 1:205 (507,83) | 3/3 | 2/6 | 2/12 | 1:148 (408,54) |
| Myc #6 | 3/3 | 7/7 | 11/21 | 4/42 | 1:125 (210,75) | 4/4 | 6/8 | 3/12 | 1:58 (121,27) |
| notch1a(ICD)#1 | 5/5 | 3/5 | 1/10 | 0/15 | 1:1066 (2130,371) | 5/5 | 4/8 | 3/12 | 1:96 (214,43) |
| notch1a(ICD)#2 | 5/5 | 2/5 | 0/10 | 0/14 | 1:2212(6428,761) | 4/4 | 1/8 | 0/11 | 1:406 (987,167) |
| notch1a(ICD)#3 | 5/5 | 2/4 | 0/10 | 0/14 | 1:1897 (5990,601) | 4/4 | 2/8 | 1/12 | 1:246 (635,95) |
| Myc+notch1a(ICD)#1 | 5/5 | 4/5 | 1/10 | 0/15 | 1:732 (1810,295) | 5/5 | 5/10 | 4/15 | 1:77 (152,39) |
| Myc+notch1a(ICD)#2 | 4/4 | 3/5 | 0/10 | 0/15 | 1:1475 (4279,509) | 4/4 | 6/10 | 5/15 | 1:69 (134,35) |
| Myc+notch1a(ICD)#3 | 5/5 | 2/5 | 0/9 | 0/5 | 1:1093 (2964,403) | 5/5 | 7/9 | 6/13 | 1:34 (66,18) |
| Myc+notch1a(ICD)#4 | 5/5 | 4/5 | 1/10 | 0/5 | 1:731 (1810,295) | 4/4 | 3/7 | 1/12 | 1:161 (419,62) |
| Myc+notch1a(ICD)#5 | 5/5 | 4/5 | 0/10 | 0/5 | 1:924 (2434,351) | 3/3 | 4/7 | 2/11 | 1:94 (221,40) |
| Myc+notch1a(ICD)#6 | 4/5 | 2/5 | 0/9 | 0/5 | 1:4743 (12290,1831) | 5/5 | 3/8 | 1/12 | 1:183 (457,73) |
| Myc+notch1a(ICD)#7 | 5/5 | 3/5 | 0/10 | 0/5 | 1:1444 (4217,494) | 4/4 | 2/8 | 0/11 | 1:308 (758,125) |
Discussion
Our work suggests that an important function of Notch signaling in T-ALL development is to expand a pre-malignant pool of T cell clones, of which a small subset acquire additional mutations to become fully transformed and capable of leukemia propagation. In this model, Notch promotes T-ALL progression through clonal expansion. An increase in the number of pre-malignant cells greatly enhances both the likelihood and speed by which these cells accumulate the necessary mutations to become fully transformed LPCs. A role for Notch in the clonal expansion of pre-malignant T cells is in agreement with work from the Kelliher group, which utilized the Tal1/Lmo2 mouse model of T-ALL to observe that spontaneous Notch mutation in pre-leukemic animals corresponded to an expansion of DN3/DN4 clones, which gave rise to T-ALL over time (13). Additional studies from Li et al. showed retroviral infection of murine lin-hematopoietic precursor cells (HPCs) with NOTCH(ICD) caused a depletion of B cells and expansion of a CD4+/CD8+ population of T cells that could spread outside of the thymus, but were non-tumorigenic (14). Similarly, we have found that Notch signaling caused an expansion of pre-malignant clones that spread outside of the thymus, and clonal analysis based on TCRβ rearrangements demonstrated that only a small fraction of these subclones were leukemogenic when introduced into transplant recipient animals, indicating Notch signaling, alone, is not sufficient to fully transform leukemia propagating cells. Further, only 4 of 9 primary Notch-induced T-ALL could engraft disease when transplanted into syngeneic recipient animals after >6 months, while the remaining five primary T-ALL did not engraft, irrespective of the number of T-ALL cells transplanted (ranging from 10 to 2.5×106 cells). These data are in agreement with work from Li et al., who demonstrated that leukemia-propagating ability arose in Notch(ICD) over-expressing murine lin-HPCs only after the up-regulation of Akt, Bmi1 and Ras (14), suggesting that the collaboration of additional oncogenes are required for transformation of Notch expressing cells. Clappier et al. recently demonstrated that xenograft of primary human T-ALL into immune-compromised mice selected for a small subset of clones found within the diagnosis leukemia (5). In Clappier’s elegant experiments, transplantation likely selected for those T-ALL cells capable of leukemia propagation, and served to identify fully malignant LPCs that were able to drive relapse. Importantly, a large number of clones found in the human primary leukemia were not detected in either transplanted animals or patients at relapse. In combination with our results from the zebrafish, these data suggest that clonal expansion of pre-malignant clones is a common feature of human T-ALL and that a large portion of genetically distinct clones found within the primary leukemia likely lack self-renewal potential and cannot remake tumor.
While Notch signaling is likely required for continued tumor growth (2,13,18), we have found that Notch may not directly promote leukemia-propagating ability. For example, Notch signaling does not collaborate with Myc to enhance the frequency of leukemia-propagating cells in T-ALL, and Notch-induced T-ALL do not have higher numbers of LPCs when compared with T-ALL that expresses Myc alone. These results are in keeping with data from McCormack et al., who showed that Notch expression in normal thymocytes does not induce serial repopulating ability or confer self-renewing capability in thymocyte reconstitution assays. (15). However, our work does not imply that Notch plays no role in LPC formation. While we have utilized the zebrafish T-ALL model to define the role of Notch in T-ALL development apart from Myc induction, an important corollary is that Myc is a critical target of Notch in mammalian T-ALL, and it is possible that Myc expression plays an important role in the ability of Notch to promote leukemia-propagation in human disease. For example, Li et al. has suggested that Ras over-expression resulting from activation of the Notch/Myc pathway axis likely contributes to LPC development in Notch(ICD) expressing HPCs (14). Further, in a recent study that utilized the TAL1/LMO1 mouse model of T-ALL, spontaneous Notch mutations were found in the Cd3ε+/+ fraction of T-ALL transplantable cells, but not the Cd3ε−/− fraction, which were unable to engraft disease in recipient animals (44). The collaboration of Notch, TAL1 and LMO1 was therefore sufficient to promote LPC formation, which the authors speculate is likely through the ability of Notch to induce Myc and repress p53. In our work, 100% of Myc-expressing T-ALL subclones could initiate disease in recipient animals, with ~1% of cells capable of leukemia- propagation, suggesting the Myc pathway is a potent initiating event in T-ALL. However, hyperplastic Myc expressing thymocytes that are initially confined to the thymus are unable to induce disease in transplanted animals, suggesting that Myc alone is also not sufficient to fully transform thymocytes. Taken together, our data strongly suggests that additional collaborating genetic events are required along with Notch and/or Myc to elicit full transformation to thymocytes into T-ALL and subsequently the creation of LPCs.
In addition to identifying an important role of Notch in expanding T-ALL subclones, our experiments also highlight the use of zebrafish as a powerful model to uncover evolutionarily conserved pathways involved in T-ALL. For example, our microarray and cross-species comparisons identified common gene signatures in zebrafish, mouse, and human T-ALL that are independent of the initiating oncogene, suggesting common molecular pathways underlie T-ALL development. Mining this data set will likely uncover important genes regulating T-ALL initiation and progression. Moreover, our microarray cross-species comparisons revealed a novel Myc signature that is found in a variety of human and mouse malignancies and identified a unique Notch signature that acts independent of Myc in T-ALL. For example, we identified hes1 and il7r as Notch target genes, both of which have been shown to be important Notch targets in human T-ALL and act independently of Myc (45,46). We also validated potential novel Notch target genes in human T-ALL cell lines including MYB, MEIS1 and BCL2, which have emerging roles in T-ALL malignancy but were previously not known to be regulated by Notch (26,47,48). Future work will determine whether these and other Notch target genes contribute to pre-malignant thymocyte expansion, altered apoptotic responses, and/or T-ALL proliferation.
In summary, we have utilized the zebrafish mosaic transgenic T-ALL model to determine that the primary role of Notch signaling is to elicit the expansion of a pool of pre-malignant thymocytes, increasing the likelihood that a subset of these cells will accumulate the necessary mutations to become malignant LPCs. Because Notch-induced clonal expansion occurs even in the presence of Myc over-expression, a model that more accurately mimics the human condition, we expect that this same cellular mechanism likely drives early clonal expansion in human T-ALL.
Supplementary Material
Acknowledgements
We thank Dr. Ravi Mylvaganam for expert advice and assistance with cell sorting, Eric Stone and Marcellino Pena for excellent animal husbandry, and Dr. Finola Moore and Tommy Jones help with data interpretation and critical review of the manuscript. JSB is supported by NIH 5T32CA09126-26. DML is supported by an American Cancer Society Research Scholar Grant, the Leukemia Research Foundation, the Alex Lemonade Stand Foundation, the Harvard Stem Cell Institute, and NIH K01AR05562190.
Footnotes
Conflict-of-interest
The authors declare no competing financial interests.
Supplementary information is available at Leukemia's website.
References
- 1.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J. Clin. Oncol. 2003 Oct 1;21(19):3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong F, Brunet de la Grange P, Gerby B, Rouyez M-C, Calvo J, Fontenay M, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009 Feb 19;113(8):1730–1740. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- 3.Gerby B, Clappier E, Armstrong F, Deswarte C, Calvo J, Poglio S, et al. Expression of CD34 and CD7 on human T-cell acute lymphoblastic leukemia discriminates functionally heterogeneous cell populations. Leukemia. 2011 Aug;25(8):1249–1258. doi: 10.1038/leu.2011.93. [DOI] [PubMed] [Google Scholar]
- 4.Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011 Jan 20;469(7330):362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 5.Clappier E, Gerby B, Sigaux F, Delord M, Touzri F, Hernandez L, et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 2011 Apr 11;208(4):653–661. doi: 10.1084/jem.20110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S-Y, Kumano K, Masuda S, Hangaishi A, Takita J, Nakazaki K, et al. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 2005 Oct;19(10):1841–1843. doi: 10.1038/sj.leu.2403896. [DOI] [PubMed] [Google Scholar]
- 7.Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 8.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol. Cell. Biol. 2000 Oct;20(20):7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 1996 May 1;183(5):2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay A-S, et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood. 2010 Dec 16;116(25):5443–5454. doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006 Jan 15;107(2):781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treanor LM, Volanakis EJ, Zhou S, Lu T, Sherr CJ, Sorrentino BP. Functional interactions between Lmo2, the Arf tumor suppressor, and Notch1 in murine T-cell malignancies. Blood. 2011 May 19;117(20):5453–5462. doi: 10.1182/blood-2010-09-309831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011 Aug 11;118(6):1579–1590. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J. Exp. Med. 2008 Nov 24;205(12):2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010 Feb 12;327(5967):879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 16.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. U.S.A. 2006 Nov 28;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma VM, Draheim KM, Kelliher MA. The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle. 2007 Apr 15;6(8):927–930. doi: 10.4161/cc.6.8.4134. [DOI] [PubMed] [Google Scholar]
- 18.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006 Aug 1;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008 Jan 10;2(1):18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Demarest RM, Dahmane N, Capobianco AJ. Notch is oncogenic dominant in T-cell acute lymphoblastic leukemia. Blood. 2011 Mar 10;117(10):2901–2909. doi: 10.1182/blood-2010-05-286351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996 Aug 1;10(15):1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 22.Pajcini KV, Speck NA, Pear WS. Notch signaling in mammalian hematopoietic stem cells. Leukemia. 2011 Oct;25(10):1525–1532. doi: 10.1038/leu.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003 Feb 7;299(5608):887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007 Mar;21(3):462–471. doi: 10.1038/sj.leu.2404546. [DOI] [PubMed] [Google Scholar]
- 25.Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2005 Apr 26;102(17):6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010 Oct 19;18(4):353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackburn JS, Liu S, Raimondi AR, Ignatius MS, Salthouse CD, Langenau DM. High-throughput imaging of adult fluorescent zebrafish with an LED fluorescence macroscope. Nat Protoc. 2011 Feb;6(2):229–241. doi: 10.1038/nprot.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assaf C, Hummel M, Dippel E, Goerdt S, Müller HH, Anagnostopoulos I, et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood. 2000 Jul 15;96(2):640–646. [PubMed] [Google Scholar]
- 29.Assaf C, Hummel M, Steinhoff M, Geilen CC, Orawa H, Stein H, et al. Early TCR-beta and TCR-gamma PCR detection of T-cell clonality indicates minimal tumor disease in lymph nodes of cutaneous T-cell lymphoma: diagnostic and prognostic implications. Blood. 2005 Jan 15;105(2):503–510. doi: 10.1182/blood-2004-06-2220. [DOI] [PubMed] [Google Scholar]
- 30.Blackburn JS, Liu S, Langenau DM. Quantifying the frequency of tumor-propagating cells using limiting dilution cell transplantation in syngeneic zebrafish. [cited 2012 Feb 29];J Vis Exp [Internet] 2011 (53) doi: 10.3791/2790. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21775966. [DOI] [PMC free article] [PubMed]
- 31.Smith ACH, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010 Apr 22;115(16):3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J.Immunol. Methods. 2009 Aug 15;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007 Jun 1;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat. Med. 2010 Nov;16(11):1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, Dose M, Kovalovsky D, Chang R, O’Neil J, Look AT, et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood. 2007 Jun 15;109(12):5463–5472. doi: 10.1182/blood-2006-11-059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JGCAM, Peters STCJM, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009 Feb;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langenau DM, Keefe MD, Storer NY, Jette CA, Smith ACH, Ceol CJ, et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene. 2008 Jul 10;27(30):4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrando AA, Look AT. Gene expression profiling in T-cell acute lymphoblastic leukemia. Semin. Hematol. 2003 Oct;40(4):274–280. doi: 10.1016/s0037-1963(03)00195-1. [DOI] [PubMed] [Google Scholar]
- 39.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009 Nov 12;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006 Jan 19;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 41.Mori S, Rempel RE, Chang JT, Yao G, Lagoo AS, Potti A, et al. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Emicro-myc model and human diffuse large B-cell lymphoma. Cancer Res. 2008 Oct 15;68(20):8525–8534. doi: 10.1158/0008-5472.CAN-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007 Apr;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JPP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br.J. Haematol. 2008 Oct;143(2):153–168. doi: 10.1111/j.1365-2141.2008.07314.x. [DOI] [PubMed] [Google Scholar]
- 44.Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hébert J, Perreault C, et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010 Jun 1;24(11):1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendorff AA, Koch U, Wunderlich FT, Wirth S, Dubey C, Brüning JC, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010 Nov 24;33(5):671–684. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Magri M, Yatim A, Benne C, Balbo M, Henry A, Serraf A, et al. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur.J. Immunol. 2009 May;39(5):1231–1240. doi: 10.1002/eji.200838765. [DOI] [PubMed] [Google Scholar]
- 47.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007 May;39(5):593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 48.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003 Jul 1;102(1):262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.