Abstract
The purpose of this work was to evaluate a previously proposed approach that aims to improve the point-spread-function (PSF) of MR spectroscopic imaging (MRSI) in order to avoid corruption by lipid signal arising from neighboring voxels. Retrospective spatial filtering can be used to alter the PSF, however, this either reduces spatial resolution or requires extending the acquisition in k-space at the cost of increased imaging time. Alternatively, the method evaluated here, PSF-Choice, can modify the PSF localization to reduce the contamination from adjacent lipids by conforming the signal response more closely to the desired MRSI voxel grid. This is done without increasing scan time or degrading SNR of important metabolites. PSF-Choice achieves improvements in spatial localization through modifications to the RF excitation pulses. An implementation of this method is reported for MRSI of the prostate, where it is demonstrated that, in 13 of 16 pilot prostate MRSI scans, intra-voxel spectral contamination from lipid was significantly reduced when using PSF-Choice. Phantom studies were also performed that demonstrate, compared to MRSI with standard Fourier phase encoding, out-of-voxel signal contamination of spectra was significantly reduced in MRSI with PSF-Choice.
Keywords: point-spread-function (PSF), MR spectroscopic imaging (MRSI), prostate imaging, truncation artifact
Introduction
PSF-Choice, a method that enables one to improve spatial localization by shaping the point-spread-function (PSF) in phase-encoding directions, was introduced previously [1]. The method involves replacing the standard RF excitation with a train of two or more sub-pulses and then modifying the amplitudes of the sub-pulses on each phase-encoding step. The pattern of the RF sub-pulse amplitudes determines the resulting PSF. In the previous work [1] and also in this study, a pattern of sub-pulse amplitudes was chosen to produce a Gaussian-like PSF. The main advantage of the method is that, even when only a very few phase encodes are acquired, as is usually the case in MR spectroscopic imaging (MRSI), there is no truncation (Gibbs ringing) artifact.
It is well known that the effective PSF can be altered by apodizing the k-space data to eliminate unwanted side lobes and ringing [2,3]. However, this post-processing procedure has the effect of reducing the spatial resolution. To maintain a specified spatial resolution while applying apodization, it would be necessary to sample further in k-space and thus increase imaging time. PSF-Choice, on the other hand may be thought of as a prospective apodization. With PSF-Choice, the sampling of higher spatial frequencies (which is necessary to achieve a sharper PSF) is done in the RF domain so that there is no increase in imaging time.
Previous work has demonstrated proof of concept of the PSF-Choice method using a conventional MR imaging sequence tested in phantoms [1] and, more recently, with a modified MRSI pulse sequence tested in animals [4]. In the work reported here and first presented in [5], PSF-Choice has been implemented by modifying an otherwise standard commercial MRSI pulse sequence and evaluated as to its ability to improve spatial localization for prostate MRSI.
Several groups have shown the utility of MRSI to detect biochemical markers in the prostate for in vivo differentiation of prostate cancer from normal tissue or from benign prostatic hyperplasia [6–9]. However, in vivo MRSI for prostate cancer diagnosis and characterization is technically challenging. One serious challenge originates with contamination of spectra from unsuppressed lipid signals [10,11]. Due to the relatively low spatial resolution of MRSI and the sinc-type shape of the PSF inherent to Fourier encoding, lipid signal contributions that ring in from distant voxels often become significant.
The goal of this work was to demonstrate the potential of PSF-Choice to reduce lipid contamination in MRSI of the prostate. To this end, the method was implemented on a clinical MRI system and was evaluated with respect to standard phase-encoded MRSI (without apodization). First, we aimed to show, from the results of phantom experiments, that SNR of important metabolite signals was not adversely impacted with PSF-Choice as compared to the standard phase-encoded MRSI (i.e. that the method does no harm). Next, we aimed to demonstrate, again through phantom experiments, that there is a significant reduction in lipid contamination with PSF-Choice and that this enables a more accurate measurement of parameters based on the content of specific metabolites relevant to the detection of prostate cancer. Finally, we wanted to demonstrate the feasibility of applying the method in prostate MRSI and to show that lipid contamination could be reduced in the in vivo context.
Methods
PSF-Choice Pulse Sequence Modifications
PROSE (PROstate imaging and Spectroscopy Exam) is a version of General Electric’s conventional Point-RESolved Spectroscopy (PRESS) pulse sequence, optimized for use in MRS of the prostate at 1.5T. The sequence was adapted for use at 3T and then further modified by us to enable PSF-Choice encoding in one of the phase-encoding dimensions.
Our pulse sequence modifications to implement PSF-Choice mainly involved a change to the 90-degree pulse by replacing it with a two-pulse RF train. The resultant pulse can be considered as a 2D spatially-selective RF pulse with slice selection along the z direction and some limited spatial selectivity along y. Our implementation also supports an interleaved mode of operation in which one phase encode with PSF-Choice is followed by a standard phase encode, permitting a comparison of PSF-Choice and standard MRSI under virtually identical conditions.
The sequence modification is the same as shown in Fig.1 of reference [1] and can be used to obtain a Gaussian-like PSF in the phase-encoding direction. Because the selection of RF weights (modified on each phase-encoding step) is what determines the exact PSF, in principle, virtually any PSF shape can be achieved with PSF-Choice depending on the number of sub-pulses, the extent of the excitation k-space, and the weighting factors used. In this work, to achieve the desired Gaussian-like PSF, 2xN RF amplitude weights were computed with 2 weights used on each of N encodes (one weight for each of the two RF pulses). The weights were computed using a windowed Gaussian function, fn = cos(pi*n/N)*exp(-a*(n/N)2) where a = 8*log(2) and n = -(N/2):(N/2 -1). Note that the cosine window is applied to remove abrupt truncation of the weights that could result in ringing in the PSF. The sum of the two RF weights used on each encode is roughly constant (within about 2%) so that there is virtually no variation in flip angle from encode to encode.
Phantom Experiments
All phantom experiments were performed on a GE 3T Signa scanner using the quadrature head coil for transmit and receive. Scan parameters were: Orientation=Axial, Spectral samples = 512, k-space matrix size = 24×24, TE/TR = 85/1000ms, Spectral Width = 2000Hz. The 24×24×512 datasets were Fourier transformed in spectral and spatial directions and zero and first order phase corrections were applied to all spectra. Windowing and zero filling were applied in the spectral dimension giving 1024 spectral points (2Hz/point). Though available, spatial saturation bands were not used in the phantom experiments so as to be able to compare the methods as to lipid contamination without any confounding effects from these saturation bands.
SNR Measurements
A small glass spherical container holding a solution of 5mM choline, 15mM creatine and 15mM citrate was used to compare SNR levels for these metabolites from MRSI experiments both with and without PSF-Choice. We aimed to show that SNR with PSF-Choice was not significantly different as compared to the standard phase-encoded MRSI method.
Using the interleaved mode of our sequence, we acquired a dataset with PSF-Choice encoding and a dataset with standard phase encoding in a single scan. Each scan produced an array of 24×24 spectra for the selected slice plane. From the 576 voxels, a subset of 62 voxel spectra was then selected for further processing according to whether there was sufficient signal. For each of the choline, creatine and citrate resonances, total signal intensity was obtained by integrating the absolute signal level over the appropriate frequency ranges.
Fat Contamination Measurements
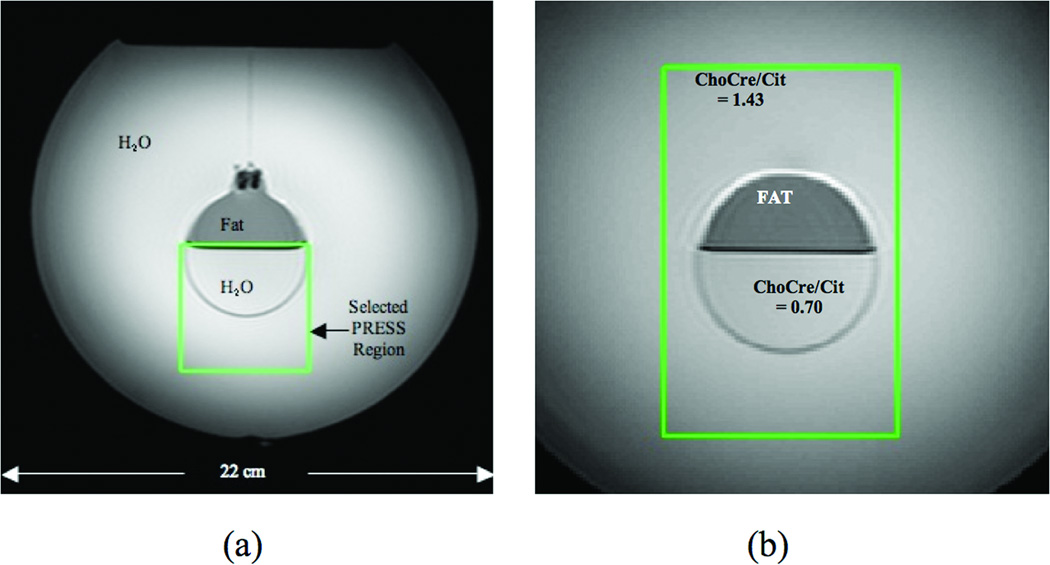
A contamination-assessment phantom was constructed using a 4-liter volume plastic spherical container holding water. A smaller spherical glass container filled with 1/2 water and 1/2 corn oil was suspended at the center of the larger sphere. Figure 1a shows an axial image through the center of the co-spherical phantom and illustrates the fat-water boundary oriented perpendicular to the gravity field with the fat layer lying above the water.
Figure 1.
Reference images of co-spherical phantoms used in experiments to compare PSF-Choice and standard phase-encoded MRSI. (a) Contamination-assessment phantom containing a fat compartment (top of inner sphere) and water (outer sphere and bottom of inner sphere). The PRESS VOI is placed with top at boundary between fat and water. (b) Diagnostic-quality phantom containing a fat compartment (top of inner sphere), a solution of choline, creatine and citrate with ChoCre/Cit = 0.70 (bottom of small sphere) and a solution with ChoCre/Cit = 1.43 (in outer sphere). The green box shows the position of the selected PRESS VOI for experiments.
A PRESS VOI (green box in Fig.1a) was selected and positioned so that the upper edge of the selected volume was at the boundary between the fat and water compartments. Scan parameters were as above with the FOV set to 120mm. From each reconstructed dataset, a subset of 9 voxel columns perpendicular to the fat-water boundary that ran from 1 voxel above to 11 voxels below the fat-water boundary was selected for analysis. The encoding direction for method comparison was set perpendicular to the fat-water boundary (i.e., in the A-P direction).
The mean signal magnitude (after baseline removal) was calculated within a spectral range where fat signal was present. The standard deviation of the signal within a spectral range where there was no significant signal from either water or fat (i.e., a range where only noise should appear – from 8.7 to 9.3 ppm) was computed as an estimate of the noise level. The ratio of the mean signal across the fat peak to the standard deviation in the noise spectral range was then computed as the measure of fat content in a voxel spectrum.
Baseline removal was used to remove the tail of the water signal, which might otherwise bias the fat measurement. An average spectrum from a water-only scan was used to derive a baseline vector. This vector was in turn used to estimate (via least-squares fitting) the component due to the water tail in the fat spectra so the bias component could then be removed by subtraction.
Metabolite Ratio Measurements
A diagnostic-quality phantom was constructed using a 4-liter volume plastic spherical container holding a solution of 2.5mM choline, 7.5 mM creatine and 7.5 mM citrate. A smaller glass spherical container, filled with corn oil in the top half and a solution of 5mM choline, 15mM creatine and 50mM citrate in the bottom half, was suspended at the center of the larger sphere. Experiments with this co-spherical phantom aimed to demonstrate that, in the presence of fat contamination, the PSF-Choice method gives better estimates of choline+creatine to citrate ratios (ChoCre/Cit).
ChoCre/Cit of the solution in the large sphere was 1.43±0.18 for the TE and TR settings used. Note that significantly elevated ChoCre/Cit ratios are thought to be associated with cancerous tissue [6,12]. In the smaller glass spherical container, the ChoCre/Cit of the solution (in the bottom half of the sphere) was 0.70±0.07. For these measurements, 64 repeated single-voxel acquisitions were averaged from a PRESS VOI placed in the respective regions.
The PRESS VOI (Fig.1b) was positioned to include the entire inner sphere (containing the fat compartment and the aqueous compartment consisting of a solution with a ChoCre/Cit ratio simulating normal tissue) plus a large contiguous region within the outer sphere (containing the solution with ChoCre/Cit ratio simulating cancer). Scan parameters were the same as in the previous phantom experiment except for the FOV, which was set at 160mm.
ChoCre/Cit was calculated by computing the mean of the intensity within the frequency ranges corresponding to choline and creatine and dividing by the mean intensity within the frequency range of citrate. Images of ChoCre/Cit were thereby produced and interpolated so as to have the same pixel size as the T2-weighted reference image of the phantom. The T2-weighted reference image of the phantom was manually segmented to separately identify regions of fat and the regions with different ChoCre/Cit values (low = 0.70 and high = 1.43).
Statistical tests
In all statistical comparisons between PSF-Choice and standard phase encoding, in terms of determining if there were significant differences in the measurements made by the two methods, the Student-t test was used and the significance level was set at 0.05.
Prostate MRSI Scans
The feasibility of PSF-Choice prostate MRSI was evaluated in a study at 3 Tesla with 16 patients, all of whom had biopsy-proven prostate cancer. All imaging was performed on a 3T GE Signa scanner. The body coil was used for transmit and endo-rectal coil for receive. Institutional Review Board approval was obtained for this study, compliant with the Health Insurance Portability and Accountability Act, and written informed consent was obtained from all patients. A single 1.5cm slice was encoded using a 14×14 matrix in a 12cm FOV (14cm FOV for two of the patients) with TE/TR = 85/1000 ms. The slice was chosen to be well within the prostate, with no special attention being made to select for areas suspected of containing tumor tissue. Saturation bands available with the product sequence were enabled for the in vivo scans. The encoding direction for comparison of the methods was R-L. The average PRESS VOI for the 16 patients contained about 18 voxels (min=8, max=24, std=5.1). Data for both the PSF-Choice method and for standard Fourier phase encoding were acquired in the interleaved mode described previously and the total scan time was 6.5 minutes. Reconstruction was performed as it was for the phantom scans, with no zero filling or filtering in the spatial dimensions.
Results from the scans were evaluated by an expert reader (an ABR certified MRI physicist) who was blinded as to which method was used in acquiring the spectra. Using a display tool (programmed in Matlab), the reader was presented with spectra (1 to 3.5 ppm) for all voxels acquired during one of the exams and was asked to decide: (1) which result had the greatest proportion of spectra contaminated by fat when looking at voxels outside the PRESS VOI (i.e., in the void where there should be no signal at all), and (2) which result had the greatest proportion of spectra contaminated by fat when looking within the prostate.
Results
Phantom Experiments
SNR Measurements
In the phantom SNR measurements, no significant difference was found between the two methods in the mean of the SNR of the Choline, Creatine and Citrate peaks (p=0.40, p=0.78, p=0.36 respectively). No significant difference was found in the values of the ChoCre/Cit ratios between the two methods (p=0.57). We also looked at the water signal intensity and found that it was almost indistinguishable using the two different methods (p=0.58).
Fat Contamination Measurements
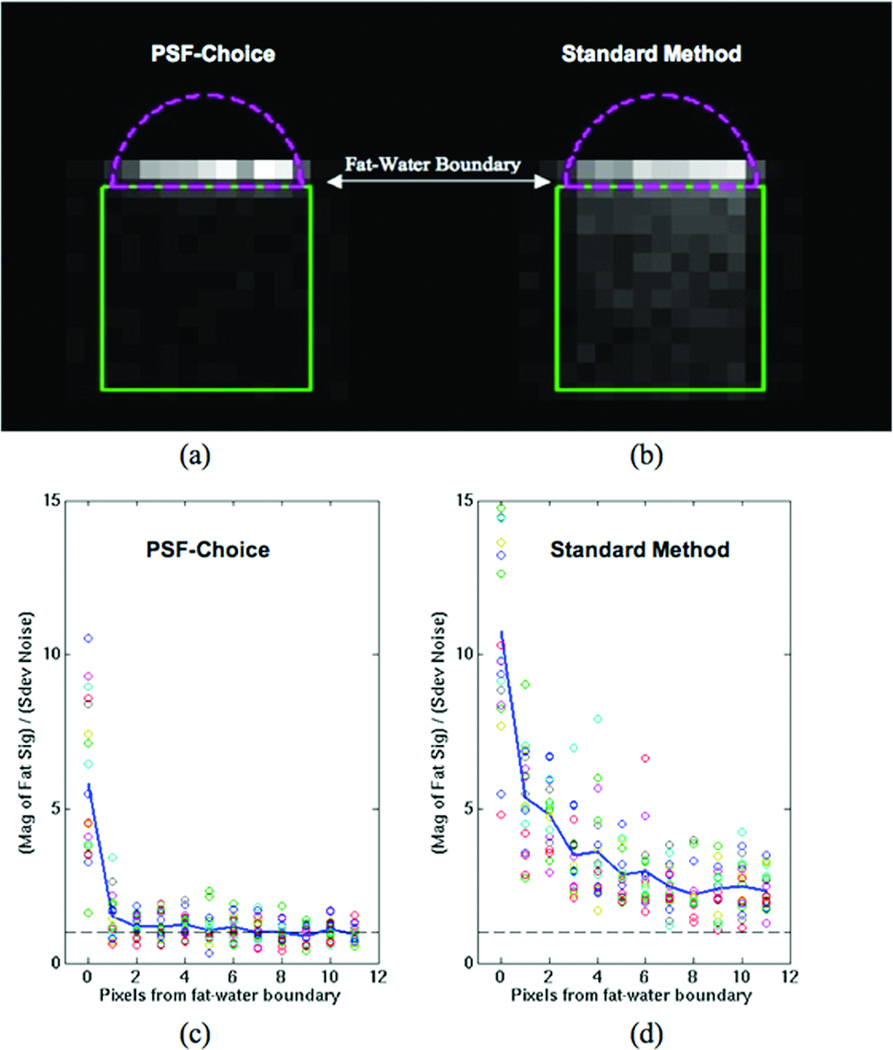
Figure 2 shows images representing the fat content (contamination) for both PSF-Choice encoding (Fig.2a) and for Fourier encoding (Fig.2b). Plots in Fig.2c and 2d show the fat content in each voxel (plotted as circles) for each distance from the fat-water boundary. A much higher degree of fat signal content for Fourier encoding (Fig.2d) compared to PSF-Choice encoding (Fig.2c) is evident at all distances from the fat-water boundary. The fat content at each distance from the fat-water boundary was compared for both methods and the fat signal content was found to be significantly greater for Fourier encoding at all pixel distances from the boundary (e.g., p=0.0005 at 1 pixel from boundary with p less than this for all other distances out to 11 pixels). At the boundary itself, as expected, the difference between the methods is not significant because of the sizeable partial volume contributions right at the boundary.
Figure 2.
Results from contamination-assessment phantom for PSF-Choice and the Standard Method (Fourier phase encoding). The images represent the fat content (contamination) for (a) PSF-Choice and (b) the Standard Method. Images are the ratio of the mean of the signal magnitude in a spectral range for fat to the standard deviation in a spectral range containing only noise. The green box shows the position of the PRESS VOI placed at the fat-water boundary. Both images are windowed identically. Encoding for comparison of methods was the vertical (A-P) direction.
The plots show the fat content for (c) PSF-Choice and (d) the Standard Method as a function of the distance from the fat-water boundary. Each circle represents the content in 1 pixel.
Metabolite Ratio Measurements
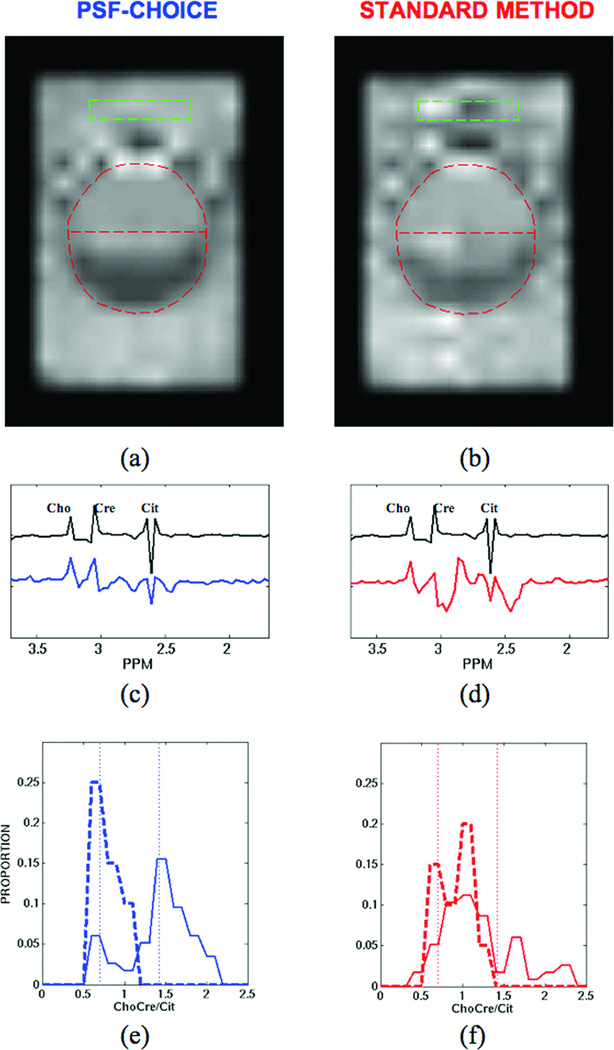
The images in Fig.3a and 3b show ChoCre/Cit ratios obtained with the diagnostic quality phantom using PSF-Choice (Fig.3a) and Standard Fourier encoding (Fig.3b). PSF-Choice was applied in the vertical direction. Note that the ChoCre/Cit image for PSF-Choice is more homogeneous than the image for Fourier encoding, both inside and outside the inner sphere (see reference image - Fig.1b).
Figure 3.
Results from diagnostic-quality phantom for PSF-Choice and the Standard Method (Fourier phase encoding): Images showing ChoCre/Cit for (a) PSF-Choice and (b) the Standard Method. The ChoCre/Cit images were interpolated and registered to the reference image shown in Fig.1b. The position of the inner sphere is indicated by the red dashed line. Black plots in (c) and (d) show a reference spectrum obtained from the compartment simulating cancer (with ChoCre/Cit = 1.43). Below the reference plots are spectra from within the simulated cancer compartment (at location indicated by green box) for both PSF-Choice (blue plot in (c)) and for standard phase encoding (red plot in (d)). Plots for (e) PSF-Choice and (f) for standard phase encoding show the distribution of metabolite ratios for the two compartments (cancer = solid line, normal = dashed line). The true values of the ratio for each of the two compartments (0.7 and 1.43 ppm) are indicated by the vertical dotted lines.
The plots in Fig.3c and 3d are of spectra obtained from the phantom. Spectra were obtained from an area (green box in images in Fig.3a and 3b) within the simulated cancer compartment for both PSF-Choice (blue plot in Fig.3c) and standard phase encoding (red plot in Fig.3d). Note the significant spectral contamination from fat for standard phase encoding results. The plot in black in Fig. 3c and 3d is a reference spectrum obtained from the compartment with the ratio measuring 1.43 (simulating ‘cancer’).
The plots in Fig.3e and 3f show the distribution of metabolite ratios for the two compartments (cancer = solid line, normal = dashed line). The true value of the ratio for each of the two compartments (0.7 and 1.43 ppm) is indicated by the dotted lines. For PSF-Choice (blue plots in Fig.3e), the distributions are reasonably well centered on the true values. For standard Fourier encoding (red plots in Fig.3f) there is a very poor correspondence between the distributions for ratios and the true values measured from the reference spectra, especially for the cancer component (solid red line in Fig.3f).
Table 1 gives the mean values and standard deviations for ChoCre/Cit measurements in the simulated normal compartment (N=20) and in the simulated cancer compartment (N=58) along with mean of error magnitude. Error magnitude was computed as magnitude of the difference between compartment measurements and the true values of 0.70 and 1.43 respectively. Note that the magnitude error in estimation of ChoCre/Cit in the ROIs with standard phase encoding is significantly higher than with PSF-Choice for the cancer compartment (p=0.00006). It is also higher for the normal compartment although the difference is not significant (p=0.2).
Table 1.
Comparison ChoCre/Cit measurement for PSF-Choice and standard phase encoding using the diagnostic-quality phantom. Error magnitude was computed as magnitude of the difference between Normal compartment measurements (N=20) and Cancer compartment measurements (N=58) and the true values of 0.70 and 1.43 respectively.
| Mean and Standard Deviation (sd) of ChoCre/Cit Ratio |
Normal Compartment True Value=0.70 sd = 0.07 |
Cancer Compartment True Value=1.43 sd = 0.18 |
Mean of Error Magnitude in Normal Compartment Measurement |
Mean of Error Magnitude in Cancer Compartment Measurement |
|---|---|---|---|---|
| PSF-Choice | 0.74 sd = 0.15 |
1.36 sd = 0.38 |
0.14 | 0.29 |
| Standard Method | 0.87 sd = 0.20 |
1.12 sd = 0.43 |
0.20 | 0.47 |
| Error (PSF-Choice) < Error (Standard Method) → | P=0.2 | P=0.0002 | ||
Prostate MRSI Scans
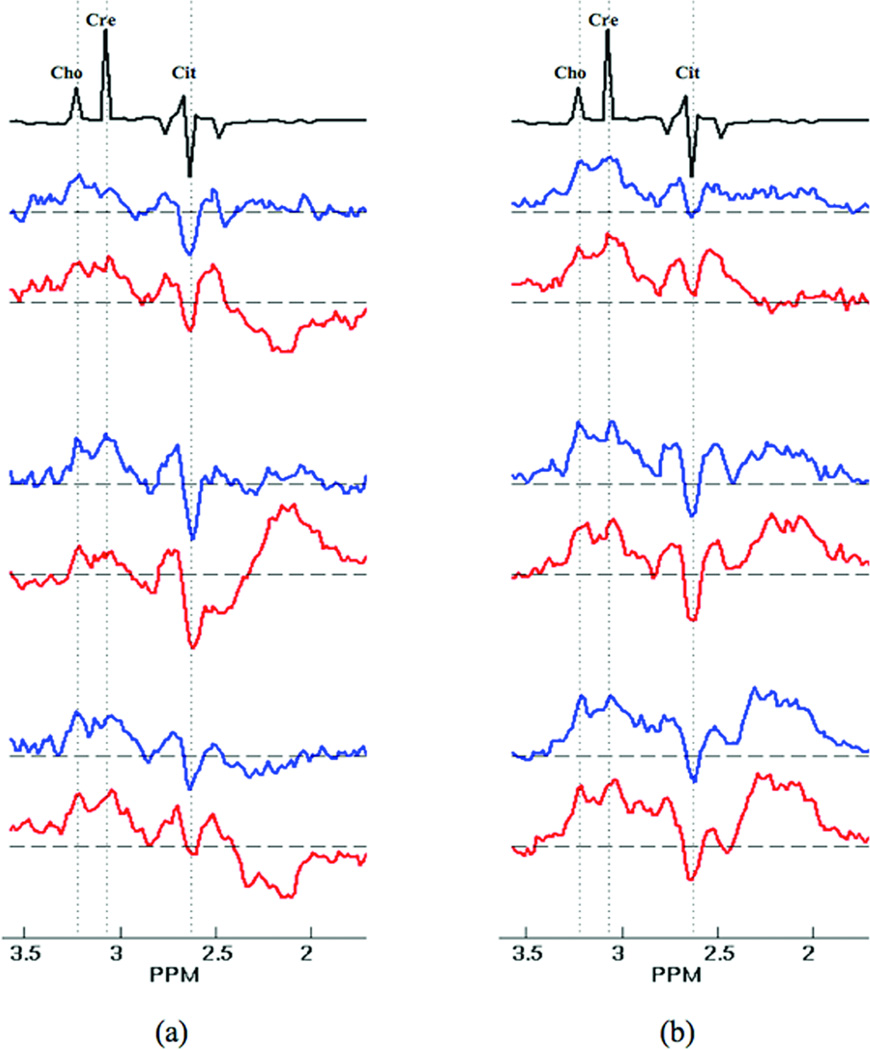
Figure 4 shows a sample of 8 voxel spectra from one of the 16 patients in the in vivo study. For reference, spectra obtained from a phantom experiment are shown (black plots) at the top of Fig.4. In Fig. 4a, the spectra obtained with PSF-Choice (blue plots) exhibit noticeably lower lipid contamination than those obtained with standard phase encoding (red plots). The voxel spectra shown in Fig.4b exhibit a roughly equivalent amount of lipid contamination for both methods.
Figure 4.
An array of spectra from 8 voxels within the prostate of one of the 16 patients along with reference spectra (plots in black) obtained from the spectral-quality phantom. (a) In one set of plots, those spectra obtained using standard phase encoding (red plots) were judged to be overly contaminated with lipid. The spectra obtained with PSF-Choice (blue plots), however, were judged to be free of this contamination. (b) In another set of spectra from the same patient, there was judged to be no significant difference between the two methods. Note that in both the phantom reference and in vivo spectra, the citrate resonance is inverted as expected for the parameters (TE=85ms) due to J-modulation of the strongly coupled citrate protons at 3T.
In the assessments by the expert reader, the spectral data obtained by standard phase encoding was judged to have more contamination in 100% of cases where a determination was made. In 13 out of the 16 patient exams, the expert reader judged that there was more fat contamination outside the selected PRESS VOI in the standard phase-encoded data as compared to the PSF-Choice data. In the remaining 3 cases, contamination was judged as equivalent in the spectra acquired by the two methods. In 10 out of the 16 patient exams, the expert reader judged that there was more fat contamination seen inside the prostate in the standard phase-encoded data as compared to the PSF-Choice data (e.g. similar to the case for the spectra in Fig.4a). In the remaining 6 cases, contamination was equivalent in the spectra acquired by the two methods (e.g. similar to the case for spectra in Fig.4b). Note that there was not a single case in which the standard phase-encoded result was considered to have less fat contamination compared to the PSF-Choice result.
Discussion
While the in vivo study with prostate patients reported here showed an advantage of PSF-Choice in terms of reducing lipid contamination, only a single slice was acquired and the in-plane resolution was relatively low compared to the current state-of-the-art in clinical prostate MRSI. Further conclusions concerning the ability of the method to improve cancer detection are limited given both the small number of patients studied and the limited volume of prostate interrogated by MRSI in each patient.
The J-modulation of strongly coupled citrate protons at 3T led to inverted citrate peaks at the TE of 85 ms used in this study [13]. Use of the MLEV-PRESS sequence has been proposed to obtain upright citrate peaks at similar TE values, which could help to improve quantification of prostate spectra at 3T [14]. However, this sequence modification was not used in this study and spectral quality is unfortunately not as high as it could be with respect to resolving and quantifying the citrate resonances.
If one defines the effective spatial resolution by the width of the PSF (e.g. full width at half maximum), we would claim that the spatial resolution with PSF-Choice remains equivalent to that obtained with standard phase encoding. The RF weights in PSF-Choice were designed so that the PSF width would be FOV/N where N is equal to the number of encodes. The width of the sinc-shaped PSF in standard phase encoding (with N encodes) is also equal to FOV/N. Although the spatial resolution remains equivalent in the two cases, because the Gaussian-like PSF obtained through PSF-Choice encoding is free of the side lobes that lead to pixel bleed, PSF-Choice can be said to improve spatial localization.
It should be noted that, although we have focused here on the negative effects of the lipid signal, the changes due to applying PSF-Choice affect all voxels and all metabolite signals. Thus, individual metabolite images reconstructed using data from standard phase encoding will also show the effects of ringing (although this may or may not be evident in those images depending on SNR). Thus, although not demonstrated in this study, PSF-Choice will also have the positive effect of improving spatial localization of metabolite signals by eliminating ringing and reducing pixel bleed.
Of course, retrospective spatial filtering (or apodization) also alters the effective PSF and could easily have been used to reduce pixel bleed and lipid contamination for the standard phase-encoded data. However, such filtering is done at the expense of a broadening of the PSF (by a factor of 2 with the Hanning filter). So, while the filtering effectively eliminates signal contributions from distant voxels and can be said to improve spatial localization, broadening the PSF width increases the effective voxel size and, therefore, can also be said to degrade spatial localization.
Approaches other than retrospective spatial filtering have been proposed that aim to reduce or eliminate truncation artifact without adversely affecting spatial resolution. In one unique approach, a standard MRS dataset, acquired at low spatial resolution and high SNR, was combined with a second MRS dataset acquired at high spatial resolution and low SNR [15–18]. These methods appear to be quite robust, however, they involve the complication of combining datasets from different scans and introduce some additional cost in imaging time. Webb et al [19] proposed a variable-tip-angle approach to create k-space weighting that has a similar effect to spatial filtering in terms of reducing pixel bleed. So as not to degrade spatial resolution from the weighing, the number of phase-encoding steps was increased. However, imaging time was not increased with this method because the TR for the higher k-space encoding steps was shortened.
PSF-Choice involves the same number of total encodes as standard phase encoding. Thus, it neither increases nor decreases total acquisition time. As was shown experimentally, PSF-Choice also maintains SNR of the metabolite peaks relative to standard phase encoding. Although the method involves RF manipulation and alteration of the spatial excitation patterns (as in standard phase encoding), the vast majority of spins are fully excited on each encode to maximize SNR. In PSF-Choice it is primarily the distribution of the phase of the transverse magnetization (rather than the magnitude) that differs from standard Fourier phase encoding.
In summary, PSF-Choice has been validated in a series of phantom experiments and its feasibility assessed in a study with 16 patients with biopsy-proven prostate cancer. Our phantom experiments quantitatively demonstrate the advantage of PSF-Choice over standard Fourier phase encoding (without retrospective spatial filtering) in terms of its ability to greatly reduce contamination of spectra due to fat signal ringing-in from neighboring voxels. This method has the potential to improve quantification of metabolite ratios important in the detection of prostate cancer. Future work is focusing on implementing the method in additional directions in order to fully exploit its benefits [20].
Acknowledgements
NIH R21-CA110092, R33-CA110092, P01-CA067165, P41-RR019703.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Panych LP, Zhao L, Mulkern RV. PSF-choice: a novel method for shaping point-spread-functions in phase-encoding dimensions. Magn Reson Med. 2005;54(1):159–168. doi: 10.1002/mrm.20525. [DOI] [PubMed] [Google Scholar]
- 2.Wood ML, Henkelman RM. Truncation artifacts in magnetic resonance imaging. Magn Reson Med. 1985;2(6):517–526. doi: 10.1002/mrm.1910020602. [DOI] [PubMed] [Google Scholar]
- 3.Vikhoff-Baaz B, Starck G, Ljungberg M, Lagerstrand K, Forssell-Aronsson E, Ekholm S. Effects of k-space filtering and image interpolation on image fidelity in (1)H MRSI. Magn Reson Imaging. 2001;19(9):1227–1234. doi: 10.1016/s0730-725x(01)00456-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen AP, Hurd RE, Cunningham CH. Proceedings of the Joint Annual Meeting ISMRM-ESMRMB 2010. Sweden: Stockholm; 2010. Non-Fourier spatial encoding for improved point-spread-functions in hyperpolarized CSI acquisitions; p. P3262. [Google Scholar]
- 5.Panych LP, Roebuck JR, Mulkern RV, Tang Y, Madore B, Chen N-K. Proceedings of the ISMRM 17th Scientific Meeting and Exhibition. Hawaii, USA: Honolulu; 2009. Improving spatial localization in MR spectroscopic imaging with PSF-Choice; p. P335. [Google Scholar]
- 6.Kurhanewicz J, Vigneron DB, Hricak H, Narayan P, Carroll P, Nelson SJ. Three-dimensional H-1 MR spectroscopic imaging of the in situ human prostate with high (0.24-0.7-cm3) spatial resolution. Radiology. 1996;198(3):795–805. doi: 10.1148/radiology.198.3.8628874. [DOI] [PubMed] [Google Scholar]
- 7.Scheidler J, Hricak H, Vigneron DB, Yu KK, Sokolov DL, Huang LR, Zaloudek CJ, Nelson SJ, Carroll PR, Kurhanewicz J. Prostate cancer: localization with three-dimensional proton MR spectroscopic imaging- clinicopathologic study. Radiology. 1999;213(2):473–480. doi: 10.1148/radiology.213.2.r99nv23473. [DOI] [PubMed] [Google Scholar]
- 8.Yuen JS, Thng CH, Tan PH, Khin LW, Phee SJ, Xiao D, Lau WK, Ng WS, Cheng CW. Endorectal magnetic resonance imaging and spectroscopy for the detection of tumor foci in men with prior negative transrectal ultrasound prostate biopsy. J Urol. 2004;171(4):1482–1486. doi: 10.1097/01.ju.0000118380.90871.ef. [DOI] [PubMed] [Google Scholar]
- 9.Hasumi M, Suzuki K, Taketomi A, Matsui H, Yamamoto T, Ito K, Kurokawa K, Aoki J, Endo K, Yamanaka H. The combination of multi-voxel MR spectroscopy with MR imaging improve the diagnostic accuracy for localization of prostate cancer. Anticancer Res. 2003;23(5b):4223–4227. [PubMed] [Google Scholar]
- 10.Verma S, Rajesh A, Futterer JJ, Turkbey B, Scheenen TW, Pang Y, Choyke PL, Kurhanewicz J. Prostate MRI and 3D MR spectroscopy: how we do it. Am J Roentgenol. 2010;194(6):1414–1426. doi: 10.2214/AJR.10.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schricker AA, Pauly JM, Kurhanewicz J, Swanson MG, Vigneron DB. Dualband spectral-spatial RF pulses for prostate MR spectroscopic imaging. Magn Reson Med. 2001;46(6):1079–1087. doi: 10.1002/mrm.1302. [DOI] [PubMed] [Google Scholar]
- 12.Kurhanewicz J, Vigneron DB, Nelson SJ, Hricak H, MacDonald JM, Konety B, Narayan P. Citrate as an in vivo marker to discriminate prostate cancer from benign prostatic hyperplasia and normal prostate peripheral zone: detection via localized proton spectroscopy. Urology. 1995;45(3):459–466. doi: 10.1016/S0090-4295(99)80016-8. [DOI] [PubMed] [Google Scholar]
- 13.Mulkern RV, Bowers JL, Peled S, Kraft RA, Williamson DS. Citrate signal enhancement with a homonuclear J-refocusing modification to double-echo PRESS sequences. Magn Reson Med. 1996;36(5):775–780. doi: 10.1002/mrm.1910360517. [DOI] [PubMed] [Google Scholar]
- 14.Chen AP, Cunningham CH, Kurhanewicz J, Xu D, Hurd RE, Pauly JM, Carvajal L, Karpodinis K, Vigneron DB. High-resolution 3D MR spectroscopic imaging of the prostate at 3 T with the MLEV-PRESS sequence. Magn Reson Imaging. 2006;24:825–832. doi: 10.1016/j.mri.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Metzger G, Sakar S, Zhang X, Heberlein K, Patel M, Hu X. A hybrid technique for spectroscopic imaging with reduced truncation artifact. Magn Reson Imaging. 1999;17(3):435–443. doi: 10.1016/s0730-725x(98)00187-8. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar S, Heberlein K, Hu X. Truncation artifact reduction in spectroscopic imaging using a dual-density spiral k-space trajectory. Magn Reson Imaging. 2002;20(10):743–757. doi: 10.1016/s0730-725x(02)00608-2. [DOI] [PubMed] [Google Scholar]
- 17.Ebel A, Maudsley AA. Comparison of methods for reduction of lipid contamination for in vivo proton MR spectroscopic imaging of the brain. Magn Reson Med. 2001;46(4):706–712. doi: 10.1002/mrm.1249. [DOI] [PubMed] [Google Scholar]
- 18.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging. 2003;21(2):113–120. doi: 10.1016/s0730-725x(02)00645-8. [DOI] [PubMed] [Google Scholar]
- 19.Webb AG, Briggs RW, Mareci TH. Volume-localized spectroscopy using selective Fourier transform with windowing by variable-tip-angle excitation. J Magn Reson. 1991;94:174–179. [Google Scholar]
- 20.Panych LP, Madore B, Hoge WS, Mulkern RV. Proceedings of the 19th Annual Meeting of the ISMRM. Montreal, Canada: 2011. Improved spatial localization in 3D MRSI with a sequence combining PSF-Choice, EPSI and a resolution-enhancement algorithm; p. P3452. [Google Scholar]