Abstract
Multidrug resistance protein 2 (MRP2, ABCC2) is an efflux membrane transporter highly expressed in liver, kidney and intestine with important physiological and pharmacological roles. The goal of this study was to investigate the functional significance of promoter region polymorphisms in ABCC2 and potential allele specific expression. Twelve polymorphisms in the 1.6 kb region upstream of the translation start site were identified by resequencing 247 DNA samples from ethnically diverse individuals. Luciferase reporter gene assays showed that ABCC2 -24C>T both alone and as part of a common haplotype (-24C>T/-1019A>G/-1549G>A) increased promoter function 35% compared to the reference sequence (P < 0.0001). No other common variants or haplotypes affected ABCC2 promoter activity. Allele specific expression was also investigated as a mechanism to explain reported associations of the synonymous ABCC2 3972C>T variant with pharmacokinetic phenotypes. In Caucasian liver samples (n=41) heterozygous for the 3972C>T polymorphism, the 3972C allele was preferentially transcribed relative to the 3972T allele (P < 0.0001). This allelic imbalance was particularly apparent in samples with haplotypes containing two or three promoter/UTR variants (-1549G>A, -1019A>G and -24C>T). The observed allelic imbalance was not associated with hepatic or renal ABCC2 mRNA expression. Additional mechanisms will need to be explored to account for the interindividual variation in ABCC2 expression and MRP2 function.
Keywords: ABC transporter, MRP2, ABCC2, pharmacogenetics, promoter, allelic imbalance
INTRODUCTION
The multidrug resistance protein 2 (MRP2), encoded by ABCC2, is a member of the ATP-binding cassette (ABC) family of membrane transporters. MRP2 is predominantly expressed in the canalicular membrane of hepatocytes, but can also be detected in the apical membranes of renal proximal tubule and intestinal epithelial cells.1, 2 MRP2 plays an important role in the pharmacokinetics of a wide variety of compounds. This transporter drives energy dependent efflux of organic anions, often conjugated to glutathione (GSH), glucuronic acid or sulfate, into the bile or urine for elimination. It can also transport neutral or basic compounds, but requires GSH for co-transport.3, 4 MRP2 substrates include endogenous compounds such as GSH, leukotriene C4,17β-D-estradiol glucuronide and the heme metabolite bilirubin glucuronide as well as xenobiotics such as the HMG-CoA reductase inhibitor pravastatin, and the anti-cancer drugs irinotecan and its glucuronide metabolite SN-38 glucuronide, cisplatin and methotrexate.5, 6
The importance of MRP2 mediated transport has been clearly demonstrated in mutant rat models lacking Mrp2 expression1 and in Dubin-Johnson syndrome patients with a hereditary deficiency of MRP2 caused by rare mutations in the coding region of ABCC2.5–7 Also, a number of common ABCC2 non-synonymous polymorphisms have been associated with alterations in ABCC2 mRNA expression and/or MRP2 function8–12 as well as with drug toxicities.13–15
The functional effects of ABCC2 coding region and promoter polymorphisms remain controversial. In particular, the mechanisms by which promoter and synonymous polymorphisms influence MRP2 expression are unclear. Currently, there is limited information about transcriptional regulation of human ABCC2. Several studies have characterized a region between 197 and 517 bases upstream from the translation start site with important basal promoter activity.16–18 This region contains consensus sequences for a number of transcription factors, including the TATA box, the liver abundant CCAAT-enhancer binding protein and HNF1/4. Single nucleotide polymorphisms (SNPs) in the promoter region may alter transcriptional activity and contribute to interindividual variability in MRP2 expression and the pharmacokinetics of MRP2 substrates.
To date, a number of SNPs have been associated with the pharmacokinetics, efficacy and toxicity of MRP2 substrates, although most findings have not been replicated19–22. For example, the ABCC2 -24C>T polymorphism has been shown to decrease,10, 19–21 increase8 as well as have no effect on ABCC2 expression.23–25 Interestingly, the ABCC2 synonymous variant 3972C>T, which is in linkage disequilibrium with a number of promoter region polymorphisms, was associated with exposure to irinotecan and its metabolite SN-38 glucuronide.22, 26 Patients who were homozygous for the 3972T allele had higher AUCs for irinotecan and SN-38 glucuronide compared to the combined group of patients having the CT or CC genotype. The higher AUC is consistent with decreased activity and/or expression of MRP2. In contrast, other studies have shown no effect of this polymorphism on ABCC2 expression and MRP2 function. 9, 10, 27–29
The current study represents a comprehensive functional analysis of genetic variants in the promoter region of ABCC2. Based on the reported associations of ABCC2 3972C>T with various phenotypes22, 30, 31, allele specific expression was also investigated as a possible mechanism conferring functional significance for this synonymous SNP.
MATERIALS and METHODS
Tissue samples
Two hundred human liver samples were provided by the Liver Tissue Procurement and Distribution System NIH Contract #N01-DK-9-2310 and by the Cooperative Human Tissue Network and were processed through Dr. Mary Relling’s laboratory at St. Jude Children’s Research Hospital (Memphis, TN); these liver samples were genotyped and used for ABCC2 allele specific expression experiments. Additional kidney (n=56) and liver (n=34) samples were provided by the Pharmacogenetics of Membrane Transporters tissue repository at the University of California San Francisco and were used to correlate ABCC2 3972C>T genotype with total ABCC2 mRNA expression.
Genetic Analysis of the 5′-region of ABCC2
A collection of 247 ethnically diverse genomic DNA samples were obtained from the Coriell Institute of Medical Research (http://coriell.umdnj.edu). This collection of samples was used to screen for genetic variations in a 1.6 kb upstream region of ABCC2. Initial screening was done using PCR, denaturing HPLC and direct sequencing as previously described.32
Plasmid Construction
A 1.6 kb region upstream of the ABCC2 translation start site was PCR amplified from genomic DNA and subcloned into pCR2.1 using the standard protocol from the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The amplified promoter DNA was digested with NheI and XhoI (New England Biolabs, Beverly, MA) and was cloned into the reporter gene expression vector pGL4.11b (Promega, Madison, WI), which contains a luciferase gene downstream from the multiple cloning site. Compared to the earlier generation pGL3 vectors, the pGL4 vectors are engineered with fewer consensus regulatory sequences and a synthetic gene which has been codon optimized for mammalian expression. Individual non-singleton variants or variants in haplotypes were introduced using specific primers (Supplemental Table 1) and the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmids were sequenced to verify the correct introduction of variants and were isolated as endotoxin-free preparations, a step that has been shown to eliminate purity bias and to significantly improve interassay variability.
Cell Culture and Reporter Gene Assay
HepG2 cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS). A day prior to transfection cells were counted and seeded on 96 well plates at a density of 5 × 104 cells per well. pGL4.11b empty vector or plasmids containing either reference or variant sequences were transiently transfected together with Renilla luciferase vector into HepG2 cells using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Renilla luciferase vector served as a transfection efficiency control. Cells were incubated for 24 hours at 37°C in humidified atmosphere with 5% CO2, after which they were washed with phosphate buffered saline and incubated with 80 μL of passive lysis buffer (Promega, Madison, WI) at room temperature on a shaker for 30 min. Aliquots were analyzed for luciferase activity in a dual luciferase reporter assay on a GloMax™ 96 luminometer (Promega, Madison,WI) according to the manufacturer’s instructions. The activities of Firefly and Renilla luciferases were measured sequentially from each sample and both reporters yield linear assays with subattomole (<10−18) sensitivities and no endogenous activity in the host cells.
Genotyping
Genomic DNA was genotyped using a 5′-nuclease assay 33 and direct sequencing. The ABCC2 5′-promoter variants -1549G>A (rs1885301) and -1019A>G (rs2804402), the UTR variant -24C>T (rs717620), the coding region variant 1249G>A (rs2273697), the intron 26 variant -34T>C (rs17216177), and the synonymous variant 3972C>T (rs3740066) were genotyped in 200 liver samples. Briefly, PCR for direct sequencing was carried out after optimization using a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA). Sequencing analysis was done on an ABI 3700 automated sequencer using the ABI PRISM BigDye system. For Taqman 5′-nuclease sequencing, PCR and sequencing were performed in one reaction using specific primers for the variant of interest, either by Assays-on-Demand (-24C>T and 3972C>T; Applied Biosystems, Foster City, CA) or Assays-by-Design (primer/probes in Supplemental Table 2). The ABCC2 3972C>T SNP was also genotyped in 56 liver and 34 kidney samples using the DMET array chip on an Illumina platform; the ABCC2 3972C>T SNP was tagged by rs7067971 SNP (r2 = 1.0). In all genotyping assays, SNPs were checked for deviations from reported minor allele frequencies and also for Hardy-Weinberg Equilibrium.
Reverse Transcription, PCR and Single Base Extension
RNA from human liver samples heterozygous for the ABCC2 3972C>T SNP were selected for reverse transcription and further analysis. One microgram of RNA was reverse transcribed using the SuperScript II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Following reverse transcription, approximately 100 bp surrounding the 3972C>T SNP site was PCR amplified from the resulting cDNA and genomic DNA (gDNA) using specific primers. Excess dNTPs and primers were removed from the amplified product by incubating in shrimp alkaline phosphatase (Promega, Madison, WI) and ExoI (New England Biolabs, Beverly, MA). PCR products then underwent single base extension using the SNaPshot mix (Applied Biosystems, Foster City, CA) and extension primers. Allele abundance was measured by an ABI 3700 sequencer. Peak heights of each allele were recorded from the resulting chromatogram.
Allele Specific Expression Analysis
An allelic imbalance ratio was quantified by the following equation:
where C1 and G1 are the peak heights of one allele in the cDNA and gDNA, respectively, and C2 and G2 are the peak heights of the second allele. The genomic DNA ratio was used as an internal normalization control to account for any fluorophore differences between the two alleles. To equalize the ratios in graphical space, the values were log-transformed. Log-transformed values were defined as follows: log R = 0, no preference for either allele; log R > 0, preference for allele 1; and log R < 0, preference for allele 2.
Quantifying mRNA Expression in Human Liver and Kidney Samples
ABCC2 mRNA expression was measured in 56 Caucasian kidney and 34 Caucasian liver samples using BioTrove Open Array technology according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA). ABCC2 mRNA expression was normalized to a geometric mean of the expression of GAPDH, beta-2 microglobin, and beta-actin and expressed as 2−ΔCt.
RESULTS
Polymorphisms in the Proximal Promoter Region
To identify polymorphisms in the ABCC2 promoter region, a 1.6 kb region upstream from the ATG start site was sequenced from 247 ethnically diverse DNA samples from the Coriell Institute. Twelve polymorphisms were found in this region (Table 1), including two SNPs in the 5′-untranslated region (−1 to −247 bp from the translation start site). All other SNPs were found more than 700 bp upstream from the translation start site. An insertion was discovered at −1059 in a single chromosome in the African American population. There were no variations found between −247 and −500 bp, the region required for basal expression [7]. Four polymorphisms (rs1885301, rs7910642, rs2804402 and rs717620) were considered cosmopolitan and were found at greater than 5% frequency in each ethnic population. The -798C>A, -1059+G, -1292A>G and -1563G>A were rare and African American specific. The -733G>A variant was specific to the Asian American population, and the -23G>A UTR SNP was Pacific Islander specific. The non-singleton variants in the Caucasian, African American and Asian American populations were organized into eight different promoter haplotypes (Table 2). In addition to the reference haplotype, a haplotype containing the -1549G>A, -1019A>G and -24C>T variants and a second containing only the -1023G>A variant were found at high frequencies in each of the ethnic groups. The combination of the -1549G>A and -1019A>G variants was common in Caucasians and African Americans (frequency of 12% and 26%, respectively) but not observed in the Asian American population. Three African American specific haplotypes were identified, specifically -1549G>A/-1292A>G/-1019A>G, 1549G>A/-1239G>A/-1019A>G and -1549G>A, at frequencies of 2%, 3% and 10%, respectively.
Table 1.
Variants Identified in the Promoter Region of ABCC2
| SNP | Position1 | Nucleotide Change | Allele Frequency2
|
||||
|---|---|---|---|---|---|---|---|
| CA | AA | AS | ME | PA | |||
| rs17222653 | −1563 | G>A | 0.000 | 0.005 | 0.000 | 0.000 | 0.000 |
| rs1885301 | −1549 | G>A | 0.430 | 0.485 | 0.150 | 0.200 | 0.357 |
| rs17222667 | −1292 | A>G | 0.000 | 0.015 | 0.000 | 0.000 | 0.000 |
| rs17222646 | −1239 | G>A | 0.005 | 0.030 | 0.000 | 0.050 | 0.000 |
| rs17216128 | −1065 | C>A | 0.000 | 0.005 | 0.000 | 0.050 | 0.000 |
| rs45593436 | −1059 | insG | 0.000 | 0.005 | 0.000 | 0.000 | 0.000 |
| rs7910642 | −1023 | G>A | 0.150 | 0.135 | 0.267 | 0.300 | 0.429 |
| rs2804402 | −1019 | A>G | 0.430 | 0.365 | 0.167 | 0.200 | 0.357 |
| rs17222533 | −798 | C>A | 0.000 | 0.005 | 0.000 | 0.000 | 0.000 |
| rs17216135 | −733 | G>A | 0.000 | 0.000 | 0.017 | 0.000 | 0.000 |
| rs717620 | −24 | C>T | 0.195 | 0.060 | 0.150 | 0.150 | 0.286 |
| rs17216156 | −23 | G>A | 0.000 | 0.000 | 0.000 | 0.000 | 0.143 |
Nucleotide positions are numbered based on the distance from the translation (+1) start site.
Frequencies were calculated for each ethnic group: CA, Caucasians (n=200); AA, African Americans (n=200); AS, Asian Americans (n=60); ME, Mexican Americans (n=20); PA, Pacific Islanders (n=14).
Table 2.
Ethnic Distribution of ABCC2 Promoter Haplotypes
| -1549G>A | -1292A>G | -1239G>A | -1023G>A | -1019A>G | -24C>T | Frequency1
|
||
|---|---|---|---|---|---|---|---|---|
| CA | AA | AS | ||||||
| G | A | G | G | A | C | 0.460 | 0.390 | 0.570 |
| A | G | 0.120 | 0.260 | 0.000 | ||||
| A | G | T | 0.200 | 0.050 | 0.160 | |||
| A | A | G | 0.000 | 0.030 | 0.000 | |||
| A | G | G | 0.000 | 0.020 | 0.000 | |||
| A | 0.000 | 0.100 | 0.000 | |||||
| A | 0.110 | 0.110 | 0.280 | |||||
Promoter haplotypes were estimated for six non-singleton 5′-variants using PHASE and the haplotype frequencies are shown for the Caucasian (CA, n=200), African American (AA, n=200) and Asian American (AS, n=60) populations. The first row is the promoter reference haplotype; nucleotide changes for each SNP site are indicated for the variant haplotypes.
Effects of ABCC2 Promoter Variants on Promoter Activity In Vitro
To assess if promoter polymorphisms have an effect on ABCC2 promoter activity a 1.6 kb fragment was amplified from genomic DNA and cloned into the multiple cloning site of the promoter-less luciferase plasmid, pGL4.11b. Single variant or promoter haplotypes were introduced by site-directed mutagenesis using specific primers. A plasmid containing the reference sequence showed approximately 70-fold higher activity than the promoter-less pGL4.11b plasmid. The ABCC2 -24C>T SNP, when introduced both as a single variant and as part of the -24C>T/-1019A>G/-1549G>A haplotype, showed a significant 35% increase in reporter activity compared to the reference ABCC2 promoter sequence (P<0.0001). Reporter activity of promoter constructs carrying other SNPs and haplotypes were not significantly different from the reference (Figure 1). There was no association of the common ABCC2 promoter variants (-1549A>G, -1023G>A and -24C>T) with ABCC2 mRNA levels in 34 Caucasian liver and 56 Caucasian kidney samples (data not shown).
Figure 1. Functional activity of ABCC2 promoter variants.
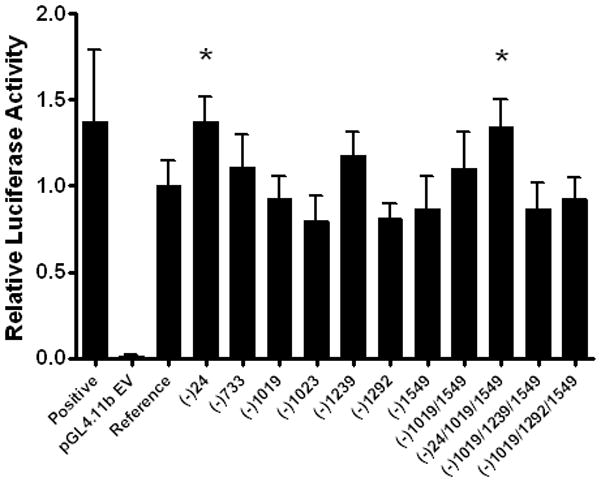
ABCC2 promoter activity was expressed as Firefly luciferase activity relative to Renilla luciferase activity; the activity of the reference sequence was set to one. Each column represents the mean ± SD of four separate transfection experiments performed in triplicate. The positive control was a liver enhancer sequence. *P < 0.001, significantly different from reference.
Allele Specific Expression
Allele specific expression was investigated as a possible mechanism for observed associations of ABCC2 3972C>T with various phenotypes.22, 30, 31 Of 200 liver samples available, the majority (n=137) were of known Caucasian descent and were used for this analysis. The sample sizes of other ethnic groups were too small for inclusion. Haplotypes were constructed using five additional polymorphisms commonly found with the 3972C>T variant and the distribution of these haplotypes in Caucasians is shown in Table 3. Ten different haplotypes were estimated for the 137 liver samples. A total of 53 Caucasian liver samples were heterozygous at the 3972C>T polymorphic site. Seven livers carried low frequency haplotypes that were not considered further and five liver RNA samples could not be reverse transcribed and were also removed from the analysis. Results of the allele specific expression analysis are presented for the most common haplotype pairs (n = 41; Figure 2).
Table 3.
Inferred ABCC2 Haplotypes of Caucasian Liver Samples
| -1549G>A | -1019A>G | -24C>T | 1249G>A | Intron 26–34T>C | 3972C>T | Frequency1 | |
|---|---|---|---|---|---|---|---|
| H1 | G | A | C | G | T | C | 0.310 |
| H2 | A | G | T | T | 0.234 | ||
| H3 | A | 0.193 | |||||
| H4 | A | G | T | 0.117 | |||
| H5 | A | G | 0.058 | ||||
| H6 | A | G | C | 0.033 | |||
| H7 | T | 0.036 | |||||
| H8 | C | 0.011 | |||||
| H9 | A | G | T | 0.004 | |||
| H10 | A | 0.000 | |||||
| H11 | A | T | 0.004 | ||||
| H12 | A | C | 0.000 |
Promoter haplotypes in 137 Caucasian liver samples were estimated for six variants using PHASE and the haplotype frequencies are shown. H1 is the promoter reference haplotype; nucleotide changes for each SNP site are indicated for the variant haplotypes.
Figure 2. Allele specific expression in Caucasian livers.
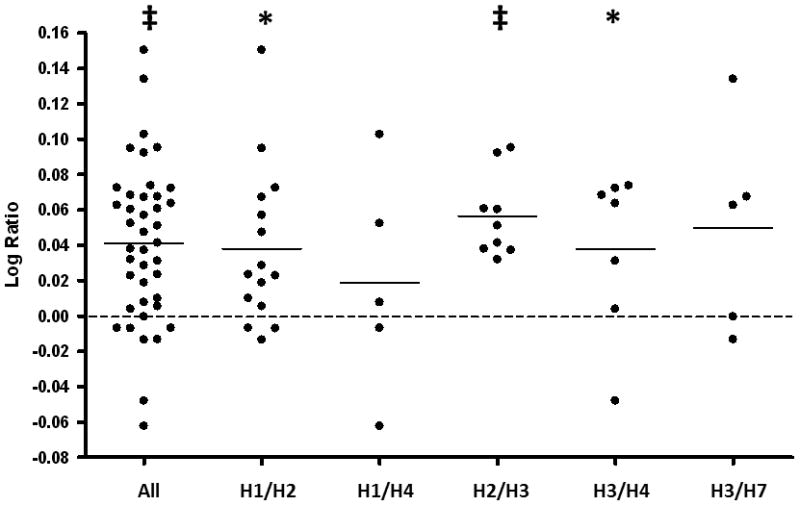
The log allelic imbalance ratio is plotted for 41 Caucasian livers. Each symbol represents an individual liver and the dashed line indicates a ratio where the C allele and T allele are equal in abundance. The mean values for each group are represented by lines. The allelic imbalance ratio is plotted for all samples and separately for each haplotype pair. Significant differences from 0 are noted (*P < 0.01; ‡P < 0.0001).
Overall, there was a statistically significant preference for the 3972C allele compared to the 3972T allele (log ratios > 0, P < 0.0001). When examining haplotype pairs, H1/H2 (P < 0.01), H2/H3 (P < 0.0001) and H3/H4 (P < 0.01) showed a significant preference for the C allele, suggesting that H2 and H4 might be driving this allelic imbalance. Samples carrying H2/H3 showed the highest mean C/T ratios of all haplotype combinations; in these samples, the C allele was 14% higher in abundance than the T allele.
Association of hepatic and renal ABCC2 mRNA expression with 3972C>T genotype
To assess if 3972C>T allelic imbalance is pronounced enough to affect total ABCC2 mRNA expression we genotyped 34 additional Caucasian liver and 56 Caucasian kidney samples for this SNP using a tagging SNP (rs7067971) and quantified ABCC2 mRNA expression. No association was observed in these samples between 3972C>T genotype and ABCC2 mRNA expression (Figure 3).
Figure 3. Association of ABCC2 3972C>T with ABCC2 mRNA expression.
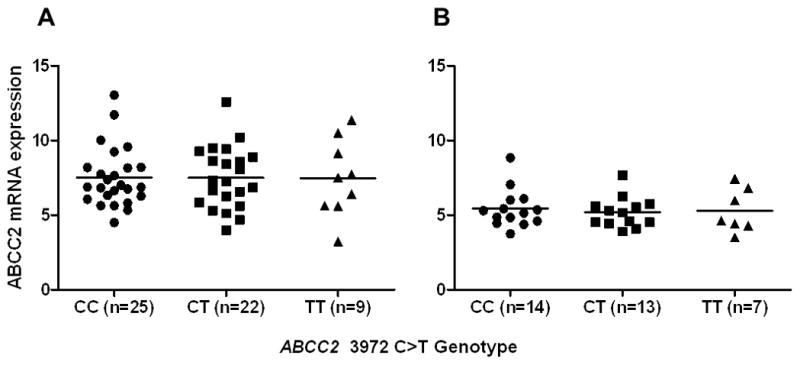
The relationship between ABCC2 3972 genotype and ABCC2 mRNA expression in kidney (A) and liver (B) samples is shown. ABCC2 mRNA expression is expressed relative to the geometric mean of the mRNA level of three housekeeping genes. Values are shown for individual samples and the mean values are indicated by the solid line.
DISCUSSION
MRP2 is one of the most important efflux transporters and can transport a large number of drugs and their conjugates. The ABCC2 gene encoding MRP2 is highly polymorphic and genetic variability is associated with interindividual differences in pharmacokinetics, drug response and toxicity. In particular, non-synonymous coding region polymorphisms and disease mutations have been associated with changes in ABCC2 expression and/or MRP2 function.8–14 The effects of ABCC2 promoter and synonymous variants are less well understood. The purpose of this study was two-fold; first, to identify and characterize the functional effect of common variants in the ABCC2 promoter and 5′-UTR. In addition, studies were carried out to determine if allelic imbalance associated with the synonymous 3972C>T allele might provide mechanistic insight into the association of this variant with irinotecan disposition and toxicity. 22, 24, 29, 34
The proximal promoter region of ABCC2 has several common polymorphisms in the 1600 bp region upstream of the transcription start site. Interestingly, the region containing the core promoter (−300 to −500 bp16, 17) is devoid of any common genetic variation, consistent with a significant role in the constitutive regulation of ABCC2 expression. The most common promoter region haplotypes contain the -1549G>A and -1019G>A polymorphisms, often together with the -24C>T UTR variant. The association of -1549G>A, -1019G>A and -24C>T polymorphisms with increased irinotecan and/or SN-38 exposure following irinotecan treatment22, 29, 34 suggests that these promoter region variants might influence ABCC2 transcription and therefore expression.
However, our in vitro studies failed to show a significant effect of the -1549G>A and -1019G>A variants, alone or in combination, with in vitro promoter activity. Interestingly, the -24C>T variant, alone and combined with the -1549G>A and -1019G>A variants, modestly increased promoter activity. If increased promoter activity translated into increased ABCC2 expression, then irinotecan and SN-38 exposure would be expected to be lower compared to patients with the reference sequence. However, ABCC2 -24C>T has been associated with increased irinotecan and SN-38 exposure, suggesting reduced transport of these substrates.22, 29
Only limited studies have been previously carried out to characterize the functional impact of genetic polymorphisms in the ABCC2 promoter and 5′-UTR region. In cell-based reporter gene assays the -24C>T/-1549G>A haplotype decreased promoter activity 39%20 and the -24C>T variant alone caused a 20% reduction in promoter activity.10 The reporter plasmids used in the current and previous studies contain different lengths of upstream DNA sequence which may account for the varying results. In cell lines with different genotypes, the -24C>T polymorphism had no effect on DNA binding or mRNA stability but was associated with reduced protein expression and corresponding effects on transport function.35 Consistent with these cell-based findings, -24C>T had no effect on the expression of ABCC2 mRNA in hippocampal, intestinal, placental or adenocarcinoma samples.9, 23, 36–38 In contrast, lower levels of ABCC2 renal mRNA were reported in patients carrying the -24T allele.10 In addition to associations of -24C>T with irinotecan and SN-38 exposure, this variant has also been associated with increased diclofenac hepatotoxicity21, greater risk of platinum hematological toxicity31 and antiepileptic drug resistance36, and increased response to irinotecan and cisplatin treatment.24 Most of these findings have not been replicated, and in the case of antiepileptic drug resistance, recent data refutes the original association.39 A possible reason for the inconsistency between studies is that ABCC2 promoter SNPs previously associated with clinical phenotypes (e.g., SN-38 pharmacokinetics and drug toxicities) are not causative and are in linkage disequilibrium with other SNPs (cis- or trans-acting) that are functional. In support of this alternative explanation, ENCODE data available on the UCSC genome browser indicates there are only two clusters of transcription factors binding in the ABCC2 promoter region and there is very little overlap of any SNPs previously associated with clinical phenotypes with this region. This publically available data is consistent with our findings that the commonly studied ABCC2 promoter SNPs are not critical for transcriptional regulation of ABCC2 and that other SNPs and transcriptional modulators may be responsible for variability in MRP2 expression and function. For example, transacting SNPs as well as epigenetic mechanisms may be involved in the regulation of ABCC2 expression. Further studies will be necessary to fully understand the functional and clinical impact of any of the ABCC2 promoter/UTR polymorphisms.
ABCC2 -24C>T is in linkage disequilibrium (LD) with a synonymous 3972C>T polymorphism, raising the possibility that previously reported associations of the 5′-UTR variant with clinical phenotypes may reflect functional changes related to the synonymous variant. In allelic imbalance studies we found a significant preference for the 3972C allele compared to the 3972T allele. This appears to be driven by a haplotype (H2) that includes polymorphisms at -1549, -1019 and -24 in ABCC2, and H4 which includes only the-1549 and -1019 SNPs. The relative effect of the observed allelic imbalance is modest, with at most a 14% average increase in transcription of the 3972C allele compared to the T allele. However, a large degree of interindividual variability was observed in this measurement and up to 45% increases were measured. The allelic imbalance did not affect total hepatic or renal ABCC2 mRNA levels, suggesting its clinical significance will be limited except perhaps in individuals with extreme degrees of allele specific expression.
A recent paper suggests that the 3972C>T variant could also be acting through a posttranscriptional mechanism. The-24C>T variant was associated with a modest decrease in MRP2 recombinant protein expression, while 3972C>T variant had much more profound effect and drove the observed decrease in protein expression with the -24T/3972T haplotype.35 Interestingly, no effect on mRNA expression was observed, suggesting these polymorphisms play an important role in MRP2 translational regulation. The contribution of genetic variation to posttranscriptional regulation of ABCC2/MRP2 expression should be further explored. Additional mechanisms for regulation of ABCC2/MRP2, including eQTLs and epigenetics, also warrant investigation. Interindividual variation in these mechanisms may contribute to variation in transport of endogenous and xenobiotic substrates of MRP2.
In summary, a number of common promoter and 5′-UTR polymorphisms in ABCC2 are found at relatively high frequency across different ethnic groups. The limited, if any, effect of these variants on promoter activity suggests that reported associations with clinical phenotypes are not a result of impaired transcription. The 3972C>T synonymous variant is associated with allelic specific expression, although the magnitude of the effect is modest. Additional mechanisms will need to be explored to account for the reported association of ABCC2 promoter variants with irinotecan disposition and other drug response phenotypes.
Acknowledgments
This work was supported by NIH grants GM61390, GM61393 and CA21765. Tan D. Nguyen and Jason M. Gow were supported in part by NIH Training Grant T32 GM007175.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
References
- 1.Paulusma CC, Oude Elferink RP. The canalicular multispecific organic anion transporter and conjugated hyperbilirubinemia in rat and man. J Mol Med. 1997;75:420–428. doi: 10.1007/s001090050127. [DOI] [PubMed] [Google Scholar]
- 2.Buchler M, Konig J, Brom M, Kartenbeck J, Spring H, Horie T, et al. cDNA cloning of the hepatocyte canalicular isoform of the multidrug resistance protein, cMrp, reveals a novel conjugate export pump deficient in hyperbilirubinemic mutant rats. J Biol Chem. 1996;271:15091–15098. doi: 10.1074/jbc.271.25.15091. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich CG, Ottenhoff R, de Waart DR, Oude Elferink RP. Role of MRP2 and GSH in intrahepatic cycling of toxins. Toxicology. 2001;167:73–81. doi: 10.1016/s0300-483x(01)00459-0. [DOI] [PubMed] [Google Scholar]
- 4.Evers R, de Haas M, Sparidans R, Beijnen J, Wielinga PR, Lankelma J, et al. Vinblastine and sulfinpyrazone export by the multidrug resistance protein MRP2 is associated with glutathione export. Br J Cancer. 2000;83:375–383. doi: 10.1054/bjoc.2000.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nies AT, Keppler D. The apical conjugate efflux pump ABCC2 (MRP2) Pflugers Arch. 2007;453:643–659. doi: 10.1007/s00424-006-0109-y. [DOI] [PubMed] [Google Scholar]
- 6.Jemnitz K, Heredi-Szabo K, Janossy J, Ioja E, Vereczkey L, Krajcsi P. ABCC2/Abcc2: a multispecific transporter with dominant excretory functions. Drug Metab Rev. 2010;42:402–436. doi: 10.3109/03602530903491741. [DOI] [PubMed] [Google Scholar]
- 7.Toh S, Wada M, Uchiumi T, Inokuchi A, Makino Y, Horie Y, et al. Genomic structure of the canalicular multispecific organic anion-transporter gene (MRP2/cMOAT) and mutations in the ATP-binding-cassette region in Dubin-Johnson syndrome. Am J Hum Genet. 1999;64:739–746. doi: 10.1086/302292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson PL, Lamba J, Aquilante CL, Schuetz E, Fletcher CV. Pharmacogenetic characteristics of indinavir, zidovudine, and lamivudine therapy in HIV-infected adults: a pilot study. J Acquir Immune Defic Syndr. 2006;42:441–449. doi: 10.1097/01.qai.0000225013.53568.69. [DOI] [PubMed] [Google Scholar]
- 9.Haenisch S, May K, Wegner D, Caliebe A, Cascorbi I, Siegmund W. Influence of genetic polymorphisms on intestinal expression and rifampicin-type induction of ABCC2 and on bioavailability of talinolol. Pharmacogenet Genomics. 2008;18:357–365. doi: 10.1097/FPC.0b013e3282f974b7. [DOI] [PubMed] [Google Scholar]
- 10.Haenisch S, Zimmermann U, Dazert E, Wruck CJ, Dazert P, Siegmund W, et al. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharmacogenomics J. 2007;7:56–65. doi: 10.1038/sj.tpj.6500403. [DOI] [PubMed] [Google Scholar]
- 11.Hirouchi M, Suzuki H, Itoda M, Ozawa S, Sawada J, Ieiri I, et al. Characterization of the cellular localization, expression level, and function of SNP variants of MRP2/ABCC2. Pharm Res. 2004;21:742–748. doi: 10.1023/b:pham.0000026422.06207.33. [DOI] [PubMed] [Google Scholar]
- 12.Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, et al. Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006;44:62–74. doi: 10.1002/hep.21214. [DOI] [PubMed] [Google Scholar]
- 13.Ranganathan P, Culverhouse R, Marsh S, Mody A, Scott-Horton TJ, Brasington R, et al. Methotrexate (MTX) pathway gene polymorphisms and their effects on MTX toxicity in Caucasian and African American patients with rheumatoid arthritis. J Rheumatol. 2008;35:572–579. [PubMed] [Google Scholar]
- 14.Izzedine H, Hulot JS, Villard E, Goyenvalle C, Dominguez S, Ghosn J, et al. Association between ABCC2 gene haplotypes and tenofovir-induced proximal tubulopathy. J Infect Dis. 2006;194:1481–1491. doi: 10.1086/508546. [DOI] [PubMed] [Google Scholar]
- 15.Wojnowski L, Kulle B, Schirmer M, Schluter G, Schmidt A, Rosenberger A, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112:3754–3762. doi: 10.1161/CIRCULATIONAHA.105.576850. [DOI] [PubMed] [Google Scholar]
- 16.Stockel B, Konig J, Nies AT, Cui Y, Brom M, Keppler D. Characterization of the 5′-flanking region of the human multidrug resistance protein 2 (MRP2) gene and its regulation in comparison withthe multidrug resistance protein 3 (MRP3) gene. Eur J Biochem. 2000;267:1347–1358. doi: 10.1046/j.1432-1327.2000.01106.x. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka T, Uchiumi T, Hinoshita E, Inokuchi A, Toh S, Wada M, et al. The human multidrug resistance protein 2 gene: functional characterization of the 5′-flanking region and expression in hepatic cells. Hepatology. 1999;30:1507–1512. doi: 10.1002/hep.510300617. [DOI] [PubMed] [Google Scholar]
- 18.Qadri I, Hu LJ, Iwahashi M, Al-Zuabi S, Quattrochi LC, Simon FR. Interaction of hepatocyte nuclear factors in transcriptional regulation of tissue specific hormonal expression of human multidrug resistance-associated protein 2 (abcc2) Toxicology and applied pharmacology. 2009;234:281–292. doi: 10.1016/j.taap.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Naesens M, Kuypers DR, Verbeke K, Vanrenterghem Y. Multidrug resistance protein 2 genetic polymorphisms influence mycophenolic acid exposure in renal allograft recipients. Transplantation. 2006;82:1074–1084. doi: 10.1097/01.tp.0000235533.29300.e7. [DOI] [PubMed] [Google Scholar]
- 20.Choi JH, Ahn BM, Yi J, Lee JH, Nam SW, Chon CY, et al. MRP2 haplotypes confer differential susceptibility to toxic liver injury. Pharmacogenet Genomics. 2007;17:403–415. doi: 10.1097/01.fpc.0000236337.41799.b3. [DOI] [PubMed] [Google Scholar]
- 21.Daly AK, Aithal GP, Leathart JB, Swainsbury RA, Dang TS, Day CP. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 2007;132:272–281. doi: 10.1053/j.gastro.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Fujita K, Nagashima F, Yamamoto W, Endo H, Sunakawa Y, Yamashita K, et al. Association of ATP-binding cassette, sub-family C, number 2 (ABCC2) genotype with pharmacokinetics of irinotecan in Japanese patients with metastatic colorectal cancer treated with irinotecan plus infusional 5-fluorouracil/leucovorin (FOLFIRI) Biol Pharm Bull. 2008;31:2137–2142. doi: 10.1248/bpb.31.2137. [DOI] [PubMed] [Google Scholar]
- 23.Moriya Y, Nakamura T, Horinouchi M, Sakaeda T, Tamura T, Aoyama N, et al. Effects of polymorphisms of MDR1, MRP1, and MRP2 genes on their mRNA expression levels in duodenal enterocytes of healthy Japanese subjects. Biol Pharm Bull. 2002;25:1356–1359. doi: 10.1248/bpb.25.1356. [DOI] [PubMed] [Google Scholar]
- 24.Han JY, Lim HS, Yoo YK, Shin ES, Park YH, Lee SY, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138–147. doi: 10.1002/cncr.22760. [DOI] [PubMed] [Google Scholar]
- 25.Miura M, Satoh S, Inoue K, Kagaya H, Saito M, Inoue T, et al. Influence of SLCO1B1, 1B3, 2B1 and ABCC2 genetic polymorphisms on mycophenolic acid pharmacokinetics in Japanese renal transplant recipients. Eur J Clin Pharmacol. 2007;63:1161–1169. doi: 10.1007/s00228-007-0380-7. [DOI] [PubMed] [Google Scholar]
- 26.Innocenti F, Ratain MJ. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping. Pharmacogenomics. 2006;7:1211–1221. doi: 10.2217/14622416.7.8.1211. [DOI] [PubMed] [Google Scholar]
- 27.Kroetz DL. Role for drug transporters beyond tumor resistance: hepatic functional imaging and genotyping of multidrug resistance transporters for the prediction of irinotecan toxicity. J Clin Oncol. 2006;24:4225–4227. doi: 10.1200/JCO.2006.07.2355. [DOI] [PubMed] [Google Scholar]
- 28.Zamboni WC, Ramanathan RK, McLeod HL, Mani S, Potter DM, Strychor S, et al. Disposition of 9-nitrocamptothecin and its 9-aminocamptothecin metabolite in relation to ABC transporter genotypes. Invest New Drugs. 2006;24:393–401. doi: 10.1007/s10637-006-6335-5. [DOI] [PubMed] [Google Scholar]
- 29.Innocenti F, Kroetz DL, Schuetz E, Dolan ME, Ramirez J, Relling M, et al. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J Clin Oncol. 2009;27:2604–2614. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoblinger A, Grunhage F, Sauerbruch T, Lammert F. Association of the c.3972C>T variant of the multidrug resistance-associated protein 2 Gene (MRP2/ABCC2) with susceptibility to bile duct cancer. Digestion. 2009;80:36–39. doi: 10.1159/000212990. [DOI] [PubMed] [Google Scholar]
- 31.Han B, Gao G, Wu W, Gao Z, Zhao X, Li L, et al. Association of ABCC2 polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Lung Cancer. 2011;72:238–243. doi: 10.1016/j.lungcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latif S, Bauer-Sardina I, Ranade K, Livak KJ, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis II: 5′-nuclease assay. Genome Res. 2001;11:436–440. doi: 10.1101/gr.156601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong FA, Scott-Horton TJ, Kroetz DL, McLeod HL, Friberg LE, Mathijssen RH, et al. Irinotecan-induced diarrhea: functional significance of the polymorphic ABCC2 transporter protein. Clin Pharmacol Ther. 2007;81:42–49. doi: 10.1038/sj.clpt.6100019. [DOI] [PubMed] [Google Scholar]
- 35.Laechelt S, Turrini E, Ruehmkorf A, Siegmund W, Cascorbi I, Haenisch S. Impact of ABCC2 haplotypes on transcriptional and posttranscriptional gene regulation and function. Pharmacogenomics J. 2011;11:25–34. doi: 10.1038/tpj.2010.20. [DOI] [PubMed] [Google Scholar]
- 36.Ufer M, Mosyagin I, Muhle H, Jacobsen T, Haenisch S, Hasler R, et al. Non-response to antiepileptic pharmacotherapy is associated with the ABCC2 -24C>T polymorphism in young and adult patients with epilepsy. Pharmacogenet Genomics. 2009;19:353–362. doi: 10.1097/fpc.0b013e328329940b. [DOI] [PubMed] [Google Scholar]
- 37.Nishioka C, Sakaeda T, Nakamura T, Moriya Y, Okamura N, Tamura T, et al. MDR1, MRP1 and MRP2 genotypes and in vitro chemosensitivity in Japanese patients with colorectal adenocarcinomas. Kobe J Med Sci. 2004;50:181–188. [PubMed] [Google Scholar]
- 38.Meyer zu Schwabedissen HE, Jedlitschky G, Gratz M, Haenisch S, Linnemann K, Fusch C, et al. Variable expression of MRP2 (ABCC2) in human placenta: influence of gestational age and cellular differentiation. Drug Metab Dispos. 2005;33:896–904. doi: 10.1124/dmd.104.003335. [DOI] [PubMed] [Google Scholar]
- 39.Hilger E, Reinthaler EM, Stogmann E, Hotzy C, Pataraia E, Baumgartner C, et al. Lack of association between ABCC2 gene variants and treatment response in epilepsy. Pharmacogenomics. 2012;13:185–190. doi: 10.2217/pgs.11.143. [DOI] [PubMed] [Google Scholar]


