Abstract
The RE1 Silencing Transcription Factor (REST) is a repressor of neuronal differentiation and its elevated expression in neural cells blocks neuronal differentiation. In the present study, we demonstrate a role for REST in the control of proliferation of medulloblastoma cells. REST expression decreased the levels of CDKNIB/p27, a cyclin-dependent kinase inhibitor and a brake of cell proliferation in these cells. The reciprocal relationship between REST and p27 was validated in human tumor samples. REST knockdown in medulloblastoma cells derepessed a novel REST-target gene encoding the deubiquitylase ubiquitin-specific peptidase 37 (USP37). Ectopically expressed wild type USP37 formed a complex with p27, promoted its deubiquitination and stabilization and blocked cell proliferation. Knockdown of REST and USP37 prevented p27 stabilization and blocked the diminution in proliferative potential that normally accompanied REST loss. Unexpectedly, wild type USP37 expression also induced the expression of REST-target neuronal differentiation genes even though REST levels were unaffected. In contrast, a mutant of USP37 carrying a site-directed change in a conserved cysteine failed to rescue REST-mediated p27 destabilization, maintenance of cell proliferation and blockade to neuronal differentiation. Consistent with these findings, a significant correlation between USP37 and p27 was observed in patient tumors. Collectively, these findings provide a novel connection between REST and the proteasomal machinery in the control of p27 and cell proliferation in medulloblastoma cells.
Keywords: REST, proliferation, p27, USP37, deubiquitylase
Introduction
The RE1 silencing transcription factor (REST) is an important regulator of neuronal differentiation (1–10). It is expressed in neural progenitors, but downregulated in most differentiated neurons (1–11). REST binds a 21–23 bp sequence called the RE1 element found in the regulatory regions of target genes through a centrally located DNA binding domain. REST has two independent repressor domains located at the amino (N) and carboxy (C) termini of the protein (3–9). The N-terminal repression domain is associated with mSin3a and HDAC1/2, whereas the C-terminal repression domain complexes with co-REST, the chromatin remodeling protein Brg1, G9a histone methyltransferase, LSD1 lysine demethylase, and HDAC1/2 (3–10). Acting through these complexes, REST represses target gene expression in neural progenitors. A number of these genes are involved in neurogenesis. The decline in REST expression at onset of neuronal specification derepresses these target genes and allows terminal neuronal differentiation (1–10).
Consistent with a role for REST in neuronal differentiation, our previous studies showed that its expression is aberrantly maintained in the undifferentiated neural tumor of childhood called medulloblastoma (12–14). V-Myc immortalized murine cerebellar progenitor cells (NSC-M) that were engineered to constitutively express human REST transgene (NSC-MR) were blocked in neuronal differentiation and formed tumors when injected into the cerebellum of mice (13). In contrast, the parental v-Myc immortalized progenitors (NSC-M) underwent neuronal differentiation in vitro and failed to form tumors in vivo (13). Importantly, constitutive REST expression provided a proliferation advantage to NSC-MR cells in vitro (13). In the study described here, we evaluated if REST played a direct role in the control of cell proliferation and also investigated the underlying molecular mechanisms.
Several studies have demonstrated the importance of the cyclin-dependent kinase inhibitor (CDKI) p27/Kip1 in the control of proliferation and cell exit in cerebellar progenitor cells (CPCs), the cells of origin of a subset of medulloblastoma (15–17). Mice that are heterozygous or nullizygous for p27 exhibit cerebellar enlargement stemming from hyperproliferation of CPCs (15–17). Cytoplasmic mis-localization of p27 is also associated with uncontrolled CPC proliferation (18). These aberrations in p27 biology contribute to medulloblastoma formation in the background of constitutive sonic hedgehog (Shh) signaling (18–21). In the present study, we provide evidence that REST-dependent effects on cell proliferation involve repression of a gene encoding a deubiquitylase (DUB), USP37 (22–24). The absence of USP37 transcript in REST-expressing medulloblastoma cells was associated with low p27 protein levels. Conversely, REST knockdown upregulated USP37 gene expression and promoted an increase in p27 protein levels. A significant correlation between p27, REST and USP37 was also seen in human tumor samples. Ectopically expressed USP37 formed a complex with p27, promoted its stabilization, blocked cell proliferation and induced the expression of REST-target neuronal differentiation genes. In contrast, ectopic expression of a USP37 transgene carrying a mutation in a conserved cysteine residue failed to rescue REST-dependent effects on p27, cell proliferation and neuronal differentiation. Since concomitant loss of REST and USP37 expression attenuated p27 stabilization and differentiation and rescued cell proliferation, our data strongly suggest that repression of USP37 and consequent p27 degradation, are important for REST-dependent maintenance of cell proliferation.
Results
REST controls cell proliferation
REST has been mostly studied in the context of its function as a regulator of neuronal differentiation genes. Our previous studies showed that elevated REST expression in v-Myc immortalized NSC-MR cells provided proliferation advantage to these cells (13). To determine if REST had a direct role in maintaining cell proliferation, we knocked down endogenous REST gene expression in DAOY and D283 medulloblastoma cell lines through transient transfection of pooled REST-specific siRNA or control scrambled (scr) siRNA. REST knockdown was confirmed by Q-RT-PCR and Western blotting analyses (Figs. 1D and 1E). A decline in REST expression promoted a 50–60% decline in total cell numbers relative to control scr-siRNA-transfected cell numbers (Fig. 1A). To determine if this decrease in cell numbers upon REST loss was mediated by an effect on cell proliferation, DAOY and D283 cells with or without REST expression were co-stained for REST and Ki-67 using specific antibodies and analyzed by immunofluorescence assay (IFA). Percent decrease in Ki-67 staining in cells lacking REST relative to control REST-expressing cells was between 50% and 70% (Fig. 1B). Flow cytometric analysis was also used to assess changes in the cell cycle distribution of DAOY cells in the presence or absence of REST expression. Loss of REST expression promoted an increase in the number of cells in the G1 phase of the cell cycle from 53% to 63%, which was associated with a corresponding decrease in the number of cells in S phase (17% to 7%) (Fig.1C). A significant change in the number of cells with a sub-G1 or G2/M DNA content was not detected, indicating that apoptosis was not a major consequence of REST loss in our assays (Fig. 1C). REST loss and the consequent increase in expression of its target gene Syn1, was also confirmed by Q-RT-PCR analyses (Fig. 1D). Western blotting also revealed REST loss to cause a decrease in the levels of the pro-proliferative marker N-Myc and an increase the levels of the inhibitor of cell proliferation, p27, in DAOY and D283 cells (Fig. 1E) (17, 19–21, 25, 26). Levels of the CDKI p21 were elevated in DAOY cells but not in D283 cells following REST loss. Interestingly, REST expression was associated with the presence of slower migrating forms of p27 in both DAOY and D283 cells. Conversely, the increase in p27 levels following REST knockdown was accompanied by the emergence of a ladder of faster migrating forms of the protein. Nuclear localization of p27, which is important for its function as a CDKI was also observed upon REST knockdown (data not shown). These data implicate REST in the control of cell proliferation and potentially through regulation of p27 protein levels.
Figure 1. REST knockdown causes a decline in medulloblastoma cell proliferation and an accumulation of p27.
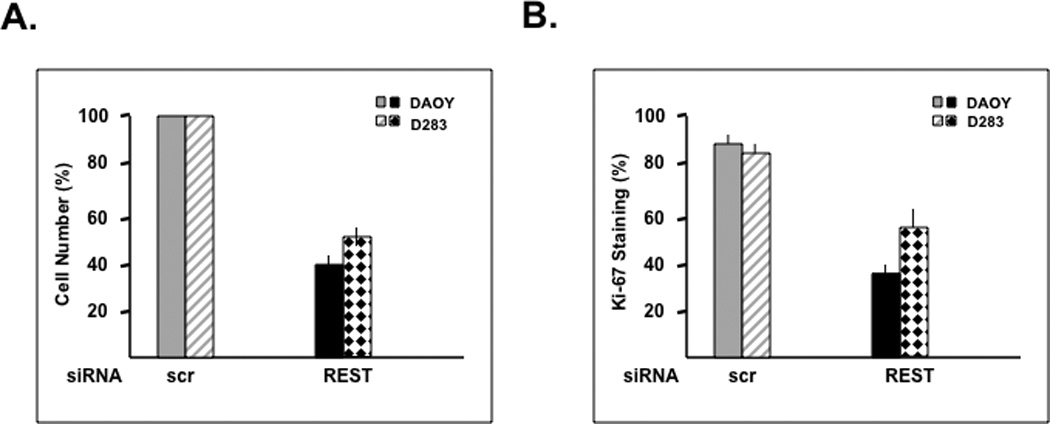
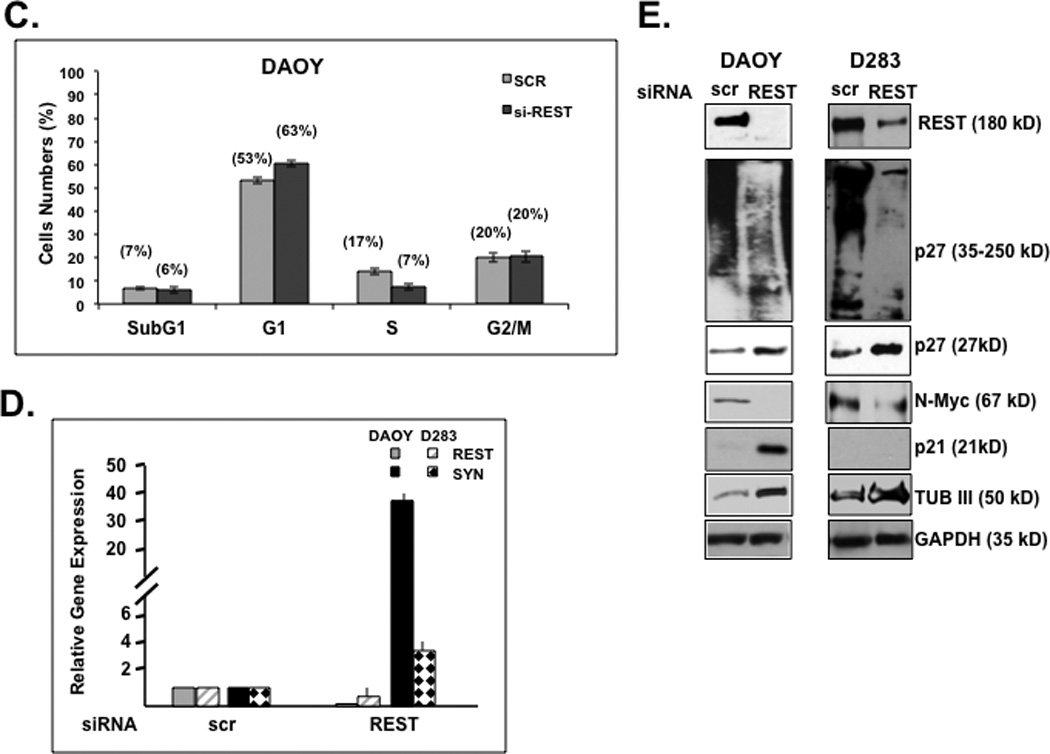
(A) REST-dependent change in the number of DAOY and D283 cells was measured 24 h post-transfection with pooled siRNA against REST or control scrambled (scr) siRNA. (B) Change in the proliferation potential of DAOY and D283 cells following transfection with REST-specific siRNA or control scr-siRNA-transfected cells was determined by co-staining for REST and Ki-67 and counting Ki67 positive cells under a fluorescence microscope. (C) DAOY cells transfected with pooled siRNA against REST or control siRNA were stained with propidium iodide and analyzed for changes in their cell cycle distribution by flow cytometry. Analysis was performed CellQuest 3.2 software. (D) SYBR Green Q-RT-PCR analysis was carried out to determine the efficiency of REST knockdown and the upregulation of its target gene Syn1 using specific primers. 18s mRNA levels were used for normalization. (E) Western blot analysis was performed to assess REST knockdown and measure changes in its target gene-product type-III beta tubulin. Levels of the pro-proliferative marker N-Myc and the anti-proliferative markers p21 and p27 following REST loss were also determined by Western blotting using specific antibodies. GAPDH and actin were measured to confirm equal loading.
REST and p27 are reciprocally expressed in human medulloblastoma samples
The relationship between REST and p27 observed in medulloblastoma cell lines was also validated in human tumor samples. Following Institutional Review Board (IRB)-approval, de-identified patient samples were co-stained for REST and p27 proteins using specific antibodies and studied by IFA. Staining in representative tumors and control normal cerebellum is shown in Fig. 2A (top panel). The corresponding hematoxylin-eosin (H&E) stained sections are included in the bottom panel (Fig. 2A). REST was expressed in all tumor samples in a focally elevated pattern, but not in the control normal cerebellum. In contrast, p27 was expressed in the normal cerebellum and either absent or present at low levels in tumors. Specifically, a total of 45 human tumors were analyzed for REST and p27 expression and scored on a scale from no (−) to high (++++) expression. Approximately, 27 of the 45 samples (60%) showing focal REST expression (+/++/+++/++++) had no detectable p27 protein. A majority (16/27) of these p27-negative tumors exhibited very high REST expression (++++) and 12/27 of p27-negative tumors exhibit lower REST staining (+/++/+++). An additional 18 of the 45 samples (40%) with REST expression also expressed p27 (+/++/+++) at levels lower or comparable to normal cerebellum (+++). The distribution of p27 protein as a function of REST levels is shown in Figures 2B and 2C. Overall, there was a statistically significant difference in level of REST protein between tumors with p27-negative and p27-positive tumors (p=0.0008, Wilcoxon rank-sum test; p=0.0006, T test).
Figure 2. REST and p27 expression are reciprocally correlated in human medulloblastoma in vitro.
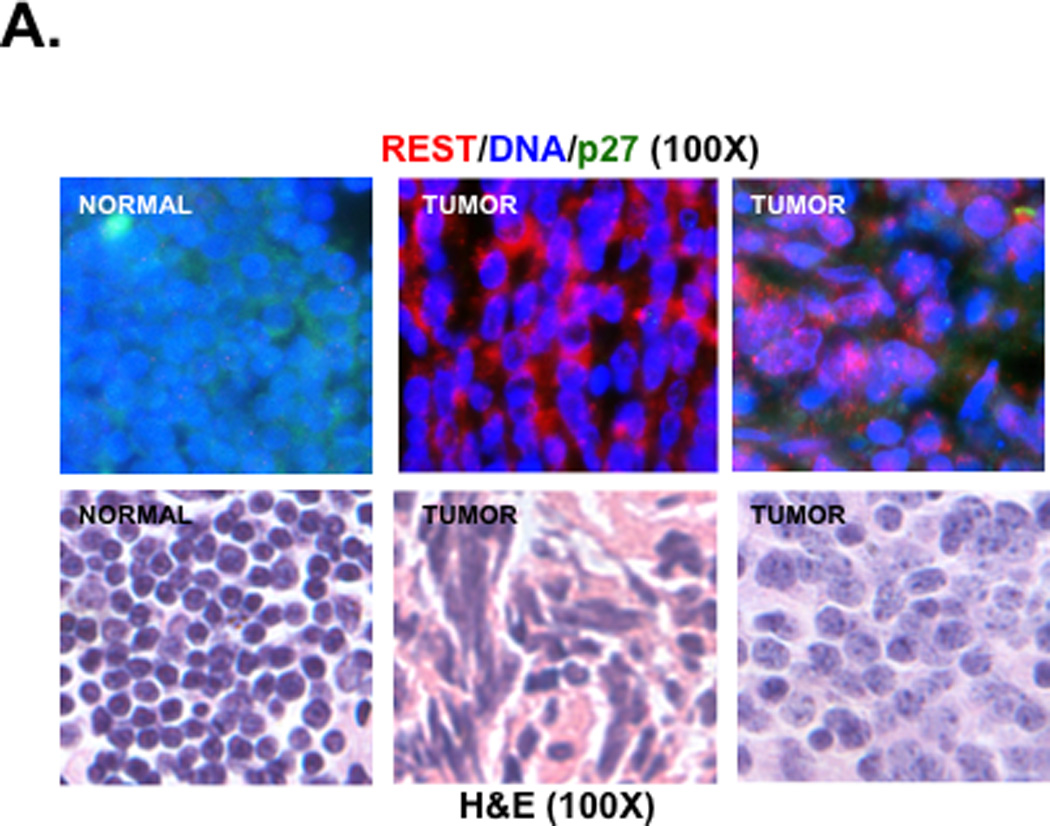
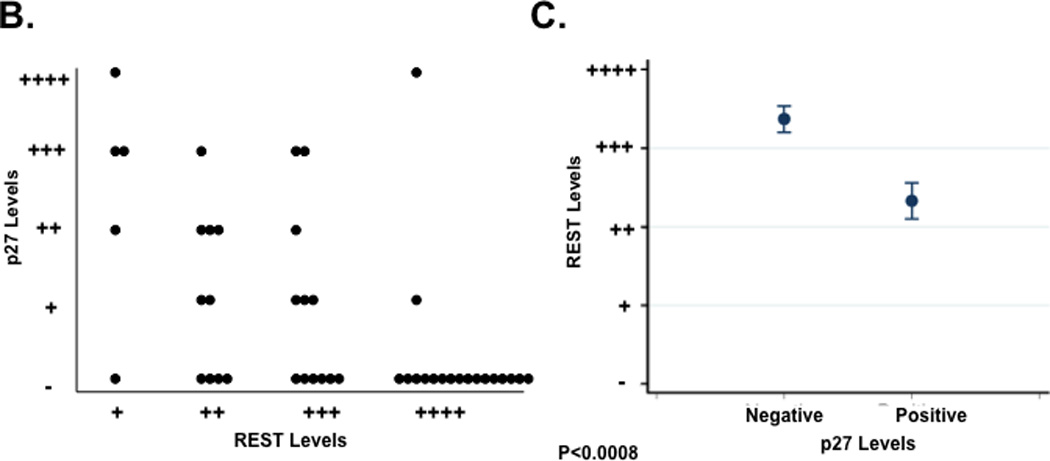
(A) co-IFA to measure REST and p27 protein levels in human medulloblastoma samples and normal cerebellum was carried out using anti-p27 (green) and anti-REST (red) primary antibodies and fluorophore-conjugated secondary antibodies. Nuclei (blue) were stained with Hoechst dye. Stained cells were visualized under a Nikon fluorescence microscope. Images were analyzed using Metamorph software (top panel). Hematoxylin-eosin (H&E) staining of these tumors and normal cerebellum is shown in the bottom panel. (B and C) Distribution of p27 expression in REST-expressing human medulloblastomas is provided. Each dot represents a tumor. Significance and correlation were measured using the Wilcoxon rank-sum test and Spearman correlation test (rank correlation=0.51 and p=0.0008).
REST represses the expression of ubiquitin-specific peptidase 37 (USP37)
The laddering appearance of p27 protein in REST-expressing cells in Fig. 1E was suggestive of post-translational regulation and modification by high molecular weight adducts. A number of studies have shown p27 levels to be post-translationally regulated by ubiquitination, which targets its for proteasomal degradation (27–37). To assess if p27 levels were post-translationally controlled in medulloblastoma cells and required the proteasome, translation of new protein synthesis was blocked by cycloheximide (CHX) treatment (0–120 minutes) in DAOY, D283 and UW228 medulloblastoma cells and p27 levels were measured by immunoblotting. As seen in Fig. 3A, there was a substantial reduction in p27 levels following CHX treatment, which could be prevented by the proteasomal inhibitor MG132. To further evaluate whether the slower migrating forms of p27 seen in REST-expressing cells were ubiquitinated and whether p27 ubiquitination changed in a REST-dependent manner, we performed immunoprecipitation (IP) assays using extracts prepared from DAOY cells transiently transfected with REST siRNA or scr siRNA. IP with anti-ubiquitin (anti-Ub) antibody followed by Western blotting with anti-p27 antibody revealed an increase in faster migrating forms of p27 in immunoprecipitates from cell extracts lacking REST compared to that from cells expressing REST. Reactions with control non-immune sera (IgG) were included as controls (Fig. 3B). Input lanes have been shown separately to highlight differences in the slower migrating forms of p27 in cells that expressed and lacked REST. These results suggest that REST loss increased p27 levels by potentially affecting its ubiquitination.
Figure 3. REST affects p27 ubiquitination by repressing the deubiquitylase USP37.
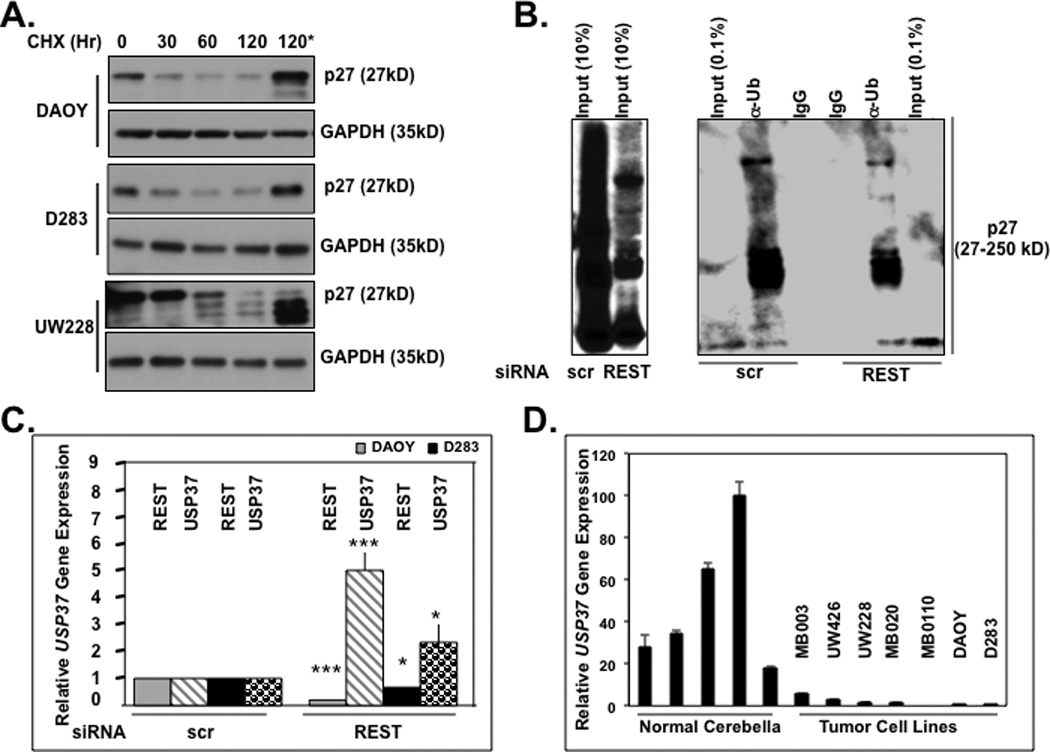
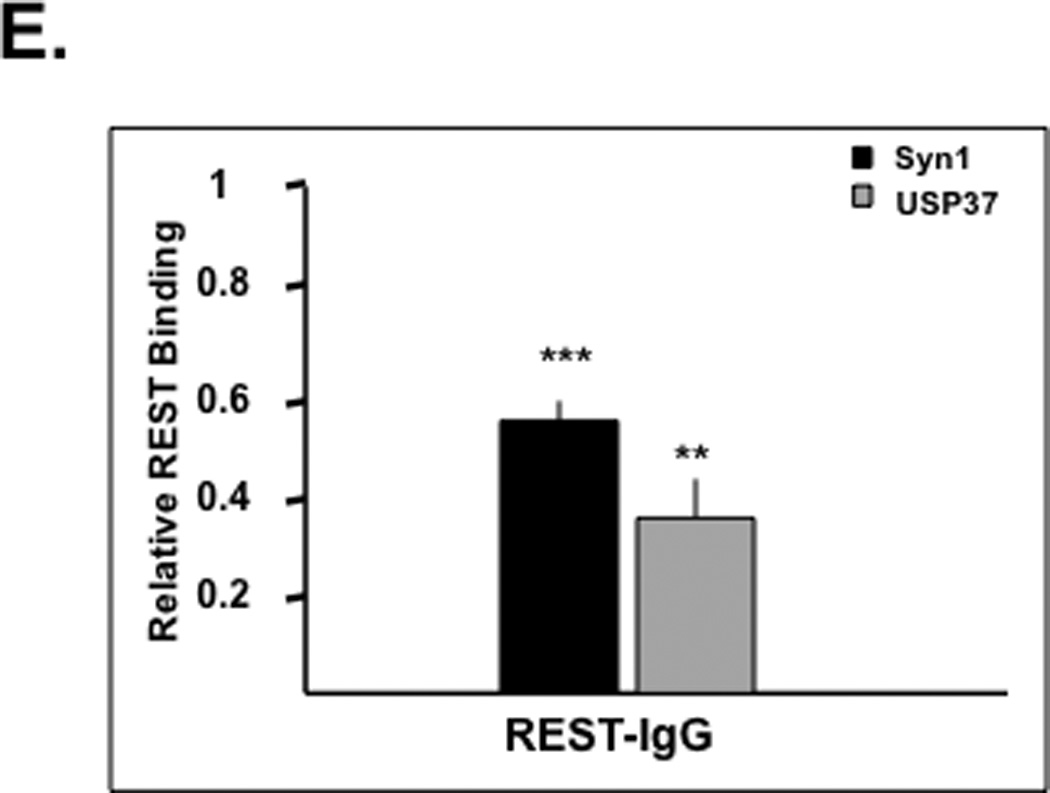
(A) Stability of p27 in DAOY, D283, and UW228 cells was measured following treatment with cycloheximide (CHX) (100 µg/ml) for various time periods (0–120 mins) or MG132 (20 µM) for 4 hrs and analyzed by Western blotting. GAPDH was used as a loading control. (B) REST-dependent changes in p27 ubiquitination were assessed by immunoprecipitation experiments following transient transfection of DAOY cells with REST-specific siRNA or scr siRNA. p27 was immunoprecipitated from whole cell extracts using anti-ubiquitin (Ub) antibody followed by Western blotting using anti-p27 antibody. Input lanes (0.5% and 10%) are included. (C) Change in USP37 gene expression in DAOY and D283 cells transfected with REST-specific siRNA or scr siRNA was determined by SYBR-Green Q-RT-PCR analysis using specific primers. Changes in REST and USP37 gene expression were plotted relative to 18S RNA. Each experiment was performed in triplicate and the standard error calculated. Significance (* p<0.1, ** p<0.05, *** p< 0.001) was calculated using Statistica 6.0 software. (D) USP37 expression in normal cerebella and established or primary medulloblastoma cultures were measured by Q-RT-PCR analyses and normalized to 18S levels. (E) REST binding to the RE1 element downstream of the USP37 gene was evaluated by chromatin immunoprecipitation assay. REST was immunoprecipitated from formaldehyde-cross-linked and sonicated nuclear DAOY cell extracts, and the associated DNA was measured by Q-RT-PCR analysis. The results shown are the average of three separate experiments. Significance (* p<0.1, ** p<0.05, *** p< 0.001) was calculated using Statistica 6.0 software.
Skp2, FBXW7 and KPC1/KPC2 are a few E3 ligases known to be important for p27 ubiquitination and proteasomal degradation (27–37). However, REST knockdown did not cause a change in the levels of these enzymes. Therefore, we asked if REST regulated the process of deubiquitylation catalyzed by a family of proteases called deubiquitylases (DUBs). A function of these enzymes is to remove ubiquitin moieties from proteins and prevent their proteasomal degradation (22–24, 38, 39). Since REST-target DUBs with activity towards p27 have not been reported in neural cells, we searched for candidates in the RE1-database that lists putative target genes based on the presence of the REST-binding RE1 element in the gene regulatory regions (40). This search identified a distal RE1 site downstream of the gene encoding the ubiquitin-specific peptidase 37 (USP37) (40). To determine whether USP37 was a REST target gene, we first assessed if USP37 expression changed in a REST-dependent manner. To this end, DAOY and D283 cells were transiently transfected with REST siRNA or scr siRNA and analyzed 24 h later for the efficiency of REST knockdown and change in USP37 expression by Q-RT-PCR analysis using 18s RNA as an internal control and for normalization. As shown in Figure 3C, both DAOY and D283 cells showed a significant increase in USP37 expression (4.5-fold and 1.6-fold, respectively) following REST knockdown. Q-RT-PCR analyses also confirmed a substantial diminution of USP37 expression in REST-positive established cell-lines (Daoy, D283) and patient-derived primary cultures (UW-228, UW-426, MB-0110, MB-020 and MB-030) compared to normal cerebella (Fig. 3D). REST binding to the RE1 motif in the USP37 regulatory region and in the positive control Syn1 in DAOY cells was confirmed by chromatin immunoprecipitation (ChIP) analyses (Fig. 3E). The signal obtained with control IgG pull-down was subtracted from that obtained with the specific REST-antibody and plotted as relative binding. Together these data indicate that REST controlled USP37 gene expression.
The relationship between USP37 and p27 was also evaluated by IFA in human medulloblastoma tumor samples. Both p27 and USP37 were expressed in the normal cerebellum whereas their expression was not detected at all or to very low levels in human tumors (Fig. 4A, top and middle panels). The corresponding H&E-stained sections are shown in the bottom panel (Fig. 4A). Of the 42 human tumor samples studied and scored, 19 (45%) showed staining for p27, while 23 (55%) did not. Of the p27-negative tumors, 18 did not express USP37, 4 expressed low levels (+) of USP37, and 1 expressed modest levels of USP37 (+++). Conversely, normal cerebellum expressed both USP37 and p27. Statistical analyses revealed that USP37 protein expression levels were significantly higher in p27-positive than in p27-negative tumors (r=0.80, p<0.0001, Spearman correlation) (Figs. 4B and 4C).
Figure 4. USP37 and p27 levels are correlated in human medulloblastoma and normal mouse cerebellum.
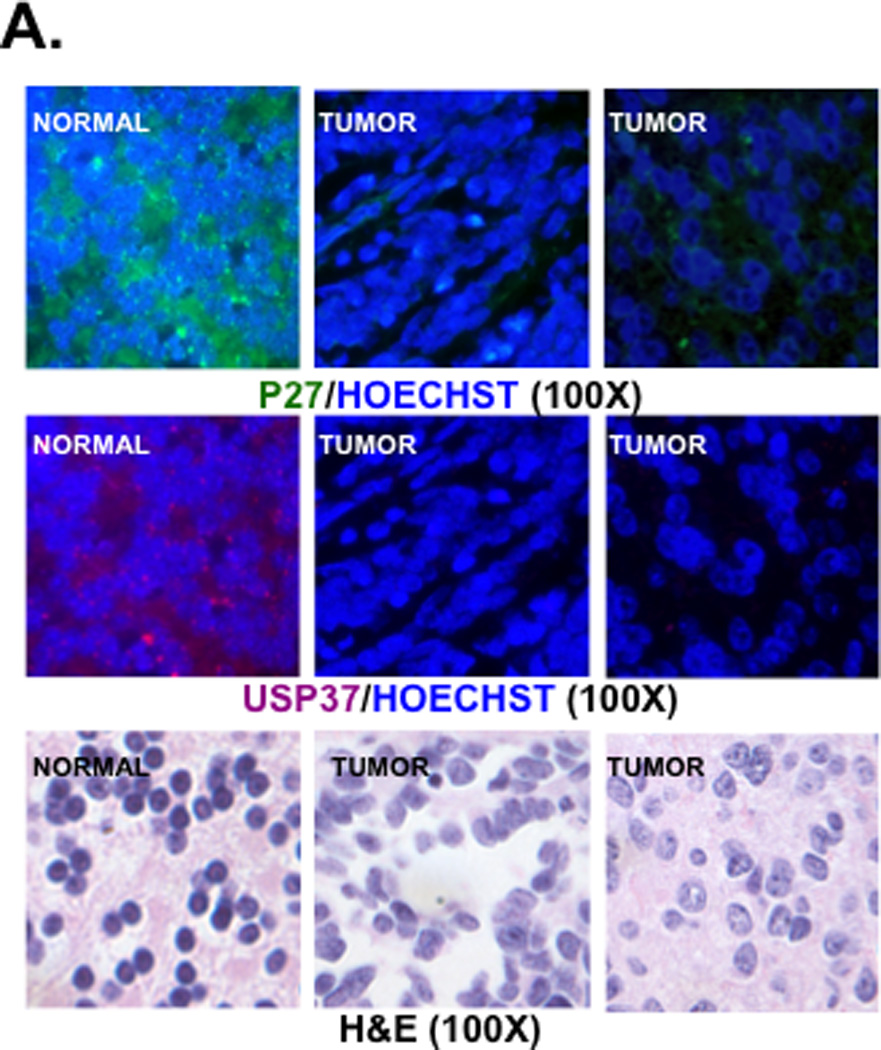
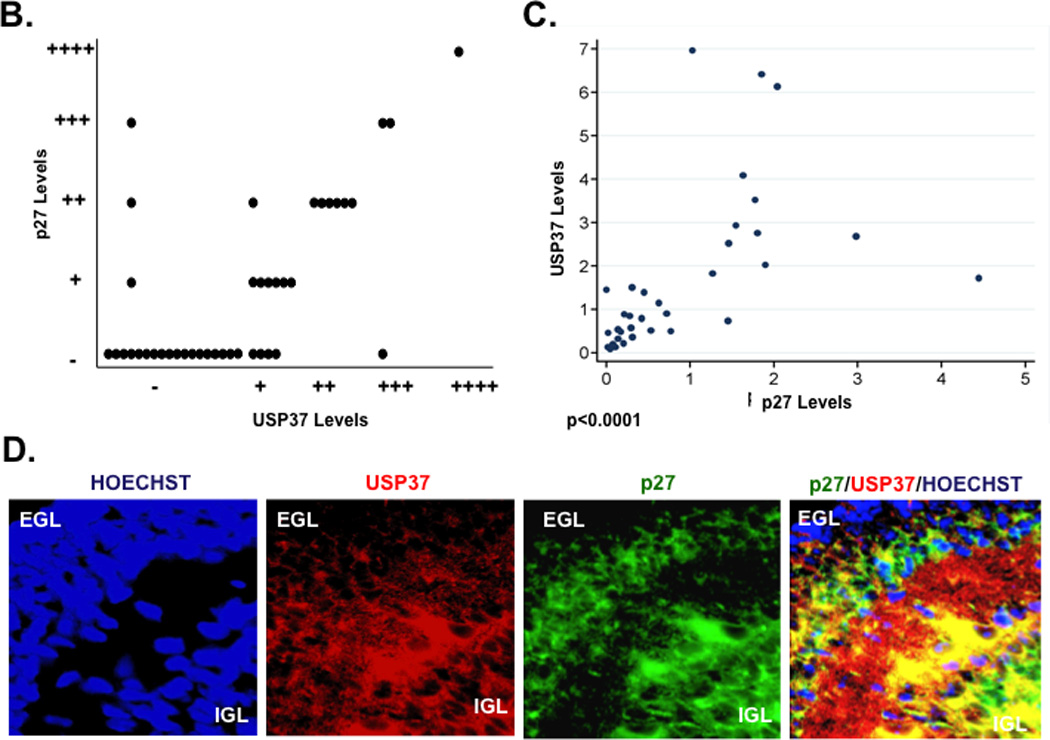
(A) Expression of USP37 (red) and p27 (green) in human medulloblastoma samples was assessed by IFA using specific antibodies and Cy3- or Alexa-488-conjugated secondary antibodies. Nuclei were stained with Hoechst dye and the images viewed and analyzed by fluorescence microscopy. (B and C) Distribution of p27 expression in USP37-expressing human medulloblastomas is provided. Significance and correlation were measured using the Wilcoxon rank-sum test and Spearman correlation test (rank correlation=0.67 and p<0.0001). (D) USP37 (red) and p27 (green) expression in cerebella of postnatal day 7 mice were evaluated by IFA as described above. EGL: external granule layer, IGL: Internal granule layer.
The expression of these proteins was also studied in the normal cerebellum of postnatal day 7 mice by IFA. USP37 expression was first seen in cells of the inner external granule layer (EGL) and was maintained in the inner granule layer (IGL) (Fig. 4D). p27 expression overlapped strongly with that of USP37 in these cells. Neither protein was detected in the outer EGL cells (Fig. 4D). These data provide strong evidence for a correlation between USP37 and p27 levels in human tumors and in the normal murine cerebellum.
Constitutive USP37 expression promotes p27 deubiquitination
To further evaluate the involvement of USP37 in the control of p27 ubiquitination, we studied the interaction between the two proteins using transiently expressed, epitope tagged USP37 (24). DAOY cells were transiently transfected with pDEST26 plasmid expressing FLAG-HA-tagged USP37 or vector alone. Transgene expression was measured by Q-RT-PCR analyses and Western blotting at various times following transfection (Figs. 5A and 5C). Interaction between USP37 and p27 was also confirmed by co-immunoprecipitation experiments using anti-p27 antibodies or control IgG followed by Western blot analysis using anti-Ub antibodies. Although slower migrating forms of p27 that are presumably ubiquitinated, were seen in the presence and absence of ectopic USP37, an increase in the lower molecular weight forms of p27 was observed in USP37 (USP37WT) expressing cells (Fig. 5B). A substantial increase in p27 protein levels accompanied by a gradual decrease in the slower migrating p27 bands over time was also apparent In USP37 transgene-expressing cells (Fig. 5C). In contrast, a mutant of USP37 carrying a cysteine to serine change at amino acid 350 (USP37C350-S), failed to stabilize p27 (Fig. 5D). Transient transfection of Myc-tagged USP1 was also ineffective in stabilizing p27 (Fig. 5E). To confirm that USP37 ubiquitinated p27, we performed in vitro DUB assays using purified epitope tagged proteins from transiently transfected 293T cells. Addition of USP37 to a reaction mix containing HA-ubiquitin and Myc-p27 caused a substantial increase in the 27kD form of p27 and a corresponding decrease in the slower migrating forms of the protein relative to reactions containing USP37 and a protease inhibitor N-ethyl maleimide (NEM) or USP37C350-S or USP1 (Fig. 6F). Collectively, these data indicate that p27 ubiquitination and stability in medulloblastoma cells is controlled by USP37.
Figure 5. Constitutive USP37 expression counters REST-mediated destabilization of p27.
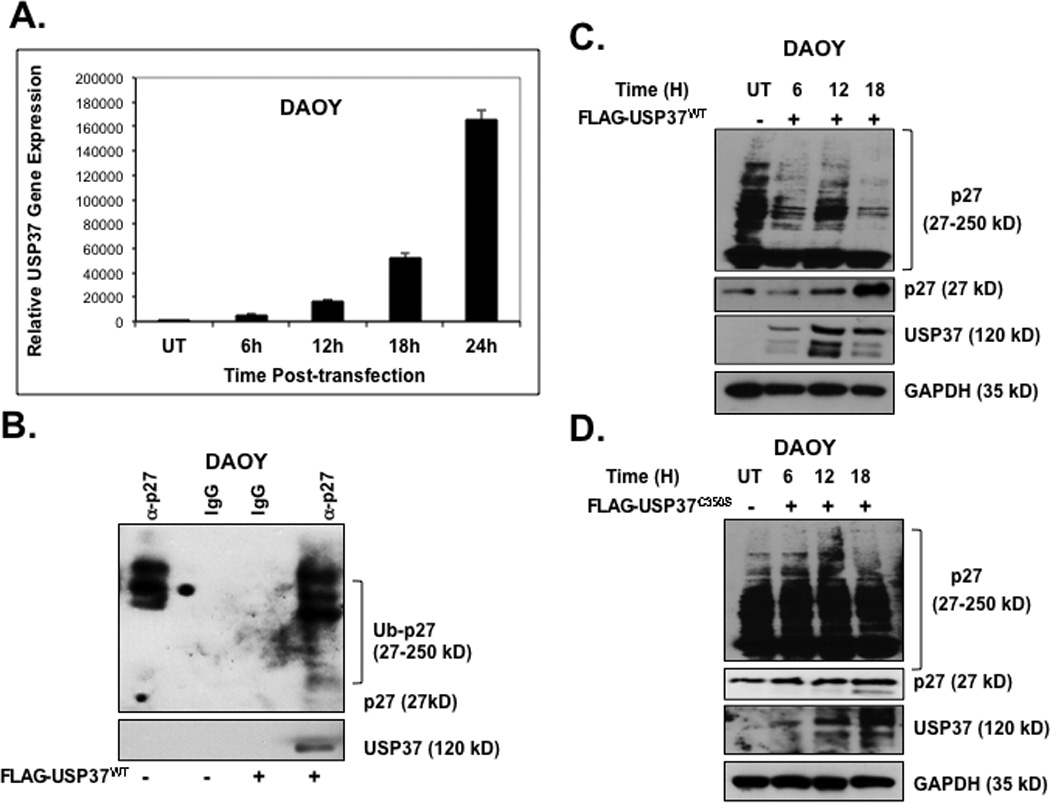
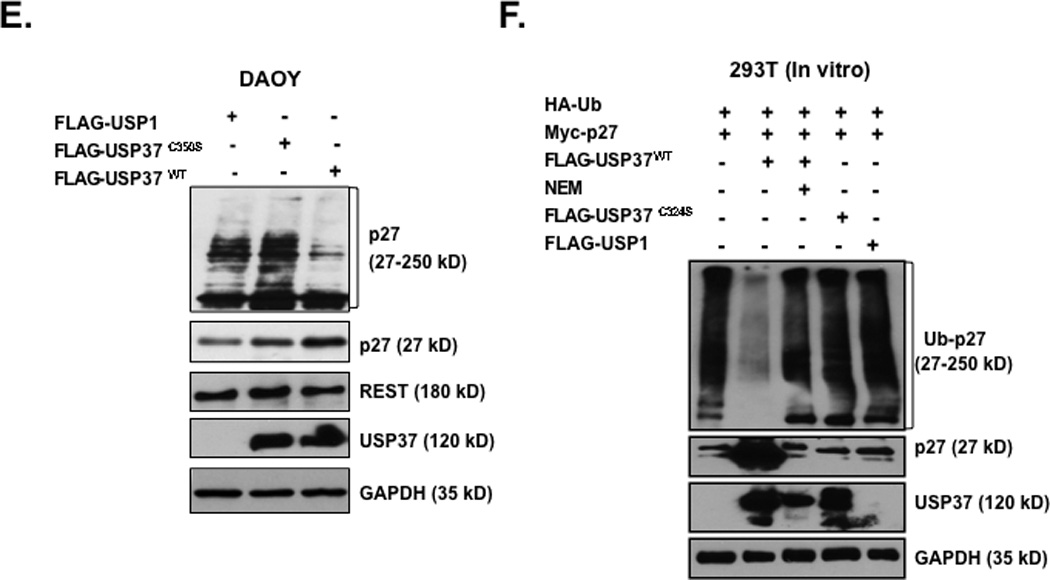
(A) DAOY cells were transiently transfected with a plasmid expressing pDEST-FLAG-HA-USP37-wildtype (WT) or vector alone (UT). Transgene expression was determined at various times post-transfection by Q-RT-PCR analysis using primers specific to USP37. Levels of 18S RNA were measured for normalization. (B) Co-immunoprecipitation assay was performed to study the association of p27 and USP37 in DAOY cells constitutively expressing FLAG-HA-USP37. Anti-p27 antibody was used to immunoprecipitate p27 from whole cell extracts prepared 16 h post-transfection. USP37 and ubiquitinated p27 pull-down was evaluated by Western blotting using anti-Ub and anti-USP37 antibodies. DAOY cells transiently expressing (C) FLAG-HA-USP37WT or (D) mutant FLAG-HA-USP37C350-S were subjected to Western blotting to assess p27 levels at various times after transfection. USP37 expression was confirmed by re-probing the blot with anti-USP37 antibodies. GAPDH was used as a loading control. (E) p27 levels in DAOY cells expressing FLAG-HA-USP1, -USP37WT -USP37C350-S were compared by Western blotting using anti-p27 antibody. Levels of REST, USP37 and GAPDH (loading control) were also measured using specific antibodies. (F) In vitro DUB assays were performed by co-incubating the substrate (immunopurified HA-Ub-Myc-p27) with FLAG-HA-USP37WT (lane 2) FLAG-HA-USP37WT in the presence of NEM (lane 3) FLAG-HA-USP37C350-S (lane 4) or FLAG-HA-USP1 (lane 5). A reaction containing substrate alone was included as a control (lane 1). Cells were treated with 20 µM MG132 for 6 hours prior to extract preparation.
Figure 6. USP37 is necessary for REST-mediated effects on p27 stability, proliferation and differentiation.
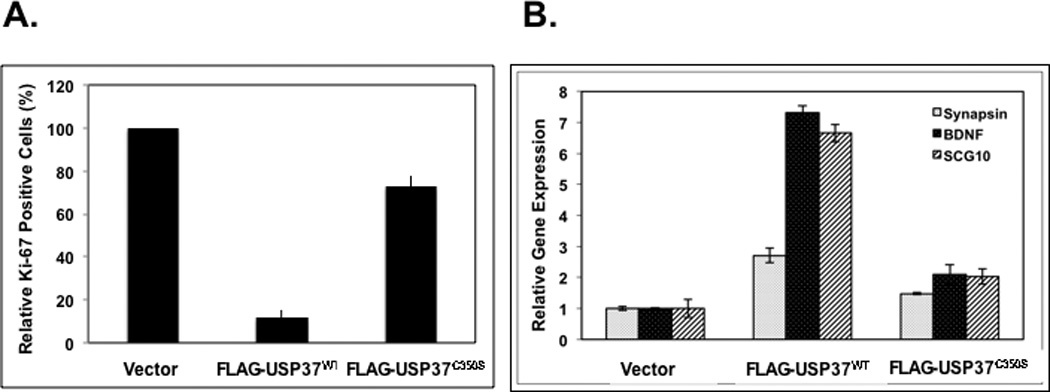
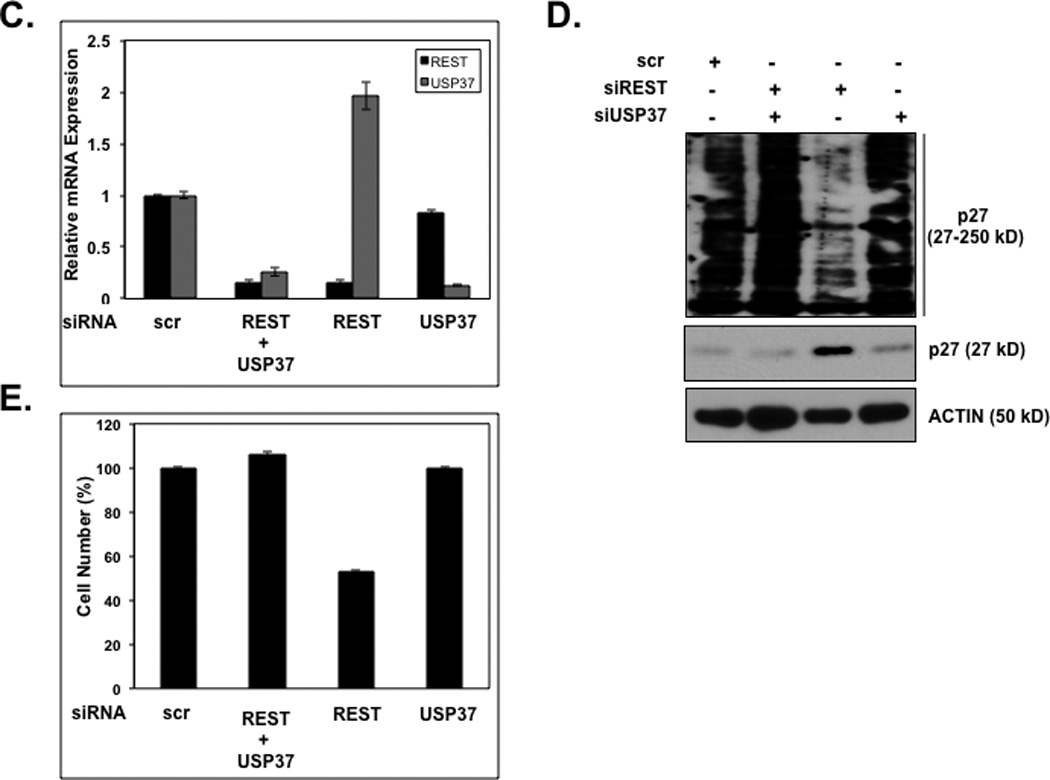
The effect of constitutive expression of USP37WT or USP37C350-S (A) on cell proliferation was assessed by measuring Ki-67 staining relative to vector-transfected cells (B) on neuronal differentiation was evaluated by Q-RT-PCR analysis using primers specific to the REST-target genes Syn1, BDNF, and SCG10. Levels of 18S RNA were measured for normalization. DAOY cells transfected with scrambled siRNA or pooled siRNA against REST or USP37 or both and (C) efficiency USP37 and REST knockdown was assessed by Q-RT-PCR analyses and normalization to 18S RNA levels (D) effect on p27 ubiquitylation and stabilization was determined by Western blotting using anti-p27 antibodies (E) and on cell numbers by trypan blue staining and counting.
REST-induced cell proliferation and blockade of neuronal gene expression is dependent on USP37
To examine the effect of USP37-dependent stabilization of p27 on proliferation and differentiation of medulloblastoma cells, we transfected DAOY cells with pDEST26 vector alone or pDEST26 expressing USP37WT or USP37C350-S. Cells were stained with anti-Ki67 antibodies and studied by IFA. Percent change in the number of Ki-67 positive cells in USP37WT or USP37C350-S-expressing cells was measured and plotted relative to vector-transfected controls (set to 100%) (Fig. 6A). Only 10% of cells expressing USP37WT were Ki-67 positive in contrast to 70% Ki-67 positivity in USP37C350-S-expressing cells (Fig. 6A). Unexpectedly, Q-RT-PCR analyses showed that WT USP37 expression promoted a 2.7–8 fold increase in expression of the REST target genes Syn1, BDNF and SCG10 respectively, relative to vector transfected cells. A 1.2–2 fold upregulation of these genes was seen in cells expressing USP37C350-S (Fig. 6B). These findings indicate that ectopic USP37 not only blocked REST-dependent cell proliferation, but also countered the blockade to neuronal differentiation even in the presence of REST.
To examine if REST-induced p27 stabilization and maintenance of cell proliferation required USP37, we transiently knocked down REST gene expression alone or REST and USP37 gene expression together in DAOY cells and the effect on p27 stabilization and cell numbers was measured. Cells transfected with USP37 siRNA or scrambled siRNA were included as controls. Knockdown efficiency was measured by Q-RT-PCR analyses (Fig. 6C). As seen in Figures 6D and 6E, REST loss promoted p27 protein stabilization whereas the concomitant loss of USP37 countered this effect. This also countered the decrease in cell numbers that was observed in cells lacking REST gene expression. Thus, our data suggest that REST-mediated destabilization of p27 and deregulation of proliferation are mediated by USP37.
Discussion
REST is expressed in embryonic stem cells (ESCs), non-neural cells, and neural progenitors, where it prevents neuronal differentiation (1–10). However, recent work from a number of groups, including our work described here, has implicated REST in the control of non-neurogenic processes (10, 41–45). We had previously shown that the aberrant maintenance of REST expression in medulloblastomas and the ectopic expression of REST in v-Myc-immortalized neural progenitors (NSC-MR) blocked neuronal differentiation (13). Elevated REST expression was also associated with sustained proliferative potential in these cells (13). A role for REST in the control of proliferation seen in our studies is consistent with its expression in the cerebellar EGL, where proliferating progenitors are housed, and its absence from post-mitotic cells in the inner EGL and IGL (46). Our findings linking REST to the control of cell proliferation are also supported by a recent report wherein elevated REST expression was shown to promote proliferation in PC12 rat pheochromocytoma cells by causing a decrease in tuberous sclerosis 2 (TSC-2) and an increase in nuclear beta-catenin levels (47).
In the current study, we suggest that the decline in cell proliferation in the absence of REST may be a consequence of p27 stabilization. The inhibitory effect of p27 on the activity of the cyclinD-CDK4 and/or cyclinE-CDK2 complex is important for cell cycle progression in both G1 and S phase (30, 48–54). Its importance in the control of proliferation of CPCs in vitro and for terminal cell cycle exit during neuronal differentiation in the murine cerebellum is well known (15–17). A decline in p27 levels or its subcellular mis-localization has also been implicated in medulloblastoma development (18–21). While these findings clearly indicate the need for tight control over p27 gene expression, protein levels and its localization in neural cells, the underlying regulatory mechanisms are only beginning to be delineated.
Studies in non-neural cells have shown p27 levels or activity to be regulated by post-translational modifications and its sub-cellular localization (30, 33, 48, 51, 52, 55). In proliferating cells, p27 is ubiquitinated by E3-ligases such as Skp2, KPC1/KPC2 and FBXW7 and targeted for proteasomal degradation (29–32, 37, 52, 56). In our studies reported here we show that the decrease in p27 levels in REST-expressing cells stemmed from the absence of the DUB, USP37. Although the levels of known p27-specific E3-ligases (Skp2, KPC1/KPC2) were unaltered in our assays, we cannot rule out a change in the activity of these enzymes at this time. Additionally, it is important to note that the introduction of a conserved cysteine residue in USP37 may have caused a change in protein folding and promoted its inactivation. To our knowledge, this is the first report of a DUB involved in p27 regulation in neural cells (22). The involvement of USP37 in cell proliferation suggested by our data is supported by a previous report in the literature by Huang and colleagues (57). This excellent study showed USP37 to be important for deubiquitylation of cyclin A and in S phase progression in 293T cells (57). However, we observed an USP37-dependent decrease in cell proliferation in neural cells. Additionally, whereas USP37 expression was found to be controlled by E2F and also subject to an auto-regulatory loop in the study by Huang et al., we showed it to be controlled by REST in neural cells (57). Whether these opposing effects on cell proliferation and cell cycle progression is a reflection of fundamental differences in the regulation and function of USP37 and or its target specificity in neural and non-neural cells is not clear at this time. In this context, it is important to note that REST has tumor suppressive and oncogenic functions in non-neural and neural cells respectively (12–14, 43, 58–61). A future line of investigation would be to assess the role of REST, if any, in the control of USP37 expression in non-neural cells. If so, it would be important to also examine its connection to E2F-dependent regulation of USP37.
In conclusion, our study has uncovered a novel role for REST and USP37 in the regulation of p27 protein and cell proliferation, although a transcriptional regulation of p27 by REST cannot be ruled out. These findings add to a growing body of literature implicating DUBs in cell cycle control (62–69). Our data suggest that REST could potentially function as a molecular switch that temporally coordinates cell proliferation with blockade of neurogenesis. This possibility needs to be evaluated in mouse models. The contribution of REST-USP37-p27 pathway to the development of neural tumors such as medulloblastoma is unclear and is a subject of ongoing investigation in our laboratory.
Materials and methods
Cell Culture
Human medulloblastoma cell lines DAOY and D283 were obtained from the American Type Culture Collection (Manassas, VA) and were maintained as previously described (70). Primary cultures MB-003, MB-020, MB-0110, UW426, and UW228 were a kind gift of Drs. James Olson, Laurence Cooper and John Silber.
Transient Transfection
DAOY and D283 cells were transiently transfected with On-Target plus Smart pool duplex siRNA against human and mouse REST, human USP37 or On-Target plus si-control non-targeting siRNA (Dharmacon, Lafayette, CO) using DharmaFect transfection reagent. Briefly, cells were grown overnight in antibiotic-free serum containing medium and transfected with 100 nM REST-specific siRNA, USP37-specific siRNA, or both siRNAs, and non-target scr siRNA and incubated for 24 hours (h) at 37°C and 5% CO2. DAOY cells were also transiently transfected with the following plasmids: pDEST-FLAG-HA-USP37WT, USP37C350-S, FLAG-HA-USP1, pcDNA3-myc3-p27 or HA-ubiquitin (Addgene, Inc). Cell numbers and viability were assessed by trypan blue staining.
Flow Cytometry
DAOY cells transfected with pooled siRNA against REST or control siRNA were resuspended in a solution containing 1 mg/mL sodium citrate, 0.1% Triton-X-100, and 0.05 mg/mL propidium iodide and incubated overnight at 4°C. Stained cells were analyzed using a Becton-Dickinson FacsScan Flow Cytometer (Franklin Lakes NJ), and the numbers of cells with sub-G1, G1, S, or G2/M DNA content were calculated using CellQuest 3.2 software (BD Bioscience, San Jose, CA).
Q-RT-PCR Analyses
RNA was isolated from DAOY and D283 cells transfected with various pooled siRNAs or wild type and mutant USP37C350-S using the RNeasy kit (Qiagen, Valencia, CA). Q-RT-PCR reactions were performed as described previously using primers for human REST, USP37, Synapsin, BDNF or SCG10 (70). Reactions were performed in triplicate and gene expression was normalized to actin, GAPDH, or 18S RNA. Relative mRNA expression was calculated using the comparative ΔΔct method (71).
Primers sequences:
-
BDNF
Forward: 5'-GCC CTG TAT CAA CCC AGA AA -3’
Reverse: 5'-CTT CAG AGG CCT TCG TTT TG-3’
-
SCG10
Forward: 5'-GAG CTG TCC ATG CTG TCA CTG-3’
Reverse: 5'-GAA GAA ACT GGA GGC TGC AGA-3’
-
Syn1
Forward: 5'- GTC TGA CAG ATA CAA GCT CTG-3’
Reverse: 5'- GAC CAC GAG CTC TAC GAT GAG-3’
-
REST
Forward: 5'- GTA GGA GCA GAA GAG GCA GAT-3’
Reverse: 5'- GCT TCA CGT TCT TCT ACT GCT-3’
-
USP37
Forward: 5'- GTG CTC TTG TCA GGC ACA AA-3’
Reverse: 5'- GCA CTC CAA CCA AGG GTA AA-3’
Western Blotting
DAOY, D283, and UW228 were treated with 100 µg/ml cycloheximide (CHX) (Sigma, St Louis, MO) or 20 µM MG132 (Calbiochem, La Jolla, CA). Extracts prepared from these cells or those transfected with expression constructs and siRNAs cells were subjected to polyacrylamide gel electrophoresis and Western blotting using one or more of the following primary antibodies: REST (Millipore, Waltham, MA); p27 (BD Biosciences, Franklin Lakes, NJ and Cell Signaling, Danvers, MA); USP37 (Bethyl Laboratories, Montgomery, TX); ubiquitin (Abcam, Cambridge, MA); type-III beta tubulin (Covance, Emeryville, CA); actin (Cell Signaling, Danvers, MA); GAPDH (Abcam, Cambridge, MA); and N-Myc (Santa Cruz Biotechnologies, Santa Cruz, CA).
Chromatin Immunoprecipitation Assay
Cross-linked DAOY cells were resuspended in sonication buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, and protease inhibitors) and sonicated and 10% of this material was saved as input DNA. The reminder of the samples were diluted 10-fold with IP buffer [16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.2 mM EDTA (pH 8.0), 1.1% Triton X-100, and protease inhibitors], precleared, and incubated with anti-REST antibody or control non-immune sera for 12 h at 4°C. Following incubation with protein A beads, washing, and elution, the cross-linking was reversed and DNA was purified with a QiaQuick PCR Purification Kit (Qiagen, Valencia, CA). The bound DNA was quantified by SYBR-Green Q-PCR analyses and analyzed using an IQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Calculations following normalization to input values were done as described previously (72). The following primers were used:
-
Human USP37
Forward: 5'-CAT CTC ACT CAG GCA GGA AAG TTG TGC-3'
Reverse: 5'-GGA CCA GGC TTC ACA GGT GAT AGG AG-3'
-
Human Syn1
As described in (12).
Immunoprecipitation and Co-immunoprecipitation Assay
Cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% Igepal, and protease inhibitors) and sonicated. Protein (1 mg) in lysis buffer was pre-cleared and incubated with anti-p27 or anti-Ubiquitin (Ub) antibody (Abcam, Waltham, MA) overnight at 4°C and then incubated with protein-G agarose. Beads were washed and sample was eluted with 2X sodium dodecyl sulfate (SDS) buffer and analyzed by Western blotting using anti-p27 or anti-Ub antibody.
Immunofluorescence Assay (IFA) and Hematoxylin Eosin staining
De-identified patient samples and normal cerebella were obtained following Institutional Review Board (IRB) approval. Samples were deparaffinized and processed for IFA using antibodies against REST (1:50, Millipore, Waltham MA), p27 (1:100, BD Transduction Laboratories, Bedford, MA) and USP37 (1:150, Bethyl Laboratories, Montgomery, TX). After washing, cells were incubated with Cy3- or Alexa 488-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) and then covered with Slowfade Gold antifade (Invitrogen, Carlsbad, CA) containing 1 µg/ml Hoechst dye to stain the nuclei. Images were visualized under a Nikon fluorescence microscope and analyzed using Metamorph software (Molecular Devices, Downington, PA). Staining for REST, p27, and USP37 expression was scored as high (++++/+++), moderate (++), low (+), or none (−). Hematoxylin-eosin staining and co-staining for REST and p27 were performed as previously described (13, 70). Brains were harvested from postnatal day 7 mice and processed for IFA using anti-p27 and anti-USP37 antibodies.
Site Directed Mutagenesis
The plasmid pDEST26-Flag-HA-USP37 expressing the human USP37 gene (Addgene, Inc) was used as a template to perform a site directed change of the conserved cysteine at position 350 to a serine using QuikChange II XL Site Directed Mutagenesis Kit (Invitrogen). The following primers were used: 5’-GGG CTT CTC CAA TTT GGG AAA TAC CTC CTA TAT GAA TGC-3’ and 5’ GCA TTC ATA TAG GAG GTA TTT CCC AAA TTG GAG AAG CCC TGC-3’. Introduction of the mutation was confirmed by sequencing.
In Vivo Deubiquitination Assay
DAOY cells were transfected with pDEST26 vector expressing Flag-HA-USP37WT, Flag-HA-USP37C350-S or Flag-HA-USP1 (Addgene, Inc). Cell extracts were prepared after addition of MG132 (20 µM) to the culture medium for a period of 6 hours prior to harvesting the cells. Cell lysates were subjected to polyacrylamide gel electrophoresis and Western blotting using anti-p27, anti-REST, anti-USP37 and anti-GAPDH antibodies.
In Vitro Deubiquitination Assay
293T cells were co-transfected with pcDNA3-myc3-p27 and HA-ubiquitin or with pDEST26-FLAG-HA-USP37WT, FLAG-HA-USP37C350-S or FLAG-HA-USP1 using Lipofectamine 2000 (invitrogen). The cells were treated with 20 µM MG132 for 6h before collection and lysed in immunoprecipitation buffer. Cell extracts from DUB-expressing cells (1 mg protein) were immunopurified using anti-FLAG M2 beads and eluted with FLAG peptide. HA-Ub-p27 substrate was purified from cell extracts (1 mg protein) using anti EZview red anti-HA affinity gel and elution with HA-peptide. DUB assays were performed by incubating equal amount of substrate with purified DUBs (USP37, USP37 C350-S or USP1) in the presence or absence of 15 mM NEM (57)). Reactions were terminated by adding 6X Laemmli buffer reactants and analyzed by Western blotting with anti-p27 and anti-Ub antibodies.
Figure 7. Model to describe the novel role for REST and USP37 in the control of p27 levels and cell proliferation.
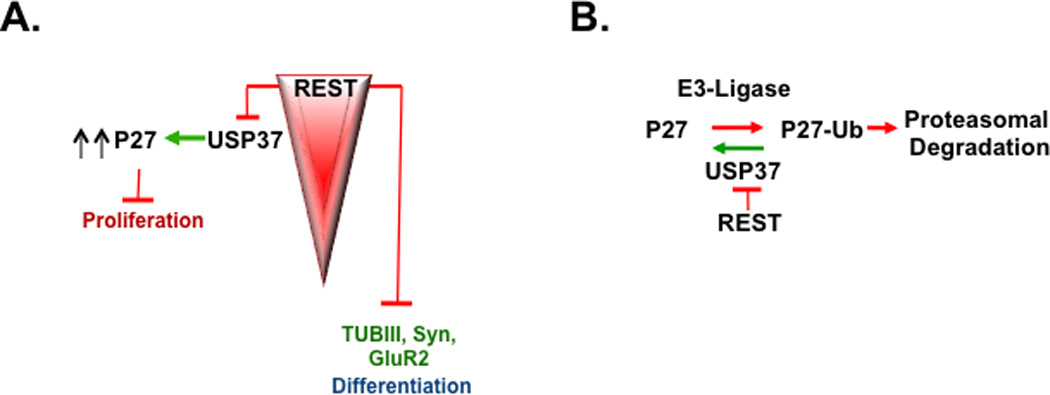
(A) REST is a known regulator of neuronal differentiation genes in neural progenitors and medulloblastoma cells. Here, we show that REST also promotes cell proliferation by repressing USP37 expression and causing p27 degradation. (B) USP37 promotes p27 deubiquitylation and prevents its proteasomal degradation, a process that is negatively regulated by REST.
Acknowledgements
This work was supported by grants from the American Cancer Center (RSG-09-273-01-DDC) and the National Institutes of Neurological Disorders and Stroke (1R03NS077021-01) to VG.
Footnotes
All authors have read and approved the manuscript.
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995 Mar 24;80(6):949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 2.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995 Mar 3;267(5202):1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 3.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005 May 20;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005 Oct;15(5):500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005 Dec;17(6):664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007 Jul;8(7):544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 7.Kagalwala MN, Singh SK, Majumder S. Stemness is only a state of the cell. Cold Spring Harb Symp Quant Biol. 2008;73:227–234. doi: 10.1101/sqb.2008.73.042. [DOI] [PubMed] [Google Scholar]
- 8.Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009;4(12):e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juliandi B, Abematsu M, Nakashima K. Chromatin remodeling in neural stem cell differentiation. Curr Opin Neurobiol. 2010 Aug;20(4):408–415. doi: 10.1016/j.conb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Gopalakrishnan V. REST and the RESTless: in stem cells and beyond. Future Neurol. 2009;4(3):317–329. doi: 10.2217/fnl.09.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't] 2011 Jun 29;31(26):9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000 Jul;6(7):826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 13.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006 Mar;26(5):1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, et al. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005 Mar;4(3):343–349. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- 15.Miyazawa K, Himi T, Garcia V, Yamagishi H, Sato S, Ishizaki Y. A role for p27/Kip1 in the control of cerebellar granule cell precursor proliferation. J Neurosci. 2000 Aug 1;20(15):5756–5763. doi: 10.1523/JNEUROSCI.20-15-05756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto T, Mitsuhashi T, Takahashi T. Altered patterns of neuron production in the p27 knockout mouse. Dev Neurosci. 2004 Mar-Aug;26(2–4):208–217. doi: 10.1159/000082138. [DOI] [PubMed] [Google Scholar]
- 17.Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci U S.A. 2006 Aug 1;103(31):11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatia B, Northcott PA, Hambardzumyan D, Govindarajan B, Brat DJ, Arbiser JL, et al. Tuberous sclerosis complex suppression in cerebellar development and medulloblastoma: separate regulation of mammalian target of rapamycin activity and p27 Kip1 localization. Cancer Res. 2009 Sep 15;69(18):7224–7234. doi: 10.1158/0008-5472.CAN-09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatia B, Malik A, Fernandez LA, Kenney AM. p27(Kip1), a double-edged sword in Shh-mediated medulloblastoma: Tumor accelerator and suppressor. Cell Cycle. 2010 Nov;9(21):4307–4314. doi: 10.4161/cc.9.21.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatia B, Nahle Z, Kenney AM. Double trouble: when sonic hedgehog signaling meets TSC inactivation. Cell Cycle. 2010 Feb;9(3):456–459. doi: 10.4161/cc.9.3.10532. [DOI] [PubMed] [Google Scholar]
- 21.Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF. Two tumor suppressors, p27Kip1 and patched-1, collaborate to prevent medulloblastoma. Mol Cancer Res. 2009 Jan;7(1):33–40. doi: 10.1158/1541-7786.MCR-08-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011 Jun 24; doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009 Jul 23;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, et al. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006 Sep 1;66(17):8655–8661. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- 26.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002 Oct 15;16(20):2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003 Feb;13(1):41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 28.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999 Aug;1(4):193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008 May 1;111(9):4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008 Jun;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara T, Kamura T, Kotoshiba S, Takahashi H, Fujiwara K, Onoyama I, et al. Role of the UBL-UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol Cell Biol. 2005 Nov;25(21):9292–9303. doi: 10.1128/MCB.25.21.9292-9303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004 Dec;6(12):1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 33.Keller UB, Old JB, Dorsey FC, Nilsson JA, Nilsson L, MacLean KH, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. Embo J. 2007 May 16;26(10):2562–2574. doi: 10.1038/sj.emboj.7601691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotoshiba S, Kamura T, Hara T, Ishida N, Nakayama KI. Molecular dissection of the interaction between p27 and Kip1 ubiquitylation-promoting complex, the ubiquitin ligase that regulates proteolysis of p27 in G1 phase. J Biol Chem. 2005 May 6;280(18):17694–17700. doi: 10.1074/jbc.M500866200. [DOI] [PubMed] [Google Scholar]
- 35.Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle. 2010 Jun 2;9(12) doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ungermannova D, Gao Y, Liu X. Ubiquitination of p27Kip1 requires physical interaction with cyclin E and probable phosphate recognition by SKP2. J Biol Chem. 2005 Aug 26;280(34):30301–30309. doi: 10.1074/jbc.M411103200. [DOI] [PubMed] [Google Scholar]
- 37.Masuda K, Ishikawa Y, Onoyama I, Unno M, de Alboran IM, Nakayama KI, et al. Complex regulation of cell-cycle inhibitors by Fbxw7 in mouse embryonic fibroblasts. Oncogene. [Research Support, Non-U.S. Gov't] 2010 Mar 25;29(12):1798–1809. doi: 10.1038/onc.2009.469. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson KD. DUBs at a glance. J Cell Sci. [Research Support, N.I.H. Extramural Review] 2009 Jul 15;122(Pt 14):2325–2329. doi: 10.1242/jcs.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. [Review] 2009 Aug;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 40.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuronrestrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun YM, Cooper M, Finch S, Lin HH, Chen ZF, Williams BP, et al. Rest-mediated regulation of extracellular matrix is crucial for neural development. PLoS One. 2008;3(11):e3656. doi: 10.1371/journal.pone.0003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. [Research Support, N.I.H. Extramural Research Support, Non-U.S. Gov't] 2008 Mar 20;452(7185):365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L, Schwarzenbach H, Meyer-Staeckling S, Brandt B, Mayr GW, Weitzel JM, et al. Expression Regulation of the Metastasis-Promoting Protein InsP3-Kinase-A in Tumor Cells. Mol Cancer Res. 2011 Apr 1; doi: 10.1158/1541-7786.MCR-10-0556. [DOI] [PubMed] [Google Scholar]
- 44.Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One. [Research Support, N.I.H. Extramural Research Support, Non-U.S. Gov't] 2009;4(11):e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, et al. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J Cell Biol. [Research Support, Non-U.S. Gov't] 2010 Apr 5;189(1):159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravanpay AC, Hansen SJ, Olson JM. Transcriptional inhibition of REST by NeuroD2 during neuronal differentiation. Mol Cell Neurosci. 2010 Jun;44(2):178–189. doi: 10.1016/j.mcn.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasoni R, Negrini S, Fiordaliso S, Klajn A, Tkatch T, Mondino A, et al. A signaling loop of REST, TSC2 and {beta}-catenin governs proliferation and function of PC12 neural cells. J Cell Sci. 2011 Sep 15;124(Pt 18):3174–3186. doi: 10.1242/jcs.087551. [DOI] [PubMed] [Google Scholar]
- 48.Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin Cancer Res. [Research Support, N.I.H. Extramural Research Support, Non-U.S. Gov't Review] 2011 Jan 1;17(1):12–18. doi: 10.1158/1078-0432.CCR-10-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. [Research Support, Non-U.S. Gov't Review] 2009 Mar;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 50.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, et al. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. [Letter] 2009 Nov;11(11):1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blain SW. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle. 2008 Apr 1;7(7):892–898. doi: 10.4161/cc.7.7.5637. [DOI] [PubMed] [Google Scholar]
- 52.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008 Apr;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 53.Kaldis P. Another piece of the p27Kip1 puzzle. Cell. [Comment Research Support, N.I.H. Intramural Review] 2007 Jan 26;128(2):241–244. doi: 10.1016/j.cell.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Pagano M. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Mol Cell. [Review] 2004 May 21;14(4):414–416. doi: 10.1016/s1097-2765(04)00268-0. [DOI] [PubMed] [Google Scholar]
- 55.Reed SI. Keeping p27(Kip1) in the cytoplasm: a second front in cancer's war on p27. Cell Cycle. [Comment Review] 2002 Nov-Dec;1(6):389–390. doi: 10.4161/cc.1.6.261. [DOI] [PubMed] [Google Scholar]
- 56.Koff A. How to decrease p27Kip1 levels during tumor development. Cancer Cell. 2006 Feb;9(2):75–76. doi: 10.1016/j.ccr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011 May 20;42(4):511–523. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. [Research Support, N.I.H. Extramural Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, Non-P.H.S.] 2008 Mar 20;452(7185):370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. [Research Support, N.I.H. Extramural Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, Non-P.H.S. Research Support, U.S. Gov't, P.H.S.] 2005 Jun 17;121(6):837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 60.Spencer EM, Chandler KE, Haddley K, Howard MR, Hughes D, Belyaev ND, et al. Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol Dis. [Research Support, Non-U.S. Gov't] 2006 Oct;24(1):41–52. doi: 10.1016/j.nbd.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 61.Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. [Review] 2005 Sep 6;15(17):R665–R668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 62.Pereg Y, Liu BY, O'Rourke KM, Sagolla M, Dey A, Komuves L, et al. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol. 2010 Apr;12(4):400–406. doi: 10.1038/ncb2041. [DOI] [PubMed] [Google Scholar]
- 63.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005 Jul 20;23(21):4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 64.Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci U S A. 2007 May 22;104(21):8869–8874. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peschiaroli A, Skaar JR, Pagano M, Melino G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene. 2010 Mar 4;29(9):1384–1393. doi: 10.1038/onc.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McFarlane C, Kelvin AA, de la Vega M, Govender U, Scott CJ, Burrows JF, et al. The deubiquitinating enzyme USP17 is highly expressed in tumor biopsies, is cell cycle regulated, and is required for G1-S progression. Cancer Res. Apr 15;70(8):3329–3339. doi: 10.1158/0008-5472.CAN-09-4152. [DOI] [PubMed] [Google Scholar]
- 67.van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008 Sep 1;7(17):2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 68.Eletr ZM, Wilkinson KD. An emerging model for BAP1's role in regulating cell cycle progression. Cell Biochem Biophys. [Review] 2011 Jun;60(1–2):3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayes SD, Harper JW. Cdc25A and Dub3 in a high-stakes balancing act. Nat Cell Biol. [Comment] 2010 Apr;12(4):311–313. doi: 10.1038/ncb2043. [DOI] [PubMed] [Google Scholar]
- 70.Aguilera DG, Das CM, Sinnappah-Kang ND, Joyce C, Taylor PH, Wen S, et al. Reactivation of death receptor 4 (DR4) expression sensitizes medulloblastoma cell lines to TRAIL. J Neurooncol. 2009 Jul;93(3):303–318. doi: 10.1007/s11060-008-9788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, et al. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TRCP) in neuroblastoma cells. Cancer. [Research Support, Non-U.S. Gov't] 2011 Nov 15;117(22):5189–5202. doi: 10.1002/cncr.26145. [DOI] [PubMed] [Google Scholar]
- 72.Das CM, Zage PE, Taylor P, Aguilera D, Wolff JE, Lee D, et al. Chromatin remodelling at the topoisomerase II-beta promoter is associated with enhanced sensitivity to etoposide in human neuroblastoma cell lines. Eur J Cancer. 2010 Oct;46(15):2771–2780. doi: 10.1016/j.ejca.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]


