Abstract
Cancer cells typically display altered glucose metabolism characterized by a preference of aerobic glycolysis, known as the Warburg effect, which facilitates cell proliferation. Hypoxia-inducible factor (HIF) and oncoprotein Myc are two prominent transcription factors that drive glycolysis. Previously we reported that the estrogen-related receptors (ERRs) act as cofactors of HIF and enhance HIF-dependent transcription of glycolytic genes under hypoxia. ERRs are orphan nuclear receptors and key regulators of energy metabolism by orchestrating mitochondrial biogenesis, fatty acid oxidation (FAO), and oxidative phosphorylation (OXPHOS). Here we show that ERRs also stimulate glycolysis under normoxia. ERRs directly bind to and activate promoters of many genes encoding glycolytic enzymes, and the ERR-binding sites in such promoters are essential for ERR-mediated transcriptional activation. ERRs interact with Myc, and the two factors synergistically activate transcription of glycolytic genes. Furthermore, overexpression of ERRs increases glycolytic gene expression and lactate production. Conversely, depletion of ERRs in cancer cells reduces expression of glycolytic genes and glucose uptake, resulting in decreased aerobic glycolysis and cell growth. Taken together, these results suggest that ERRs are important transcriptional activators of the glycolytic pathway and contribute to the Warburg effect in cancer cells.
Keywords: aerobic glycolysis, Warburg effect, nuclear receptor, Randle cycle
Introduction
Glycolysis is the metabolic process through which cells metabolize glucose to pyruvate. In normal differentiated cells under well oxygenated conditions, pyruvate is primarily directed into mitochondria where it is oxidized into acetyl-CoA by the pyruvate dehydrogenase (PDH) complex. Acetyl-CoA then feeds into the tricarboxylic acid (TCA) cycle to generate reducing equivalents, which produce ATP through oxidative phosphorylation (OXPHOS). In hypoxic/anaerobic conditions, pyruvate is shifted from the mitochondrial oxidative pathway and instead is reduced to lactate by lactate dehydrogenase (LDH) in cytoplasm. Glucose metabolism in cancer cells is typically characterized by a shift from aerobic, oxidative metabolism to a glycolytic pathway. Compared to normal cells, cancer cells take up much more glucose and even in the presence of oxygen, most pyruvate derived from glycolysis is diverted into lactate production instead of mitochondrial oxidation. This aerobic glycolysis phenotype is known as the Warburg effect and is a hallmark of cancer cell metabolism (1). The altered glucose metabolism facilitates cancer cell proliferation, which requires not only ATP but also synthesis of biomass (2). Many glycolytic intermediates are important external carbon sources and precursors to generate nucleotides, amino acids, and lipids. Increased glucose uptake and glycolysis should increase glycolytic ATP production and flux through these synthetic pathways, allowing cancer cells to accommodate both bioenergetic and anabolic demands for assembling new cells (2). The importance of a proliferative metabolic program in malignancy is exemplified by recently discovered amplification of the PHGDH gene in human cancer (3,4). PHGDH encodes phosphoglycerate dehydrogenase, which directs a glycolytic metabolite into synthesis of serine and glycine, and contributes to cancer cell proliferation (3,4).
Metabolic switch to aerobic glycolysis in cancer cells is driven primarily by oncogenic signaling pathways involving kinases such as PI3K and Akt, and transcription factors, most notably, hypoxia-inducible factor (HIF) and Myc (5–9). Either due to an intratumoral hypoxic microenvironment or as a result of genetic defects, HIF is stabilized in cancer cells. HIF directly binds to and activates transcription of glucose transporter and nearly every gene in the glycolytic pathway (10). Meanwhile, HIF upregulates pyruvate dehydrogenase kinase (PDK) 1, which in turn inhibits the PDH complex, a rate-limiting enzyme for glucose oxidation (11–13). Therefore, HIF induces a dramatic reprogramming of cancer cell metabolism involving increased glucose uptake and glycolytic flux, and concomitantly decreased glucose oxidation. Many genes encoding glycolytic enzymes are also direct targets of Myc (14). Myc enhances glycolysis without hypoxia. Furthermore, HIF and Myc, both of which are highly expressed in most tumor types, collaborate to direct a transition to glycolytic metabolism during cell proliferation or tumorigenesis (7).
We recently identified the estrogen-related receptors (ERRs) α, β, and γ (NR3B1, 2, and 3) as coactivating factors of HIF (15). ERRs interact with HIF and enhance HIF-induced glycolytic and angiogenic gene expression under hypoxia (15). ERRs are orphan nuclear receptors that are constitutively active without exogenously added ligands, although their transcription activity is further augmented in the presence of coactivator proteins, in particular the PGC-1 family of coregulatory proteins (16,17). Expressed mostly in tissues with high metabolic demands, ERRs play a predominant role in orchestrating mitochondrial biogenesis and cellular energy metabolism such as oxidative phosphorylation (OXPHOS), tricarboxylic acid (TCA) cycle, fatty acid oxidation (FAO), and ATP synthesis (16). ERRs directly activate transcription of numerous genes involved in mitochondrial oxidative metabolism. Consistently, engineered ablation of ERRα or ERRγ in mice results in impaired mitochondrial biogenesis and oxidative capacity in heart muscle, fat cells, and macrophages (16).
Glucose and fatty acids compete for their oxidation, which is described as the Randle cycle (18). While promoting FAO, ERRs inhibit glucose oxidation by upregulating PDK4 (19–21). Like PDK1, PDK4 inactivates PDH and decreases glucose carbon flux into TCA. The similar activity of ERRs and HIF in blocking glucose oxidation and their collaboration in hypoxic gene transcription prompted us to examine whether ERRs might also directly regulate glycolysis. Accumulating evidence implicates ERRs in the glycolysis pathway. Genome-wide chromatin immunoprecipitation (ChIP)-based binding studies in mouse and human cells revealed the occupancy of ERRs not only at genes of oxidative metabolism but also at glycolytic gene loci (22–24). Moreover, the Drosophila ortholog of ERR, dERR, is required for induction of glycolysis to support cell proliferation during mid-embryonic development (25). The fly Pfk glycolytic gene is a direct transcriptional target of dERR, and most glycolytic genes and lactate levels are significantly downregulated in dERR mutants (25).
ERRα has been identified as a negative prognosticator in breast and other cancers, and its expression generally correlates with advanced tumor stage and histological grading (26,27). ERRγ mRNA was found to be overexpressed in 75% of the breast tumors compared to normal mammary epithelial cells, and its overexpression is associated with estrogen receptor (ER)-positive status, and thus, anti-estrogen sensitivity and a favorable prognosis (26). Depletion of ERRα in the MDA-MB-231 breast cancer cells decreases the growth rate of tumor xenografts (28). Given that altered metabolism is vital for tumor growth and that ERRs are global metabolic regulators, we investigated whether ERRs might regulate glucose metabolism in cancer. In the present study, we confirmed that ERRs bind to promoters of many glycolytic genes and activate their expression through the ERR-binding sites. ERRs interact and synergize with Myc in activation of glycolytic genes. Overexpression of ERRs increases glycolytic gene expression and lactate production; conversely, depletion of ERRs in cancer cells downregulates the aerobic glycolytic phenotype and cell growth. Collectively, these findings suggest that ERRs promote glycolysis, and together with well-established glycolysis-driving transcription factors Myc and HIF, contribute to the metabolic transformation of cancer cells.
Results
ERRs bind to promoters of glycolytic enzyme genes
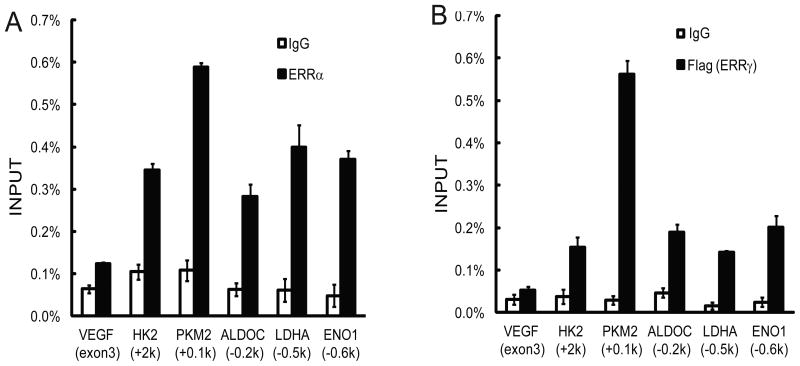
By examining the ChIP-seq-based global transcription factor binding database from the ENCODE Consortium, we found that ERRα occupies the transcription start sites (TSSs) and/or extended promoter regions of most glycolytic genes such as ENO1, PKM2, and LDHA in HepG2 human liver cells (Fig. S1; Table S1). An Enhancer Elements Locator (EEL) program identified putative ERR-binding motifs (ERREs) in the ERRα-bound glycolytic gene promoters. To confirm the binding of ERR, we designed corresponding primers and performed ChIP analysis in MCF7 human breast cancer cells. Compared to control immunoglobin G (IgG), the ERRα antibody significantly enriched genomic DNA fragments of HK2, ALDOC, ENO1, PKM2, and LDHA promoters (Fig. 1A). Because antibodies for ERRγ suitable for ChIP analysis were not available, MCF7 cells were transduced with lentivirus expressing TetON-inducible Flag-tagged ERRγ. Upon induction with a low concentration of Doxcyclin, cells were subjected to ChIP analysis with anti-Flag antibody. Significant binding of Flag-ERRγ was observed at these glycolytic gene promoters as well (Fig. 1B). These results confirm that ERRs bind to promoter regions of glycolytic genes in cancer cells.
Figure 1. ChIP analysis of the binding of ERRs to promoters of glycolytic genes in cancer cells.
Real-time PCR was performed with primers corresponding to the 5′ region of the indicated genes. The exon 3 of the VEGFa gene serves as a negative control for background binding.
(A) ChIP analysis of ERRα binding to glycolytic genes in MCF7 cells with an anti-ERRα antibody.
(B) ChIP analysis of ERRγ binding in MCF7 cells stably expressing Flag-ERRγ with an anti-Flag antibody.
ERRs activate glycolytic gene promoters through ERREs
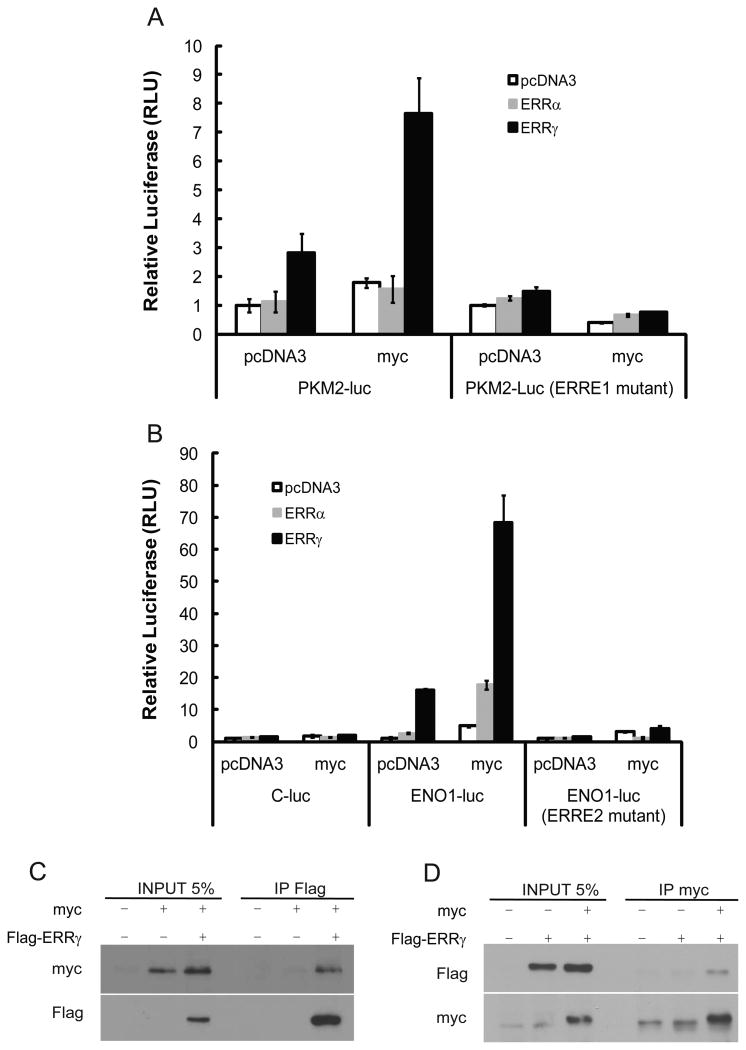
The presence of ERRs at glycolytic genes prompted us to examine their effect on transcription of these genes. The PKM2 proximal promoter contains two putative ERREs (Fig. 2A). When this promoter was linked to a Luciferase reporter, ERRγ significantly activated the reporter when co-transfected into HEK293 cells (Fig. 2A). This activation was further enhanced in the presence of exogenous PGC-1 proteins which are coactivators of ERRs (Fig. 2A). ERRα only possessed weak transcriptional activities in vitro, and did not significantly stimulate reporter activity. To verify if ERRγ activated through direct binding to the promoter, we individually mutated the two ERREs in the PKM2 promoter. While one ERRE was dispensable, the other was essential for ERRγ-mediated reporter activation (Fig. 2B).
Figure 2. Activation of luciferase reporters driven by glycolytic gene promoters by ERRα and ERRγ.
(A) Activation of the PKM2-luciferase by ERRs and PGC-1 coactivator proteins. (top) schematic representation of the positions of two putative ERREs in the PKM2 promoter.
(B) ERRγ-mediated activation of reporters carrying mutated PKM2 promoters lacking individual ERRE.
(C) Synergistic activation of ENO1 promoter-dependent reporter by ERRs and PGC-1. (top) The locations of two ERREs in the ENO1 promoter are indicated. C-luc represents the control reporter without the ENO1 promoter.
(D) ERRγ activation of the reporters driven by the ENO1 promoters with mutations in ERREs.
Similarly, we analyzed the promoter of ENO1, which also harbors two potential ERREs (Fig. 2C). A control reporter carrying the basal promoter of PDK4 was not responsive to ERRs under all experimental conditions (Fig. 2C). When an ENO1 5′ upstream fragment was inserted in front of the basal promoter, the resulting reporter was activated mildly by ERRα but robustly by ERRγ (Fig. 2C). Addition of PGC-1 proteins further markedly augmented activation by both ERRα and ERRγ (Fig. 2C). Mutation of one ERRE (site 1) in the ENO1 promoter substantially impaired ERRγ-mediated activation, and mutation of the other ERRE (site 2) completely abolished induction of reporter by ERRγ (Fig. 2D), suggesting both ERREs are required for ERR-dependent activation of the ENO1 promoter. The reporter assays together with ChIP analyses suggest that glycolytic genes are direct transcriptional targets of ERRs.
Interplay between ERRs and Myc in activation of glycolytic genes
Many genes encoding enzymes in the glycolytic pathway are transcriptional targets of Myc. The ENCODE ChIP analysis of Myc binding in Hela tumor cells uncovered the remarkable presence of Myc at glycolytic gene promoters (Fig. S1). Intriguing, the occupancy of Myc overlaps with that of ERRα (Fig. S1). We tested whether Myc may influence ERR’s activation of glycolytic genes in reporter-based assays. Myc alone exerted only very mild effect on the reporter construct carrying either PKM2 or ENO1 promoter (Fig. 3A and 3B). Remarkably, in the presence of exogenous ERRγ, Myc and ERRγ synergistically activated both reporters (for example, 70 fold induction of the ENO1 promoter-driven reporter) (Fig. 3A and 3B). Myc also synergized with ERRα on the ENO1 reporter, albeit to a milder extent. Mutation of the critical ERREs in these reporters absolutely abrogated such synergy (Fig. 3A and 3B), suggesting that binding of ERRs to DNA is essential for the cooperative activation by ERRs and Myc.
Figure 3. Myc potentiates ERR-mediated transcriptional activation of glycolytic genes.
(A) Myc enhances ERRγ-mediated activation of the luciferase reporter carrying the wild type, but not the ERRE mutant PKM2 promoter in transiently transfected HEK293 cells.
(B) Myc stimulates both ERRα- and ERRγ-dependent activation of the wild type but not the ERRE mutant ENO1 reporters.
(C) Exogenous Myc and ERRγ proteins form a complex in cells. Whole cell lysates from HEK293 cells transfected with Myc and Flag-ERRγ singly or in combination were subjected to co-immunoprecipitation (CoIP) assay with anti-Flag antibody. The presence of exogenous Myc and ERRγ in the precipitates and lysates was shown by immunoblotted with anti-Myc and anti-Flag antibodies.
(D) Reciprocal CoIP assay detects ERRγ-Myc association. CoIP of cellular lysates as in (C) with anti-Myc antibody, followed by immunoblotting with anti-Myc and anti-Flag antibodies.
Given their overlapping binding pattern and synergy in reporter activation, we wondered whether ERR might interact with Myc. Myc and Flag-tagged ERRγ were co-transfected into HEK293 cells. Immunoprecipitation (IP) of ERRγ from cellular lysates with anti-Flag antibody pulled down exogenous Myc (endogenous Myc was not detectable) (Fig. 3C). Similarly, a reciprocal assay with anti-Myc antibody for IP also detected ERR-Myc association (Fig. 3D). These data suggest that ERR and Myc form a complex and co-regulate glycolytic genes.
Overexpression of ERRs activates endogenous glycolytic genes and enhances aerobic glycolysis
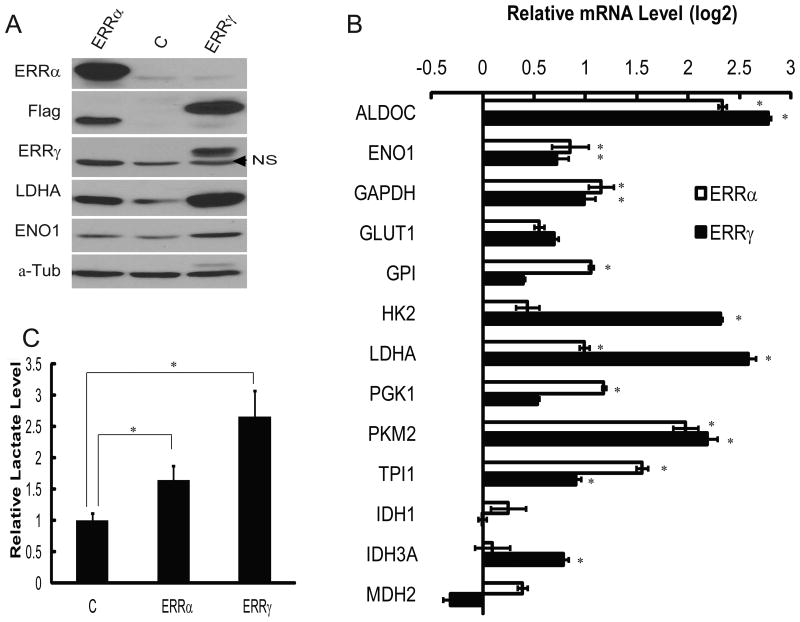
To investigate the effect of ERRs on transcription of endogenous glycolytic genes, we generated lentivirus expressing either Flag-tagged ERRα or ERRγ, and transduced MCF7 cells. Expression of ERRs was validated by immunoblotting analysis. Exogenous ERRα was expressed at much higher levels than the endogenous one (Fig. 4A). Overexpression of ERRα evidently increased protein expression of LDHA (Fig. 4A). Forced expression of ERRγ led to an even stronger effect. In ERRγ-overexpressing cells, LDHA protein level was dramatically increased, and ENO1 expression was also elevated as compared to control cells (Fig. 4A).
Figure 4. Overexpression of exogenous ERRs increases glycolytic gene expression and aerobic glycolysis in cancer cells.
(A) Immunoblotting analysis of ERRs and glycolytic enzymes ENO1 and LDHA in MCF7 cells transduced with control (“C”), or Flag-ERR-expressing lentivirus. Tubulin serves as a loading control. The ERRγ antibody only recognized exogenous protein. Arrowhead denotes a non-specific band (NS).
(B) Quantitative determination of glycolytic gene expression by RT-PCR in control, ERRα- and ERRγ-overexpressed MCF7 cells. * represents significant changes (p<0.05).
(C) Comparison of lactate levels in culture media from control, ERRα- and ERRγ-expressing MCF7 cells. * represents significant changes (p<0.05).
To quantify the induction of glycolytic genes by forced expression of ERRs in MCF7 cells, realtime RT-PCR was performed to measure mRNA levels of glycolytic genes including LDHA, ENO1, HK2, ALDOC, PKM2, TPI1, and GAPDH. A few known ERR target genes such as IDH1, IDH3a and MDH2, which encode enzymes involved in mitochondrial metabolism, were included for comparison. Both ERRα and ERRγ stimulated expression of these glycolytic genes to varying degrees (Fig. 4B). Activation of endogenous glycolytic genes by ectopic expression of ERRs was also observed in MDA-MB-231 human breast cancer cells (Fig. S2A).
Consistent with elevated expression of enzymes in the glycolytic pathway, in particular LDHA, lactate concentrations in media of MCF7 cells were also stimulated by overexpression of ERRα and ERRγ, the latter had a more potent effect (Fig. 4C). ERRs similarly increased lactate production in MDA-MB-231 cells (Fig. S2B).
Depletion of ERRs reduces glycolytic gene expression, glucose uptake, ATP and lactate levels, and growth rate in cancer cells
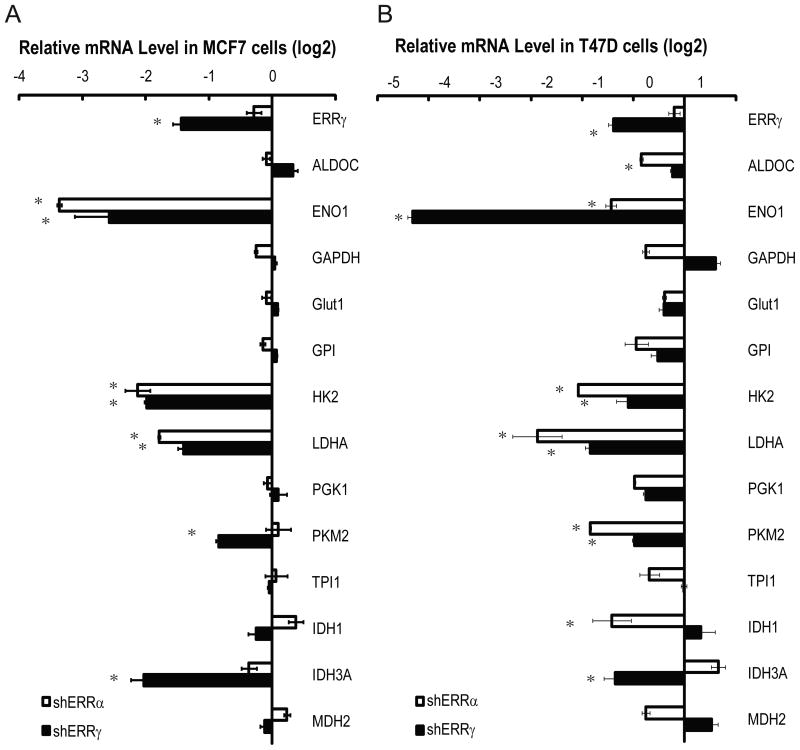
Cancer cells exhibit increased glycolysis even under normoxic conditions. Because exogenous ERRs enhanced glycolysis, we asked whether the glycolytic phenotype of cancer cells might be (in part) attributable to the endogenous ERRs. ERRα or ERRγ was each depleted in MCF7 cells with two independent lentiviral short hairpin RNAs (shRNAs). Efficient knockdown of endogenous ERRα was confirmed by immunoblotting analysis (not shown), and depletion of ERRγ was verified by quantitative RT-PCR (Fig. 5A). Depletion of either ERRα or ERRγ significantly downregulated RNA expression of HK2, ENO1, and LDHA (Fig. 5A). PKM2 RNA levels decreased only in ERRγ-depleted, but not ERRα-depleted MCF7 cells (Fig. 5A). We also examined other cancer cell lines for gene regulation by ERR and confirmed that expression of endogenous HK2, ENO1, LDHA, and PKM2 was largely dependent on both ERRs in T47D and MDA-MB-435 cells as well (Fig. 5B and Fig. S3A, respectively).
Figure 5. Endogenous ERRs are required for glycolytic gene expression in cancer cells.
Quantitative RT-PCR analysis of glycolytic gene mRNA levels in MCF7 (A) and T47D (B) breast cancer cells depleted of ERRα or ERRγ by lentiviral shRNAs. Cells infected with empty virus were used as control. * represents significant changes (p<0.05).
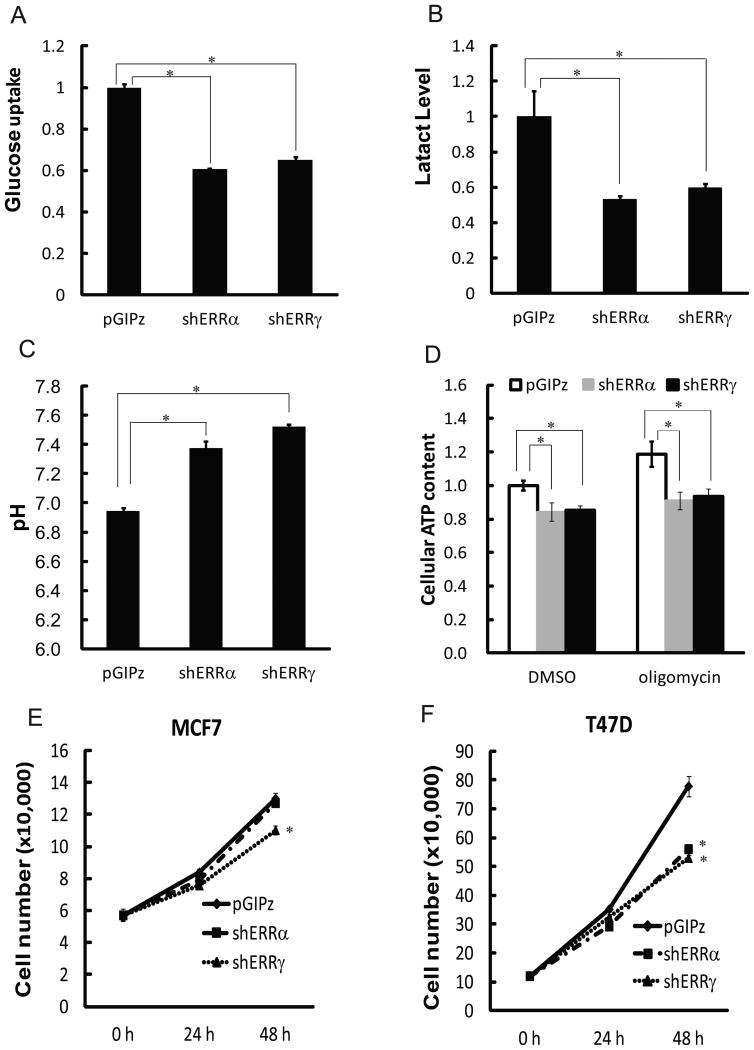
Reduced expression of glycolytic enzymes is expected to lead to reduced glycolysis rates. Therefore, we determined the effect of depletion of ERRs on aerobic glycolysis. Compared to control MCF7 cells, cells depleted of ERRs exhibited markedly decreased glucose consumption (Fig. 6A) and lactate production (Fig. 6B). In addition to generation of energy and metabolites, cellular metabolism converts substrates into byproducts lactate and CO2, which are released into the environment and cause extracellular acidification. Under typical cell culture conditions, extracellular acidification by cancer cells is dominated by lactic acid derived from glycolysis. Consistent with reduced lactate production, ERR-depleted MCF7 cells displayed weaker capacity to decrease medium pH values than control cells (Fig. 6C). Elevated aerobic glycolysis represents an integral source for energy production in cancer cells. As expected, in ERR-depleted cells in which glycolysis was impaired, cellular ATP levels also declined (Fig. 6D). While glucose oxidation is generally attenuated in cancer cells, OXPHOS is still functional to certain extent and contributes to ATP generation. Oligomycin, an inhibitor of the F1F0 ATP synthase, suppresses OXPHOS and mitochondrial ATP synthesis. In response to oligomycin treatment, cancer cells upregulate glycolysis as a compensatory mechanism for ATP generation to sustain ATP levels (29). When MCF7 cells were exposed to oligomycin, cellular ATP levels in ERR-depleted cells were significantly lower than those in control cells (Fig. 6D). Collectively, these bioenergetic measurements suggest that endogenous ERRs contribute to the aerobic glycolysis phenotype of cancer cells.
Figure 6. Depletion of endogenous ERRs reduces aerobic glycolysis and cell growth.
Culture media from control (pGIPz), ERRα- and ERRγ-depleted MCF7 cells were analyzed to measure glucose uptake (A), lactate levels (B), pH value (C), and ATP contents (D). Data were normalized to cell number and are presented as mean ± SEM (n=3). Cell growth curves from control, ERRα- and ERRγ-knockdown MCF7 cells (E) and T47D (F) cells are presented as mean ± SEM (n=3). * indicates significant change (p<0.05).
Increased glucose consumption and glycolysis facilitate cell proliferation by meeting both bioenergetic and anabolic needs of dividing cells. As depletion of ERRs reduced glycolysis, it was anticipated that cell growth would be affected as well. MCF7 cells depleted of ERRγ demonstrated modestly yet significantly reduced growth rate compared to control cells (Fig. 6E), but depletion of ERRα did not have a clear effect (Fig. 6E). In T47D cells, depletion of either ERRα or ERRγ significantly impaired cell growth (Fig. 6F). Similar phenomenon was also observed in MDA-MB-435 cells (Fig. S3B). These results suggest that ERRs are modulators of cancer cell growth.
Discussion
Aerobic glycolysis, or the Warburg effect, is a characteristic feature of cancer cells. The altered glucose metabolic profile satisfies the unique bioenergetic and anabolic requirement of proliferating cancer cells. HIF and Myc are two prominent drivers of the Warburg effect. Here we identified the orphan nuclear receptor ERRs as global activators of glycolysis and important contributors to the glycolytic phenotype in cancer cells. ERRs recognize the promoter regions of many genes encoding enzymes in the glycolytic pathway. ERRs activate glycolytic genes through the ERR-binding motifs in these promoters. Overexpression and depletion of ERRs enhance and reduce glycolytic gene expression and aerobic glycolysis in cancer cells, respectively. Together with our previous report (15), the current study suggests that ERRs can collaborate with both Myc and HIF in activating glycolytic gene expression, reinforcing the important role of ERRs in induction of glycolysis and metabolic transformation of cancer. ERRs have been implicated in tumor growth and progression (26,27). ERRs may contribute to malignant development in part by conferring metabolic advantages to tumor cells.
According to the Randle cycle (18), oxidation of glucose and fatty acids is reciprocally inhibitory. FAO produces acetyl-CoA, which enters the TCA cycle to generate NADH. Both acetyl-CoA and NADH can inhibit the PDH complex, uncoupling glycolysis from subsequent oxidation (18). ERRs are known to promote oxidative metabolism of fatty acids by activating genes critical for mitochondrial uptake and β-oxidation of fatty acids (16). By enhancing FAO and inhibiting glucose oxidation through upregulation of PDK4, ERRs may be part of the regulatory mechanism underlying the Randle cycle and metabolic substrate selectivity. However, the two FAO products do not directly inhibit glycolysis. Consequently, unlike glucose oxidation, glycolysis is compatible with FAO. Indeed, inhibition of glucose oxidation by FAO should further diverts pyruvate to lactate, and hence enhance aerobic glycolysis. Therefore, stimulation of glycolysis by ERRs does not necessarily conflict with ERRs’ existing role in FAO. This metabolic control is reminiscent of AMPK, a key kinase in energy metabolism. AMPK orchestrates cellular energy conservation by activating catabolic pathways, including glucose uptake, glycolysis, and FAO (30).
Cancer cells generally retain intact mitochondrial function. While they exhibit a shift from glucose oxidation to glycolysis, cancer cells may have varying degrees of oxidative metabolism, probably depending availability of oxygen and fuel types (e.g. glutamine and fatty acids) (31). Oxidative metabolism also feeds the energetic and anabolic demands of dividing cells: the TCA cycle supplies important intermediates for lipid and amino acid biosynthesis, and OXPHOS efficiently generates ATP (2,9). ERRs are master regulators of mitochondrial oxidative metabolism including the TCA cycle and OXPHOS (16). The finding that ERRs also promote glycolysis suggests that the two main metabolic pathways can be integrated under the control of a common transcription factor, which may offer growth advantages to cancer cells. Although different ERR members may function differently (e.g. depending on cell types) (32), ERR-expressing cancer cells may rely on different metabolic programs to adapt to nutrient supply, oxygen availability, and energy demand. In this regard, Myc stimulates aerobic glycolysis as well as mitochondrial respiration and glutaminolysis (33,34). Anti-glycolysis treatment as monotherapy so far only results in a limited effect on tumorigenesis (8), which might be attributed to tumor’s ability to switch from glucose dependence to a reliance on oxidative metabolism of other fuels.
Materials and Methods
Plasmids, Antibodies, and Mice
Full-length human ERRα, ERRγ, PGC-1α, and PGC-1β cDNAs were amplified by PCR and cloned into pcDNA3 (Invitrogen) with a Flag epitope tag at the amino-terminus. Lentiviral expression vectors for Flag-ERRα-IRES-Puro and Flag-ERRγ-IRES-Puro were made in the pGIPz vector (OpenBiosystems) by replacing GFP with ERRα or ERRγ. Tet-on inducible lentiviral expression vector for ERRα and ERRγ were made in pTriPz (OpenBiosystems) by replacing RFP and shRNA with ERRα and ERRγ. The human PKM2 promoter (-284bp to +271bp) was PCR-amplified and cloned into the pGL4.22 vector (Promega) for luciferase reporter assays. The rat PDK4 basal promoter region (−302bp to +42bp) was PCR amplified and cloned into pGL4.22, giving rise to a control reporter called “C-luc”; Subsequently, the ENO1 promoter (−1067bp to −328bp) was amplified and inserted in front of the PDK4 basal promoter in C-luc. Mutagenesis of the putative ERR-binding sites in the PKM2 and ENO1 luciferase reporters was achieved using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). pGIPz-based lentiviral shRNA constructs targeting ERRα and ERRγ were from OpenBiosystems. The following antibodies were used in this study: anti-ERRα (Santa Cruz and GeneTex); anti-ERRγ (GeneTex); anti-Flag (Sigma); anti-LDHA and anti-ENO1(Cell Signaling); anti-Tubulin (Sigma). ERRα knockout mutant mice were obtained from the Jackson Laboratory.
Cell Culture, Transfection, Viral Infection, Luciferase Assay, and RT-PCR
The human cell lines HEK293, MCF7, MDA-MB-231, T47D, and MDA-MB-435 cells were grown in DMEM medium (Cellgro) supplemented with 10% bovine calf serum (BCS). All transfections were done using the Turbofect transfection reagent (Fermentas) according to the manufacturer’s instructions. For the reporter assay, HEK293 cells were transfected with the Luciferase reporter and TK-driven Renilla reporter plasmids, and harvested for Luciferase assays 24 hours later. Luciferase activities were determined using the dual Luciferase Reporter Assay System (Promega). Renilla luciferase was used as a reference to normalize transfection efficiency. Data are shown in relative luciferase units (RLUs) and are presented as mean ± standard deviations (S.D.) from three independent experiments. For RNA isolation and realtime RT-PCR assays, ERRα-IRES-Puro and ERRγ-IRES-Puro expressing lentiviruses were used to infect MCF7 and MDA-MB-231 cells; pGIPz ERRα shRNA, ERRγ shRNA and empty vector lentiviruses were used to transduce MCF7, T47D, and MDA-MB-435 cells. The transduced cells were selected by puromycin (1ug/ml) for 48h. Total RNA from cells was extracted with TRIzol reagent (Invitrogen) following the manufacturer’s instruction, and cDNA was synthesized using M-MuLV reverse transcriptase with random primers. The expression levels of selected genes were determined by quantitative PCR. Primer sequences were listed in Table S3.
Co-immunoprecipitation and Chromatin Immunoprecipitation (ChIP)
Co-immunoprecipitation assays were performed as previously described (15). For ChIP assay, cells were cross-linked with 1% formaldehyde for 10 min. The reaction was stopped by the addition of glycine. Cross-linked cells were washed in 1× PBS and collected. Cell pellets were washed in washing buffer (0.25% TritonX-100, 10mM EDTA, 0.5mM EGTA, 10mM Tris pH8.0), resuspended in sonication buffer (1mM EDTA, 0.5mM EGTA, 10mM Tris pH8.0), mixed with glass beads, and subjected to sonication. The sonicated samples were diluted in ChIP buffer (0.01% SDS, 1.0% TX-100, 1.0mM EDTA, 20mM Tris pH 8.1, 150 mM NaCl) and incubated with specific antibodies and protein A slurry (Invitrogen). The immunoprecipitates were subjected to a series of washing steps to remove non-specific binding. After reverse-crosslinking, the DNA samples were purified and then analyzed by real-time quantitative PCR. Primers used in ChIP assay were listed in Table S2. Final results represent percentage of input chromatin and error bars indicate S.D. from triplicate experiments.
Measurement of Lactate Concentrations
Lactate concentration was measured using the Lactate assay kit (Biovision) following manufacturer’s instruction. Lactate levels in culture media were normalized to cell number, and lactate levels in mice were normalized to protein concentration. Data are presented as mean value ± S.D. from triplicate experiments.
Measurement of Glucose uptake and pH
Cells grown at 70% confluence in 6-well plates were washed once with fresh DMEM containing 10% fetal bovine serum, and then incubated in 2ml of the fresh medium for 48h. The culture medium was collected and analyzed for glucose levels by Glucose Assay Kit (Eton Bioscience). Glucose uptake was determined as the difference between glucose level of fresh medium and that of conditioned medium. The cultured medium was also analyzed for pH value using a pH meter.
Measurement of ATP
Cellular ATP changes were measured by Cell Titer-Glo reagent according to the instructions (Promega). Cells grown at 40% confluence in 12-well plates were washed once with fresh DMEM containing 10% bovine calf serum, and then incubated in 500μl of the fresh medium with or without 5μM oligomycin for 4 h. The cells were trypsinized and collected to measure ATP by Cell Titer-Glo reagent immediately.
Cell Growth Assay
MCF7, T47D, and MDA-MB-435 cells were infected with pGIPz lentivirus and were selected by puromycin (1ug/ml) for 48h. Cells were seeded in 12 wells plates, and later trypsinized and counted by TC10™ Automated Cell Counter (BioRad) every 24h.
Statistical Analysis
For all experiments with quantitative results, the data are expressed as the mean and 95% confidence interval (CI) from three identical experiments carried out independently. Means and 95% confidence intervals were calculated using Microsoft Office Excel 2007 software.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute (R01CA137021) and Florida Bankhead-Coley Cancer Research Program (09BN-12-23092 and 2BT01).
Footnotes
Conflict of interest
The authors declare there is no competing financial interests in relation to this work.
References
- 1.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 8.Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 9.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 10.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim Biophys Acta. 2011;1813:1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci USA. 2008;105:7821–7826. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 17.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 18.Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297:E578–591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, Wang F, et al. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem. 2006;281:39897–39906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- 21.Araki M, Motojima K. Identification of ERRalpha as a specific partner of PGC-1alpha for the activation of PDK4 gene expression in muscle. FEBS J. 2006;273:1669–1680. doi: 10.1111/j.1742-4658.2006.05183.x. [DOI] [PubMed] [Google Scholar]
- 22.Dufour BJ, Wilson JM, Huss DP, Kelly WA, Alaynick M, Downes RM, et al. Genone-wide orchestration of cardiac functions by orphan nucler receptors ERRα and γ. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Deblois JA, Hall MC, Perry J, Laganière M, Ghahremani M, Park M, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor α as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 24.Charest-Marcotte CR, Dufour BJ, Wilson AM, Tremblay LJ, Eichner DH, Arlow VK, et al. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev. 2010;24:537–542. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr Top Med Chem. 2006;6:203–215. doi: 10.2174/1568026610606030203. [DOI] [PubMed] [Google Scholar]
- 27.Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell. 2011;20:500–510. doi: 10.1016/j.ccr.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein RA, Chang CY, Kazmin DA, Way J, Schroeder T, Wergin M, et al. Estrogen-related receptor α is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–8812. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 30.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. mir-378* mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.