Katz et al. present a novel mRNA tethering assay as a method for understanding the role of mRNA compartmentalization in the control of cellular processes. In this study, they combine this tethering assay with advanced imaging techniques to show that the β-actin mRNA-binding protein ZBP1 directs β-actin mRNA to focal adhesions, where it stabilizes focal adhesions in an actin protein-dependent manner and affects the directionality of cell movement.
Keywords: cell directionality, focal adhesion, mRNA localization, post-transcriptional control, single-particle tracking, translation control
Abstract
Directed cell motility is at the basis of biological phenomena such as development, wound healing, and metastasis. It has been shown that substrate attachments mediate motility by coupling the cell's cytoskeleton with force generation. However, it has been unclear how the persistence of cell directionality is facilitated. We show that mRNA localization plays an important role in this process, but the mechanism of action is still unknown. In this study, we show that the zipcode-binding protein 1 transports β-actin mRNA to the focal adhesion compartment, where it dwells for minutes, suggesting a means for associating its localization with motility through the formation of stable connections between adhesions and newly synthesized actin filaments. In order to demonstrate this, we developed an approach for assessing the functional consequences of β-actin mRNA and protein localization by tethering the mRNA to a specific location—in this case, the focal adhesion complex. This approach will have a significant impact on cell biology because it is now possible to forcibly direct any mRNA and its cognate protein to specific locations in the cell. This will reveal the importance of localized protein translation on various cellular processes.
mRNA localization is a highly conserved method for the subcellular spatial and temporal control of gene expression (Lecuyer et al. 2007, 2009; Besse and Ephrussi 2008; Martin and Ephrussi 2009). As one of the initial examples of mRNA localization, β-actin mRNA was shown to be distributed to the leading edge of migrating fibroblasts (Kislauskis et al. 1997). Directionality of cell movement was shown to be dependent on the zipcode-binding protein 1 (ZBP1), which binds the zipcode sequence in mRNA and is necessary for mRNA localization (Kislauskis et al. 1997; Ross et al. 1997). Introducing antisense oligos to the zipcode sequence in the 3′ untranslated region (UTR) of β-actin mRNA or truncating the C-terminal mRNA-binding domains in ZBP1 exhibited significant decreases in the directionality of fibroblasts (Shestakova et al. 2001; Farina et al. 2003). ZBP1 binding acts to repress β-actin mRNA translation until a Src kinase phosphorylates the 396 tyrosine, a site proximal to the KH3–KH4 domains responsible for binding the mRNA zipcode (Huttelmaier et al. 2005; Chao et al. 2010). This was the first indication of a tightly regulated mechanism involving ZBP1 for not only localization of an mRNA target, but also its translational control.
Newly synthesized β-actin within the leading edge would be expected to supply a small fraction of the actin monomer concentration known to contribute to all filaments within the leading edge (Chan et al. 1998; Condeelis and Singer 2005). Therefore, it was hypothesized that local actin synthesis must be targeted to a specific compartment, presumably one that would impact cell directionality. For this reason, we focused our experimental methods on focal adhesions within the leading edge, a site known to have active Src kinase. Additionally, previous studies have shown that other RNA-binding proteins can concentrate near adhesion complexes (Babic et al. 2009; Lee et al. 2009) and may play a role in adhesion maturation (Chicurel et al. 1998; de Hoog et al. 2004; Vikesaa et al. 2006). Here we used single-particle tracking of endogenous β-actin mRNA to identify a dwelling population of mRNA within the leading edge of live fibroblasts. We found increased mRNA dwelling as the distance to adhesions decreased, indicating a compartment around adhesions that corralled mRNA. Furthermore, we showed that localizing β-actin mRNA to the adhesion compartment was dependent on ZBP1 and that ZBP1 knockout cells exhibited decreased focal adhesion stability. Development of an mRNA tethering assay illustrated that an increased concentration of β-actin mRNA at adhesions altered adhesion dynamics and cell motility statistics. This effect on adhesions and motility was found to be translation-dependent. Taken together, these data elucidate the dynamic role of ZBP1-mediated β-actin mRNA localization to leading edge compartments in order to asymmetrically control adhesion stability and regulate the directionality of fibroblast movement through local β-actin synthesis.
Results and Discussion
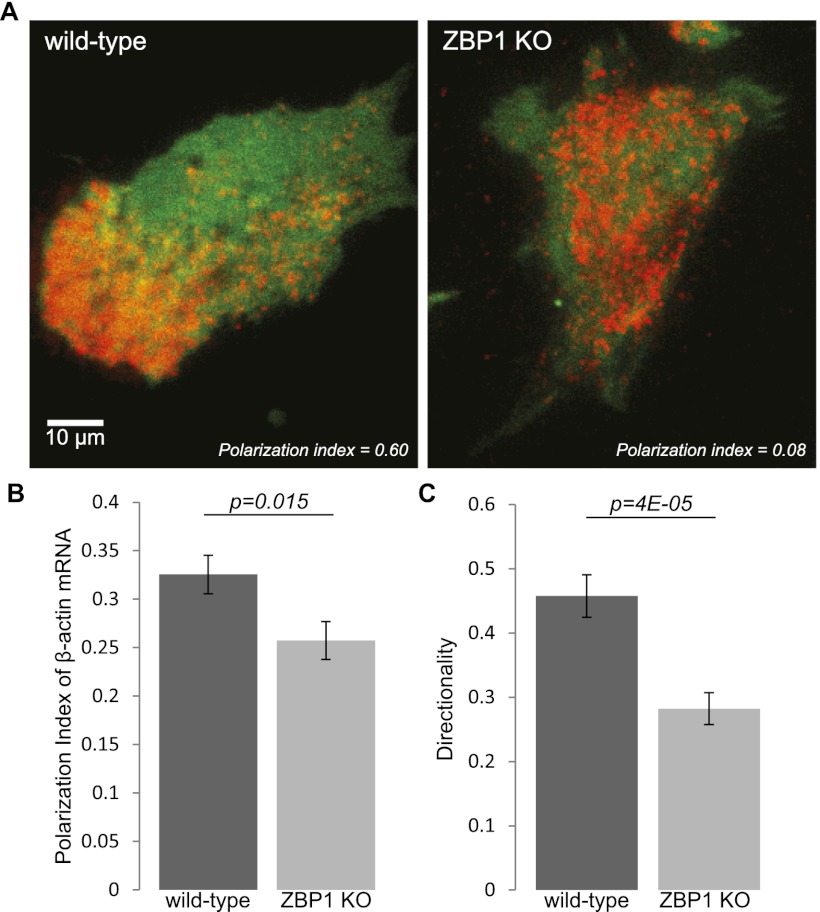
With the recent advent of the β-actin-MS2-binding site knock-in (MBS) mouse, with all β-actin mRNA labeled with MS2 stem loops in the 3′ UTR, we now can visualize and track endogenous mRNAs in real time (Lionnet et al. 2011). Crossing the MBS mouse with our ZBP1 knockout mouse enabled us to investigate mRNA dynamics in a ZBP1-null environment (Supplemental Fig. S1). This allowed direct investigation of mRNA localization dependent on ZBP1 expression. We first investigated whether a significant localization deficiency could be measured in ZBP1−/− cells. Fibroblasts derived from littermates of a ZBP1+/−MBS+/+ cross were immortalized and stably infected with the MS2 coat protein (MCP) fused to TagRFPt, a red fluorescent protein. Live-cell images of MCP-TagRFPt-labeled β-actin mRNA were taken in total internal reflection fluorescence (TIRF), and the mRNA distribution was quantified against GFP as a cytoplasmic marker (Fig. 1A). As expected, the polarization index, computed through a recently reported algorithm (Park et al. 2012), was significantly reduced in MBS–ZBP1 knockout fibroblasts when compared with MBS wild type (Fig. 1B). Additionally, we investigated whether the localization defect translated into a cell motility defect. Motility analysis over 12 h revealed that ZBP1 knockout fibroblasts had a significantly reduced directionality (Fig. 1C; Supplemental Movie 1; Supplemental Table 1). This confirmed previous findings that showed that a deficiency in cytoplasmic localization of β-actin mRNA resulted in defective directionality of migrating fibroblasts (Shestakova et al. 2001; Farina et al. 2003).
Figure 1.
ZBP1 is necessary for β-actin mRNA localization and directionality in mouse embryonic fibroblasts (MEFs). (A) Live-cell TIRF images of wild-type and ZBP1 knockout fibroblasts with TagRFPt-labeled β-actin mRNA and free GFP as the cytoplasmic marker. Corresponding polarization indices are shown, based on a reported algorithm that assesses asymmetry by computing the intensity-weighted centroids of mRNA and cytoplasmic GFP (Park et al. 2012). (B) The average polarization index of β-actin mRNA distribution was significantly lower in cells without ZBP1. n = 60 for each condition, and bars represent polarization index averages ± SEM. (C) The directionality of fibroblasts (computed as the net path divided by the total path) was also found to be significantly reduced in fibroblasts without ZBP1. n = 50 for each condition ±SEM.
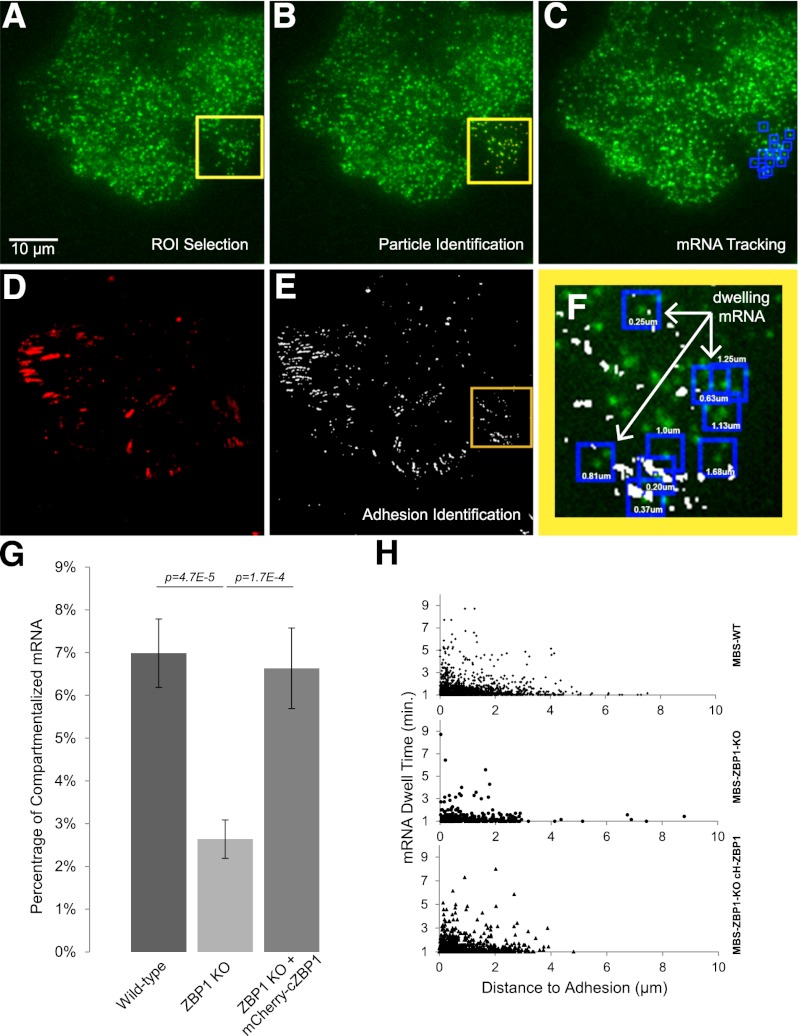
In order to directly investigate the difference in β-actin mRNA localization dynamics between MBS wild-type and MBS–ZBP1 knockout cells, we created an analysis approach to track single mRNA particles that dwell in the leading edge of live fibroblasts. Images were taken every 10 sec for a total of 10 min, and TIRF excitation ensured a single plane of focus on the ventral surface. Endogenous β-actin mRNA was labeled with MCP fused to GFP, while cell adhesions were labeled with paxillin-mCherry to enable coordinate analysis of dwelling mRNAs to the nearest focal adhesion (Fig. 2A–F). Movies were then analyzed by a previously described single-particle tracking program (Larson et al. 2005). The slower acquisition rate allowed specific observation of mRNAs that dwelled for long time periods within a confined distance on the ventral surface of the cell. Tracked mRNA particles that persisted >1 min were categorized as “dwelling” mRNAs and counted as a single tracked data set (Fig. 2F; Supplemental Movie 2). This analytical approach produced two critical results for explaining the importance of localizing β-actin mRNA in motile cells. First, the percentage of dwelling mRNAs in the cell periphery was significantly greater in wild-type cells than in ZBP1 knockout cells (Fig. 2G), although mRNA and adhesion densities within the analyzed regions were the same (Supplemental Fig. S2A). Expressing exogenous mCherry-cZBP1, sorted to express at endogenous levels, rescued the defect that we observed in MBS–ZBP1 knockout cells (Fig. 2G). Second, we measured the distance from each dwelling mRNA (coordinates were determined by the track centroid) to the nearest focal adhesion and found an increased proportion of mRNA dwelling as the distance to adhesions decreased (Fig. 2H). This trend was observed in all cell lines (Supplemental Fig. S3A), and we therefore concluded that the focal adhesion environment was responsible for corralling β-actin mRNA. We rarely observed mRNA dwelling directly on adhesions; instead, a majority of mRNAs dwelled within 1 μm of an adhesion, suggesting that the environment around adhesions retains mRNA. It is reasonable to hypothesize that these mRNAs are translating, since they would be less mobile when associated with polysomes (multiple ribosomes). We tested this hypothesis by the addition of puromycin, an antibiotic that causes premature translation termination and dissociation of ribosomes from mRNA and hence would be expected to eliminate the dwelling (translating) mRNAs. This experiment yielded a clear result. We found that the percent of dwelling mRNAs was significantly reduced after the addition of 100 ug/mL puromycin (Supplemental Fig. S3B; Supplemental Movie 3). This showed that dwelling mRNAs were likely translating.
Figure 2.
ZBP1 localizes β-actin mRNA to the adhesion compartment, where it dwells for minutes. (A) Endogenous β-actin mRNA labeled with 2xMCP-GFP bound to 24 MS2 stem loops in the 3′ UTR was imaged in TIRF every 10 sec for 10 min. (B) For each frame, mRNA particles were identified within a chosen region of interest (ROI) (yellow square) in the cellular periphery with Localize software (Larson et al. 2005), as indicated by the red dots. (C) Identified mRNAs that persisted for >1 min (seven consecutive frames) with less than five pixels of displacement between each frame were tracked and categorized as dwelling mRNAs (identified by the blue boxes). (D,E) The first and last frame included acquisition of paxillin-mCherry labeling to coordinate dwelling mRNAs to the nearest adhesion. The centroid coordinates of each tracked dwelling mRNA was coordinated to the nearest adhesion identified by thresholding (shown in E). (F) Distances (shown for each dwelling mRNA in the figure) were calculated for each dwelling mRNA centroid to the nearest adhesion (masked in white) and used to analyze the significance of ZBP1 on β-actin mRNA compartmentalization within the leading edge. (G) We found that ZBP1 expression in MEFs significantly increased the probability of β-actin mRNA dwelling for >1 min in the cell periphery. Data shown represents dwelling mRNA tracks found within each ROI divided by the average number of mRNA particles found per minute ± SEM, with the following n-values: wild type (WT) = 60; ZBP1 knockout (KO) = 46; ZBP1 KO + mCh-ZBP1 = 33. β-actin mRNA particles near the perinuclear region were not found to dwell for >1 min for all samples (data not shown). (H) All dwelling mRNAs for each condition as a factor of dwell time versus distance to the nearest adhesion were plotted. More dwelling mRNA was observed as distance to the nearest adhesion decreased.
The distribution of mRNA dwell times within 1 μm of an adhesion was found to be consistent among samples, even though mRNAs dwell in ZBP1-expressing cells more frequently (Supplemental Fig. S3C). Therefore, the decreased number of longer-dwelling mRNAs in ZBP1 knockout cells reflected a transport deficiency of β-actin mRNA to the compartment around adhesions. We predict that this was directly related to ZBP1's mechanism of binding to β-actin mRNA in the perinuclear region and asymmetrically transporting its target along the cytoskeleton to the cellular periphery (Latham et al. 2001; Oleynikov and Singer 2003), a site known to have increased Src activity near adhesions (Zaidel-Bar et al. 2007). Once ZBP1 is phosphorylated and β-actin mRNA is no longer translationally repressed, newly synthesized β-actin protein can directly integrate at the interface between the actin filament and the adhesion, thereby helping to stabilize adhesions at the leading edge.
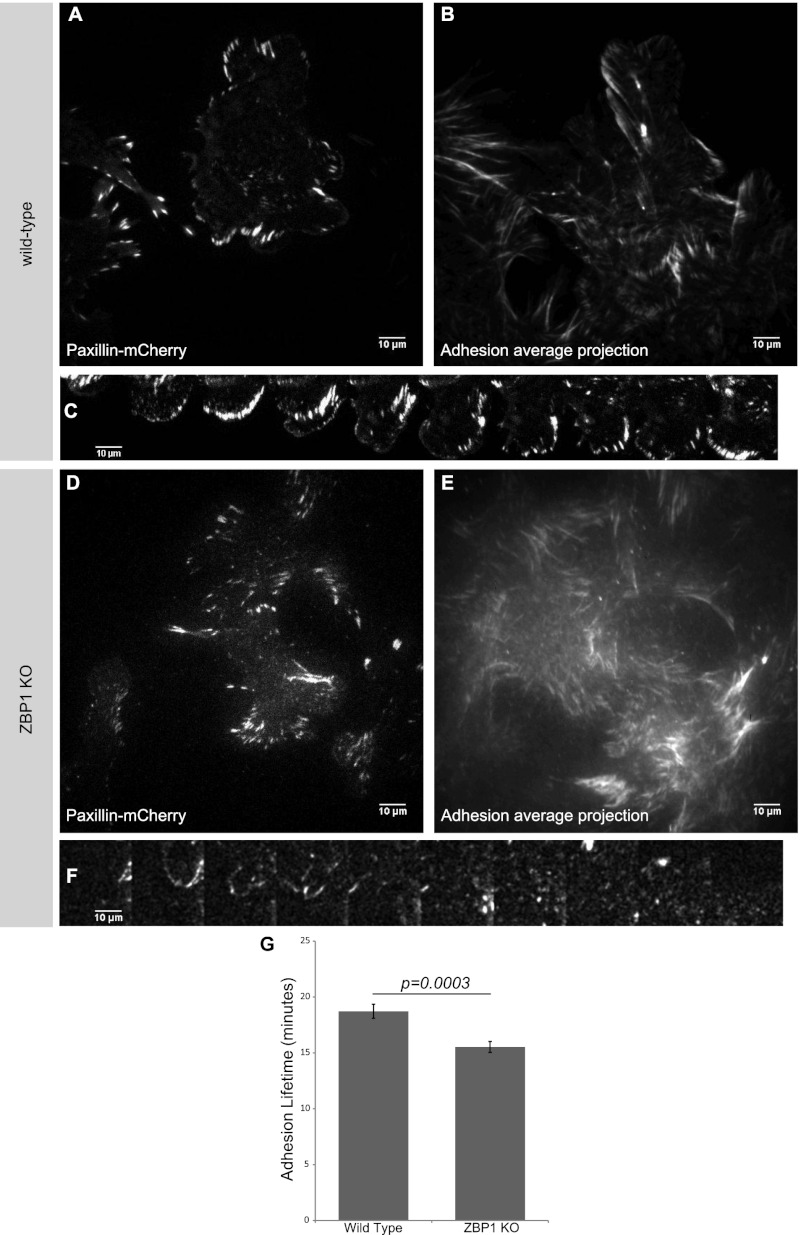
Since localization of β-actin mRNA to the adhesion compartment was found to be reduced in ZBP1 knockout cells, we sought to understand the role of β-actin mRNA localization on adhesion dynamics. To assess focal adhesion dynamics, we measured paxillin-mCherry lifetimes in motile fibroblasts by tracking adhesions in TIRF for 6 h (Fig. 3; Supplemental Movie 4). We found a significant decrease in focal adhesion stability (measured by tracking the lifetime of each adhesion) in ZBP1 knockout cells (Fig. 3G), although the average adhesion size of all adhesions was not significantly different between wild-type and ZBP1 knockout fibroblasts (data not shown). Analysis of individual protrusions allowed for closer visualization of each adhesion life cycle within the cellular periphery. In wild-type fibroblasts, we observed a progressive maturation of adhesions as the leading edge extended (Fig. 3C), whereas protrusions in ZBP1 knockout fibroblasts showed very little adhesion stability within a protrusion over the same time scale (Fig. 3F). Averaged projections over the 6-h acquisition showed clear protruding waves of adhesions in wild-type fibroblasts, while a disarray of adhesions in ZBP1 knockout fibroblasts did not show a persistent front of maturing adhesions (Fig. 3B,E). Re-expressing exogenous ZBP1 at wild-type levels rescued the adhesion stability defect measured in ZBP1 knockout cells (Supplemental Fig. S4). We suspect that ZBP1's ability to asymmetrically localize β-actin mRNA to specific adhesion compartments within the cell periphery enabled focal complex maturation at the leading front of migrating fibroblasts and dictated the directionality of cell movement. ZBP1 knockout fibroblasts had a decreased probability of localizing β-actin mRNA to specified adhesion compartments, and therefore adhesion stability was not asymmetrically coordinated, resulting in reduced directionality in migrating cells.
Figure 3.
Focal adhesion stability is directly related to ZBP1 expression in migrating MEFs. Paxillin-mCherry was expressed in wild-type MEFs (A–C) and ZBP1 knockout MEFs (D,E) and imaged every minute for 6 h in TIRF (Supplemental Movie 3). (B,E) Average projections of adhesion movement movies depict progressive waves of adhesions in the direction of migration in wild-type MEFs but a disarray of adhesion movement in ZBP1 knockout MEFs. (G) We measured a significant reduction in average adhesion lifetimes in ZBP1 knockout cells, although adhesion size was not significantly different (data not shown). The time series of individual protrusions in C and F depict a coordinated protrusion and adhesion maturation paradigm in wild-type MEFs, while no clear adhesion stability is seen in the ZBP1 knockout protrusion over the same time course (each montage frame is 15 min). Data shown for wild type and ZBP1 knockout represent averages with n-values of >18 fields over three independent experiments ±SEM.
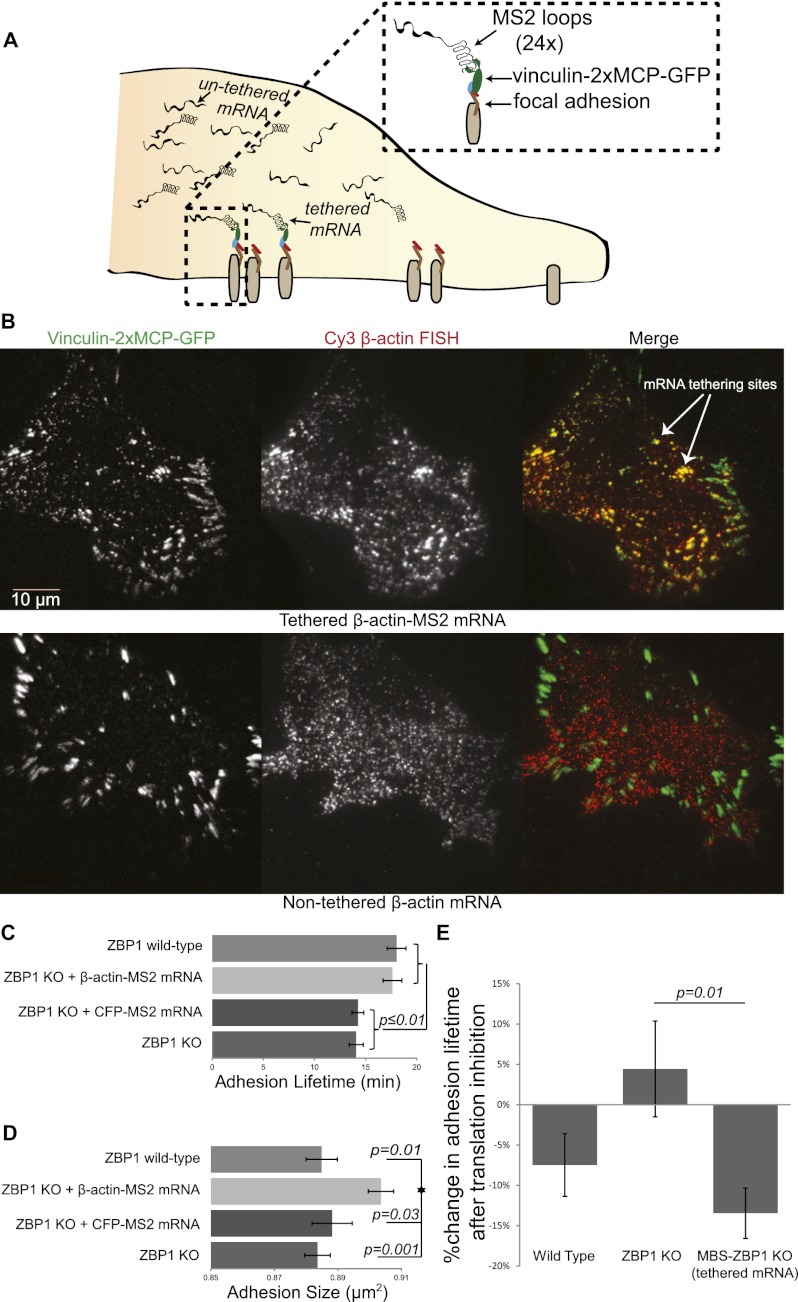
If increased dwelling of β-actin mRNA at the adhesion compartment directs cell movement through adhesion stabilization, then artificially tethered β-actin mRNA at adhesions would elicit an exaggerated response. We therefore exploited the MS2 system to tether mRNA to adhesion complexes in order to directly correlate β-actin mRNA locality at adhesions with adhesion lifetimes and motility statistics (Fig. 4A). We stably infected cells with vinculin fused to the MS2 capsid protein dimer and GFP (denoted as vinculin-2xMCP-GFP), which enabled us to tether endogenous β-actin mRNA to adhesions when put into MBS fibroblasts. We confirmed this result by fluorescent in situ hybridization (FISH) of β-actin mRNA (in MBS and non-MBS cells) and colocalized this signal with the GFP signal of the vinculin construct (Fig. 4B). We observed increased colocalized signal with mature adhesions further back from the leading edge, which may relate to MS2-β-actin mRNA accumulating on an adhesion as it matures over time (Supplemental Fig. S5). As expected, cells that did not express MS2-β-actin mRNA showed reduced colocalization of β-actin mRNA with vinculin-2xMCP-GFP (Supplemental Fig. S6). Next, we investigated focal adhesion dynamics, as done previously, by tracking vinculin-2xMCP-GFP fluorescence in wild-type, ZBP1 knockout, and MBS-ZBP1 knockout cells. We found that cells expressing vinculin-2xMCP-GFP in a non-MBS background (no MS2-β-actin mRNA tethered to adhesions) acted similarly to before, with significantly reduced adhesion lifetimes in ZBP1 knockout cells compared with wild type. However, when we tested MBS–ZBP1 knockout cells expressing vinculin-2xMCP-GFP, we observed a noticeable difference in their adhesion profile (Supplemental Movie 5). MBS–ZBP1 knockout cells with β-actin mRNA tethered to adhesions exhibited a significant increase in adhesion lifetimes, comparable with levels that we measured in wild-type cells (Fig. 4C). Additionally, we were able to measure a significant increase in the average adhesion size in cells with β-actin mRNA tethered to vinculin (Fig. 4D). These observations appear to be specific to β-actin mRNA, as the same effect was not found when we expressed CFP-MS2 mRNA in ZBP1 knockout cells infected with vinculin-2xMCP-GFP (CFP mRNA tethered to adhesions).
Figure 4.
β-Actin mRNA tethering to focal adhesions specifically increases adhesion size and lifetimes. (A) Schematic for the mRNA tethering assay. mRNA with multiple MS2 stem–loops in the 3′ UTR binds with high affinity to the vinculin construct fused to the MCP dimer and GFP. As adhesions mature, more vinculin-2xMCP-GFP accumulates, and tethered mRNA increases in concentration at the adhesion. (B) Cy3 FISH probes were used to confirm that β-actin mRNA was tethered to the vinculin-2xMCP-GFP construct. (Top) Tethered cells show dense colocalizing signal in TIRF. (Bottom) Non-MBS ZBP1 knockout cells (nontethered) showed no overlay of FISH signal with vinculin-2xMCP-GFP. Tethering β-actin mRNA to adhesions in ZBP1 knockout cells increased adhesion lifetimes (C) to wild-type values but also significantly increased the adhesion size (D). CFP mRNA with MS2 stem–loops in the 3′ UTR was used as a control tethered mRNA in ZBP1 knockout cells and produced no increase in adhesion size or lifetime. All data shown represent averages of fields analyzed with n-values as follows: ZBP1 knockout (KO) = 30; ZBP1 KO + CFP-MS2 = 17; MBS-ZBP1 KO = 30; wild type (WT) = 24. (E) To test the influence of translation, adhesions were tracked before and after cycloheximide (100 ug/mL) addition. The percent change in adhesion lifetime after dosage was calculated for each experimental group. n-values are as follows: wild type = 19; MBS-ZBP1 KO = 18; ZBP1 KO = 17. Error bars represent SEM, and P-values above data bars are from a two-tailed Student's t-test compared between experimental groups.
To investigate how actin stress fibers may differ in cells with β-actin mRNA tethered to adhesions, we stained filaments with rhodamine phalloidin and imaged in TIRF (Supplemental Fig. S7). Actin filament ends integrated into adhesions similarly in all experimental groups (blue arrows in Supplemental Fig. S7). However, we observed adhesions on actin filaments in the TIRF field further into the interior of cells when β-actin mRNA was tethered in MBS cells (purple arrows in Supplemental Fig. S7). This was consistent with our real-time observation that adhesions with tethered β-actin mRNA did not turn over normally and appeared to track along filaments into the interior of a cell (Supplemental Movie 4). Therefore, we tracked cell movement, as done previously, and found that ZBP1 knockout cells with tethered β-actin mRNA had a significantly decreased cell velocity (Supplemental Table S2), which correlated with our predicted increase in adhesiveness. Because MBS cells expressing vinculin-2xMCP-GFP cannot localize β-actin mRNA, they would exhibit an equal ability to tether the mRNA at all adhesions, without establishing a polarity. Therefore, each adhesion would be equally stabilized, and as a consequence, cells with tethered β-actin mRNA exhibited no increase in directionality in the absence of an asymmetric force (Supplemental Table S2; Supplemental Movie 6).
Implicit in these results is the implication that increased translation of β-actin protein at adhesions strengthens the adhesive force of the ventral surface on the extracellular matrix. To test whether these results were translation-dependent, we applied cycloheximide, an inhibitor of translation elongation, to cells expressing vinculin-2xMCP-GFP. Consistent with our hypothesis, a significant reduction in adhesion lifetime was observed after translation inhibition only in MBS–ZBP1 knockout cells (those that tethered β-actin mRNA to all adhesions). Cells that did not tether mRNA did not show a significant reduction (Fig. 4E; Supplemental Movie 7), although wild-type cells showed a trend of reduced adhesion stability after translation inhibition. This indicated that local translation of β-actin mRNA increased adhesion stability in cell lines with increased mRNA around adhesions.
These results have elucidated the location where local β-actin mRNA regulates cellular guidance. Focal adhesion complexes are known sites of active Src, the kinase necessary for relieving the translational control that ZBP1 exerts on its mRNA target (Huttelmaier et al. 2005; Zaidel-Bar et al. 2007). Recent evidence from adhesion proteome analysis revealed that several RNA-binding proteins (such as hnRNP K, PABP1, and hnRNP E2) and many ribosomal proteins were present at focal adhesions (Kuo et al. 2011). Once ZBP1 transports β-actin mRNA to the adhesion compartment, Src would enable its release, and additional RNA-binding proteins could ensure its retention near the adhesion site. Locally synthesized β-actin would integrate into a maturing nascent adhesion to stabilize the leading edge protrusion. Recent cryo-electron microscopy evidence suggested that only two to three actin filaments integrate into an individual adhesion particle (Patla et al. 2010), while live-cell imaging revealed that short, thin actin filaments emanated from maturing nascent adhesions (Choi et al. 2008), implying that tight spatial control over actin filament assembly greatly alters the stability of a maturing adhesion plaque. Newly synthesized β-actin was also shown to be post-translationally modified through N-terminal arginylation, a known mechanism specific to the β-actin isoform that promotes proprietary incorporation into filaments (Saha et al. 2010; Zhang et al. 2010). Furthermore, actin isoform-specific antibodies have revealed the spatial distribution of β-actin protein to the ventral surface within the myosin-dependent contractile network (Dugina et al. 2009). This evidence supports the necessity of localized β-actin protein synthesis within distinct cytoplasmic compartments. Understanding β-actin mRNA spatial regulation through ZBP1 has enabled a greater understanding of how mRNA localization can elicit significant control over cell dynamics. Whether other mRNA targets of ZBP1 act in a way similar to β-actin mRNA has yet to be fully determined, although there is recent evidence to support a conserved mechanism (Patel et al. 2012). ZBP1 expression has implications for cancer prognosis (Wang et al. 2004; Vainer et al. 2008), neural regulation (Donnelly et al. 2011; Welshhans and Bassell 2011), and development (Hansen et al. 2004). Determining how, where, and for what purpose ZBP1 regulates its mRNA targets will have significant implications for understanding cell dynamics.
This work introduces a novel approach to study the effects of increased protein synthesis at specified cellular locations through forcibly localizing mRNA. Results from tethering β-actin mRNA to focal adhesion complexes supported the role that ZBP1 plays in facilitating β-actin mRNA localization to adhesions. Because the mRNA is permanently associated with all adhesions, rather than dissociating and diffusing elsewhere, these results do not phenocopy the mechanism of ZBP1-facilitated localization. Thus, the change in adhesion properties (e.g., slower turnover) as a result gives us insight into how constantly restricting β-actin mRNA to adhesions affects how a cell can move. Evidence suggests that other mRNAs important for motility are likewise ZBP1-regulated (Mingle et al. 2005; Gu et al. 2012) and may alter cell movement if tethered to adhesions. Further use of mRNA tethering with any MS2-labeled mRNA will enable us to test their impact on multiple cellular processes in addition to cell motility. Tethering mRNA to spatially restricted complexes would also alter the localization status of any desired mRNA and hence its cognate protein. For instance, an mRNA could be forcibly localized to neuronal processes to enable direct demonstration of this mRNA on a specific cellular function (e.g., spine stability or synaptic plasticity). Conversely, mRNAs with mutant coding regions can be forcibly localized to specific structures to test dysfunctional protein effects on cell dynamics. mRNA tethering therefore presents a novel way for understanding the necessity of mRNA compartmentalization, or lack of it, in the control of cellular processes.
Materials and methods
Live-cell imaging
All experiments were performed on an Olympus IX-71 TIRF station customized for laser-illuminated TIRF with an environmental control chamber and an Andor EMCCD camera. mRNA tracking was performed with an Olympus 150× 1.45 NA TIRFM objective. Focal adhesion tracking and mRNA localization experiments were performed with an Olympus 60× 1.45 NA TIRFM objective. Cell motility analysis was performed with an Olympus 10× 0.30 NA phase1 objective. Further explanation of acquisition procedures for each experimental approach can be found in the Supplemental Material.
mRNA tethering assay
The chicken vinculin ORF was cut from a GFP-vinculin construct (gift from Louis Hodgson) and inserted into our 2xMCP-GFP lentiviral backbone. The vinculin-2xMCP-GFP construct enabled stable expression of our tethering apparatus in MBS cells and non-MBS cells. MBS cells have β-actin mRNA labeled with 24 MS2 stem–loops in the 3′ UTR and therefore tethered β-actin mRNA to all labeled adhesions. A list of the cy3 FISH probes used to confirm β-actin mRNA tethering may be found in the previously published Supplemental Material for Lionnet et al. (2011). As a control, ZBP1 knockout cells (non-MBS background) were nucleofected with a CFP construct with MS2 stem–loops in the 3′ UTR. For the translation inhibition assay, cycloheximide (Sigma) was applied approximately half-way through the adhesion tracking experiment at 100 uM. Construct maps for all experiments can be found in Supplemental Figure S8.
Statistical tests
Statistical significance between experimental conditions was determined by using a two-tailed Student's t-test.
Acknowledgments
We thank Xiuhua Meng and Carolina Eliscovich for help with constructs. We thank Chiso Nwokafor and Melissa Lopez-Jones for breeding and care of mice. We thank Timothee Lionnet for the use of MatLab code, and Gal Haimovich for discussions that led to the tethering work. This work was supported by NIH grants T32 GM007491 (to Z.B.K.) and GM84364 (to R.H.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.190413.112.
References
- Babic I, Sharma S, Black DL 2009. A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol Cell Biol 29: 5564–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Ephrussi A 2008. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 9: 971–980 [DOI] [PubMed] [Google Scholar]
- Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci 111: 199–211 [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicurel ME, Singer RH, Meyer CJ, Ingber DE 1998. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392: 730–733 [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR 2008. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol 10: 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH 2005. How and why does β-actin mRNA target? Biol Cell 97: 97–110 [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M 2004. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117: 649–662 [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, et al. 2011. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J 30: 4665–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugina V, Zwaenepoel I, Gabbiani G, Clement S, Chaponnier C 2009. β- and γ-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci 122: 2980–2988 [DOI] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Katz Z, Wu B, Park HY, Li D, Lin S, Wells AL, Singer RH 2012. Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J Cell Sci 125: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC 2004. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH 1997. β-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol 136: 1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR 3rd, Waterman CM 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol 13: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Johnson MC, Webb WW, Vogt VM 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci 102: 15453–15458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH 2001. A Rho-dependent signaling pathway operating through myosin localizes β-actin mRNA in fibroblasts. Curr Biol 11: 1010–1016 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Krause HM 2009. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol 21: 409–415 [DOI] [PubMed] [Google Scholar]
- Lee JH, Rangarajan ES, Yogesha SD, Izard T 2009. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure 17: 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 8: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci 118: 2425–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Trcek T, Wells AL, Chao JA, Singer RH 2012. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Reports 1: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. 2012. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patla I, Volberg T, Elad N, Hirschfeld-Warneken V, Grashoff C, Fassler R, Spatz JP, Geiger B, Medalia O 2010. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat Cell Biol 12: 909–915 [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, Kashina A 2010. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol Biol Cell 21: 1350–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci 98: 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainer G, Vainer-Mosse E, Pikarsky A, Shenoy SM, Oberman F, Yeffet A, Singer RH, Pikarsky E, Yisraeli JK 2008. A role for VICKZ proteins in the progression of colorectal carcinomas: Regulating lamellipodia formation. J Pathol 215: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikesaa J, Hansen TV, Jonson L, Borup R, Wewer UM, Christiansen J, Nielsen FC 2006. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J 25: 1456–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 64: 8585–8594 [DOI] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ 2011. Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci 31: 9800–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B 2007. Functional atlas of the integrin adhesome. Nat Cell Biol 9: 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, Kashina A 2010. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329: 1534–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]